Abstract
Purpose
The Tamoxifen and Exemestane Adjuvant Multinational (TEAM) trial included a prospectively planned pathology substudy testing the predictive value of progesterone receptor (PgR) expression for outcome of estrogen receptor–positive (ER-positive) early breast cancer treated with exemestane versus tamoxifen.
Patients and Methods
Pathology blocks from 4,781 TEAM patients randomly assigned to exemestane versus tamoxifen followed by exemestane for 5 years of total therapy were collected centrally, and tissue microarrays were constructed from samples from 4,598 patients. Quantitative analysis of hormone receptors (ER and PgR) was performed by using image analysis and immunohistochemistry, and the results were linked to outcome data from the main TEAM trial and analyzed relative to disease-free survival and treatment.
Results
Of 4,325 eligible ER-positive patients, 23% were PgR-poor (Allred < 4) and 77% were PgR- rich (Allred ≥ 5). No treatment-by-marker effect for PgR was observed for exemestane versus tamoxifen (PgR-rich hazard ratio [HR], 0.83; 95% CI, 0.65 to 1.05; PgR-poor HR, 0.85; 95% CI, 0.61 to 1.19; P = .88 for interaction). Both PgR and ER expression were associated with patient prognosis in univariate (PgR HR, 0.53; 95% CI, 0.43 to 0.65; P < .001; ER HR, 0.66; 95% CI, 0.51 to 0.86; P = .002), and multivariate analyses (P < .001 and P = .001, respectively). A trend toward a treatment-by-marker effect for ER-rich patients was observed.
Conclusion
Preferential exemestane versus tamoxifen treatment benefit was not predicted by PgR expression; conversely, patients with ER-rich tumors may derive additional benefit from exemestane. Quantitative analysis of ER and PgR expression provides highly significant information on risk of early relapse (within 1 to 3 years) during treatment.
INTRODUCTION
For decades, tamoxifen remained the gold standard for adjuvant endocrine therapy for estrogen receptor–positive (ER-positive) and/or progesterone receptor–positive (PgR-positive) early breast cancer. Patients with ER-positive tumors who received tamoxifen for 5 years experienced significantly reduced rates of recurrence and death.1 However, a significant group with hormone receptor–positive (HRec-positive) tumors exhibited resistance, either acquired or de novo, to tamoxifen therapy.2,3 Several resistance mechanisms are hypothesized, including signaling via human epidermal growth factor receptor (EGFR) homolog 2 (HER2), EGFR/HER1, and HER34–7 signaling pathways,8,9 or loss of ER-mediated gene regulation.10,11 Differential sensitivity to endocrine agents (eg, aromatase inhibitors [AIs] v tamoxifen) has been linked to bypassing resistance pathways.12–14
The third-generation AIs—anastrozole, exemestane, and letrozole—induce nearly complete suppression of circulating estrogens in postmenopausal women. In randomized clinical trials in postmenopausal women with HRec-positive early breast cancer,13–15 AIs significantly improved disease-free survival (DFS) compared with tamoxifen. In the Intergroup Exemestane Study (IES), patients switching to exemestane after 2 to 3 years of tamoxifen experienced significant improvements in outcome compared with those remaining on tamoxifen.16 Additionally, patients with PgR-negative tumors in the Arimidex, Tamoxifen, Alone or in Combination (ATAC) trial appeared to gain significant additional benefit from anastrozole versus tamoxifen.17
The Tamoxifen and Exemestane Adjuvant Multinational (TEAM) trial compared the efficacy of exemestane versus sequential therapy of tamoxifen followed by exemestane for 5 years of total adjuvant endocrine therapy.18 TEAM provided an ideal setting to test hypotheses regarding primary tamoxifen resistance. PgR is considered “time-dependent” because the impact of low PgR expression on relapse risk in ER-positive, tamoxifen-treated patients with breast cancer is confined to the first 3 years of endocrine therapy.10 The study presented here addresses the hypothesis that low PgR expression predicts clinical benefit for patients receiving initial AI treatment rather than tamoxifen in the first 2.75 years of therapy.17 The translational aspect of TEAM is unique because the hypotheses were prospectively defined and powered at the time of study design.
PATIENTS AND METHODS
Design
The TEAM trial is a multinational, randomized, open-label, phase III trial in postmenopausal women with HRec-positive early breast cancer. Women were randomly assigned to receive exemestane 25 mg once daily or tamoxifen 20 mg once daily for the first 2.5 to 3 years followed by exemestane (for a total of 5 years of treatment). Coprimary end points were DFS at 2.75 and 5 years. This study complied with the Declaration of Helsinki, individual independent ethics committee guidelines, and the International Conference on Harmonization and Good Clinical Practice guidelines. All patients provided informed consent.
One hypothesis tested in the TEAM pathology substudy was that initial therapy with exemestane is superior to tamoxifen in patients with ER-positive and PgR-negative or PgR-poor tumors. The prospectively powered outcome analysis compared high PgR expressers (approximately 75% of tumors) with low PgR expressers (approximately 25%). This analysis was planned for 2.75 years after last patient enrollment and was not event driven. By using a two-sided α = .05 and assuming a hazard ratio (HR) of 1.9 and PgR-poor prevalence of 25%, a sample size of ≥ 4,000 patients gave > 90% power to detect a treatment-biomarker interaction.
Patients
Inclusion and exclusion criteria differed slightly by participating country.18 Eligible patients were postmenopausal with HRec-positive resectable breast cancer (defined locally), with histologically or cytologically confirmed T1-3 N0-2 M0 breast adenocarcinoma and adequate surgical resection followed by radiotherapy and/or adjuvant chemotherapy if indicated. Five of nine countries that participated in the TEAM trial (United Kingdom/Ireland, the Netherlands, Belgium, Germany, and Greece) provided tumor samples for this substudy.
Staining Methodology
Briefly, tissue blocks were received at the central laboratory, logged, reviewed for sufficient tissue, subjected to pathology quality assurance and tissue marking, and had six cores extracted and placed on six replicate tissue microarrays (TMAs). Standard immunohistochemical techniques were used to stain TMAs for ER (Clone 6F11; Novocastra, Newcastle, United Kingdom) and PgR (Clone PgR636, DAKO, Cambridge, United Kingdom). Assays were performed by using a single batch of antibody and reagents, with incubations rigorously controlled for temperature. Each run included a quality-control, six-sample TMA with various ER and PgR expression, and runs were accepted only if histoscore inter-run variation was < 15%. Staining and scoring of ER and PgR by using TMAs and the Ariol SL-50 image analysis system (Genetix, New Milton, United Kingdom) were previously validated.19 Samples were stained in sextuplet (6 × 0.6 mm2 cores). Slides were counterstained; each TMA section was scanned, mapped to positional map, and individually assessed for tumor content (tumor areas were marked and checked and a second pathology quality-assurance assessment was performed). The actual number and percentage of cells in each category were recorded, with a minimum of 100 tumor nuclei per patient required for eligibility. Nuclear HRec staining was assessed by using a previously validated scoring algorithm,19 with nuclei grouped into categories of negative, weak (1+), moderate (2+), and strong (3+) nuclear staining, to generate a continuous histoscore (range, 0 to 300). The histoscore was calculated by multiplying the sum of the percentage of weakly stained cells times 1, the percentage of moderately stained cells times 2, and the percentage of strongly stained cells times 3.
Statistical Analysis
All analyses were performed for DFS (censored at 2.75 years), defined as time from random assignment to earliest documentation of disease relapse (ie, primary tumor recurrence [locoregional or distant] and ipsilateral/contralateral breast cancer) or death from any cause. Cox proportional hazards regression models were used to assess prognostic and predictive values of PgR and ER expression. Interactions between treatment arms and PgR or ER expression levels were evaluated by examining the log likelihood ratio statistic. Both PgR and ER were included as binary covariates for Allred score (PgR-poor ≤ 4 v PgR-rich ≥ 5; ER-poor ≤ 6 v ER-rich ≥ 7). The PgR cutoff was informed by our previous data10; the ER cutoff was informed by the TransATAC study.20 Adjusted analyses were performed for patient age (continuous variable), tumor size (< 2 cm v > 2 cm), nodal status (negative v positive), and histologic grade on an intent-to-treat basis. Continuous HRec expression data were checked for nonlinearity by applying a log transformation and assessing changes in Akaike information criterion between univariate Cox proportional hazards models of transformed and untransformed data.21 The best-fitting model was used to plot the predicted log HR and 95% CI values against quantitative HRec expression. All data were analyzed by using SAS/STAT statistical software (SAS Institute, Cary, NC).
A prognostic (risk) score (scale, 0.0 to 4.0) was derived based on prognostic modeling of ER and PgR expression as continuous variables; tumor grade, tumor size, and nodal status as dichotomous variables; and age as a continuous variable. Each variable was assessed for nonlinearity by using log transformations and fractional polynomials; factors considered significant at α = .1 were included in the model. Individual patients' prognostic factors were multiplied by the coefficients defined in the multivariate prognostic model as follows: score = (PgR histoscore × −0.00372) + (ER histoscore × −0.00227) + ([age-38] × 0.67) + (grade × −0.38992) + (T size × −0.45302) + (nodal status × −0.61221), where age-38 represents age at diagnosis minus 38 years, and T size represents pathologic tumor size.
Patients' scores were plotted against their survival estimate as calculated by the Cox proportional hazards model, and results were split by randomized treatment. The number needed to treat (NNT) to prevent one recurrence was calculated as described previously.22
RESULTS
Study Population
Eligible samples (n = 4,781) were received and analyzed by country (samples received/randomly assigned patients) from United Kingdom/Ireland (1,097/1,275; 86.0%), the Netherlands (2,722/2,753; 98.9%), Belgium (122/414; 29.5%), Germany (745/1,471; 50.6%), and Greece (95/207; 45.9%). In total, 4,598 samples were suitable for TMA construction and ER and PgR staining. Following staining, 219 samples contained < 100 stained tumor nuclei, 42 samples were ER-negative (of which 28 were PgR-positive), and 12 samples were ineligible for other reasons. For the remaining samples, a median of > 1,200 cells per tumor were analyzed. Only 14 ER-negative/PgR-negative patients (0.3%) were identified in the pathology cohort, and 4,325 eligible ER-positive patients (87% of target population) were analyzed. Patient tumor characteristics were similar among samples from the pathology subset and all patients from countries participating in the pathology substudy, but there were slight differences between these two groups and the entire TEAM population regarding tumor size, nodal status, and grade (Table 1). Most tumors (87.3%) in the pathology substudy had ER Allred scores of 7 to 8, with only 12.7% having Allred scores ≤ 6. In total, 3,341 tumors (77.2%) were PgR-rich (Allred score, 5 to 8) and 984 (22.8%) were PgR-poor (Allred score, 0 to 4).
Table 1.
Patient and Tumor Characteristics
| Characteristic | Subset of Pathology Samples Received(n = 4,325) |
Pathology Substudy Population(n = 6,120)* |
TEAM Study ITT Database(n = 9,766) |
|||
|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | |
| Age, years | ||||||
| < 60 | 1,393 | 32 | 2,085 | 34 | 3,348 | 34 |
| ≥ 60 | 2,932 | 68 | 4,035 | 66 | 6,418 | 66 |
| Nodal status | ||||||
| Positive | 2,602 | 60 | 3,494 | 57 | 4,585 | 47 |
| Negative | 1,713 | 40 | 2,604 | 43 | 5,113 | 52 |
| Unknown | 10 | < 1 | 22 | < 1 | 68 | 1 |
| Tumor size, cm | ||||||
| ≤ 2 | 2,120 | 49 | 3,028 | 50 | 5,697 | 58 |
| > 2 | 2,197 | 51 | 3,027 | 50 | 4,047 | 41 |
| Unknown | 8 | < 1 | 20 | < 1 | 20 | < 1 |
| Grade | ||||||
| 1 | 486 | 11 | 616 | 10 | 1,677 | 19 |
| 2 | 2,199 | 51 | 3,166 | 52 | 4,795 | 54 |
| 3 | 1,370 | 32 | 1,835 | 30 | 2,438 | 27† |
| ER Allred score | ||||||
| 0-4 | 153 | 3.5 | N/A | N/A | ||
| 5-6 | 398 | 9.2 | N/A | N/A | ||
| 7-8 | 3,774 | 87.3 | N/A | N/A | ||
| PgR Allred score | ||||||
| 0-4 | 984 | 22.8 | N/A | N/A | ||
| 5-8 | 3,341 | 77.2 | N/A | N/A | ||
Abbreviations: TEAM, Tamoxifen and Exemestane Adjuvant Multinational; ITT, intent-to-treat; ER, estrogen receptor; N/A, not available; PgR, progesterone receptor.
Randomly assigned patients from the United Kingdom, Ireland, the Netherlands, Belgium, Germany, and Greece were included in the pathology substudy.
Grade 3 to 4.
Efficacy
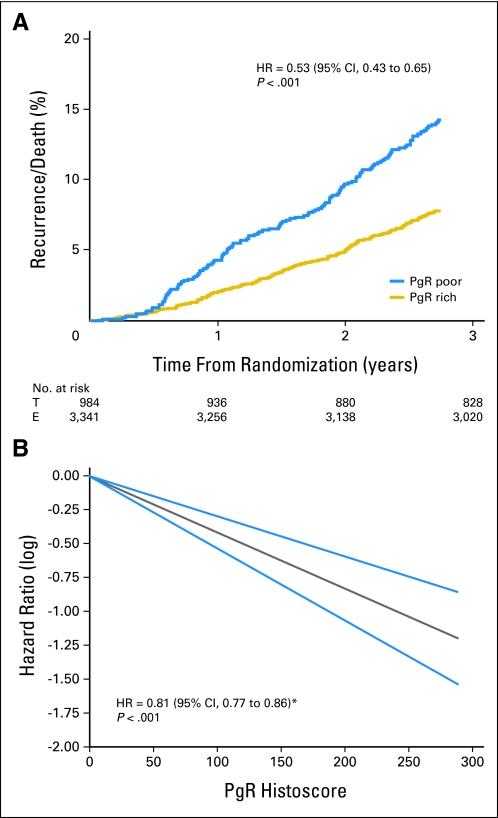
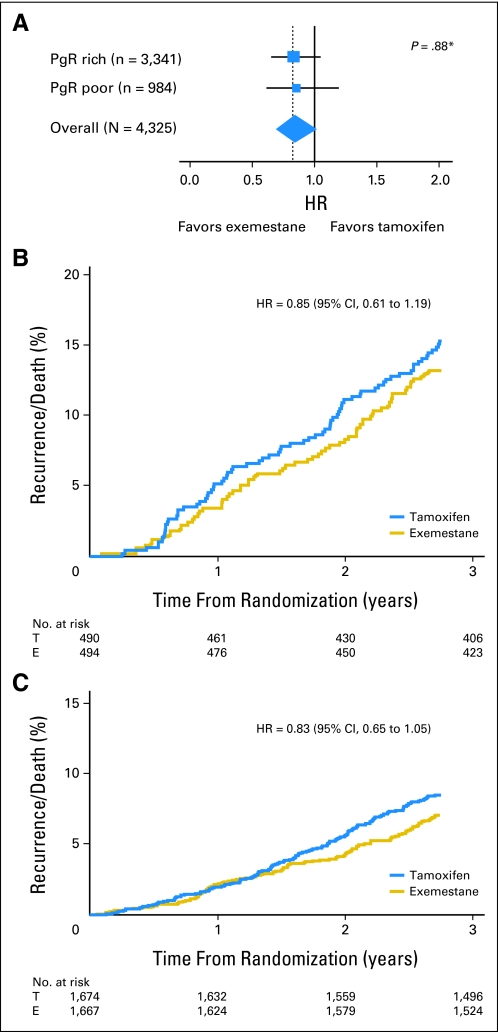
In the pathology substudy, 397 DFS events were recorded within 2.75 years of random assignment with a trend toward a DFS benefit of exemestane compared with tamoxifen (HR, 0.84; 95% CI, 0.69 to 1.02; P = .078). The HR for the DFS benefit of exemestane within the whole trial was similar to that observed for the pathology subgroup (HR, 0.89; 95% CI, 0.77 to 1.03; P = .12). Overall, the pathology substudy population was well matched to the patient population from which the samples were collected, but the patients were of higher risk than those in the United States (Table 1). Expression of PgR was a significant prognostic indicator in univariate analyses (PgR-rich v PgR-poor; HR, 0.53; 95% CI, 0.43 to 0.65; P < .001; Fig 1A) and adjusted multivariate analyses (HR, 0.54; 95% CI, 0.44 to 0.68; P < .001). Univariate Cox regression with PgR expression as a continuous variable yielded an HR of 0.81 (95% CI, 0.77 to 0.86; P < .001) per 50-unit increase of PgR expression (eg, PgR histoscore of 50 was associated with a 19% reduction in DFS compared with histoscore of 0). Nonlinearity between PgR expression and DFS was investigated but not confirmed, and PgR was therefore modeled under the assumption of linearity (Fig 1B). Multivariate Cox regression analysis, including a treatment-by-marker analysis, showed no evidence that PgR expression was predictive of a differential DFS benefit for exemestane compared with tamoxifen (PgR-poor HR, 0.85; 95% CI, 0.61 to 1.19; PgR-rich HR, 0.83; 95% CI, 0.65 to 1.05; test for interaction P = .88; Fig 2). Analyses were also performed by using the actual switch point rather than 2.75 years. The interaction for differential benefit of treatment was not statistically significant (PgR-poor HR, 0.94; 95% CI, 0.65 to 1.35; PgR-rich HR, 1.12; 95% CI, 0.85 to 1.49; test for interaction P = .7); hence the conclusions remain unchanged.
Fig 1.
Progesterone receptor (PgR) as a prognostic marker of disease-free survival by (A) Allred score (≤ 4 v ≥ 5) and (B) quantitative PgR expression plotted versus log hazard ratios (HRs); gray line represents PgR histoscore versus hazad ratio; blue lines represent 95% CI of estimate. T, tamoxifen; E, exemestane. (*) Per 50-unit increase in PgR.
Fig 2.
Progesterone receptor (PgR) is not predictive of exemestane (E) versus tamoxifen (T) benefit. (A) Hazard ratio (HR) by Allred score (≤ 4 [PgR poor] v ≥ 5 [PgR rich]); (B) exemestane versus tamoxifen in PgR-poor, and (C) PgR-rich tumors. (*) Test for interaction by PgR staining.
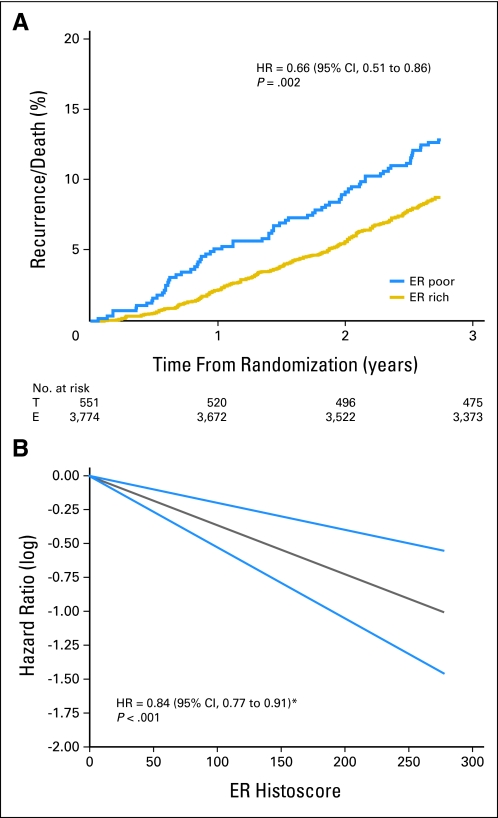
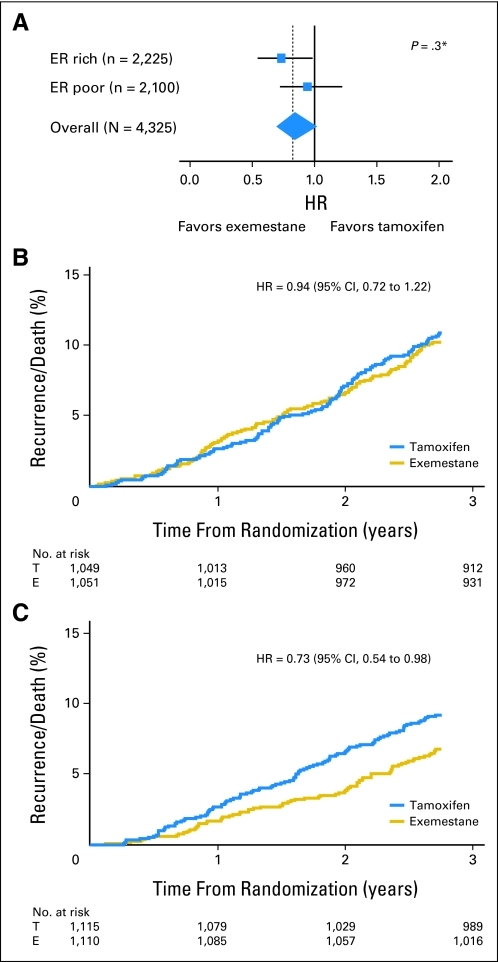
A similar analysis confirmed that moderate to poor ER expression (Allred score < 6) was associated with a significantly worse prognosis than ER-rich tumors (Allred score > 7) in both univariate (ER-rich v ER-poor HR, 0.66; 95% CI, 0.51 to 0.86; P = .002; Fig 3A) and multivariate regression analysis (adjusted HR, 0.65; 95% CI, 0.50 to 0.85; P = .001). Again, after investigating the nonlinearity assumption of ER expression, univariate analysis for linearly increasing ER histoscores as a continuous variable found a 17% risk reduction related to a 50-unit increase in histoscore (HR, 0.84; 95% CI, 0.77 to 0.91; P < .001; Fig 3B). In an unplanned analysis of the predictive value of ER expression, a trend toward benefit in favor of exemestane was observed (Fig 4A). Tumors with ER histoscores ≤ 190 (median, ER-poor) had an HR for exemestane versus tamoxifen of 0.94 (95% CI, 0.72 to 1.22; Fig 4B), whereas the HR in favor of exemestane for ER-rich tumors (histoscore > 190) was 0.73 (95% CI, 0.54 to 0.98; Fig 4C). However, adjustment for other prognostic factors did not confirm a significant treatment-by-marker effect (P = .2). Analyses performed by using the actual switch point showed that the interaction for differential benefit of treatment was not statistically significant (ER-poor HR, 1.13; 95% CI, 0.84 to 1.51; ER-rich HR, 0.97; 95% CI, 0.69 to 1.36; test for interaction P = .5).
Fig 3.
Estrogen receptor (ER) as a prognostic disease-free survival marker by (A) Allred score (≤ 6 v ≥ 7) and (B) quantitative ER expression plotted against log hazard ratios (HRs); gray line represents ER histoscore versus hazard ratio; blue lines represent 95% CI of estimate. T, tamoxifen; E, exemestane. (*) Per 50-unit increase in ER.
Fig 4.
Estrogen receptor (ER) as a predictive marker for exemestane (E) versus tamoxifen (T) treatment. (A) Hazard ratio (HR) by histoscore (≤ 190 [ER-poor tumors] v > 190 [ER-rich tumors]); (B) exemestane versus tamoxifen in ER-poor and (C) ER-rich tumors. (*) Test for interaction by ER staining.
Prognostic Model
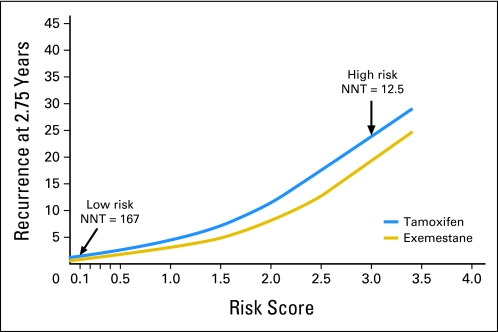
By combining and weighting prognostic variables shown to independently influence risk of early relapse (within 2.75 years), a risk score integrating ER and PgR expression level, age, tumor grade, tumor size, and nodal status was derived and plotted against risk of recurrence in each treatment arm (Fig 5). In this model, lower levels of ER and PgR expression correspond with a higher risk score, as do increased tumor size and grade, node positivity, and patient age. In the highest-risk population (risk score, 3.0), the NNT with exemestane to prevent one recurrence was 12.5. Conversely, in patients with a low risk of early recurrence (risk score, 0.1), the absolute benefit derived with initial exemestane was minimal (NNT = 167).
Fig 5.
Risk score is prognostic for disease-free survival and predicts greater benefit from exemestane in patients with a higher risk score. NNT, number needed to treat.
DISCUSSION
The TEAM pathology substudy included the largest prospective biomarker analysis within an adjuvant AI trial, rejecting the a priori hypothesis that low PgR expression might predict improved benefit from AI treatment. Both PgR and ER expression levels were quantitatively associated with DFS. The relative risk of relapse increased proportionally with decreasing expression of either receptor, demonstrating a concentration effect of HRec expression on prognosis (Figs 1B and 3B). However, PgR status did not predict additional benefit of exemestane versus tamoxifen (P = .88 for PgR-rich v PgR-poor interaction with treatment). This prospective analysis demonstrates that PgR is strongly prognostic for patients treated with endocrine agents (either AIs or tamoxifen) but does not provide a marker to select patients who derive additional benefit from AIs over tamoxifen when analyzed as a dichotomous variable. These data are consistent with the analysis of PgR in the TransATAC23 and Breast International Group (BIG 1-98)24–26 studies, but contrast with the primary ATAC trial report.17 The prospectively powered and planned analysis within the TEAM study provides high-level evidence to refute the original observation from the primary ATAC report that low PgR expression is a predictive biomarker for initial therapy selection with AIs versus tamoxifen.17 When viewed in the context of other analyses and translational studies, current evidence supports the conclusion that the relative benefit from AIs is similar for tumors that express low versus high levels of PgR.
Nonetheless, ER and PgR are significant prognostic factors in postmenopausal patients with early breast cancer who are receiving adjuvant endocrine therapy.10,27–30 Recent evidence, including our study, suggests that PgR as a prognostic marker is both time- and concentration-dependent.10 The current study produced highly consistent quantitative HRec expression data by using image analysis and robustly quality-assured immunohistochemistry, allowing us to demonstrate a significant link between recurrence risk and quantitative changes in HRec expression, irrespective of endocrine treatment modality. These and similar analyses31,32 argue strongly against the current practice of dichotomizing patient risk of recurrence relative to specific HRec expression cutoffs. Allred scores of 2 to 4 are widely used to discriminate between patients who are likely to respond to hormone therapy and those who are not, but little attention is paid to markedly varying risk of relapse across the 80% to 85% of ER-positive patients or the > 75% of PgR-rich patients demonstrated herein. Although patients derive approximately equally proportional benefit from exemestane in PgR-poor and PgR-rich tumors, the strong prognostic influence of HRec expression may affect the absolute benefit observed in different subgroups of HRec-positive patients. A 10-fold reduction in risk of relapse for ER-moderate versus ER-rich patients with a constant proportional benefit from AIs versus tamoxifen suggests a 10-fold increase in absolute NNTs to prevent one recurrence. Such exploratory analyses, which were also performed in the BIG 1-98 and ATAC trials, imply that many more patients in low-risk groups with high HRec expression are exposed to the risk of adverse effects relative to the numbers that benefit compared with higher-risk groups with low HRec expression. This observation clearly underpins the continuing use of patient risk stratification in many local and national guidelines for AI use. By modeling the relative contribution of HRec expression with clinical and pathologic variables (eg, age, nodal status) within the TEAM pathology cohort, we extended this risk stratification to produce a risk estimate for individual patients within the first 2.75 years of treatment with either exemestane or tamoxifen. At higher risk scores, the absolute DFS benefit of exemestane versus tamoxifen was more apparent, whereas at lower risk scores, the absolute benefit was modest.
In an unplanned analysis, quantitative image analysis that used median ER histoscores, following the categorization used in previously published TransATAC data, suggested that high expression of ER is predictive of preferential benefit for exemestane compared with tamoxifen. Although this interaction was not significant (P = .3), the HRs and 95% CIs (HR, 0.72; 95% CI, 0.53 to 0.97 and HR, 0.92; 95% CI, 0.70 to 1.20) for ER-rich and ER-poor patients, respectively, indicated a potential clinical significance of quantitative ER levels for selection of endocrine therapies. Similar results were reported in the TransATAC study, in which a significant relationship between increasing ER expression and time to relapse was reported only for patients treated with anastrozole (P < .001) but not tamoxifen (P = .78).23 These results conflict with observations from neoadjuvant studies,33 which showed that AIs appeared to provide additional benefit in HRec-poor patients. The observations in the TEAM and ATAC studies suggest that more effective endocrine agents could have proportionally greater impact on highly endocrine-responsive tumors rather than on less endocrine-responsive tumors as previously believed.23 Therefore, current data confirm a strong prognostic effect (ie, decreased risk of relapse as HRec expression increases) and a potential predictive effect of HRec expression (ie, increased benefit of exemestane as ER expression increases). Statistical modeling of this potential effect is complicated by possible interactions between ER and PgR expression and because the fewest events (ie, low recurrence risk) are observed at the point of potential maximum divergence between exemestane and tamoxifen treatment. These results, however, were derived from retrospective, unplanned analyses and are not sufficient to alter current clinical practice but must be regarded as hypothesis generating. Future modeling, perhaps in a meta-analysis of all AI trial pathology archives, is required to explore this potential interaction between the prognostic and predictive impact of HRec expression.
The TEAM pathology substudy provides, to the best of our knowledge, the only prospectively planned and powered analysis of the interaction between PgR and efficacy of exemestane versus tamoxifen as initial endocrine therapy. By using expression scores derived from quantitative image analysis for HRec expression in analyses that were not preplanned, this study further demonstrated that patients derive benefit from endocrine therapies directly proportional to their relative risk, which is strongly influenced by quantitative changes in ER and PgR expression as well as other clinical and pathologic features. In an unplanned analysis, patients with breast tumors expressing high ER levels were shown to potentially benefit preferentially from initial treatment with exemestane over tamoxifen. Validation of these observations in a meta-analysis of data from all AI translational studies may provide evidence of a clinically useful marker approach to patient stratification for early versus delayed treatment with AIs.
Acknowledgment
We thank Chris A. Kirk, PhD, Complete Healthcare Communications, and Jeffrey S. Riegel, PhD, AccuVerus, Beachwood, OH, for their editorial support (funded by Pfizer). We also thank all pathologists and also the patients, who consented to provide paraffin blocks for the study, for their support.
Footnotes
See accompanying editorial on page 1504
Supported by unrestricted research grants from Pfizer and funding from the Breast Cancer Institute and Cancer Research UK.
Presented at the 31st Annual San Antonio Breast Cancer Symposium, San Antonio, TX. December 14-18, 2008.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Clinical trial information can be found for the following: NCT00036270.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: John M.S. Bartlett, Pfizer (C), Roche (C), Novartis (C), Bayer Pharmaceuticals (C), Abbott Laboratories (C), DAKO (C); Dirk G. Kieback, Pfizer (C); Daniel Rea, Pfizer (C), Novartis (C) Stock Ownership: None Honoraria: John M.S. Bartlett, Pfizer, Roche, Abbott Laboratories, DAKO, Bayer Pharmaceuticals; Fiona M. Campbell, Roche; Annette Hasenburg, Pfizer; Dirk G. Kieback, Pfizer; Christos Markopoulos, Pfizer; Daniel Rea, Pfizer Research Funding: John M.S. Bartlett, Pfizer, Roche, AstraZeneca, Novartis, GlaxoSmithKline; Dirk G. Kieback, Pfizer; Christos Markopoulos, Pfizer; Daniel Rea, Pfizer, Novartis, AstraZeneca Expert Testimony: None Other Remuneration: John M.S. Bartlett, Pfizer, Roche
AUTHOR CONTRIBUTIONS
Conception and design: John M.S. Bartlett, Hein Putter, Daniel Rea
Administrative support: John M.S. Bartlett
Provision of study materials or patients: John M.S. Bartlett, Fiona M. Campbell, Annette Hasenburg, Dirk G. Kieback, Christos Markopoulos, Luc Dirix, Caroline Seynaeve, Daniel Rea
Collection and assembly of data: John M.S. Bartlett, Tammy Robson, Fiona M. Campbell, Margaret Grant, Annette Hasenburg, Elysée T.M. Hille, Charlene Kay, Dirk G. Kieback, Christos Markopoulos, Elma Meershoek-Klein Kranenbarg, Elizabeth A. Mallon, Luc Dirix
Data analysis and interpretation: John M.S. Bartlett, Cassandra L. Brookes, Tammy Robson, Lucinda J. Billingham, Annette Hasenburg, Hein Putter, Luc Dirix, Daniel Rea
Manuscript writing: John M.S. Bartlett, Cassandra L. Brookes, Tammy Robson, Cornelis J.H. van de Velde, Lucinda J. Billingham, Fiona M. Campbell, Margaret Grant, Annette Hasenburg, Elysée T.M. Hille, Charlene Kay, Dirk G. Kieback, Hein Putter, Christos Markopoulos, Elma Meershoek-Klein Kranenbarg, Elizabeth A. Mallon, Luc Dirix, Caroline Seynaeve, Daniel Rea
Final approval of manuscript: John M.S. Bartlett, Cassandra L. Brookes, Tammy Robson, Cornelis J.H. van de Velde, Lucinda J. Billingham, Fiona M. Campbell, Margaret Grant, Annette Hasenburg, Elysée T.M. Hille, Charlene Kay, Dirk G. Kieback, Hein Putter, Christos Markopoulos, Elma Meershoek-Klein Kranenbarg, Elizabeth A. Mallon, Luc Dirix, Caroline Seynaeve, Daniel Rea
REFERENCES
- 1.Tamoxifen for early breast cancer. An overview of the randomised trials—Early Breast Cancer Trialists' Collaborative Group. Lancet. 1998;351:1451–1467. [PubMed] [Google Scholar]
- 2.Berstein LM, Wang JP, Zheng H, et al. Long-term exposure to tamoxifen induces hypersensitivity to estradiol. Clin Cancer Res. 2004;10:1530–1534. doi: 10.1158/1078-0432.ccr-0433-03. [DOI] [PubMed] [Google Scholar]
- 3.Fisher B, Dignam J, Bryant J, et al. Five versus more than five years of tamoxifen for lymph node-negative breast cancer: Updated findings from the National Surgical Adjuvant Breast and Bowel Project B-14 randomized trial. J Natl Cancer Inst. 2001;93:684–690. doi: 10.1093/jnci/93.9.684. [DOI] [PubMed] [Google Scholar]
- 4.Schiff R, Massarweh S, Shou J, et al. Breast cancer endocrine resistance: How growth factor signaling and estrogen receptor coregulators modulate response. Clin Cancer Res. 2003;9:447S–454S. [PubMed] [Google Scholar]
- 5.Witton CJ, Reeves JR, Going JJ, et al. Coexpression of EGFr, HER2, HER3, and HER4 in primary human breast carcinoma. Presented at the 24th Annual San Antonio Breast Cancer Symposium; December 10-13, 2001; San Antonio, TX. abstr 32. [Google Scholar]
- 6.Witton CJ, Reeves JR, Going JJ, et al. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J Pathol. 2003;200:290–297. doi: 10.1002/path.1370. [DOI] [PubMed] [Google Scholar]
- 7.Massarweh S, Osborne CK, Creighton CJ, et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68:826–833. doi: 10.1158/0008-5472.CAN-07-2707. [DOI] [PubMed] [Google Scholar]
- 8.Osborne CK, Coronado-Heinsohn EB, Hilsenbeck SG, et al. Comparison of the effects of a pure steroidal antiestrogen with those of tamoxifen in a model of human breast cancer. J Natl Cancer Inst. 1995;87:746–750. doi: 10.1093/jnci/87.10.746. [DOI] [PubMed] [Google Scholar]
- 9.Kirkegaard T, McGlynn LM, Campbell FM, et al. Amplified in breast cancer 1 in human epidermal growth factor receptor–positive tumors of tamoxifen-treated breast cancer patients. Clin Cancer Res. 2007;13:1405–1411. doi: 10.1158/1078-0432.CCR-06-1933. [DOI] [PubMed] [Google Scholar]
- 10.Tovey S, Dunne B, Witton CJ, et al. Can molecular markers predict when to implement treatment with aromatase inhibitors in invasive breast cancer? Clin Cancer Res. 2005;11:4835–4842. doi: 10.1158/1078-0432.CCR-05-0196. [DOI] [PubMed] [Google Scholar]
- 11.Gross GE, Clark GM, Chamness GC, et al. Multiple progesterone receptor assays in human breast cancer. Cancer Res. 1984;44:836–840. [PubMed] [Google Scholar]
- 12.Johnston SR. Combinations of endocrine and biological agents: Present status of therapeutic and presurgical investigations. Clin Cancer Res. 2005;11:889s–899s. [PubMed] [Google Scholar]
- 13.Miller WR, Bartlett JM, Canney P, et al. Hormonal therapy for postmenopausal breast cancer: The science of sequencing. Breast Cancer Res Treat. 2007;103:149–160. doi: 10.1007/s10549-006-9369-7. [DOI] [PubMed] [Google Scholar]
- 14.Spears M, Kenicer J, Munro AF, et al. Type I receptor tyrosine kinases as predictive or prognostic markers in early breast cancer. Biomark Med. 2008;2:397–407. doi: 10.2217/17520363.2.4.397. [DOI] [PubMed] [Google Scholar]
- 15.Arimidex, Tamoxifen, Alone or in Combination (ATAC) Trialists' Group. Forbes JF, Cuzick J, et al. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 2008;9:45–53. doi: 10.1016/S1470-2045(07)70385-6. [DOI] [PubMed] [Google Scholar]
- 16.Coombes RC, Kilburn LS, Snowdon CF, et al. Survival and safety of exemestane versus tamoxifen after 2-3 years' tamoxifen treatment (Intergroup Exemestane Study): A randomised controlled trial. Lancet. 2007;369:559–570. doi: 10.1016/S0140-6736(07)60200-1. [DOI] [PubMed] [Google Scholar]
- 17.Dowsett M, Cuzick J, Wale C, et al. Retrospective analysis of time to recurrence in the ATAC trial according to hormone receptor status: An hypothesis-generating study. J Clin Oncol. 2005;23:7512–7517. doi: 10.1200/JCO.2005.01.4829. [DOI] [PubMed] [Google Scholar]
- 18.Jones SE, Seynaeve C, Hasenburg A, et al. Results of the first planned analysis of the TEAM (tamoxifen exemestane adjuvant multinational) prospective randomized phase III trial in hormone sensitive postmenopausal early breast cancer. Presented at the 31st Annual San Antonio Breast Cancer Symposium; December 10-14, 2008; San Antonio, TX. abstr 15. [Google Scholar]
- 19.Faratian D, Kay C, Campbell F, et al. Automated image analysis for high-throughput quantitative detection of ER and PgR expression levels in large-scale clinical studies: The TEAM trial experience. Presented at the 30th Annual San Antonio Breast Cancer Symposium; December 13-16, 2007; San Antonio, TX. [Google Scholar]
- 20.Dowsett M, Ebbs SR, Dixon JM, et al. Biomarker changes during neoadjuvant anastrozole, tamoxifen, or the combination: Influence of hormonal status and HER-2 in breast cancer—a study from the IMPACT trialists. J Clin Oncol. 2005;23:2477–2492. doi: 10.1200/JCO.2005.07.559. [DOI] [PubMed] [Google Scholar]
- 21.Collett D. Modelling Survival Data in Medical Research. London, United Kingdom: Chapman & Hall; 1994. [Google Scholar]
- 22.Altman DG, Andersen PK. Calculating the number needed to treat for trials where the outcome is time to an event. BMJ. 1999;319:1492–1495. doi: 10.1136/bmj.319.7223.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowsett M, Allred C, Knox J, et al. Relationship between quantitative estrogen and progesterone receptor expression and human epidermal growth factor receptor 2 (HER-2) status with recurrence in the Arimidex, Tamoxifen, Alone or in Combination trial. J Clin Oncol. 2008;26:1059–1065. doi: 10.1200/JCO.2007.12.9437. [DOI] [PubMed] [Google Scholar]
- 24.Coates AS, Keshaviah A, Thürlimann B, et al. Five years of letrozole compared with tamoxifen as initial adjuvant therapy for postmenopausal women with endocrine-responsive early breast cancer: Update of study BIG 1-98. J Clin Oncol. 2007;25:486–492. doi: 10.1200/JCO.2006.08.8617. [DOI] [PubMed] [Google Scholar]
- 25.Breast International Group (BIG) 1-98 Collaborative Group. Thürlimann B, Keshaviah A, et al. A comparison of letrozole and tamoxifen in postmenopausal women with early breast cancer. N Engl J Med. 2005;353:2747–2757. doi: 10.1056/NEJMoa052258. [DOI] [PubMed] [Google Scholar]
- 26.Viale G, Regan MM, Maiorano E, et al. Prognostic and predictive value of centrally reviewed expression of estrogen and progesterone receptors in a randomized trial comparing letrozole and tamoxifen adjuvant therapy for postmenopausal early breast cancer: BIG 1-98. J Clin Oncol. 2007;25:3846–3852. doi: 10.1200/JCO.2007.11.9453. [DOI] [PubMed] [Google Scholar]
- 27.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 28.Harvey JM, Clark GM, Osborne CK, et al. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 29.Osborne CK, Schiff R. Estrogen-receptor biology: Continuing progress and therapeutic implications. J Clin Oncol. 2005;23:1616–1622. doi: 10.1200/JCO.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 30.Bardou VJ, Arpino G, Elledge RM, et al. Progesterone receptor status significantly improves outcome prediction over estrogen receptor status alone for adjuvant endocrine therapy in two large breast cancer databases. J Clin Oncol. 2003;21:1973–1979. doi: 10.1200/JCO.2003.09.099. [DOI] [PubMed] [Google Scholar]
- 31.McCabe A, Dolled-Filhart M, Camp RL, et al. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst. 2005;97:1808–1815. doi: 10.1093/jnci/dji427. [DOI] [PubMed] [Google Scholar]
- 32.Mohsin SK, Weiss H, Havighurst T, et al. Progesterone receptor by immunohistochemistry and clinical outcome in breast cancer: A validation study. Mod Pathol. 2004;17:1545–1554. doi: 10.1038/modpathol.3800229. [DOI] [PubMed] [Google Scholar]
- 33.Ellis MJ, Tao Y, Luo J, et al. Outcome prediction for clinical stage II and III ER+ breast cancer based on treatment response, pathological stage, tumor grade, Ki67 proliferation index, and estrogen receptor status after neoadjuvant endocrine therapy. Presented at the 30th Annual San Antonio Breast Cancer Symposium; December 13-16, 2007; San Antonio, TX. abstr 62. [Google Scholar]