Abstract
Purpose
Merkel cell carcinoma (MCC) is a polyomavirus-associated skin cancer that is frequently lethal and lacks established prognostic biomarkers. This study sought to identify biomarkers that improve prognostic accuracy and provide insight into MCC biology.
Patients and Methods
Gene expression profiles of 35 MCC tumors were clustered based on prognosis. The cluster of genes overexpressed in good-prognosis tumors was tested for biologic process enrichment. Relevant mRNA expression differences were confirmed by quantitative polymerase chain reaction and immunohistochemistry. An independent set of 146 nonoverlapping MCC tumors (median follow-up, 25 months among 116 living patients) was employed for biomarker validation. Univariate and multivariate Cox regression analyses were performed.
Results
Immune response gene signatures were prominent in patients with good prognoses. In particular, genes associated with cytotoxic CD8+ lymphocytes were overexpressed in tumors from patients with favorable prognoses. In the independent validation set, cases with robust intratumoral CD8+ lymphocyte infiltration had improved outcomes (100% MCC-specific survival, n = 26) compared with instances characterized by sparse infiltration (60% survival, n = 120). Only stage and intratumoral CD8 infiltration (but not age, sex, or CD8+ lymphocytes localized to the tumor-stroma interface) were significant in both univariate and multivariate Cox regression analyses. Notably, traditional histologic identification of tumor-infiltrating lymphocytes was not a significant independent predictor of survival.
Conclusion
Intratumoral CD8+ lymphocyte infiltration can be readily assessed on paraffin-embedded tissue, is independently associated with improved MCC-specific survival, and therefore, may provide prognostic information that enhances established MCC staging protocols.
INTRODUCTION
Merkel cell carcinoma (MCC) is a neuroendocrine skin cancer associated with advanced age, ultraviolet exposure, and immune suppression (approximately 10% of this patient population is chronically immune suppressed).1,2 Recently, MCC has gained attention for two reasons. The first is its rapidly increasing reported incidence (incidence rate in the United States tripled from 0.2 cases per 100,000 in 1986 to 0.6 per 100,000 in 2006).2,3 Second, MCC has been linked to the recently discovered Merkel cell polyomavirus, a finding that has been validated by multiple groups worldwide.4–10
MCC can be aggressive and has a mortality rate more than twice that of melanoma. Indeed, the MCC-attributable mortality is 46% at 5 years.11 Furthermore, little is known about factors associated with MCC pathogenesis and progression.12 The current staging system for MCC is based on two factors: size (largest dimension) of the primary tumor and extent of disease spread at diagnosis.13 Tumor dimension is of limited prognostic value, providing only a 15% relative survival difference at 5 years between patients with small local (≤ 2 cm) and large local (> 2 cm) disease.11 This means that patients in the best prognostic category for MCC (stage Ia, local tumor ≤ 2 cm with pathologically proven negative nodal status) still have a disease-associated mortality of 21% at 5 years. This statistic contrasts with the best prognosis (stage Ia) patients with melanoma who have 4.7% cancer-associated mortality at 5 years.14 Currently, there are no established biomarkers that can improve the prognostic accuracy of the MCC staging system.
We hypothesized that an unbiased mRNA profiling approach comparing MCC tumors from patients with good and poor outcomes might reveal important aspects of MCC pathogenesis and identify new prognostic markers. In this study, we employed transcriptome-wide mRNA profiling followed by gene set enrichment analyses to isolate factors that differentiate patients with MCC with excellent clinical outcomes from those with rapidly progressive disease. In order to determine prognostic utility, array findings were validated by immunohistochemistry on an independent set of 146 clinically annotated MCC tumors.
PATIENTS AND METHODS
Patients and Tumors
Studies were performed in accordance with Helsinki principles and approved by the institutional review board of the Fred Hutchinson Cancer Research Center and the University Clinic of Wuerzburg. Patient materials and clinical information used in this study were obtained from the MCC Tissue and Data Repository. Patients were diagnosed with MCC between 1980 and 2009. MCC diagnosis was confirmed by at least two pathologists. Fresh tumor tissues originated from three centers in Europe, Australia, and the United States. Paraffin blocks from patients enrolled in this repository were obtained from more than 100 distinct pathology laboratories, and were sectioned and stained in a central study–associated laboratory.
Determination of Virus Status
Patients with available DNA (n = 80) were characterized as Merkel cell polyomavirus (MCPyV or MCV) positive or MCPyV negative using real-time polymerase chain reaction (PCR), as published.15 Virus status on some patients has been previously reported.15 The lower limit of detection was approximately 1 copy per 1,000 cells.
mRNA Profiling and Analysis
Fresh MCC tumors from Europe and Australia were flash frozen. MCC tumors from the United States were preserved in RNA-Later (Ambion, Austin, TX). All tumors were macrodissected from surrounding stroma. RNA isolation was performed with RNeasy or Allprep Mini kits (Qiagen, Alameda, CA). RNA was quantified by RiboGreen RNA Quantitation Reagent (Invitrogen, Carlsbad, CA) and its quality assessed by Agilent RNA 6000 Pico Kit (Agilent, Santa Clara, CA) in an Agilent 2100 Bioanalyzer. Samples were amplified and labeled using Ovation WB protocol (NuGEN Technologies, San Carlos, CA). Resulting cDNAs were hybridized to the Human Rosetta Custom Affymetrix 2.0 Chip (Affymetrix, Santa Clara, CA) in a single batch at Rosetta Inpharmatics. Images were analyzed by Affymetrix GeneChip Operating Software and processed further to derive sequence-based intensities by the robust multichip average algorithm. Although 40 tumors were profiled, five tumors failed standard Rosetta quality control guidelines and were excluded from analysis; 35 tumors from 34 patients were included for gene expression analysis. No country-of-origin specific patterns were observed, and samples from different continents readily admixed on two-dimensional unbiased clustering. Expression data has been made publicly available in the GEO database (accession number GSE22396).
Patient Stratification for mRNA Profiling Studies
Because long-term follow-up was not available for all patients and because primary, recurrent, and metastatic lesions were studied, a prognostic stratification was used. Cases were separated into the following clinical/prognostic groups: poor prognosis (MCC presented with or progressed to distant metastasis), moderate prognosis (recurrent local disease, development of nodal metastasis, or nodal disease at presentation with no progression during follow-up of fewer than 24 months), or favorable prognosis (local disease presentation with no subsequent recurrence or nodal disease at presentation with no progression during follow-up of longer than 24 months).
Reverse Transcriptase PCR
Adequate RNA remained after gene expression profiling for subsequent studies on 33 (94%) of 35 samples. Reverse transcription was performed using a random-primed reverse transcription kit (Applied Biosystems, Carlsbad, CA). Taqman real-time PCR was performed on an ABI 7900 HT machine with commercially available assays and cycling conditions recommended by the manufacturer (Applied Biosystems). Transcripts for the following genes were analyzed: CD8a, CXCL10, GZMB, and IFNG, with 18s RNA serving as input control (Applied Biosystems). Relative quantities of RNA were determined using the ΔΔCT method.
CD8 Immunohistochemistry
Among 35 patients in the microarray set, 20 (57%) had available formalin-fixed paraffin-embedded material corresponding to the same lesion. An additional 146 patients with MCC comprised the validation set. In the event a patient had multiple tissues available, data from only one specimen was included according to the following hierarchy for selection: primary (most cases) > nodal metastasis or presenting node > recurrence > skin metastasis > distant metastasis. CD8a immunohistochemistry was performed with antibody 4B11 (Novocastra, Bannockburn, IL) at a 1:200 dilution. Epitope retrieval was heat induced and unmasking performed in pH 8 buffer.
CD8 scoring was performed by an observer who was blinded to patient characteristics, including outcome. Peritumoral and intratumoral CD8+ infiltrates were each semiquantitatively scored on a 0 to 5 scale with 0 representing no CD8 cells and 5 representing a strong CD8 infiltrate (Data Supplement, online only). The scoring system was established before data collection in the validation set, and the 0 to 5 score was used as a continuous variable in Cox regression survival analyses (details below).
To determine intraobserver variability, an independent pathologist who was blinded to both patient outcome and scores from the initial observer scored a random sample of 41 patients. A weighted κ statistic was calculated, with identical scores being weighted as 1, scores 1 bin apart weighted as 0.8, and all others counted as 0. The observed agreement was 85% and κ calculated was 0.65, consistent with substantial agreement between observers.16
Tumor-Infiltrating Lymphocyte Scoring and Staging
Tumor-infiltrating lymphocytes (TILs) were scored on hematoxylin and eosin–stained sections as recommended for MCC,17 by a pathologist who was blinded to CD8+ score and patient outcome.
MCC stage was determined using 2010 American Joint Committee on Cancer criteria.11,13
Statistical Analysis
Gene set enrichment analysis was performed with Resolver software (version 6.0, Rosetta Biosoftware, Seattle, WA). Linear regression was employed to determine correlation between mRNA array and corresponding reverse transcriptase PCR or immunohistochemistry. Disease-specific survival effects were tested with Cox regression and utilized robust SEs. Multivariate models included stage, age older than 65 (yes/no), and sex. Separate models were considered for TILs and CD8 analyses due to concerns of collinearity. CD8 analyses considered intratumoral and peritumoral CD8 scores as continuous variables and did not employ a cut point. Regression analyses were performed with Stata software (StataCorp, College Station, TX). Kaplan-Meier survival curves were generated for data visualization purposes. For these curves, a preselected cut point of intratumoral CD8 score of 3 was employed. This point corresponded to the score of moderate, and split the six bins into two groups of three bins each.
RESULTS
Good-Prognosis Expression Signature Is Enriched for Immune Response Genes
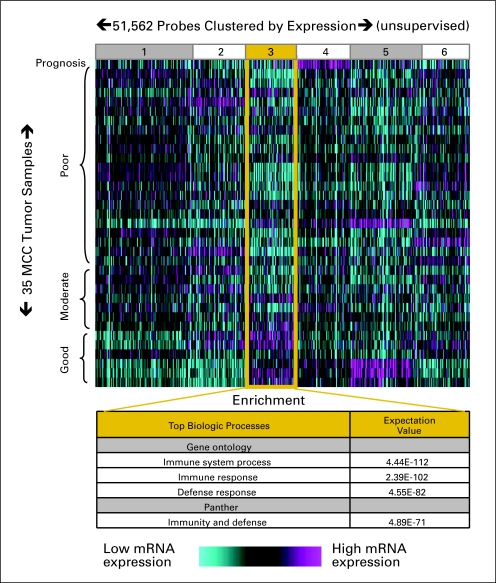
mRNA expression profiles were obtained from 35 MCC tumors, the clinical details of which are summarized in Table 1. Profiles were clustered by prognostic category (poor, moderate, or favorable) as well as by gene expression level (relative to the average among the MCC samples). Clustering was performed in an unbiased manner and 51,562 gene probes were grouped into six bins based on pattern of expression (Fig 1). One cluster of genes (bin 3) contained mRNAs overexpressed in favorable prognosis tumors as compared with moderate and poor prognosis tumors. Both Gene Ontology18 and Panther19,20 gene set enrichment analyses of this bin found that genes involved in the immune response were greatly over-represented (expectation values between 10−71 and 10−112; Fig 1).
Table 1.
Patient and Tumor Characteristics
| Characteristic | mRNA Array(n = 34) |
Validation Set(n = 146) |
||
|---|---|---|---|---|
| No. | % | No. | % | |
| Age at diagnosis, years | ||||
| Mean | 68 | 66 | ||
| Range | 44-90 | 31-92 | ||
| Male sex | 25 | 74 | 96 | 66 |
| Stage at MCC presentation* | ||||
| I | 10 | 29 | 47 | 33 |
| II | 3 | 9 | 31 | 22 |
| III | 18 | 53 | 60 | 42 |
| IV | 3 | 9 | 6 | 4 |
| Lesion type studied* | ||||
| Primary | 9 | 31 | 113 | 80 |
| Regional metastasis/rec | 15 | 52 | 25 | 18 |
| Distant metastasis | 5 | 17 | 5 | 4 |
| MCPyV DNA detectable? | ||||
| Yes | 19 | 70 | 40 | 75 |
| No | 8 | 30 | 13 | 25 |
NOTE. Patients represented in the validation set were entirely nonoverlapping with patients represented in the mRNA array set. Stage information was not available for seven patients; lesion type information was not available for eight patients; and MCPyV DNA status was not available for 100 patients. Patients with nodal presentation and unknown primary are represented in the regional metastasis category (n = 14).
Abbreviations: MCC, Merkel cell carcinoma; rec, recurrence; MCPyV, Merkel cell polyomavirus.
Significant differences (P < .05) between array and validation set.
Fig 1.
Unbiased gene expression analysis reveals association between immune response and Merkel cell carcinoma (MCC) prognosis. Thirty-five tumors were analyzed from 34 patients who had been categorized by prognosis as described in Patients and Methods. Probes were grouped into six bins by the K means algorithm based on expression pattern. Cluster bin 3 displayed the expression pattern of interest (relatively highly expressed in good prognosis patients) and was further investigated through gene set enrichment analysis as indicated.
The 30 genes in this cluster (bin 3) most overexpressed in good- (versus poor-) prognosis patients are listed in Table 2. Prominently represented are genes encoding components of cytotoxic granules (granzymes A, 5.4-fold, B, 6.0-fold, H, 6.3-fold, and K, 5.8-fold), chemokines (CCL19, 4.6-fold), lymphocyte activation genes (SLAMF1, 6.4-fold and NKG2D, 6.2-fold),21,22 and α chain of the CD8 receptor (CD8a, 5.0-fold). Affymetrix array data reproducibility was assessed by quantitative reverse transcription PCR for four immune response genes (CD8a, CXCL10, GZMB, IFNG). For all four genes, reverse transcription quantitative PCR findings correlated closely with the mRNA array data (R2 values: 0.48 to 0.81).
Table 2.
Genes Most Highly Upregulated in Good Prognosis Tumors
| Gene Abbreviation | Gene's Full Name |
|---|---|
| ALDH1A1 | Aldehyde dehydrogenase 1 family, member A1 |
| AMICA1 | Adhesion molecule, interacts with CXADR antigen 1 |
| BHLHE41 | Basic helix-loop-helix family, member e41 |
| CCL19 | Chemokine (C-C motif) ligand 19 |
| CCR2 | Chemokine (C-C motif) receptor 2 |
| CD8a | CD8a molecule |
| CGA | Glycoprotein hormones, alpha polypeptide |
| CHI3L1 | Chitinase 3-like 1 |
| CHIT1 | Chitinase 1 |
| CHRNA9 | Cholinergic receptor, nicotinic, alpha 9 |
| FAM46C | Family with sequence similarity 46, member C |
| FBP1 | Fructose-1,6-bisphosphatase 1 |
| GZMA | Granzyme A |
| GZMB | Granzyme B |
| GZMH | Granzyme H |
| GZMK | Granzyme K |
| HLA-DPA1 | Major histocompatibility complex, class II, DP alpha 1 |
| HLA-DRB5 | Major histocompatibility complex, class II, DR beta 5 |
| IGJ | Immunoglobulin J polypeptide |
| IGKC | Immunoglobulin kappa constant |
| ITGBL1 | Integrin, beta-like 1 |
| KLRK1 | Killer cell lectin-like receptor subfamily K, member 1 (NKG2D) |
| LYZ | Lysozyme |
| MMP7 | Matrix metallopeptidase 7 |
| POU2AF1 | POU class 2 associating factor 1 |
| PROM1 | Prominin 1 |
| SLAMF1 | Signaling lymphocytic activation molecule family member 1 |
| TMEM200A | Transmembrane protein 200A |
| TNFRSF17 | Tumor necrosis factor receptor superfamily, member 17 |
| TRBC1 | T cell receptor beta constant 1 |
NOTE. The 30 genes in cluster bin 3 (Fig 1) most highly upregulated in good prognosis patients as compared with poor prognosis patients are listed in alphabetical order. Fold overexpression ranged from five- to 13-fold.
Overexpression of RNAs for four granzymes and CD8a (Table 2) suggested that CD8+ lymphocytes contribute to the signature associated with good prognosis. Consistent with this hypothesis, known lymphocyte attractant chemokines (CCL5, 4.1-fold),23 cytokines produced by T-cell activation (IFNG, 4.2-fold), and T-cell receptor– associated genes (CD3E, 4.0-fold, CD8B, 4.2-fold) were also represented in the favorable prognosis signature with increased expression in good prognosis tumors.
Immunohistochemical Corroboration of CD8+ Lymphocyte mRNA Signature
Archival formalin-fixed, paraffin-embedded materials were available for 20 of 35 specimens represented on the array. The CD8+ lymphocyte infiltrate was scored on a 0 to 5 scale (0 absent to 5 strong) both in the tumor center (intratumoral) and at the tumor periphery (peritumoral). Expression of CD8a on the mRNA array was correlated with the combined peritumoral and intratumoral score for CD8 infiltration (R2 = 0.69; Fig 2).
Fig 2.
CD8+ lymphocytic infiltration correlates with mRNA expression of CD8a. (A) Top row: hematoxylin and eosin (H/E) –stained sections; arrows indicate tumor-infiltrating lymphocytes. Bottom row: immunohistochemistry (IHC) for CD8 on corresponding serial sections. Peritumoral and intratumoral CD8+ lymphocytic infiltrates were each scored on a 0 to 5 scale (Data Supplement). (B) Correlation between CD8a mRNA expression and immunohistochemistry. Twenty samples had available archival materials.
Characteristics of 146 Additional Patients With MCC (validation set)
To test whether CD8 infiltrate is a useful prognostic marker in MCC, we employed a validation set of 146 patients with MCC whose clinical details are summarized in Table 1. These patients were entirely nonoverlapping with the mRNA array group. Patients were annotated using disease-specific survival information with a median follow-up of 25 months among living patients (n = 116). The validation and array sets differed in terms of tumor lesion studied (validation set was mostly primary lesions) and stage at presentation (validation set patients tended to present at earlier stage).
Validation set patients were similar to national registry data11 in terms of stage at presentation (stage I, II, III, IV: 33%, 22%, 42%, 4%, respectively, in validation v 36%, 22%, 33%, 10% in registry [99% or 101% due to rounding]) and sex (66% male v 61% in registry). However, validation patients were younger than the national average (median 66 years in validation set v 76 years in registry).
Strong Intratumoral CD8+ Infiltration Observed in 18% of MCC Tumors
Intratumoral and peritumoral CD8 infiltrates were scored separately and semiquantitatively (see Patients and Methods). Of 146 patients with MCC, 39% had no CD8 intratumoral infiltrate, 33% had a score of 1, 10% a score of 2, 12% a score of 3, 3% a score of 4, and 2% received a maximum score of 5 (99% due to rounding). Among the 72% of patients with no or very low CD8 intratumoral infiltrate (intratumoral score of 0 or 1, n = 105), 46% exhibited a prominent stalling phenomenon with high numbers of peritumoral CD8+ cells localized along the tumor-stroma border (peritumoral CD8 scores of 3 to 5).
Consistent with other published studies,4–6,24 MCPyV was detectable by real-time PCR in 75% of validation set tumors with available DNA (n = 53). No relationship was observed between intratumoral CD8 infiltration and virus status (data not shown).
TILs, As Assessed by Standard Histology, Are Not an Independent Prognostic Factor in MCC
Corresponding hematoxylin and eosin slides from 129 of 146 cases were scored for TILs; the remaining 17 could not be assessed due to crush artifact or difficulty distinguishing pyknotic nuclei from lymphocytes. 44 tumors (34%) lacked TILs, 50 tumors (39%) had an infiltrate that was not brisk, and 35 tumors (27%) had a brisk infiltrate.
Consistent with prior reports,25 TILs were significant on univariate but not multivariate analyses. On univariate analysis, the hazard ratio (HR) associated with the presence of TILs was 0.4 (P = .03; 95% CI, 0.2 to 0.9). MCC-specific survival among patients with TIL-positive tumors was modestly improved at 5 years (70% v 55%; Fig 3A). However, in a multivariate Cox model considering stage, age at diagnosis, sex, and TILs, only stage was significant.
Fig 3.
T-cell infiltration and Merkel cell carcinoma (MCC)–specific survival in an independent set of 146 patients. (A) Tumor infiltrating lymphocytes (TILs) analysis by routine histology among 129 patients. (*) TILs were prognostically significant on univariate (P = .03) but not multivariate (P = .12) analysis. (B) Intratumoral (IT) CD8+ lymphocyte infiltration. Brisk CD8s were defined as an intratumoral CD8 score of 3 to 5 (corresponding to approximately 60 or more CD8s per typical 40× high power field), sparse as 0 to 2. (†) IT CD8 infiltration was a statistically significant predictor of outcome on univariate (P < .01) and multivariate (P = .01) regression analyses (Table 3). (C) Subgroup breakdown of (B), by extent of disease at presentation (as indicated). Extent of disease at presentation was not known for two patients. Statistical analysis was not performed on subgroups; instead, multivariate Cox regression is listed in Table 3.
Intratumoral CD8+ Lymphocyte Infiltration Independently Predicts MCC-Specific Survival
In contrast to standard histologic TILs, intratumoral CD8+ lymphocyte infiltration was a statistically significant predictor of outcome on both univariate and multivariate analyses (Table 3). The HR associated with each one point increase on the 0 to 5 intratumoral CD8 infiltration scale was 0.5 and was statistically significant (95% CI, 0.5 to 0.7, P < .01). The 26 patients with MCC (18%) with intratumoral CD8 infiltrate scores of 3 to 5 on the scale (greater than approximately 60 CD8+ cells per typical high powered field) had 100% disease-specific survival at 5 years after diagnosis. This compared with 60% survival among the remaining 120 patients with intratumoral CD8 scores of 0 to 2 (Fig 3B). Furthermore, in both multivariate Cox regression and subgroup Kaplan-Meier analyses, CD8+ infiltrate distinguished outcomes among patients with MCC of the same stage (Table 3 and Fig 3C). Other factors predictive of survival on univariate and multivariate analysis included stage III disease (regional) versus stage I (local, ≤ 2 cm), and stage IV disease (metastatic) versus stage I. Lacking prognostic significance in this cohort were sex, age at diagnosis, and stage II disease (local > 2 cm, versus stage I). Additionally, the degree of peritumoral CD8 infiltration was also not significantly associated with outcome (Table 3).
Table 3.
Multivariate Cox Regression Analysis Demonstrates Intratumoral CD8 Score Is an Independent Predictor of Merkel Cell Carcinoma Outcome
| Characteristic | Univariate |
Multivariate |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Stage | ||||||
| II v I | 0.8 | 0.1 to 5.0 | .86 | 1.1 | 0.2 to 6.6 | .92 |
| III v I | 6.5 | 1.9 to 22.6 | < .01 | 5.5 | 1.4 to 21.2 | .02 |
| IV v I | 18.8 | 3.4 to 104.5 | < .01 | 31.5 | 6.8 to 147.0 | < .01 |
| Female sex | 0.4 | 0.1 to 1.0 | .06 | 0.6 | 0.2 to 1.7 | .31 |
| Age at diagnosis ≥ 65 years | 1.1 | 0.5 to 2.3 | .87 | 0.8 | 0.4 to 1.8 | .62 |
| CD8, per increase on 0 to 5 scale | ||||||
| Peritumoral | 0.8 | 0.6 to 1.0 | .06 | 0.9 | 0.6 to 1.4 | .79 |
| Intratumoral | 0.5 | 0.5 to 0.7 | < .01 | 0.5 | 0.3 to 0.9 | .01 |
NOTE. CD8+ scoring scale is described in Methods and in the scoring guide provided as a Data Supplement. All variables listed in this table were included in the multivariate analysis.
Abbreviation: HR, hazard ratio.
DISCUSSION
The current staging for the aggressive skin cancer MCC relies solely on disease size and extent of spread, with no biomarkers recommended for collection.11,13 To develop new prognostic markers, as well as gain insight into MCC biology, we undertook an unbiased expression profiling approach. The set of genes highly expressed in good-prognosis tumors was enriched for immune response genes, particularly those expressed by CD8+ lymphocytes. In an independent set of 146 tumors, intratumoral but not peritumoral CD8+ infiltration was independently predictive of MCC-specific survival.
The association of immune response genes and prognosis is concordant with recent studies in other cancers such as melanoma26 and colon cancer.27 Furthermore, this observation is consistent with the clinical association between cellular immune suppression and MCC: persons with any of several forms of T-cell immune suppression are at greater than 10-fold increased risk of MCC.1,28,29
Overexpression of CD8β and CD3E in tumors from patients with good prognoses suggests that CD8+ T cells (rather than natural killer [NK] cells) are the major infiltrating cell type. Direct assessment of NK infiltration was not feasible because greater than 90% of MCC tumors express CD56,30,31 the best immunohistochemical marker for NK cells. Interestingly, a CD8+ T lymphocyte cellular defense is critical for eliminating mouse tumors persistently expressing the SV40 polyomavirus T-antigen oncoprotein32 (this model is relevant to MCC biology since most MCC tumors persistently express MCPyV oncoproteins).33
In the validation set, patients presenting with local-only disease tended to have brisk intratumoral CD8+ lymphocytes more often than those presenting with regional disease (23% v 10%, not significant). However, CD8+ infiltration did not merely track with stage, but also added additional prognostic information to both of these subgroups and was significant on multivariate analyses that included the current MCC stages.13
In contrast, TILs as assessed by routine histology did not independently predict disease-specific survival in two studies of more than 100 patients with MCC25 (and this study). At least two factors may be relevant in explaining these differences. The first relates to the current definition of TILs for both MCC and melanoma, which includes both peritumoral and intratumoral lymphocytes.17 Indeed, the findings in this study suggest that intratumoral lymphocytes are relevant for MCC outcome, whereas peritumoral lymphocytes are not. This is also true for other cancers, such as ovarian cancer and colon cancer.27,34 Second, immunohistochemical CD8+ evaluation may be more sensitive and specific for identification of TILs than routine histology. This is because T cells can sometimes be indistinguishable from MCC tumor cells using hematoxylin and eosin staining.
This study has several limitations despite the fact that it is both the largest molecular and immunohistochemical examination of MCC yet reported, to our knowledge. The median age of the patient population (66 years) was younger than that for MCC nationally (76 years).3,11 This may in part reflect the fact that patients in this cohort were ascertained because they sought specialty care or information/research participation via the Internet. In these regards, this population is not fully representative of the overall population of patients with MCC; however, it is not clear how these biases would affect the observed survival benefit of CD8 infiltration. A second limitation is that our study was designed and powered to address whether CD8 infiltration adds prognostic information to a four-category staging system rather than to the new staging system with eight substages.11,13 These new substages include microscopic versus clinical-only evaluation of nodal status. Although a larger prospective follow-up study will be required to resolve this issue, a subset analysis of validation set patients with sufficient data (n = 115) suggests that CD8 infiltration is a significant predictor of outcome even when substage is considered (Appendix Table A1, online only).
These results demonstrate a strong association between MCC prognosis and the extent of intratumoral CD8+ infiltration, a readily assessed biomarker that provides useful additional survival information beyond the newly adopted MCC staging system.11,13 Future studies may extend these observations by further characterizing the effector lymphocyte response against MCC and investigating rational immunotherapy for this cancer.
Acknowledgment
We thank Betsy Williams and Sue Montgomery for excellent administrative support; Brenda Kurland for statistical advice; and James S. Hardwick for his extensive support relating to the microarray experiments.
Appendix
Table A1.
Multivariable Cox Regression Model Considering the Subset of Patients With Available Substaging Information (n = 115)
| Variable | Hazard Ratio | 95% CI | P |
|---|---|---|---|
| Stage | |||
| IB v IA | 16.0 | 1.0 to 247.3 | .05 |
| IIA v IA | Not calculable | ||
| IIB v IA | 24.0 | 2.1 to 273.2 | .01 |
| IIIA v IA | Not calculable | ||
| IIIB v IA | 22.8 | 2.5 to 203.2 | < .01 |
| IV v IA | 166.9 | 14.2 to 1965.9 | < .01 |
| Intratumoral CD8 score per increase on 0 to 5 scale | 0.4 | 0.2 to 0.7 | < .01 |
| Female sex v male | 0.8 | 0.3 to 2.0 | .60 |
| Age at diagnosis > 65 years | 0.2 | 0.1 to 0.7 | < .01 |
NOTE. Substaging as per 2010 American Joint Commission on Cancer staging manual. All listed variables were considered in the model. Hazard ratios for stage IIA and IIIA were not calculable because there were no events among the patients in these substages. There were no patients studied with stage 0 (in situ) or stage IIC (in transit nodal metastases) disease.
Footnotes
Supported by American Cancer Society Grant No. RSG-08-115-01-CCE, National Institutes of Health (NIH) Grants No. RC2CA147820 and NIH K24 CA139052-0 (P.N.), and NIH T32 CA80416-10 and F30ES017385 (K.P.), ULRR025014 to UW CTSA, TL1RR025016 (A.T.), Deutsche Forschungsgemeinschaft KFO124 (J.C.B., D.S.), and the MCC Patient Gift Fund at the University of Washington. Transcriptional profiling was generously provided by Rosetta Inpharmatics/Merck.
Presented in part in preliminary form at the Molecular Biology of Immune Escape Keystone Symposium, February 7-12, 2010, Keystone, CO, and at the Society for Investigative Dermatology Meeting, May 5-8, 2010, Atlanta, GA.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: Michele A. Cleary, Merck & Co (C) Consultant or Advisory Role: None Stock Ownership: None Honoraria: None Research Funding: None Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Kelly G. Paulson, Juergen C. Becker, Michele A. Cleary, Paul Nghiem
Financial support: Kelly G. Paulson, Michele A. Cleary, Paul Nghiem
Administrative support: Paul Nghiem
Provision of study materials or patients: Jayasri G. Iyer, Renee Thibodeau, Shinichi Koba, David Schrama, Bianca D. Lemos, David R. Byrd, J. Helen Leonard, Juergen C. Becker, Paul Nghiem
Collection and assembly of data: Kelly G. Paulson, Jayasri G. Iyer, Andrew R. Tegeder, Janell Schelter, William T. Simonson, Zsolt B. Argenyi, Juergen C. Becker, Michele A. Cleary, Paul Nghiem
Data analysis and interpretation: Kelly G. Paulson, Jayasri G. Iyer, Andrew R. Tegeder, Janell Schelter, David M. Koelle, Denise A. Galloway, Margaret M. Madeleine, Zsolt B. Argenyi, Mary L. Disis, Juergen C. Becker, Michele A. Cleary, Paul Nghiem
Manuscript writing: Kelly G. Paulson, Jayasri G. Iyer, Andrew R. Tegeder, Renee Thibodeau, Janell Schelter, Shinichi Koba, David Schrama, William T. Simonson, Bianca D. Lemos, David R. Byrd, David M. Koelle, Denise A. Galloway, J. Helen Leonard, Margaret M. Madeleine, Zsolt B. Argenyi, Mary L. Disis, Juergen C. Becker, Michele A. Cleary, Paul Nghiem
Final approval of manuscript: Kelly G. Paulson, Jayasri G. Iyer, Andrew R. Tegeder, Renee Thibodeau, Janell Schelter, Shinichi Koba, David Schrama, William T. Simonson, Bianca D. Lemos, David R. Byrd, David M. Koelle, Denise A. Galloway, J. Helen Leonard, Margaret M. Madeleine, Zsolt B. Argenyi, Mary L. Disis, Juergen C. Becker, Michele A. Cleary, Paul Nghiem
REFERENCES
- 1.Heath M, Jaimes N, Lemos B, et al. Clinical characteristics of Merkel cell carcinoma at diagnosis in 195 patients: The AEIOU features. J Am Acad Dermatol. 2008;58:375–381. doi: 10.1016/j.jaad.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hussain SK, Sundquist J, Hemminki K. Incidence trends of squamous cell and rare skin cancers in the Swedish National Cancer Registry point to calendar year and age-dependent increases. J Invest Dermatol. 2010;130:1323–1328. doi: 10.1038/jid.2009.426. [DOI] [PubMed] [Google Scholar]
- 3.Albores-Saavedra J, Batich K, Chable-Montero F, et al. Merkel cell carcinoma demographics, morphology, and survival based on 3870 cases: A population based study. J Cutan Pathol. doi: 10.1111/j.1600-0560.2009.01370.x. [epub ahead of print on July 21, 2009] [DOI] [PubMed] [Google Scholar]
- 4.Feng H, Shuda M, Chang Y, et al. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker JC, Houben R, Ugurel S, et al. MC polyomavirus is frequently present in Merkel cell carcinoma of European patients. J Invest Dermatol. 2009;129:248–250. doi: 10.1038/jid.2008.198. [DOI] [PubMed] [Google Scholar]
- 6.Kassem A, Schopflin A, Diaz C, et al. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res. 2008;68:5009–5013. doi: 10.1158/0008-5472.CAN-08-0949. [DOI] [PubMed] [Google Scholar]
- 7.Katano H, Ito H, Suzuki Y, et al. Detection of Merkel cell polyomavirus in Merkel cell carcinoma and Kaposi's sarcoma. J Med Virol. 2009;81:1951–1958. doi: 10.1002/jmv.21608. [DOI] [PubMed] [Google Scholar]
- 8.Houben R, Shuda M, Weinkam R, et al. Merkel cell polyomavirus infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010;84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter JJ, Paulson KG, Wipf GC, et al. Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma. J Natl Cancer Inst. 2009;101:1510–1522. doi: 10.1093/jnci/djp332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busam KJ, Jungbluth AA, Rekthman N, et al. Merkel cell polyomavirus expression in merkel cell carcinomas and its absence in combined tumors and pulmonary neuroendocrine carcinomas. Am J Surg Pathol. 2009;33:1378–1385. doi: 10.1097/PAS.0b013e3181aa30a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemos BD, Storer B, Iyer JG, et al. Pathologic nodal evaluation improves prognostic accuracy in Merkel cell carcinoma: Analysis of 5,823 cases as the basis of the first consensus staging system for this cancer. J Am Acad Dermatol. 2010;63:751–761. doi: 10.1016/j.jaad.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemos B, Nghiem P. Merkel cell carcinoma: More deaths but still no pathway to blame. J Invest Dermatol. 2007;127:2100–2103. doi: 10.1038/sj.jid.5700925. [DOI] [PubMed] [Google Scholar]
- 13.Edge SE, Byrd DR, Carducci MA, et al., editors. AJCC Cancer Staging Manual (ed) New York, NY: Springer; 2010. Merkel cell carcinoma; pp. 315–324. [Google Scholar]
- 14.Balch CM, Buzaid AC, Soong SJ, et al. Final version of the American Joint Committee on Cancer staging system for cutaneous melanoma. J Clin Oncol. 2001;19:3635–3648. doi: 10.1200/JCO.2001.19.16.3635. [DOI] [PubMed] [Google Scholar]
- 15.Garneski KM, Warcola AH, Feng Q, et al. Merkel cell polyomavirus is more frequently present in North American than Australian Merkel cell carcinoma tumors. J Invest Dermatol. 2009;129:246–248. doi: 10.1038/jid.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 17.Rao P, Balzer BL, Lemos BD, et al. Protocol for the examination of specimens from patients with merkel cell carcinoma of the skin. Arch Pathol Lab Med. 2010;134:341–344. doi: 10.5858/134.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: Tool for the unification of biology: The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas PD, Kejariwal A, Campbell MJ, et al. PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas PD, Kejariwal A, Guo N, et al. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cocks BG, Chang CC, Carballido JM, et al. A novel receptor involved in T-cell activation. Nature. 1995;376:260–263. doi: 10.1038/376260a0. [DOI] [PubMed] [Google Scholar]
- 22.Groh V, Rhinehart R, Randolph-Habecker J, et al. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol. 2001;2:255–260. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- 23.Lapteva N, Huang XF. CCL5 as an adjuvant for cancer immunotherapy. Expert Opin Biol Ther. 2010;10:723–733. doi: 10.1517/14712591003657128. [DOI] [PubMed] [Google Scholar]
- 24.Bhatia K, Goedert JJ, Modali R, et al. Merkel cell carcinoma subgroups by Merkel cell polyomavirus DNA relative abundance and oncogene expression. Int J Cancer. 2010;126:2240–2246. doi: 10.1002/ijc.24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andea AA, Coit DG, Amin B, et al. Merkel cell carcinoma: Histologic features and prognosis. Cancer. 2008;113:2549–2558. doi: 10.1002/cncr.23874. [DOI] [PubMed] [Google Scholar]
- 26.Harlin H, Meng Y, Peterson AC, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69:3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 28.Engels EA, Frisch M, Goedert JJ, et al. Merkel cell carcinoma and HIV infection. Lancet. 2002;359:497–498. doi: 10.1016/S0140-6736(02)07668-7. [DOI] [PubMed] [Google Scholar]
- 29.Koljonen V, Kukko H, Pukkala E, et al. Chronic lymphocytic leukaemia patients have a high risk of Merkel-cell polyomavirus DNA-positive Merkel-cell carcinoma. Br J Cancer. 2009;101:1444–1447. doi: 10.1038/sj.bjc.6605306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurokawa M, Nabeshima K, Akiyama Y, et al. CD56: a useful marker for diagnosing Merkel cell carcinoma. J Dermatol Sci. 2003;31:219–224. doi: 10.1016/s0923-1811(03)00029-x. [DOI] [PubMed] [Google Scholar]
- 31.McNiff JM, Cowper SE, Lazova R, et al. CD56 staining in Merkel cell carcinoma and natural killer-cell lymphoma: Magic bullet, diagnostic pitfall, or both? J Cutan Pathol. 2005;32:541–545. doi: 10.1111/j.0303-6987.2005.00378.x. [DOI] [PubMed] [Google Scholar]
- 32.Lowe DB, Shearer MH, Jumper CA, et al. Tumor immunity against a simian virus 40 oncoprotein requires CD8+ T lymphocytes in the effector immune phase. J Virol. 2010;84:883–893. doi: 10.1128/JVI.01512-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shuda M, Arora R, Kwun HJ, et al. Human Merkel cell polyomavirus infection I: MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int J Cancer. 2009;125:1243–1249. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Conejo-Garcia JR, Katsaros D, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]