Abstract
Purpose
Medulloblastomas are heterogeneous tumors that collectively represent the most common malignant brain tumor in children. To understand the molecular characteristics underlying their heterogeneity and to identify whether such characteristics represent risk factors for patients with this disease, we performed an integrated genomic analysis of a large series of primary tumors.
Patients and Methods
We profiled the mRNA transcriptome of 194 medulloblastomas and performed high-density single nucleotide polymorphism array and miRNA analysis on 115 and 98 of these, respectively. Non-negative matrix factorization–based clustering of mRNA expression data was used to identify molecular subgroups of medulloblastoma; DNA copy number, miRNA profiles, and clinical outcomes were analyzed for each. We additionally validated our findings in three previously published independent medulloblastoma data sets.
Results
Identified are six molecular subgroups of medulloblastoma, each with a unique combination of numerical and structural chromosomal aberrations that globally influence mRNA and miRNA expression. We reveal the relative contribution of each subgroup to clinical outcome as a whole and show that a previously unidentified molecular subgroup, characterized genetically by c-MYC copy number gains and transcriptionally by enrichment of photoreceptor pathways and increased miR-183∼96∼182 expression, is associated with significantly lower rates of event-free and overall survivals.
Conclusion
Our results detail the complex genomic heterogeneity of medulloblastomas and identify a previously unrecognized molecular subgroup with poor clinical outcome for which more effective therapeutic strategies should be developed.
INTRODUCTION
Advancements in genome scale technologies have led to an evolving molecular classification of me dulloblastomas. Previous gene expression studies have suggested up to five molecular subtypes of this disease,1–4 which include a subgroup with Sonic Hedgehog (SHH) pathway activation1,5; another characterized by β-catenin mutations and monosomy chromosome 64,6,7; and others expressing neuronal differentiation markers, photoreceptor transcriptional markers, or both.3
Numerous genetic alterations have also been reported but have not been extensively detailed for the various molecular subgroups of medulloblastoma.3,8–10 Similarly, miRNA studies have revealed their differential expression in medulloblastomas but have largely focused on the SHH pathway–activated subgroup without appreciating the heterogeneity reported at the mRNA level.11–14
Here, we present a genomic classification of medulloblastoma that is based on an analysis of 194 tumor samples. We identify distinct molecular subgroups of medulloblastoma with unique combinations of copy number alterations that globally influence mRNA and miRNA expression. We correlate clinical behavior of medulloblastomas in the context of these molecular subgroups and reveal a newly identified molecular subgroup with a particularly aggressive course. We validate our findings in three independently published data sets and, in a separate study, integrate our knowledge of molecular subtypes into a novel outcome prediction model.14b
PATIENTS AND METHODS
Biologic Samples
Tumors were serially accrued from Children's Oncology Group (COG) (ACNS02B3; n = 89; 2004 to 2007), Children's Hospital Boston (n = 55; 1996 to 2007), University of Washington (n = 27; 2001 to 2005), Texas Children's Hospital (n = 24; 2000 to 2004), and Johns Hopkins Medical Center (n = 10; 2000 to 2001). Normal cerebellum (n = 11) was obtained from University of Washington and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Brain and Tissue Bank for Developmental Disorders, University of Maryland, Baltimore, MD. All samples were collected with approval from respective institutional review boards, and informed consent and/or assent was obtained from all patients or their parents.
Central histopathologic review was performed on patients from Children's Hospital Boston and COG. For samples obtained from University of Washington, Johns Hopkins, and Texas Children's, hematoxylin and eosin was not available; therefore, histopathology reports provided by the contributing institutions were reviewed.
Gene Expression, SNP Array, and miRNA Data
DNA and total RNAs were extracted as described in the Appendix (online online). Gene expression data were generated by using Affymetrix_HT-HG-U133A chips (Affymetrix, Santa Clara, CA). Microarray data from Kool et al,3 Fattet et al,7 and Thompson et al4 were obtained from the Gene Expression Omnibus (GSE10327 and GSE12992) and from http://www.stjuderesearch.org/data/medulloblastoma/, respectively. Copy number data were generated from Affymetrix_250K_Sty and Affymetrix_6.0 arrays (Affymetrix). miRNA profiling was performed as previously described.15,16
Computational Methods
Detailed methods are provided in the Appendix (online only). Briefly, non-negative matrix factorization (NMF)2; silhouette width analysis17,18; gene set enrichment analysis (GSEA)19,20; genomic identification of significant targets in cancer (GISTIC) analysis21,22; and metagene projection23 were performed as previously described.
Immunohistochemistry and Fluorescent In Situ Hybridization
Anti-CRX and GRM8 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and Sigma-Aldrich (St Louis, MO), respectively. Probes for c-MYC and MYCN were obtained from Abbott Molecular. (Abbott Park, IL). GLI2 BAC clones were obtained from CHORI (http://www.chori.org). Immunohistochemistry and fluorescent in situ hybridization (FISH) were performed on formalin-fixed, paraffin-embedded samples as previously described24,25 (Appendix, online only).
Survival Analysis
Kaplan-Meier curves and log-rank statistics were generated by using GraphPad Prism (GraphPad Software, La Jolla, CA). Clinical annotations are provided in the Appendix (online only).
RESULTS
Unsupervised Clustering of mRNA Expression Data Identifies Six Medulloblastoma Subgroups
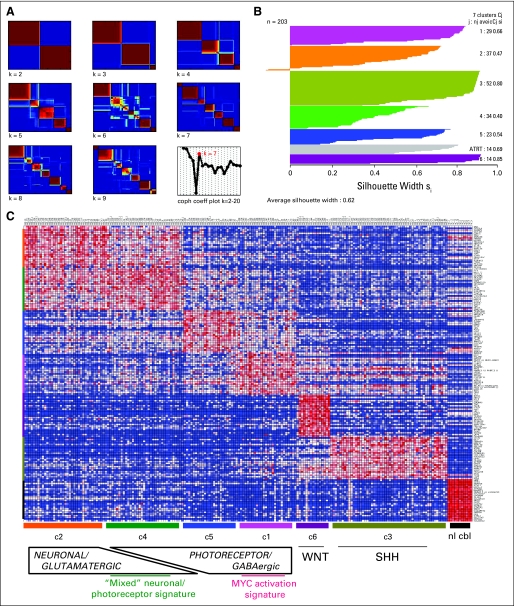
To identify molecular subtypes of medulloblastoma, we utilized an unsupervised clustering algorithm that was based on NMF.2 NMF identifies metagenes, or aggregate patterns of gene expression, which are then used to determine the most stable clustering by calculating a cophenetic coefficient for each number of clusters. When applied to our gene expression microarray data consisting of 194 primary medulloblastomas and, for comparison, nine atypical teratoid/rhabdoid tumors (ATRTs), the optimal number of clusters in our data set is k = 7. These represent six medulloblastoma subgroups (designated c1 through c6) plus one ATRT subgroup (Fig 1A).
Fig 1.
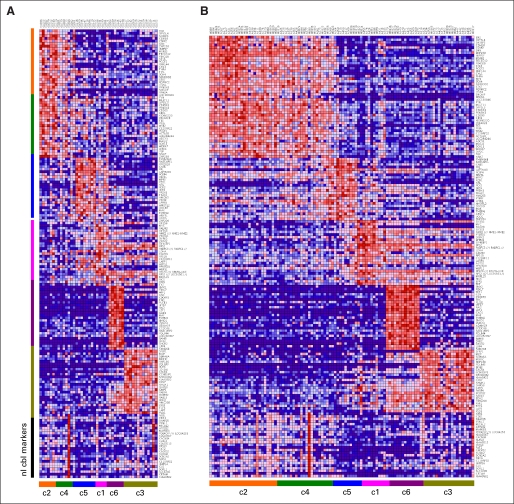
(A) Non-negative matrix factorization consensus clustering of mRNA expression array data from 194 primary medulloblastomas and nine atypical teratoid/rhabdoid tumors (ATRTs) reveals seven (k = 7) stable subgroups (six medulloblastoma subgroups, c1 through c6, plus an additional ATRT subgroup). (B) Silhouette widths indicate a strong similarity of samples to others within their subgroup relative to samples from other subgroups. (C) Heat map of the top 25 upregulated genes for each subgroup shows significant overlap between the c2 and c4 subgroups, which are characterized by enrichment of neuronal/glutamatergic signatures, and overlap to a lesser extent between c1 and c5 subgroups, which both have enrichment of photoreceptor transcriptional signatures; normal cerebellum samples are included to assess for the degree of stromal contamination. coph coeff, cophenetic coefficient; WNT, Wingless signaling pathway; SHH, Sonic Hedgehog signaling pathway.
Silhouette width analysis17 was performed to assess the similarity of each sample to its assigned subgroup compared with samples from other subgroups (Fig 1B). The average silhouette width for NMF subgroups was 0.62 (range, 0.4 to 0.85), suggesting samples within each subgroup are highly coherent (Fig 1B). The c2 and c4 subgroups displayed the lowest overall silhouette scores; at the gene expression level, significant overlap was seen between these subgroups (Fig 1C).
Functional Annotation of Medulloblastoma Subgroups
To identify biologic pathways associated with each NMF subgroup, we performed gene set enrichment analysis (GSEA)19 with 2,163 annotated pathways. The c1 subgroup shows marked enrichment of MYC and related translational/ribosomal signatures. Also enriched are photoreceptor transcriptional programs, and expression of GABRA5, features prominent in c5 as well (Data Supplement). A pairwise GSEA between c1 and c5, however, substantiates the marked upregulation of MYC-related signatures as specific to c1 (Data Supplement).
c2 and c4 are characterized by neuronal differentiation markers, including increased expression of the metabotropic glutamatergic receptors GRM1 and GRM8 (Data Supplement). Despite considerable overlap between these subgroups, a pairwise GSEA revealed enrichment of gene sets related to tumor necrosis factor (TNF) -α/interferon(IFN) -β/nuclear factor-κB in c2, whereas signatures related to both MYC and neuronal development were enriched in c4 (Data Supplement).
Although c4 tumors predominantly express neuronal/glutamatergic signatures, they also express GABRA5 and the photoreceptor transcriptional program to some degree (Data Supplement). To investigate whether concomitant expression of both signatures represented distinct subclonal populations within the tumor, we performed immunostaining for CRX and GRM8, surrogate markers for the photoreceptor/GABAergic and neuronal/glutamatergic signatures, respectively. We identify nests of CRX-immunopositive cells that were distinct from GRM8-immunopositive cells, confirming the presence of subpopulations within c4 tumors and explaining their mixed gene expression signature (Data Supplement).
The c3 subgroup shows enrichment of gene sets associated with SHH signaling, as observed previously.1 c6 was enriched with TGF-β, WNT/β-catenin, and epithelial mesenchymal transition (EMT) gene sets, consistent with previous reports3,7 (Data Supplement).
Unique Combinations of Copy Number Alterations Define Each Molecular Subgroup
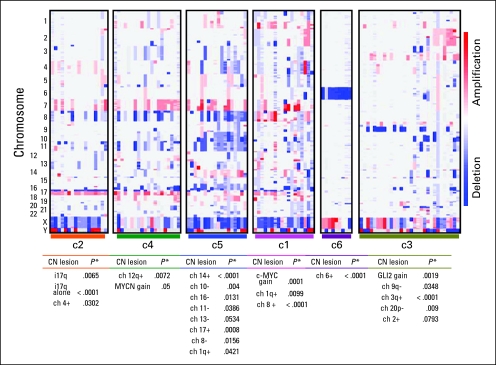
GISTIC analysis on our full cohort identified several genetic lesions that have been previously reported (Data Supplement).8,10,21,26 To assess subgroup-specific copy number alterations and to evaluate their intersubgroup significance, we performed GISTIC analysis on NMF subgroups individually (Figs 2A and 2B; Data Supplement). As previously reported,4,7,27 a strong correlation of monosomy chromosome 6 with the c6/WNT subgroup was observed (nine of nine; P < .001; Figs 2A and 2B).
Fig 2.
Copy number (CN) profiles of samples arranged by non-negative matrix factorization subgroup reveals statistically significant CN alterations associated with each. Detailed files of significant CN lesions within each subgroup, as determined by genomic identification of significant targets in cancer (GISTIC) analysis, are provided in the Data Supplement. ch, chromosome.
We found c-MYC copy number gain exclusively in c1 (13 of 18; P < .001; Fig 2B; Data Supplement) along with frequent broad gains of 1q (12 of 18; P = .0099; Fig 2B; Data Supplement). Several c-MYC gains appeared focal and high level, which was suggestive of amplifications. Thus, we performed FISH for c-MYC and MYCN to confirm focal amplification of c-MYC in several c1 tumors and, less commonly, focal MYCN amplification in the rare samples without c-MYC gains (Data Supplement). The one sample without c-MYC or MYCN copy number gains on SNP analysis had no evidence for subclonal amplification by FISH but had polysomy of chromosome 2, on which the MYCN locus resides.
Given the similarities between c1 and c5, we also performed FISH for c-MYC and MYCN on c5 tumors. Within the limits of this technique, we found no evidence of c-MYC amplifications, even subclonally. However, c5 tumors had numerous other copy number alterations, which were predominantly arm-level or whole-chromosome changes. The notable exception was focal PTEN and/or 10q loss (14 of 18; P = .004). Gain of 14 (11 of 18; P < .001), loss of 16 or 16q (17 of 18; P = .0131), and broad loss of 11 (13 of 18; P = .0386) were common (Figs 2A and 2B; Data Supplement). Isochromosome 17q (i17q) was notably underrepresented in c5 (five of 18) compared with c1 (11 of 18), c2 (15 of 17; P = .0065), and c4 (13 of 20); however, numerical gain of 17 (six of 18; P = .0008) was noted in c5. Neither c3 nor c6 had evidence for chromosome 17 alterations (Figs 2A and 2B; Data Supplement).
Differences between c2 and c4 become more apparent at the DNA level when compared with gene expression. In particular, c2 tumors had fewer genetic lesions than c4 tumors (Data Supplement). Core c2 tumors, as defined by the samples with the highest silhouette scores (Fig 1B), often had no more than an i17q (P < .001) and, occasionally, gain of chromosome 4 (P = .0302; Figs 2A and 2B; Data Supplement). Specific to c4 was gain of 12q (seven of 20; P = .0072). MYCN gain, often focal and high level, was also seen in 45% (nine of 20; P = .05) of c4 tumors, whereas low-level c-MYC gain was observed in 25% of the c4 subgroup (five of 20). Additional copy number alterations shared by other subgroups, such as gain of 7 and 18 and deletions of 8, 11, and 16q, were displayed by c4 tumors (Figs 2A and 2B; Data Supplement).
Notable genetic heterogeneity was observed in c3 tumors. Broad loss of 9q (13 of 30; P = .0348) and 20p (five of 30; P = .009) and gains of 3q (12 of 30; P < .001) were specific for this subgroup (Figs 2A and 2B; Data Supplement). Loss of 10q (seven of 30) and 16q (10 of 30) were frequent features of c3 that were also seen in other subgroups. Loss of 9q and 10q appeared to be mutually exclusive except in one sample. Both broad and focal gains were noted on chromosome 2 (P = .0783). In particular, focal gains of GLI2 (five of 30; P = .0019), MYCN (three of 30), or—interestingly—both (two of 30) were observed (Data Supplement). FISH analysis for GLI2 and MYCN confirmed their high-level gains in a subset of SHH-activated medulloblastomas (Data Supplement). We obtained analyzable FISH results on 19 c3/SHH tumors. We identified one sample with high-level MYCN amplification, one with focal GLI2 amplification (plus polysomy for chromosome 2), and several with polysomy for chromosome 2 (n = 8). We did not find evidence for subclonal amplification of either GLI2 or MYCN but did detect samples with subclonal (< 5% nuclei) polysomy for chromosome 2.
To assess the influence of DNA copy number alterations on gene expression, we performed GSEA on each NMF subgroup by using sets of genes that were based on their physical chromosomal location (ie, positional). We observed statistically significant gene expression changes concordant with chromosomal gains and losses identified in our subgroup-specific GISTIC analyses (Data Supplement). However, in c3/SHH tumors, a correlation between GLI2 copy number gain and GLI2 mRNA expression was not observed (Data Supplement).
Medulloblastoma Subgroups Have Distinct miRNA Profiles
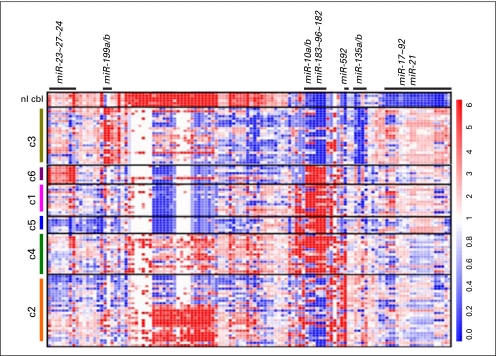
As previously reported,13,14 the miR-17∼92 cluster is upregulated across medulloblastoma samples relative to normal cerebellum (Appendix Fig A1, online only; Data Supplement). The most robust expression of these miRNAs was noted in c1, c3, c5, and c6; lower levels of expression were noted in c2 and c4 (Fig 3). miR-21, a known oncomir,28 was also upregulated across all tumors relative to normal cerebellum (Data Supplement). Conversely, several miRNAs were highly expressed in normal cerebellum relative to primary medulloblastomas. c2 and c4 tumors appeared to have the most overlap with normal cerebellum miRNA expression consistent with the differentiated neuronal phenotype suggested by GSEA.
The miR-183∼96∼182 cluster is specific for medulloblastomas expressing photoreceptor transcriptional genes (c1, c5 and c4; P < .001), whereas miR-592 expression is observed primarily in c2 and c4 (P < .001). Thus, c4 tumors express both miR-592 and miR-183∼96∼182 (P < .001), which additionally corroborates the mixed subpopulations in these tumors (Data Supplement).
Finally, downregulation of miR-135a/b, miR-338, miR-124, and miR-138 and upregulation of miR-199b, miR-378, miR-28, miR-95 and miR-625 were noted in c3 tumors (Data Supplement). c6 tumors are distinctive in their marked upregulation of the miR-23∼27∼24 cluster (Fig A1; P < .001).
Demographic Characteristics of Molecular Subgroups
The general characteristics of our study population were consistent with those previously reported (Table 1).29,30 A male predominance of 1.5 to 1 was noted for the full cohort and was largely driven by the c1 subgroup, which had a 3.8-to-1 male-to-female ratio. The median age for the cohort was 6.5 years. However, age distribution varied across subgroups; the c3/SHH subgroup contained the highest percentage of patients younger than 3 years of age (34%; P = .0142) and all patients older than 25 years of age (n = 4).
Table 1.
Clinicohistologic Characteristics of Individual NMF Subgroups and of the Overall Study Cohort
| Variable | NMF Subgroup |
Overall(N = 189) | |||||
|---|---|---|---|---|---|---|---|
| c1: MYC (n = 29) | c2: Neuronal (n = 37) | c3: SHH (n = 52) | c4: Mixed (n = 34) | c5: Photoreceptor (n = 23) | c6: WNT (n = 14) | ||
| % of overall patients | 15.34 | 19.58 | 27.51 | 17.99 | 12.17 | 7.40 | |
| Sex, No. | |||||||
| Male | 23 | 25 | 25 | 21 | 16 | 3 | 113 |
| Female | 6 | 12 | 27 | 13 | 7 | 10 | 75 |
| Male-to-female ratio | 3.83 | 2.08 | 0.93 | 1.61 | 2.29 | 0.3 | 1.51 |
| Age, months | |||||||
| Median | 64 | 103 | 67 | 84 | 64 | 96 | 79 |
| Range | 7-178 | 48-227 | 11-527 | 12-264 | 12-144 | 48-159 | 7-527 |
| Age group by years, % | |||||||
| < 3 | 25 | 0 | 34* | 14.71 | 21.74 | 0 | 18.09 |
| 3-7 | 42.86 | 37.14 | 22 | 41.48 | 56.52 | 30.77 | 35.64 |
| 7-12 | 17.86 | 51.43 | 20 | 23.53 | 21.74 | 53.85 | 28.19 |
| 12-18 | 14.29 | 8.57 | 16 | 17.65 | 0 | 15.38 | 12.23 |
| 18-25 | 0 | 2.86 | 2 | 2.94 | 0 | 0 | 1.60 |
| > 25 | 0 | 0 | 6 | 0 | 0 | 0 | 1.60 |
| Histologic subtype, % | |||||||
| Classic | 73 | 85.7 | 51 | 79.4 | 61.9 | 85.71 | 71 |
| Nodular/desmoplastic | 4 | 11.4 | 38.3* | 8.8 | 4.76 | 0 | 15.3 |
| Large cell/anaplastic | 23 | 2.86 | 10.6 | 11.8 | 33.33 | 7.1 | 13.6 |
| M+ disease | |||||||
| No. | 7 | 4 | 8 | 4 | 0 | 1 | 24 |
| No. evaluated | 25 | 30 | 42 | 31 | 21 | 12 | 163 |
| % | 28 | 13.33 | 18.18 | 12.90 | 0 | 8.33 | 14.72 |
| Relapse-free survival, %† | 21.01 | 51.94 | 57.65 | 76.35 | 68.38 | 81.82 | 60.56 |
| Overall survival, %† | 21.01‡ | 87.67 | 64.99 | 95 | 74.07 | 90.91 | 73.57 |
Abbreviations: NMF, non-negative matrix factorization; SHH, Sonic Hedgehog.
Statistically significant P value (< .05) for association of subgroup v all other subgroups.
Kaplan-Meier adjusted 6-year survival (EFS and OS) for patients greater than 36 months of age.
Statistically significant difference between survival curves for subgroup patients v all other patients (log-rank test).
Nodular/desmoplastic histology was associated with c3 (P < .001), and large cell or anaplastic (LC/A) histologies were noted in all NMF subgroups. However, c5 and c1 had the highest percentages of LC/A (33.33% and 23%, respectively; P = .0518 and .2404, respectively). Interestingly, all patients with LC/A histology in the c3/SHH subgroup were older than 7 years of age, whereas patients with LC/A tumors in c5 were all younger than 7 years of age.
Aggressive Clinical Behavior Was Associated With c1 Subgroup
We analyzed event-free survival (EFS) and overall survival (OS) for 115 and 112 patients in our cohort, respectively. To minimize treatment heterogeneity, we excluded patients younger than 3 years of age (n = 16) and confirmed that all patients older than 3 years of age received treatment with radiation and chemotherapy. Individuals who died as a result of reasons other than primary disease (n = 3) were excluded.
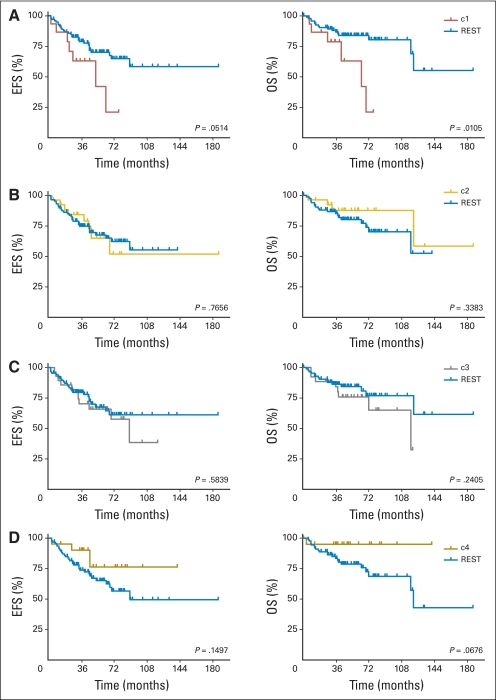
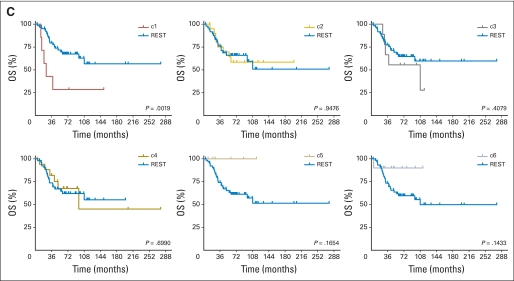
Kaplan-Meier analysis revealed that poor clinical outcome was driven by c1 tumors (P = .0514 and .0105 for EFS and OS, respectively; Table 1; Appendix Fig A2A, online only). Patients with c3/SHH tumors also had relatively poor prognoses. Indeed, c1 and c3, together, were responsible for the majority of relapses and death caused by medulloblastoma as a whole. Intermediate rates of EFS and OS were seen in c5 and c2. Patients with c6 tumors appeared to have good clinical outcome, consistent with previous reports (Data Supplement).7,27
To investigate whether our results were biased by the exclusion of patients younger than 3 years of age, we analyzed survival from the whole patient cohort without filtering for age, and we also analyzed the age group younger than 3 years old independently (Data Supplement). Although the small number of patients in the age group younger than 3 years old limited statistical significance, patients with c1 tumors still fared poorly irrespective of age, whereas patients with c4 tumors in the age group younger than 3 years old fared worse relative to the older age groups (Data Supplement). Patients with c3 tumors appeared to have equivalent survival trends in both age groups.
Validation of Findings in Three Independent Medulloblastoma Data Sets
We analyzed data sets published by Thompson et al,4 Kool et al,3 and Fattet et al7 by building a classifier that was based on metagene projection23 to assign samples from each data set to the NMF subgroups identified in this study (Data Supplement). A heatmap of the top marker genes for each subgroup again revealed overlap between c2 and c4 (Figs A3A and A5B, online only). GSEA of positional gene sets in these subgroups confirmed copy number–driven gene expression changes similar to the GISTIC and positional GSEA results obtained on our data set (Data Supplement). Importantly, the equivalent c1 subgroups showed enrichment of genes at the c-MYC locus (8q24). Accordingly, genes along chromosome 14 were enriched in the c5 subgroup, whereas genes at the PTEN locus (10q23) were enriched in nonc5 tumors relative to c5, suggesting gain of chromosome 14 and loss of chromosome 10/10q, respectively (Data Supplement).
In a separate analysis, we performed consensus NMF clustering on the validation data sets by using parameters that identified the six subgroups in our data set. In a combined data set of Kool et al3 and Fattet et al7, NMF clustering identified six subgroups similar to those in our data set. In the data set of Thompson et al4, however, eight subgroups were identified because of splits in the c3/SHH and c6/WNT subgroups. Silhouette analysis revealed one of the additional c6/WNT subgroups had a low silhouette width (Si, 0.03) and that the extra c3/SHH subgroup had a notable degree of stromal contamination signature that was based on the signature from our normal cerebellum samples.
Validation of Poor Survival Associated With c1 Subgroup Tumors
Survival data from a combined total of 102 patients from the three validation cohorts was analyzed. Again, patients younger than 3 years of age (n = 21) and patients older than 3 years of age who received no radiation therapy (n = 8) were analyzed independently.
Kaplan-Meier analysis confirmed that poor clinical outcome was associated with c1 tumors (P = .0019; Appendix Fig A3C, online only). Statistically significant differences in survival curves were not achieved for other NMF subgroups; however, c5 and c6 appeared to have the best overall survival (P = .1654 and .1433, respectively; Fig A3C).
In patients younger than 3 years of age (and in patients older than 3 years of age who received no radiotherapy), c1 tumors were again associated with decreased survival (Data Supplement); however, the size of these cohorts limited their statistical significance. Younger patients with c5 tumors also fared worse relative to their older counterparts (Data Supplement).
DISCUSSION
Our integrated genomic analysis identified six distinct subgroups of medulloblastoma. However, given the similarities between c1 and c5 subgroups and the c2 and c4 subgroups, one could imagine four broad medulloblastoma subgroups: photoreceptor/GABAergic (composed of c1 and c5), neuronal/glutamatergic (composed of c2 and c4), SHH, and WNT/β-catenin. Yet, key differences exist between these subgroups (Data Supplement). As first described by Kool et al,3 there are several medulloblastomas that have both neuronal/glutamatergic and photoreceptor/GABAergic features. We have provided evidence that this mixed signature, seen in our c4 subgroup, results from distinct subclonal populations within these tumor (Data Supplement). This also explains overlaps in our miRNA data, in which tumors expressed either miR-592 alone (c2), miR-183∼96∼182 alone (c1, c5), or both miR-592 and miR-183∼96∼182 together (c4; Data Supplement). Interestingly, miR-592 is physically located within an intron of the GRM8 gene, which suggests a common transcriptional regulation; conversely, miRNA-96∼182∼183 has been implicated in retinal development and stem-cell maintenance.31–33
The distinction between c1 and c5 is less subtle, and our findings clearly implicate MYC activation, predominantly via genomic amplification, as highly specific for c1 tumors. From a clinical standpoint, distinguishing c1 from c5 is critically important, as patients with c1 tumors in our cohort and three independent cohorts had poor survival (Figs A2A and A3C). Whether this warrants more aggressive up-front therapy for patients with c1 tumors will need to be determined. Nonetheless, our results emphasize that, if treatment stratification is based on four molecular subgroups rather than six, and if patients with c1 and c5 tumors are treated similarly, an unacceptable number of patients would be overtreated on the basis of an errant assumption that c1 and c5 tumors share the same clinical behavior.
Other than the c1 subgroup (and perhaps c6), however, there are no other strong associations with outcome and NMF subgroup. We also acknowledge that our results merely associate clinical outcome with molecular subgroups rather than actively predict outcome for individual patients. Therefore, in a concurrent study, we describe an outcome prediction model that incorporates molecular subtypes into risk stratification by identifying factors that influence outcome within each subgroup.14b
Finally, within the c3/SHH subgroup, we observe notable genetic heterogeneity, which has tremendous implications on the efficacy of targeted therapies currently entering clinical trials (eg, smoothened inhibitors). In particular, MYCN and GLI2, both downstream mediators of the SHH signaling pathway, are amplified in some c3 tumors. Given that smoothened inhibitors target the SHH pathway upstream of GLI2 and MYCN, one might predict GLI2- or MYCN–amplified tumors to be refractory to these therapies.34,35 Thus, our results emphasize the importance of having genomic data to inform clinical trials, either up front or in post hoc analyses.
Supplementary Material
Acknowledgment
We thank the Children's Oncology Group (T. Zhou and A. Buxton) and the Children's Hospital Tumor Network for provision of samples and outcomes data. We thank N. Schouten-van Meeteren (Amsterdam), S. Fattet, F. Doz (France), and C. Haberler (Austria) for sharing genomic and clinical outcome data. We also thank L. Ambrogio, J. Wang, J. Lu, and G. Getz (Broad Institute); Natalie Vena (Dana-Farber Cancer Institute); and J. Mariani (Children's Hospital Boston) for technical assistance.
Appendix
Methods
Biologic samples/nucleic acids extraction.
Snap-frozen tumors were obtained from Children's Oncology Group (ACNS02B3; n = 89) and from Children's Hospital Boston (n = 55), University of Washington Medical Center (n = 27), Texas Children's Hospital (n = 24), and Johns Hopkins Medical Center (n = 10). Normal cerebellum (n = 11) was obtained from University of Washington Medical Center and NICHD Brain and Tissue Bank for Developmental Disorders, University of Maryland, Baltimore, MD. All samples were collected with approval from respective institutional review boards, and proper informed consent and/or assent was obtained from all patients or their parents.
Tumors were processed at Children's Hospital Boston except for samples from Texas Children's, where similar protocols were used. DNA was prepared by using the Puregene DNA Extraction Kit (Gentra Systems, Minneapolis, MN), and total RNA was extracted by using Trizol (Invitrogen, Carlsbad, CA), both per manufacturers' instructions.
Gene expression, SNP array, and miRNA data.
Gene expression data was generated by hybridizing labeled RNAs to Affymetrix HT HG-U133A arrays (Affymetrix, Santa Clara, CA). Samples meeting a minimum of 37% p-calls were used in this analysis. A number of samples with borderline 3′/5′ GAPDH and ACTIN ratios were identified but did not contribute to any batch effects in our non-negative matrix factorization (NMF) analysis; these remained in our subsequent analyses. Gene expression data from Kool et al3 and Fattet et al7 were obtained from the Gene Expression Omnibus (GSE10327 and GSE12992, respectively). Gene expression data from Thomson et al4 was downloaded from http://www.stjuderesearch.org/data/medulloblastoma/. All data sets were preprocessed with the Robust Multichip Average (RMA) algorithm to generate the .gct files used in subsequent analyses (Irizarry RA, et al: Biostatistics 4:249-264, 2003).
DNA copy number data was generated by using Affymetrix 250K Sty arrays or Affymetrix 6.0 arrays to obtain signal intensities and genotype calls. Genomic DNA was digested, adaptor ligated, and amplified by polymerase chain reaction to achieve fragments ranging from 200 base pairs to 1,100 base pairs. These fragments were pooled, concentrated, and additionally fragmented with DNaseI, which was followed by labeling, denaturing, and hybridization to arrays in batches of 96 samples. Arrays were then scanned by using the GeneChip Scanner 3000 7G (Affymetrix). Signal intensities were normalized against a reference set of normal genomic DNA data, as previously described.22
miRNA profiling was performed by using a bead-based detection system containing 582 miRNA probes, representing 435 individual miRNAs, as previously described.15,16 Raw signal intensities were transformed from log2 to linear values, and each miRNA probe was then normalized to the median and median polishing applied when necessary with a threshold cutoff for signal intensities less than 4. Samples were assigned to NMF subgroups on the basis of their mRNA expression data, and an analysis of variance was performed to identify statistically significant miRNAs associated with each subgroup (false discovery rate, 0.0001). The miRNA data were preprocessed and visualized with the Genspring GX7.3.1 software package (Agilent Technologies, Santa Clara, CA).
NMF clustering.
We used a two-step clustering technique to define biologic subtypes of our samples by using microarray expression data. First, we reduced the dimensionality of the expression data from thousands genes to a few metagenes by applying NMF.2 We computed multiple factorizations of an expression matrix A that contained our samples. For each factorization rank k between 2 and 20, we computed 100 decompositions A ∼ W × H. We assessed the stability of the factorizations for each rank k and selected the best factorization (in terms of estimation error) from each of the most stable three ranks for additional analysis.
The second step involved the clustering of the samples in the much smaller metagene space represented by the matrix H by a technique similar to consensus clustering (Monti S, et al: Machine Learning 52:91-118, 2003). For each number of clusters n from 2 to 20, we clustered a random sample of the data containing 85% of the samples by using the Partitioning Around Medoids algorithm (Kaufman L, et al: Finding Groups in Data: An Introduction to Cluster Analysis. Wiley, John, and Sons, Hoboken, NJ, 2005). We repeated this process 500 times and measured the stability of the clustering solutions for each number of clusters by computing a consensus matrix and computing its cophenetic coefficient.2
By using this method described, we clustered a data set containing the expression data for 198 primary medulloblastomas, 11 normal cerebellum samples, and 46 cell lines and other tumor types. Factorizations with four, seven, and 11 metagenes were the most stable, but we performed the clustering step on the representation of the data in seven metagenes space and not four because of the diversity of the samples process. The most stable solutions were achieved for partitioning of the samples into 11, six, and 15 clusters. All of the normal cerebellum samples were assigned to the same cluster (in the most stable clustering solution into 11 clusters). Medulloblastoma samples that were assigned to the cluster of the normal samples (n = 4) were removed from additional analysis.
Next, we performed a similar analysis on the remaining medulloblastoma samples (n = 194) and a comparison group of atypical teratoid/rhabdoid tumors (ATRT; n = 9). The most stable factorizations were achieved using 12, eight, and five metagenes. After the factorization using 12 metagenes, the most stable clustering solution was achieved when grouping into eight clusters; however, we identified a metagene specific for the samples/RNA obtained from Texas Children's Hospital, indicating a batch effect. Therefore, we removed this metagene and reclustered the data set into seven clusters by using the remaining metagenes, which resulted in the final clustering solution. All ATRT samples were assigned to the same cluster, and medulloblastoma samples that were assigned to the rhabdoid cluster were removed from additional medulloblastoma specific analyses (n = 5). The six clusters of medulloblastoma left are those we use to define disease subtypes c1 through c6. The top 50 marker genes for each disease subtype are provided as a Data Supplement.
Silhouette width analysis.
As a means to validate the stability of subgroups identified by NMF clustering and to assess the similarity of each sample to other samples within its subgroup and dissimilarity to samples from other subgroups, we utilized an algorithm described by Rouseeuw17 that calculates silhouette widths for each sample. A silhouette width close to 1 indicates a sample highly similar to other samples within its subgroup; a width of 0 indicates that the sample is equally similar to samples within its subgroup as it is to samples of one another subgroup; conversely, a negative silhouette score indicates the existence of another subgroup in which members are more similar to the current sample than members of the sample's subgroups are. Silhouette widths are provided in the Data Supplement.
Gene set enrichment analysis and single same gene set enrichment analysis: gene set databases.
We utilized both single sample gene set enrichment analysis (ssGSEA) and conventional gene set enrichment analysis (GSEA), as previously described, with a reference database of 2,163 gene sets: 1,891 gene sets from the MSigDB-C2 database (http://www.broad.mit.edu/gsea/msigdb) and 272 additional manually curated gene sets representing oncogene activation/tumor suppressor deregulation (Oncogene Pathway Activation Model [OPAM] database v3).19,20 To all gene sets we added sets that combined up and down regulated sets derived from the same experimental condition or publication. The final total was 2,599 signatures.
We also used the MSigDB positional gene sets (MSigDB, C1.all.v2.5 [positional]) in an independent analysis (Data Supplement) to evaluate the effect of DNA copy number–driven gene expression changes. This database is comprised of gene lists that are based on chromosomal loci/bands (http://www.broad.mit.edu/gsea/msigdb).
Pathway activation features and ssGSEA procedure.
For ssGSEA (Data Supplement), we used the same procedure as described by Barbie et al.20 First, we preprocessed the datasets by mapping from probe identification to gene symbols by using the probe collapse by max procedure as is done in GSEA analysis (www.broad.mit.edu/gsea).19 Then, for each of the 2,165 gene sets in the database, we define an enrichment score that represents the degree of absolute enrichment of a given gene set in each of the samples in the train or test data set. The gene expression values for a given sample are first rank normalized, and then an enrichment score is produced by evaluating an enrichment statistic that is a function of the differences in the empirical cumulative distribution functions (ECDFs) of the genes in the gene set and the remaining genes. This procedure is similar to the one used in GSEA,19 but instead of using a gene list ranked by differential expression, the list is ranked by absolute expression (in one sample). The enrichment score is obtained, not by a weighted Kolmogorov-Smirnov statistic like in GSEA, but by an integration of the difference between the ECDFs. For a given gene set G of size NG and single sample S, of the train or test data set of N genes, the genes are replaced by their ranks according the their absolute expression from high to low: L = {r1, r2, r3,…,rN}. An enrichment score ES(G, S) is obtained by a weighted sum (integration) of the difference between the ECDF of the genes in the gene set and the ECDF of the remaining genes:
This calculation is repeated for each signature and each sample in the data set. Notice that this quantity is signed and that the exponent α = 3/4 adds a weight to the rank. This quantity is slightly more robust and more sensitive to differences in the tails than the Kolmogorov-Smirnov statistic. It is particularly well suited to represent the activation score of gene sets on the basis of a relatively small subset of the genes attaining high expression values. For gene sets with both upregulated (UP) and downregulated (DN) versions, in addition to their independent UP and DN enrichment scores, a combined score is computed by ES(GUP, S) – ES(GDN,S).
Genomic identification of significant targets in cancer analysis.
Genomic identification of significant targets in cancer (GISTIC) analysis was performed on segmented copy number data by using default parameters in Genepattern (www.genepattern.org; Reich M et al: Nat Genet 38:500-501, 2006), except for samples in the c6 subgroup, for which the q-value threshold was decreased to 0.05 to allow for smaller sample size and lack of high-level focal copy number alterations in these tumors.
Metagene projection.
To predict which NMF subgroup each sample from three independent validation data sets (ie, Kool et al,3 Fattet et al,7 and Thompson et al4) belonged to, we used a metagene projection algorithm23 to project the Thompson et al4 data set and a merged Kool et al3/Fattet et al7 data set onto the metagenes used in our clustering solution. We then built a classifier that was based on a support vector machine to classify each of the samples in the validation datasets into one of the six subgroups identified in our data set (subgroup assignments and confidence values provided with clinical annotations in Data Supplement).
In addition, we performed cluster analysis of the validation datasets using their metagene projection described earlier in the Appendix Methods section. We used the method described in the NMF clustering section to cluster random samples of the validation data sets into two to 20 clusters. For the data set from Thompson et al,4 the most stable clustering solution was achieved at n = 8. The clustering solution corresponded well to the NMF subgroups observed in our data set, except that the c3 (SHH) and c6 subgroups (WNT) each split into two subgroups each. For the merged data set, the most stable clustering solution was also at n = 8, except that the corresponding c1 and c4 subgroups were additionally split into two subgroups each. The split subgroups seen in the validation data sets appeared largely technical, given the lack of significant changes in gene expression, copy number, or demographics between the split subgroups. However, information on the methods used to obtain the samples or how the data sets were generated was not readily available for us to analyze.
Immunohistochemistry.
Four-micron tissue sections were mounted on standard glass slides, and heat-induced antigen retrieval was performed with pressure cooker treatment at 120°C for 30 seconds at 15 PSI (range, 10 to 20 PSI) in citrate buffer (pH, 6.0). Dual immunohistochemistry was performed by using a polyclonal rabbit antibody to anti-GRM8 (#G2045; Sigma-Aldrich, St Louis, MO) at a dilution of 1:50 incubated 45 minutes at room temperature followed by labeled polymer-horseradish peroxidase anti-rabbit incubation for 30 minutes and visualization with 3,3′-diaminobenzidine (brown color; Envision System, Dako, Carpinteria, CA). Subsequently, a rabbit polyclonal antibody to Crx (sc-30150; Santa Cruz Biotechnology, Santa Cruz, CA) was applied at a dilution of 1:150 (overnight incubation at 4°C) followed by alkaline phosphatase polymer incubation for 30 minutes at room temperature with visualization by using Dako permanent red.24
Fluorescent in situ hybridization.
Four-micron tissue sections were mounted on standard glass slides and were baked at 60°C for at least 2 hours. Then, they were deparaffinized and digested for 5 minutes, as described previously.25
The following three DNA probes were cohybridized: RP11-57i19 (labeled in SpectrumGreen; Abbott Molecular, Abbott Park, IL), which maps to 2q14.2; RP11-1115a22 (labeled in SpectrumGreen), which also maps to 2q14.2; and NMYC (labeled in SpectrumOrange; Abbott Molecular), which maps to 2p24. Alternatively, the CMYC Dual-Color, Break Apart Rearrangement Probe (Abbott Molecular), which maps to 8q24, and the D8Z2 centromeric probe (labeled in SpectrumAqua; Abbott Molecular) were cohybridized. The GLI2 BAC clones were obtained from CHORI (http://www.chori.org). DNA probes were direct-labeled by using nick translation and were precipitated by using standard protocols. The final probe concentration was approximately 100 ng/μL. The final concentration used for the commercial probes followed manufacturer's recommendations.
The tissue sections and probes were codenatured at 80°C for 5 minutes, were hybridized at least 16 hours at 37°C in a darkened humid chamber, were washed in 2× saline sodium citrate at 70°C for 10 minutes, were rinsed in room temperature 2× saline sodium citrate, and were counterstained with 4′,6-diamidino-2-phenylindole (Abbott Molecular). Slides were imaged with an Olympus BX51 fluorescence microscope (Olympus America, Center Valley, PA). Individual images were captured by using an Applied Imaging system running CytoVision Genus, version 3.92 (Applied Imaging, Grand Rapids, MI).
Fig A1.
miRNA expression profiles of samples arranged by non-negative matrix factorization subgroup reveal statistically significant enrichment of the miR-17∼92 cluster and miR-21 in all tumors relative to normal cerebellum; miR-183∼96∼182 in c1, c5, and c4 tumors; miR-23∼27∼24 in c6 (WNT pathway) tumors; miR-592 in c2 and c4 tumors; and miR-199a/b in c3 (SHH) tumors. P value less than .001 for each miRNA-subgroup association.
Fig A2.
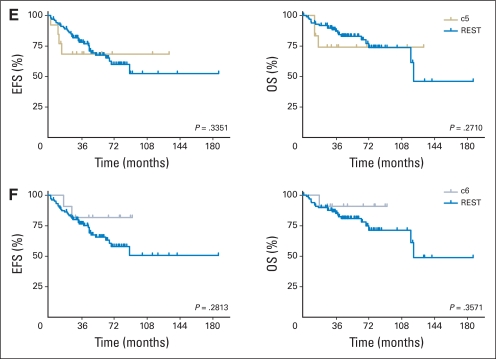
(A-F) Kaplan-Meier survival analysis of patients in each non-negative matrix factorization subgroup reveals (A) decreased survival (event-free survival [EFS] and overall survival [OS]) of patients having c1 subgroup tumors relative to all other tumors (REST). (F) Increased OS is noted for c6 subgroup tumors. Survival analyses were based on clinical risk, histologic subtype, and M stage and are provided as figures in the Data Supplement.
Fig A3.
Validation of molecular subgroups in three independent data sets. Samples were assigned to corresponding non-negative matrix factorization (NMF) subgroups by using metagene projection. (A) Heat map of the top 25 upregulated genes for each NMF subgroup in the Thompson et al4 data set and (B) a merged data set containing the Kool et al3 and Fattet et al7 expression array data. (C) Kaplan-Meier survival analysis of patients from data sets by Thompson et al,4 Kool et al,3 and Fattet et al7 confirms poor overall survival associated with the c1 NMF subgroup. REST, all other tumors.
Footnotes
Supported by Grants No. NIH-R01-109467, NIH-R33-CA97556-01, NIH-6R01-CA-121941-04, NIH-R01-GM074024, NIH-P50-CA112962, and NIH-R01-NS055089.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Yoon-Jae Cho, Pablo Tamayo, Jill P. Mesirov, Scott L. Pomeroy
Financial support: Matthew Meyerson, Jill P. Mesirov,Scott L. Pomeroy
Provision of study materials or patients: Yoon-Jae Cho, Liliana Goumnerova, Charles G. Eberhart, Ching C. Lau, James M. Olson, Richard J. Gilbertson, Amar Gajjar, Marcel Kool, Keith Ligon,Scott L. Pomeroy
Collection and assembly of data: Yoon-Jae Cho, Aviad Tsherniak, Pablo Tamayo, Sandro Santagata, Azra Ligon, Heidi Greulich, Rameen Berhoukim, Vladimir Amani, Liliana Goumnerova, Charles G. Eberhart, Ching C. Lau, James M. Olson, Richard J. Gilbertson, Amar Gajjar, Olivier Delattre, Keith Ligon, Matthew Meyerson, Jill P. Mesirov,Scott L. Pomeroy
Data analysis and interpretation: Yoon-Jae Cho, Aviad Tsherniak, Pablo Tamayo, Sandro Santagata, Azra Ligon, Heidi Greulich, Rameen Berhoukim, Matthew Meyerson, Jill P. Mesirov, Scott L. Pomeroy
Manuscript writing: Yoon-Jae Cho, Aviad Tsherniak, Pablo Tamayo, Sandro Santagata, Azra Ligon, Heidi Greulich, Rameen Berhoukim, Vladimir Amani, Liliana Goumnerova, Charles G. Eberhart, Ching C. Lau, James M. Olson, Richard J. Gilbertson, Amar Gajjar, Olivier Delattre, Marcel Kool, Keith Ligon, Matthew Meyerson, Jill P. Mesirov, Scott L. Pomeroy
Final approval of manuscript: Yoon-Jae Cho, Aviad Tsherniak, Pablo Tamayo, Sandro Santagata, Azra Ligon, Heidi Greulich, Rameen Berhoukim, Vladimir Amani, Liliana Goumnerova, Charles G. Eberhart, Ching C. Lau, James M. Olson, Richard J. Gilbertson, Amar Gajjar, Olivier Delattre, Marcel Kool, Keith Ligon, Matthew Meyerson, Jill P. Mesirov, Scott L. Pomeroy
REFERENCES
- 1.Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 2.Brunet JP, Tamayo P, Golub TR, et al. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kool M, Koster J, Bunt J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson MC, Fuller C, Hogg TL, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 5.Kim JY, Nelson AL, Algon SA, et al. Medulloblastoma tumorigenesis diverges from cerebellar granule cell differentiation in patched heterozygous mice. Dev Biol. 2003;263:50–66. doi: 10.1016/s0012-1606(03)00434-2. [DOI] [PubMed] [Google Scholar]
- 6.Clifford SC, Lusher ME, Lindsey JC, et al. Wnt/Wingless pathway activation and chromosome 6 loss characterize a distinct molecular sub-group of medulloblastomas associated with a favorable prognosis. Cell Cycle. 2006;5:2666–2670. doi: 10.4161/cc.5.22.3446. [DOI] [PubMed] [Google Scholar]
- 7.Fattet S, Haberler C, Legoix P, et al. Beta-catenin status in paediatric medulloblastomas: Correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. J Pathol. 2009;218:86–94. doi: 10.1002/path.2514. [DOI] [PubMed] [Google Scholar]
- 8.Lo KC, Rossi MR, Eberhart CG, et al. Genome wide copy number abnormalities in pediatric medulloblastomas as assessed by array comparative genome hybridization. Brain Pathol. 2007;17:282–296. doi: 10.1111/j.1750-3639.2007.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo KC, Rossi MR, Burkhardt T, et al. Overlay analysis of the oligonucleotide array gene expression profiles and copy number abnormalities as determined by array comparative genomic hybridization in medulloblastomas. Genes Chromosomes Cancer. 2007;46:53–66. doi: 10.1002/gcc.20388. [DOI] [PubMed] [Google Scholar]
- 10.Northcott PA, Nakahara Y, Wu X, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferretti E, De Smaele E, Po A, et al. MicroRNA profiling in human medulloblastoma. Int J Cancer. 2009;124:568–577. doi: 10.1002/ijc.23948. [DOI] [PubMed] [Google Scholar]
- 12.Ferretti E, De Smaele E, Miele E, et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. Embo J. 2008;27:2616–2627. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uziel T, Karginov FV, Xie S, et al. The miR-17∼92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci U S A. 2009;106:2812–2817. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Northcott PA, Fernandez-L A, Hagan JP, et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14b.Tamayo P, Cho Y-J, Tsherniak A, et al. Predicting relapse in patients with medulloblastoma by integrating evidence from clinical and genomic features. J Clin Oncol. doi: 10.1200/JCO.2010.28.1675. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Lu Y, Ryan SL, Elliott DJ, et al. Amplification and overexpression of Hsa-miR-30b, Hsa-miR-30d and KHDRBS3 at 8q24.22-q24.23 in medulloblastoma. PLoS One. 2009;4:e6159. doi: 10.1371/journal.pone.0006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:12. [Google Scholar]
- 18.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beroukhim R, Getz G, Nghiemphu L, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy number alterations across multiple human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamayo P, Scanfeld D, Ebert BL, et al. Metagene projection for cross-platform, cross-species characterization of global transcriptional states. Proc Natl Acad Sci U S A. 2007;104:5959–5964. doi: 10.1073/pnas.0701068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santagata S, Maire CL, Idbaih A, et al. CRX is a diagnostic marker of retinal and pineal lineage tumors. PLoS One. 2009;4:e7932. doi: 10.1371/journal.pone.0007932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Firestein R, Bass AJ, Kim SY, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boon K, Eberhart CG, Riggins GJ. Genomic amplification of orthodenticle homologue 2 in medulloblastomas. Cancer Res. 2005;65:703–707. [PubMed] [Google Scholar]
- 27.Ellison DW, Onilude OE, Lindsey JC, et al. Beta-catenin status predicts a favorable outcome in childhood medulloblastoma: The United Kingdom Children's Cancer Study Group Brain Tumour Committee. J Clin Oncol. 2005;23:7951–7957. doi: 10.1200/JCO.2005.01.5479. [DOI] [PubMed] [Google Scholar]
- 28.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 29.Central Brain Tumor Registry of the United States. Hinsdale, IL: Central Brain Tumor Registry of the United States; 2009. Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2004-2005. [Google Scholar]
- 30.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loscher CJ, Hokamp K, Kenna PF, et al. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol. 2007;8:R248. doi: 10.1186/gb-2007-8-11-r248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loscher CJ, Hokamp K, Wilson JH, et al. A common microRNA signature in mouse models of retinal degeneration. Exp Eye Res. 2008;87:529–534. doi: 10.1016/j.exer.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viswanathan SR, Mermel CH, Lu J, et al. MicroRNA expression during trophectoderm specification. PLoS One. 2009;4:e6143. doi: 10.1371/journal.pone.0006143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romer JT, Kimura H, Magdaleno S, et al. Suppression of the SHH pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1(+/-)p53(-/-) mice. Cancer Cell. 2004;6:229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 35.Sasai K, Romer JT, Lee Y, et al. SHH pathway activity is down-regulated in cultured medulloblastoma cells: Implications for preclinical studies. Cancer Res. 2006;66:4215–4222. doi: 10.1158/0008-5472.CAN-05-4505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.