Abstract
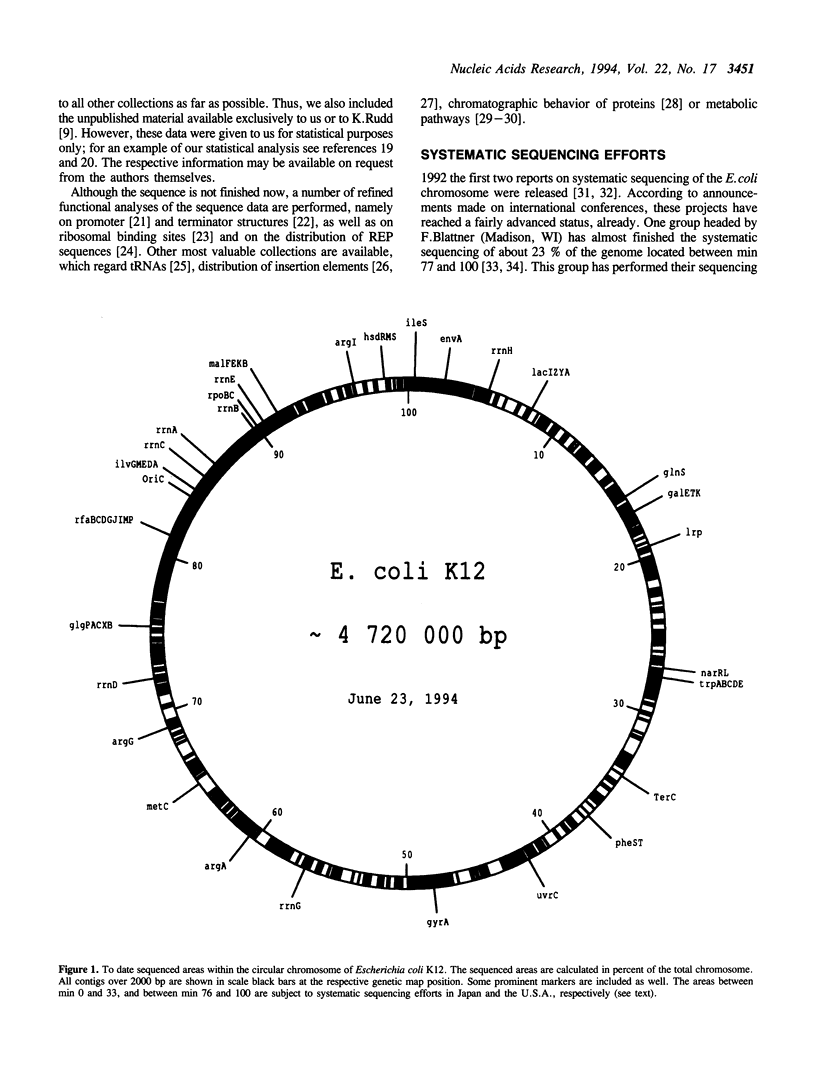
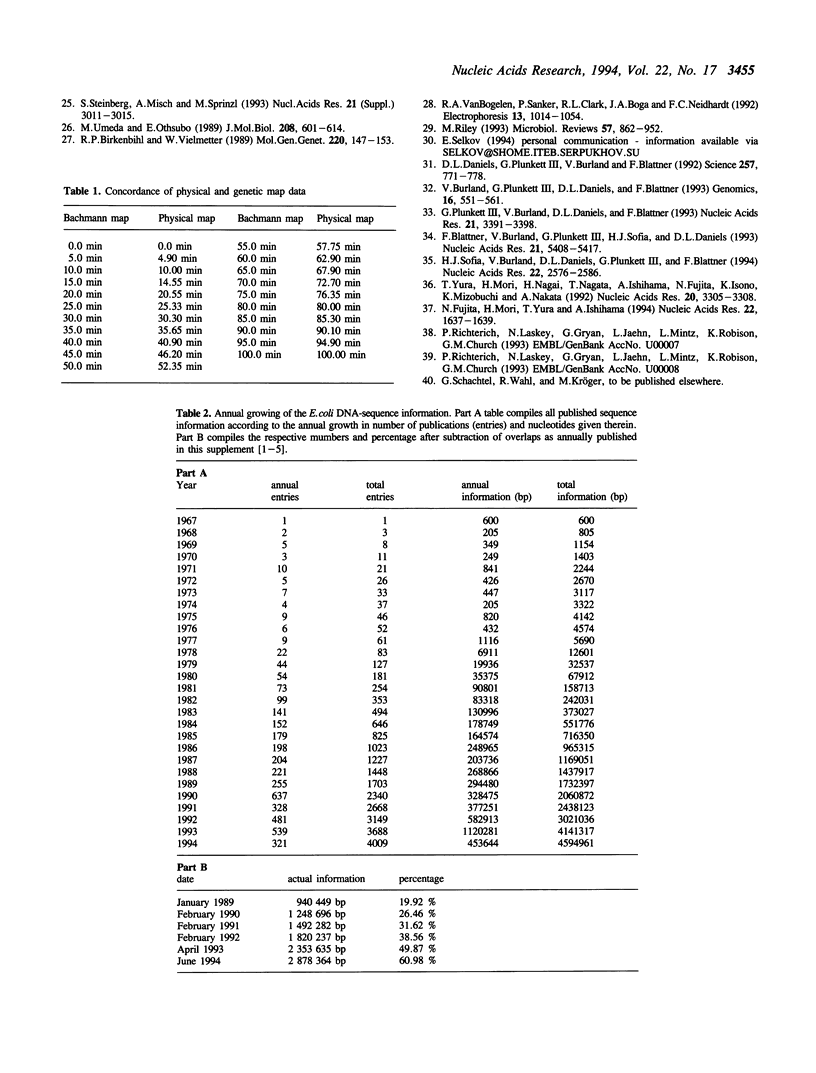
We have compiled the DNA sequence data for E. coli available from the GENBANK and EMBL data libraries and independently from the literature. Starting with this update of our Escherichia coli database (ECD release 20) we provide major changes compared to previous issues. This update not only represents another substantial increase in sequence information, it also allows now to find the exact physical location of each individual gene or regulatory region, even regarding discrepancies in nomenclature. In order to save space this printed version does not contain the database itself anymore, but we provide several examples. The complete database is publically available in electronic form together with a self explaining application program or as a flat file. The complete compilation including a full set of genetic map data and the E. coli protein index can be obtained in machine readable form from the EMBL data library as a part of the CD-ROM issue of the EMBL sequence database, released and updated every three months. After deletion of all detected overlaps a total of 2,878,364 individual bp is found to be determined till the end of June 1994. This corresponds to a total of 60.98% of the entire E. coli chromosome consisting of about 4,720 kbp. This number may actually be higher by 9161 bp derived from other strains of E. coli.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl R. P., Vielmetter W. Complete maps of IS1, IS2, IS3, IS4, IS5, IS30 and IS150 locations in Escherichia coli K12. Mol Gen Genet. 1989 Dec;220(1):147–153. doi: 10.1007/BF00260869. [DOI] [PubMed] [Google Scholar]
- Birkenbihl R. P., Vielmetter W. Cosmid-derived map of E. coli strain BHB2600 in comparison to the map of strain W3110. Nucleic Acids Res. 1989 Jul 11;17(13):5057–5069. doi: 10.1093/nar/17.13.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F. R., Burland V., Plunkett G., 3rd, Sofia H. J., Daniels D. L. Analysis of the Escherichia coli genome. IV. DNA sequence of the region from 89.2 to 92.8 minutes. Nucleic Acids Res. 1993 Nov 25;21(23):5408–5417. doi: 10.1093/nar/21.23.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burland V., Plunkett G., 3rd, Daniels D. L., Blattner F. R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics. 1993 Jun;16(3):551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- Daniels D. L., Plunkett G., 3rd, Burland V., Blattner F. R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992 Aug 7;257(5071):771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- Fujita N., Mori H., Yura T., Ishihama A. Systematic sequencing of the Escherichia coli genome: analysis of the 2.4-4.1 min (110,917-193,643 bp) region. Nucleic Acids Res. 1994 May 11;22(9):1637–1639. doi: 10.1093/nar/22.9.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott V., Blake D. J., Brownlee G. G. Completion of the detailed restriction map of the E. coli genome by the isolation of overlapping cosmid clones. Nucleic Acids Res. 1989 Aug 11;17(15):5901–5912. doi: 10.1093/nar/17.15.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kröger M., Wahl R., Rice P. Compilation of DNA sequences of Escherichia coli (update 1990). Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2549–2587. doi: 10.1093/nar/18.suppl.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger M., Wahl R., Rice P. Compilation of DNA sequences of Escherichia coli (update 1991). Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2023–2043. doi: 10.1093/nar/19.suppl.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger M., Wahl R., Rice P. Compilation of DNA sequences of Escherichia coli (update 1993). Nucleic Acids Res. 1993 Jul 1;21(13):2973–3000. doi: 10.1093/nar/21.13.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger M., Wahl R., Schachtel G., Rice P. Compilation of DNA sequences of Escherichia coli (update 1992). Nucleic Acids Res. 1992 May 11;20 (Suppl):2119–2144. doi: 10.1093/nar/20.suppl.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisawa T., Nakamura M., Watanabe H., Otsuka J., Tsugita A., Yeh L. S., George D. G., Barker W. C. Escherichia coli K12 genomic database. Protein Seq Data Anal. 1990 Jun;3(2):157–162. [PubMed] [Google Scholar]
- Lisser S., Margalit H. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 1993 Apr 11;21(7):1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Bhagwat A. S. Biased DNA repair. Nature. 1992 Feb 13;355(6361):595–596. doi: 10.1038/355595b0. [DOI] [PubMed] [Google Scholar]
- Merkl R., Kröger M., Rice P., Fritz H. J. Statistical evaluation and biological interpretation of non-random abundance in the E. coli K-12 genome of tetra- and pentanucleotide sequences related to VSP DNA mismatch repair. Nucleic Acids Res. 1992 Apr 11;20(7):1657–1662. doi: 10.1093/nar/20.7.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médigue C., Bouché J. P., Hénaut A., Danchin A. Mapping of sequenced genes (700 kbp) in the restriction map of the Escherichia coli chromosome. Mol Microbiol. 1990 Feb;4(2):169–187. doi: 10.1111/j.1365-2958.1990.tb00585.x. [DOI] [PubMed] [Google Scholar]
- Médigue C., Hénaut A., Danchin A. Escherichia coli molecular genetic map (1000 kbp): update I. Mol Microbiol. 1990 Sep;4(9):1443–1454. [PubMed] [Google Scholar]
- Médigue C., Viari A., Hénaut A., Danchin A. Colibri: a functional data base for the Escherichia coli genome. Microbiol Rev. 1993 Sep;57(3):623–654. doi: 10.1128/mr.57.3.623-654.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médigue C., Viari A., Hénaut A., Danchin A. Escherichia coli molecular genetic map (1500 kbp): update II. Mol Microbiol. 1991 Nov;5(11):2629–2640. doi: 10.1111/j.1365-2958.1991.tb01972.x. [DOI] [PubMed] [Google Scholar]
- Plunkett G., 3rd, Burland V., Daniels D. L., Blattner F. R. Analysis of the Escherichia coli genome. III. DNA sequence of the region from 87.2 to 89.2 minutes. Nucleic Acids Res. 1993 Jul 25;21(15):3391–3398. doi: 10.1093/nar/21.15.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Fuchs R., Higgins D. G., Stoehr P. J., Cameron G. N. The EMBL data library. Nucleic Acids Res. 1993 Jul 1;21(13):2967–2971. doi: 10.1093/nar/21.13.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M. Functions of the gene products of Escherichia coli. Microbiol Rev. 1993 Dec;57(4):862–952. doi: 10.1128/mr.57.4.862-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd K. E., Miller W., Ostell J., Benson D. A. Alignment of Escherichia coli K12 DNA sequences to a genomic restriction map. Nucleic Acids Res. 1990 Jan 25;18(2):313–321. doi: 10.1093/nar/18.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofia H. J., Burland V., Daniels D. L., Plunkett G., 3rd, Blattner F. R. Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 minutes. Nucleic Acids Res. 1994 Jul 11;22(13):2576–2586. doi: 10.1093/nar/22.13.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S., Misch A., Sprinzl M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1993 Jul 1;21(13):3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M., Ohtsubo E. Mapping of insertion elements IS1, IS2 and IS3 on the Escherichia coli K-12 chromosome. Role of the insertion elements in formation of Hfrs and F' factors and in rearrangement of bacterial chromosomes. J Mol Biol. 1989 Aug 20;208(4):601–614. doi: 10.1016/0022-2836(89)90151-4. [DOI] [PubMed] [Google Scholar]
- VanBogelen R. A., Sankar P., Clark R. L., Bogan J. A., Neidhardt F. C. The gene-protein database of Escherichia coli: edition 5. Electrophoresis. 1992 Dec;13(12):1014–1054. doi: 10.1002/elps.11501301203. [DOI] [PubMed] [Google Scholar]
- Watanabe H., Kunisawa T. Computer-assisted analysis of chromosomal locations and transcriptional directions of Escherichia coli genes. Protein Seq Data Anal. 1990 Jun;3(2):149–156. [PubMed] [Google Scholar]
- Yura T., Mori H., Nagai H., Nagata T., Ishihama A., Fujita N., Isono K., Mizobuchi K., Nakata A. Systematic sequencing of the Escherichia coli genome: analysis of the 0-2.4 min region. Nucleic Acids Res. 1992 Jul 11;20(13):3305–3308. doi: 10.1093/nar/20.13.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Aubenton Carafa Y., Brody E., Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990 Dec 20;216(4):835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]



