Abstract
Importance to the field
Virtual screening is a computer-based technique for identifying promising compounds to bind to a target molecule of known structure. Given the rapidly increasing number of protein and nucleic acid structures, virtual screening continues to grow as an effective method for the discovery of new inhibitors and drug molecules.
Areas covered in this review
We describe virtual screening methods that are available in the AutoDock suite of programs, and several of our successes in using AutoDock virtual screening in pharmaceutical lead discovery.
What the reader will gain
A general overview of the challenges of virtual screening is presented, along with the tools available in the AutoDock suite of programs for addressing these challenges.
Take home message
Virtual screening is an effective tool for the discovery of compounds for use as leads in drug discovery, and the free, open source program AutoDock is an effective tool for virtual screening.
Keywords: virtual screening, computer-aided drug design, computational docking, AutoDock
Introduction
AutoDock is a suite of software for predicting the optimal bound conformations of ligands to proteins [1, 2]. The initial applications of AutoDock were in the analysis of binding modes and catalytic properties of protein and nucleic acid complexes [3, 4], and a typical study would include results from several dozen docking simulations. More recently, however, enhancements in the performance of AutoDock combined with the availability of high speed computers and clusters of computers has allowed much larger experiments, where entire compound libraries are screened against pharmaceutically-relevant targets [5]. In this report, we will describe the methods that are available within AutoDock to perform virtual screening experiments, and describe some of the successes in virtual screening with AutoDock.
Computational Docking
Computational docking is used to predict the binding modes of two or more molecules. Building on two decades of research, many successful methods for docking of ligands to macromolecular targets have been developed [6–13]. Computational docking relies on two methods: first, a force field to estimate the free energy of binding of the complex, typically estimated based on a particular bound conformation, and second, a search method to explore the conformational space available to the ligand and target. Often, many approximations must be built into the method, both in the force field and in the conformation search, to allow docking with a reasonable computational effort. These may include use of simplified force fields, restriction of the search space, or limitations to the conformational flexibility of the ligand and/or target.
The current version of AutoDock, AutoDock4.2, relies on a number of approximations to predict the conformation and free energy of binding during a docking simulation. The ligand is treated as flexible, but unlike traditional molecular mechanics methods, only torsional degrees of freedom are explored, holding bond angles and bond lengths constant. This allows very rapid transformations of coordinates during the search, but may cause problems if the complex requires significant distortion of the ligand upon binding. In addition, the simple tree-like structure of the data representation used for the ligand does not allow direct modeling of flexibility in rings, although several methods to reclose ring structures during a docking experiment are currently available in AutoDock.
The empirical free energy force field is based on a molecular mechanics force field, which includes typical terms for dispersion/repulsion, hydrogen bonding, electrostatics, desolvation, and torsional entropy. The force field has been calibrated against a large database of complexes with known structure and binding constant, allowing the force field to predict binding free energies. During the docking simulation, a grid-based method is used for energy evaluation, where interaction energies are precalculated around the target structure and then used as look-up table to allow rapid evaluation of ligand-protein interaction. However, the use of this grid-based method requires that the target molecule is treated as rigid, unless specific sidechains are treated explicitly outside the grid.
Several search methods are available in AutoDock, including genetic algorithms, simulated annealing, and local search. All of these methods are stochastic, so repeated docking simulations are often used to validate the exhaustiveness of the search and the solution.
Virtual Screening
Today, virtual screening is widely used to predict the binding of a large database of ligands to a particular target, with the goal of identifying the most promising compounds from the database for further study [10, 14–20]. Hundreds of thousands of compounds may evaluated in a virtual screen, so two aspects of the search are critical. First, we must be confident that the docking method will find a relevant conformation. Docking methods are typically validated by “redocking” experiments, where a series of known complexes are separated and then redocked, ensuring that the docking algorithm can reproduce the observed binding mode. From this type of validation study, we have found that the current version of AutoDock will consistently dock “drug-like” molecules with up to about 10 degrees of torsional freedom [1]. Second, the predicted free energy of binding must be accurate enough to allow ranking of compounds, ensuring that compounds that are predicted to bind most strongly actually do bind when tested experimentally. Most computational docking techniques, including AutoDock, have an accuracy of free energy prediction of about 2–3 kcal/mol standard deviation [21]. This is not sufficient, unfortunately, to provide confident ranking. Rather, we typical refer to the process of “enrichment,” where the set of compounds that are predicted to bind tightly are enriched in compounds that actually show strong binding upon testing.
The two-step docking process used in AutoDock, where a map of interaction energies is calculated first and then used during the docking simulation, is particularly effective for virtual screening, since the map need only be calculated once at the beginning of the screen. A variety of map modification methods are available for extending the basic capabilities of AutoDock. In these methods, the grid maps are modified prior to docking to incorporate a new physical or chemical property. Examples include energy-weighted averaging to model protein flexibility (described below) [22], covalent maps for prediction of covalently-linked complexes or metal coordination [1], mutable atom approaches for de novo design of ligands [23], and water maps for the prediction of bridging water positions (manuscript in preparation).
Choice of Ligand Libraries
A variety of ligand libraries are available for use in virtual screening. The most important criterion, of course, is the ability to obtain samples from the library for testing—so the general rule of thumb is: use what you can get! Several large databases are available, such as the NCBI PubChem (pubchem.ncbi.nlm.nih.gov), eMolecules (www.emolecules.com) and ZINC [24], and most of them include commercially available compounds. ZINC now distributes AutoDock input files for several of these different libraries, from vendors including ChemBridge, Otava, and Asinex.
Other libraries are targeted for specific needs, such as the lead-like compounds [25], nutraceuticals [26], natural products [27], and metabolome [28] libraries, which bring together compounds that might be expected to have good biological properties. In addition, the library of FDA-approved drugs (www.epa.gov/ncct/dsstox) may be of use for repositioning compounds that have already shown biological activity and acceptable safety/toxicity profiles. Finally, virtual screening may be used in tandem to combinatorial chemistry, evaluating the set of molecules that are synthetically accessible within a given combinatorial scheme.
Large databases are often prefiltered to create smaller databases that capture the diversity of the entire set, while reducing the computational demands of the virtual screen campaign [29]. The NCI Diversity Set (dtp.nci.nih.gov/branches/dscb/diversity_explanation.html) is a popular example, which includes 1990 compounds that represent the diversity of 140,000 compounds available at NCI. A new NCI Diversity Set II is also now available (dtp.nci.nih.gov/branches/dscb/div2_explanation.html), which contains a similar number of compounds, but chosen to have more desirable physicochemical properties than the first set. These types of filtered libraries, or diversity subsets, are often particularly effective in a two-stage study. The best ranking compounds from the screen of the diversity set are tested, and the actives are then used as seeds to perform a similarity search on the entire database, generating a focused library of second-generation compounds with chemotypes that structurally resemble the first-generation active compounds. In several of our own applications (described below), this second generation resulted in higher success rates and better activities upon testing.
Several sets of guidelines have been proposed to improve the sampling of the available chemical space, which has been estimated to include 1060 unique compounds [30]. Most notably, Lipinski and coworkers [31] identified common chemical properties that recurr in FDA-approved molecules, presenting the familiar “rule of 5” for drug-like molecules (5 hydrogen bond donors and 10 acceptors, less than 500 daltons, and logPoctanol/water coefficient lower than 5). A similar set of guidelines has been proposed for identifying suitable fragments for drug development [32]. These guidelines are useful for pruning ligand libraries to remove potentially undesirable molecules, however, care must be taken in their application. Many of the most successful drugs do not fit these guidelines, and would have been pruned by a strict application of the guidelines. Like many other laboratories, we have often used a stepwise approach, where a less stringent pruning is used before the virtual screen, and the more stringent pruning is used in combination with manual inspection after the screen.
Choice of Target
In many cases, the target molecule has a well-defined active site. In these cases, it is ideal to have a target structure with an inhibitor or substrate bound, thus forcing the target to adopt a conformation that is more relevant to binding of new compounds. Of course, we often do not have this luxury and must begin with unbound target molecules, homology models, or other target coordinate sets. In these cases, issues of flexibility and protonation state (see below) or errors in modeling must be addressed, and results must be interpreted in this light.
In some cases, we are faced with a completely new target molecule, with no knowledge of potential binding sites for ligands. In these cases we can do a blind docking to the entire protein, to identify sites that bind tightly to ligands. Limitations in the size of the precalculated grid maps in AutoDock pose challenges for blind docking. The maps are typically limited to about 128 grid points in each dimension, otherwise the computation time and file sizes become prohibitive. We have taken two approaches to solve this problem. First, a large grid spacing of 1 Å may be used, allowing the entire target to fit into the map space. However, this may cause problems with accuracy, since the dispersion/repulsion and hydrogen bonding potentials are very steep at short intermolecular distances (i.e., the distances that are most interesting). Alternatively, we have had success with creating separate maps at the typical 0.375 Å spacing, each centered at a different place on the protein surface. Separate docking simulations are then performed within the separate search spaces and the results are combined for analysis.
It is also possible to predict the optimal binding sites on a target molecule, identifying likely candidates for drug-binding sites and using them as the targets for docking analysis, using programs such as AutoLigand [33 and references therein]. AutoLigand analyzes the affinity values around the protein structure, identifying the contiguous volume with the best interaction energy for a given size of ligand molecule. In tests, AutoLigand is successful in identifying the active sites of known complexes and predicting the size of the optimal ligand that will bind to the site. We have applied it to several cases of blind docking, including development of compounds to stabilize protein dimerization in transcription factors (Figure 1, described in more detail below). We have also used results from AutoLigand to define a reduced volume for screening, limiting an AutoDock virtual screen to the area predicted to provide the strongest binding potential.
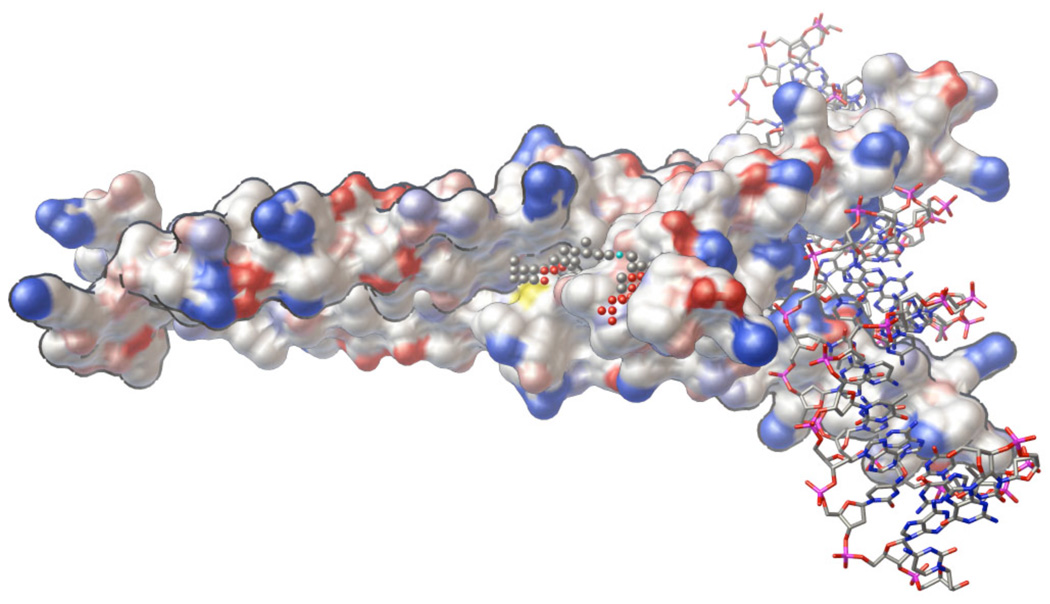
Figure 1.
AutoLigand was used to identify potential binding sites to stabilize the homodimer of the transcription factor Max. The illustration shows the homodimer of Max (surface representation) bound to DNA (ball-and-sticks) and the small spheres show the optimal binding site identified by AutoLigand. The gray spheres have the highest affinity for carbon, the red spheres for hydrogen bond acceptors, and the cyan spheres for hydrogen bond donors. The search was done on a 1 Å grid spacing using only the protein coordinates (illustration created with PMV [56]).
Preparation of Coordinates
Of course, a virtual screen is only as good as the coordinates that are used. For ligand coordinates, a number of processing methods are available, including the ZINC protocols and Corina, which can successfully calculate energetically meaningful three dimensional coordinates starting from the two dimensional representations often provided with compound libraries. Then, AutoDockTools may be used to convert coordinates into the form needed for AutoDock calculations, often adding hydrogen atoms and charges, merging non-polar hydrogens onto their respective heavy atoms, and assigning atom types in the process. It pays to be critical, however: incorrect calculation of the starting conformation, protonation state, and partial charges can dramatically influence docking results, particularly in the case of rigid ligands containing macrocyclic rings or exotic chemical groups. In addition, errors in the crystallographic coordinates or protein-induced distortions from standard geometry may require energy minimization of ligand coordinates before and/or after it is used in docking calculations. Often, preparation of difficult ligands may be improved using sophisticated tools such as Marvin from ChemAxon (www.chemaxon.com) or Avogadro (avogadro.openmolecules.net) to generate starting coordinates for unusual conformations or protonation states.
Tautomers and protonation states pose problems for automated preparation of coordinates. Ideally, we would like to test all possible tautomeric and protonation states of a given ligand and target, to ensure that the state with optimal interaction is included in the screening process. A recent study, however, questions the utility of tautomer and protomer enumeration for improving the enrichment of active molecules, compared to use of a single predicted form of each molecule. A retrospective virtual screening was performed using AutoDock on 19 drug targets with a publicly available data set, and the authors propose that with respect to efficiency, the use of the most probable tautomer/protomer is better than docking the entire enumeration ensemble, since the scoring functions are generally not accurate enough to discriminate among them [34]. On the other hand, other work suggests that tautomer/protomer enumeration should be more suitable when limited information is available for the target structure, or when standard protonation methods do not perform satisfactorily [35].
Flexibility in the Target
In biological systems, we are often faced with heterogeneity in targets. Most often, this is a consequence of flexibility, ranging from small motions of side-chains to flexing of entire domains. Polymorphism in the target, in which multiple distinct, fairly rigid conformational states exist in equilibrium, can add another dimension of complexity to the target’s landscape. Also, we may have a target that can undergo resistance mutations and desire to find compounds that bind to a range of different primary structures. With AutoDock, we have explored several ways to approach this issue of diversity in the target.
Incorporation of target flexibility into AutoDock is tricky, since the grid-based method used for energy evaluation limits us to a rigid model for the target. The current version of AutoDock allows explicit modeling of flexibility in selected sidechains, but for larger motions, other methods must be used. The most obvious approach to incorporating target flexibility into docking is to generate a representative ensemble of structures, and then to perform docking simulations against each one. This ensemble may be a collection of experimental structures (such as from NMR spectroscopy), or in the case of the “relaxed complex” scheme, they may be a collection of conformations harvested from a molecular dynamics or a Monte Carlo simulation [36, 37].
The proper representation of protein flexibility can determine the difference between success and failure. For instance, as part of the FightAIDS@Home project, we recently reexamined nine compounds that did not perform well when they were docked against 77 different crystal structures of HIV protease, but when tested they displayed anti-HIV activity in the standard FRET-based protease inhibition assay. We then used the relaxed complex scheme, docking these nine false negative compounds against 2,000 different snapshots of a wild type HIV-1b protease harvested from 20 ns of molecular dynamics [38]. These AutoDock calculations were surprisingly successful (Table 1)—all 9 compounds scored better than the threshold of −7.0 kcal/mol that was established in the early FightAIDS@Home experiments [39], allowing us to retrospectively re-classify all nine of these compounds as actual hits. A comparison with the original virtual screen that was performed against the rigid crystal structures indicated that clashes and less favorable electrostatic interactions with the crystallographic conformations of a mobile arginine sidechain were the likely cause of the false negative results. Looking to other applications of the relaxed complex method, we see that the method typically yields a good success rate, so these gains in correct scoring of false negatives are not accompanied by a significant increase in false positives [40, 41].
Table 1.
Comparison of Ensemble Docking Methods
| Ligand | Average Energy(1) | Relaxed Complex Energy(2) |
|---|---|---|
| 007223 | −6.58 | −12.91 |
| 065828 | −6.65 | −11.52 |
| 119886 | −5.82 | −11.51 |
| 119889 | −2.88 | −10.27 |
| 119911 | −5.43 | −9.19 |
| 119913 | −4.72 | −10.39 |
| 172033 | −4.35 | −9.50 |
| 270718 | −4.56 | −10.87 |
| 402959 | −6.41 | −15.89 |
Compounds were docked to 77 crystallographic conformations of HIV protease, and the best energy from each complex averaged. Values are kcal/mol.
Compounds were docked against snapshots from a molecular dynamics simulation of one HIV protease structure. Energies are from the conformation with the best energy from the largest cluster. Values are kcal/mol.
A number of methods have been developed to reduce the computation burden of docking to multiple target structures. These have the advantage of both decreasing the computational cost and reducing the amount of human time needed for visual analysis of the docking results. For instance, the “in situ cross docking method” [42] performs all the docking simulations simultaneously, by placing several instances of the binding site within the search space and letting the ligand choose the most favorable one during the docking simulation (Figure 2). The only limitation resides in the number of conformations that can fit within the search space.
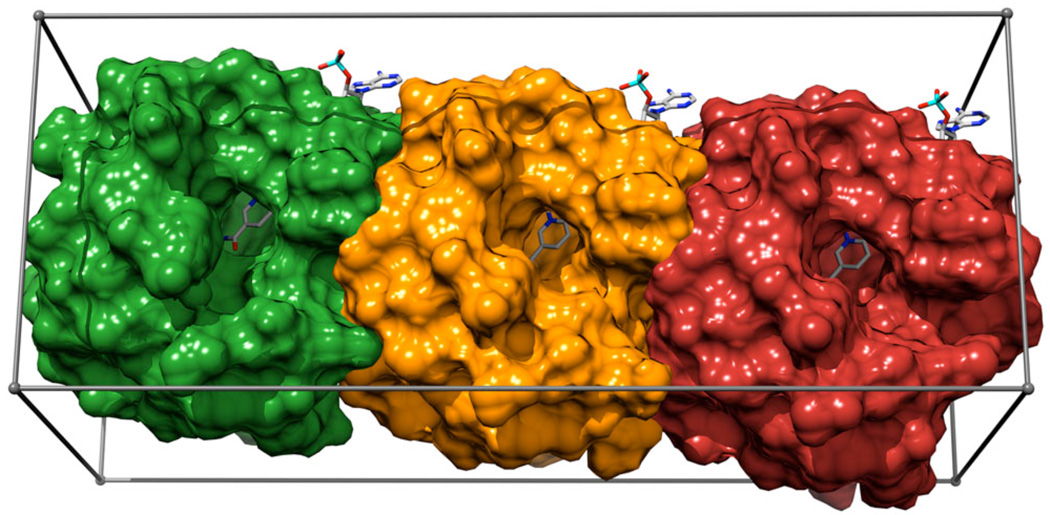
Figure 2.
Aldose reductase construct used for virtual screening with the in situ cross docking approach. 2pdk, 1us0, and 2fzd subsites are represented as red, orange, and green surfaces, respectively. The gray box represents the search area (illustration created with Chimera [57]).
We have also explored methods to define a single representation of the target in a way that captures the structural features of the entire ensemble. One method is to overlap all of the structures in the ensemble, and then to create an averaged map that incorporates features from all the structures. We found that an energy-weighted average map was able to improve docking of a series of HIV protease inhibitors, where the conformation of a mobile arginine was critical to binding of larger ligands [22]. We have also used this method in a virtual screen of the NCI datatabase against an ensemble of x-ray structures β-secretase. Docking analysis was performed individually with each structure, and the different conformations of β-secretase were also combined to generate a unified description of the proteins conformational ensemble. The enrichment factors from these two approaches showed similar predictive power in identifying the positive controls [43].
More recently, we have used principal component analysis to analyze a large cross-docking experiment with 1771 ligands docked to 268 HIV protease structures [39]. The analysis was able to identify a small collection of “spanning” protease structures that capture the energetic features of the entire set. These spanning structures may then be used in future studies in place of the entire set of protease structures.
Analysis of Results
One of the most difficult and subjective steps of virtual screening is the process of analyzing the docking results and choosing the compounds that will actually be ordered and tested. The process is tricky because of the inaccuracies of the scoring functions, which result in errors in ranking. We have used a number of different techniques to help improve the success rate.
Prior to virtual screening calculations, it is often useful to test docking performance on the studied system. Typically, this is done by redocking the co-crystallized conformation of a ligand, if present. This provides many benefits, including validation of the target preparation, tuning of parameters for the docking calculation, and the validation of the method for predicting the known binding pose. If a set of known active compounds exists, these may also be docked against the target protein, and the results used to define a baseline energy value for the selection of virtual screening results that will be considered for further study.
The simplest method for ranking is to use the predicted free energy found for each compound. An additional refinement is to look at the consistency of a particular solution in reiterated docking calculations, often evaluated by clustering the docked conformations based on the RMSD of coordinates. We and others have found that this consistency is related to the conformational entropy of the system, and solutions that are found many times in reiterated docking experiments typically correspond to compounds with better free energy of binding [44]. We often use a combination of these two metrics, requiring conformations to have both favorable predicted free energy and consistent clustering of docking conformations [45]. Several variations on this approach are described in the applications below. A further measure used by some workers is the ligand efficiency [46], the free energy of binding per non-hydrogen atom in the ligand. The ligand efficiency is designed to counter the strong bias of virtual screening towards large compounds, since the predicted binding affinity is often closely proportional to the number of atoms in the ligand.
In specialized cases, negative design elements may also be incorporated in the selection of ligands for testing, to try to increase specificity of binding and potentially reduce the possibility of toxic side effects. This approach may include results from docking to competing targets, to competing sites on the receptor, or to undesired/decoy conformations of the target site. The Myc-Max virtual screen described below is an example.
These types of quantitative measures, based on the predicted free energies and clustering of docking poses, are often followed by a set of more subjective techniques. Visual inspection is one of the most critical steps in virtual screening, as it can greatly help to increase the success rate. Given that methods like AutoDock have a typical error of ±2 kcal/mol in the prediction of free energies of binding, estimated free energy values should not be used as the sole criterion for selecting the ligands that will eventually be tested.
Ideally, many criteria may be used to prescreen and remove undesirable compounds, such as those that contain reactive groups, insoluble compounds, compounds that are too large/less extendable, or highly flexible compounds. Docking simulations are so fast, however, and these properties are often difficult to evaluate computationally, so prescreening is often only performed in a rudimentary way, and compounds are screened manually for these properties after docking. Several aspects of the docked conformation may also be used to filter the set of compounds, such as the presence of key contacts with critical amino acids in the target (identified by mutagenesis), similarity to known positions of ligands or waters in the active site, or the presence of unpaired hydrogen bond donors or acceptors in the ligand-receptor complex. In addition, the ligands may be filtered based on similarity to known actives (such as by calculating “Tanimoto coefficients”), particularly if the screen is a second generation screen [47].
Automation and Scripting
Virtual screening is largely a bookkeeping exercise, and careful planning will ensure an orderly study and effective, efficient analysis of results. We are currently developing graphical user interfaces to streamline the entire process of virtual screening. The first step has been to automate the workflow for preparing the initial input files. The usual virtual screening calculations involve docking a large number of ligands against a single protein (or nucleic acid) structure. This implies the generation of AutoDock ligand format files, the relative parameter files for calculating the affinity maps, and the file with the run parameters used during docking. More sophisticated approaches can include partial target flexibility and multiple target conformations or mutations of the same protein. All these required steps can be performed by using Raccoon, a graphical user interface for AutoDock virtual screening (autodock.scripps.edu/resources/raccoon). Raccoon can split multiple-molecule ligand files, convert them into the AutoDock format, and filter them by using common criteria (e.g., Lipinski's rules, fragment-like “rule of 3”, and drug-likeness). A validation check of the input files is performed at every step, which includes checking for the presence of non-standard atom types and ensuring that parameters, input filenames, and grid maps have a coherent format.
Virtual screening rapidly becomes a major computational effort, particularly if flexibility and/or multiple targets are required. For our largest experiments, which involve virtual screening of libraries of hundreds of thousands of ligands against multiple molecular dynamics snapshots of a series of different mutant targets, we have created the distributed computing system FightAIDS@Home, which is now part of IBM’s “World Community Grid” (fightaidsathome.scripps.edu). This system has allowed the application of over 100,000 CPU years of AutoDock effort devoted to virtual screens of HIV drug-resistance strains.
ACAR Transformylase Inhibitors
In collaboration with Ian Wilson, our first successful virtual screen was targeted against ACAR transformylase [48], which is required for cell division and tissue growth in mammals and is a potential target for cancer chemotherapy. We screened the NCI Diversity set against the crystal structure of human ACAR transformylase bound to the BW1540 inhibitor (PDB entry 1p4r). A simple selection procedure based on the binding energy was used to select 44 compounds for testing. As is often the case with compounds from the NCI Diversity Set, 10 were insoluble, 18 precipitated, and the remaining 16 had properties that allowed testing. Of these 16, 8 showed inhibition better than 250 µM. A second generation screen on the NCI-3D set (213,628 compounds), using compounds with >70% similarity to the best leads, yielded 138 compounds. Of these 138, 12 compounds were tested, and 11 showed inhibition of 50 µM or better.
Protein phosphatase 2C Inhibitors
In collaboration with Paul Greengard, we used virtual screening to identify inhibitors of protein phosphatase 2C [49]. After initial validation with three known ligands, the NCI Diversity Set was screened. Docked compounds were ranked by the predicted free energy of binding, and the best 100 compounds were ordered for testing. Of these, 4 compounds showed >30% inhibition at 100 µM, with the best two displaying 5–10 and 20–30 µM IC50 values. A second generation screen was performed using a similarity search of the Open NCI Database. This yielded 6,000 compounds that were docked using AutoDock. Based on the predicted free energies in these complexes, 156 compounds were ordered and tested. 11 of these compounds showed >30% inhibition at 100µM.
APS reductase inhibitors
In collaboration with Kate Carroll, we have performed virtual screens to identify inhibitors of APS reductase [50], a critical enzyme in bacterial sulfate metabolism and an attractive target for tuberculosis therapy. AutoDock was used to screen the NCI Diversity Set against the P. aeruginosa APS reductase crystal structure (PDB code 2goy). The results were sorted on the basis of their predicted free energies of binding, which ranged from −3.16 to −13.76 kcal/mol, and according to the cluster size for each docked conformation. Solutions with a predicted binding free energy higher than −8.0 kcal/mol and a cluster size lower than 20 out of 100 individuals were discarded. The remaining 192 compounds were visually inspected for interactions with three positively charged residues lining the active site. After this final step, 42 compounds corresponding to 2% of the original NCI Diversity Set were selected for biological evaluation. Five compounds exhibited more than 50% inhibition at 100 µM.
A second generation virtual screen was performed using 890 compounds with least 80% Tanimoto similarity to the first generation compounds, chosen from 250,000 compounds in the Open NCI database. Similarity was evaluated using default settings at the Enhanced NCI Database Browser (http://cactus.nci.nih.gov/ncidb2/). After docking these 890 compounds, the 40 highest-scoring solutions, ranked according to the criteria outlined above, were experimentally evaluated. Five compounds were identified that displayed dissociation constants lower than 50 µM. The dihydrophenanthrendione 23180 (Figure 3) was the most potent inhibitor identified in this study, with a dissociation constant of less than 10 µM.
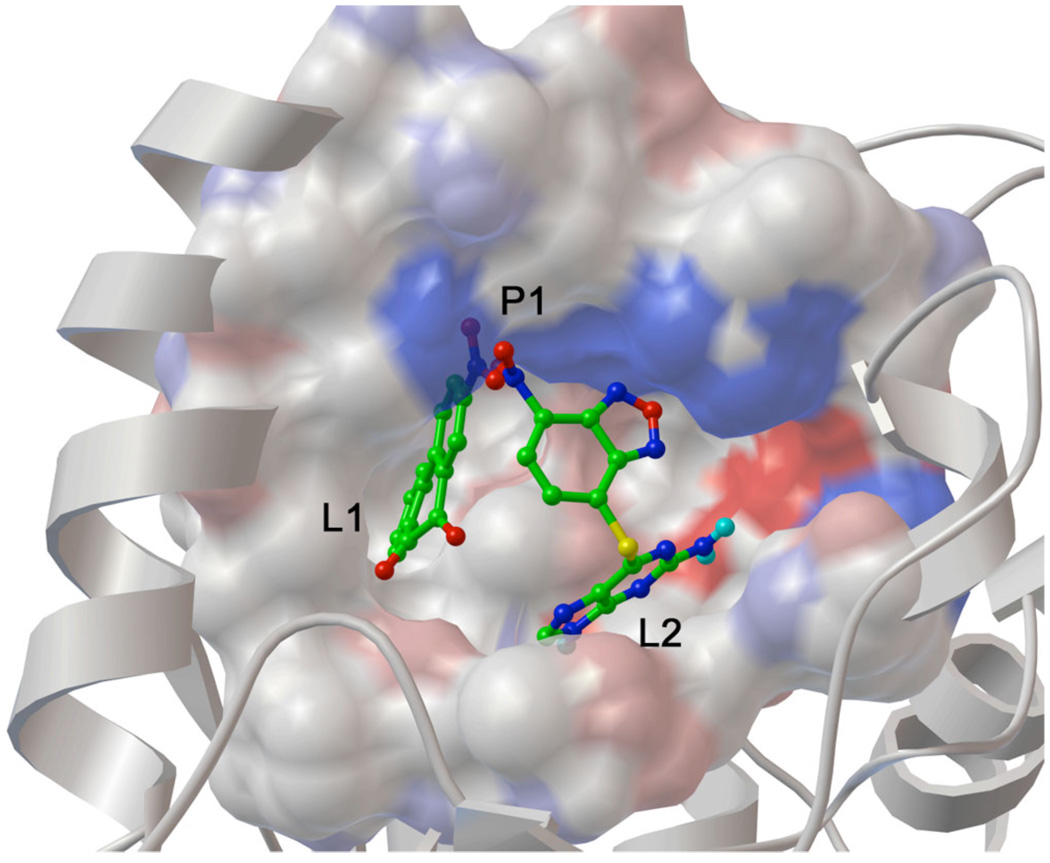
Figure 3.
Binding pose of both NSC23180 and NSC348401 in the APS reductase binding site. The two compounds bind to three separate sites in the protein, termed L1, L2, and P1. These observations suggest that a ligand that occupies both L1 and L2 clefts and that displays polar hydrogen bond accepting groups to interact with the P1 site may exhibit sub-µM activity against APS reductase (illustration created with PMV [56]).
Max Homodimer Stabilizers
In collaboration with Peter Vogt, we performed virtual screens to identify stabilizers of a transcription factor interface [51]. Myc is an oncogene that mediates progression of the cell cycle; thus, it is a potential target for cancer chemotherapy. It forms a series of homodimers and heterodimers with the similar protein Max. The goal of the virtual screen was to block dimerization between Myc and Max by specifically stabilizing the normally weak Max-Max homodimer. This is an unusual application for drug design, since the goal is to stabilize a protein-protein interaction rather than to block formation of the biologically-relevant complex.
Two crystallographic structures were used: Myc-Max (PDB entry 1nkp) and the structure we sought to stabilize, Max-Max (PDB entry 1an2). We screened the NCI Diversity Set using a larger grid spacing than normal (1 Å), in order to provide a blind search of the entire protein. A longer search was also employed to ensure that the larger search space was adequately sampled. All results were clustered to identify 12 potential binding sites on the protein surface. Three of these clusters contained 85% of the docked compounds and included all of the compounds with the lowest predicted free energy of binding. Consequently, the compounds bound to these three sites were chosen for experimental testing. These sites were also identified using an early version of AutoLigand (Figure 1).
Forty compounds were chosen for testing from the Myc-Max docking results, and forty from the Max-Max results. After duplicates were removed, 68 compounds were obtained and tested by FRET analysis. At 10 µM, 13 compounds showed stabilization of Max-Max and compounds predicted to bind to Myc-Max did not stabilize Max-Max. Further in vivo testing with the best compound showed that the stabilization of Max-Max was not a function of DNA binding, and that stabilization of Max-Max inhibited formation of Myc-Max. Tests in cell culture with the best compound showed that it interferes with Myc-induced transformation, Myc-dependent cell growth, and Myc-mediated transcriptional activation.
Cobratoxin Antitoxin
In collaboration with Opa Vajragupta and Palmer Taylor, we used virtual screening to identify inhibitors of cobratoxin, for potential use as snakebite antidotes [52]. We used several sets of coordinates, including a complex of α-cobratoxin with acetylcholine-binding protein (PDB entry 1yi5) and two structures of α-cobratoxin alone (PDB entries 1ctx and 2ctx). We screened the NCI Diversity Set against all three coordinate sets and saved the top 175 compounds from each screen. The sets were then compared, and choosing the 77 compounds that in all three screens for a more thorough analysis. An additional filtering step based on ligand efficiency < −0.30 and manual inspection for “drug-like properties” yielded 19 compounds for testing. A second protocol for filtering, which looked simply at the predicted free energy value and the cluster size, yielded an additional 20 compounds. These 39 compounds were tested for their ability to block binding between α-cobratoxin and acetylcholine-binding protein. Four of these compounds were active and were able to displace the antagonists epibatidine and α-bungarotoxin with µM to nM dissociation constants. The best case showed a 13.8 nM dissociation constant. The best compounds were tested in vivo, showing increased survival time in mice challenged with cobratoxin. Three compounds increased survival if given 30 minutes before the toxin, and two showed increased survival if given as an antidote immediately after toxin administration.
DNA Quadruplex Groove Binders
In collaboration with Ettore Novellino and Antionio Randazzo, we used virtual screening to find DNA quadruplex groove binders [53]. G-quadruplexes are four-stranded helical DNA or RNA structures involved in a number of medically-relevant biological processes, such as replication, recombination, transcription, and translation, and they form the teleomere structure. So far, distamycin A is the only agent for which a pure groove binding mode has been demonstrated.
We screened a diversity set of the commercially available Life Chemicals database (6000 compounds) against the simple quadruplex [d(TGGGGT)]4 (PDB entry 1s45, Figure 4). The virtual screen results were sorted on the basis of their predicted binding free energies, which ranged from −0.95 to −9.55 kcal/mol. Solutions with a predicted binding free energy greater than −6.0 kcal/mol and a cluster size lower than 10 out of 100 individuals were discarded. Based on these criteria, 137 individuals were retained for further consideration. The binding poses calculated for these compounds were then visually inspected to discard compounds that did not establish tight interactions with the groove of the quadruplex structure. More precisely, compounds that were not able to form H-bonds with any of the guanine bases and/or to establish an electrostatic interaction with the backbone phosphate groups were not considered for subsequent tests. After this final step, 30 compounds corresponding to 0.5% of the original Life Chemicals database were selected and purchased for further analysis.
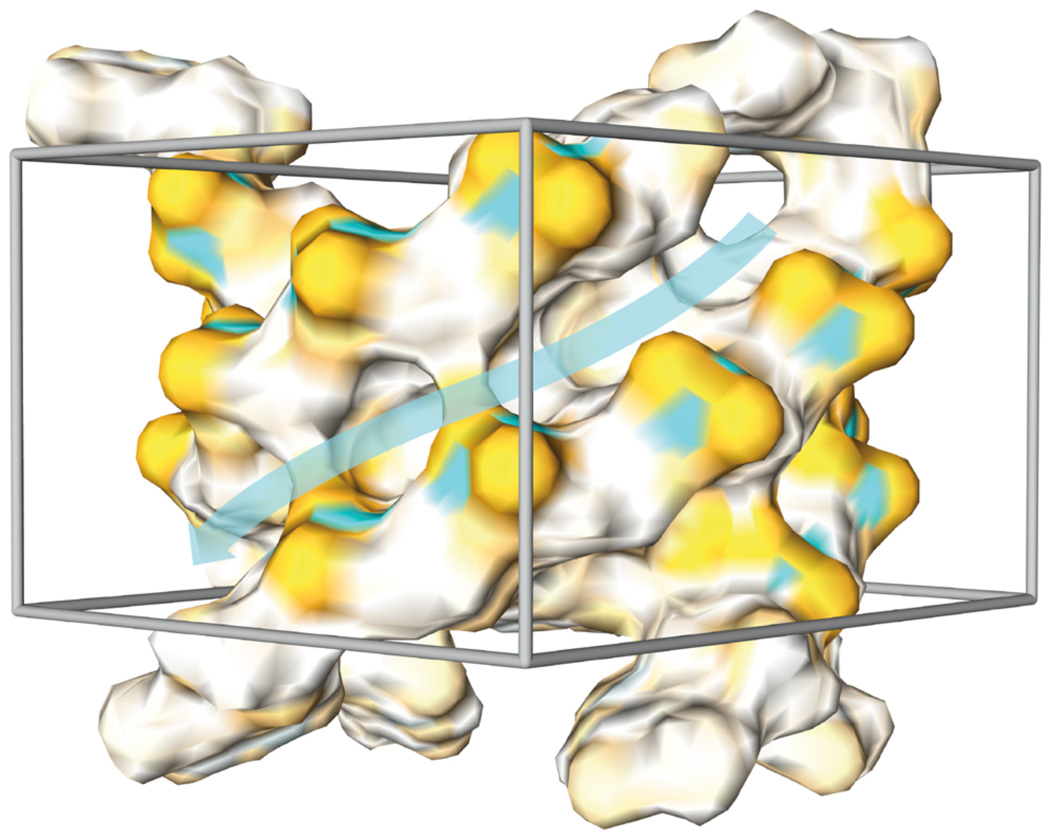
Figure 4.
Surface representation of the quadruplex structure. Orange, white and cyan areas represent negatively, neutrally and positively charged regions, respectively. The grey box represents the searched area in virtual screening calculations. The cyan transparent arrow outlines the groove area taken into consideration (illustration created with Chimera [57]).
The experimental testing was performed by NMR titrations. By monitoring resonance chemical shift changes of DNA, we measured whether a given compound is able to interact with the quadruplex and determined the binding site. Six molecules were found to cause an appreciable chemical shift of the G3 and G4 resonances signals, indicative of a groove binding interaction.
Aldose Reductase Inhibitors
In collaboration with Ettore Movellino, we performed virtual screens against aldose reductase (ALR2), a potential target for preventing the onset, progression, and severity of diabetic complications [54]. So far, the known ALR2 inhibitors failed in clinical trials because of poor pharmacokinetic properties and unexpected side effects. From the structural point of view, ALR2 represents one of the most striking examples of the induced fit effect upon ligand binding. X-ray studies have demonstrated that at least three different binding site conformations exist, depending on the bound ligand, which poses a great challenge for virtual screening. We used “in situ cross-docking” (described above) to combine the three most divergent X-ray structures of human ALR2 (PDB entries 2pdk, 1us0, and 2fzd) into one grid map for docking. The combined grids were then used to perform a preliminary docking calculation on a set of 141 active ligands, which showed good agreement with the experimental data.
We then screened the commercially available Maybridge HitFinder database (14,400 compounds) against the combined grids. Virtual screen results were sorted on the basis of the predicted free energy values, which ranged from −3.49 to −13.87 kcal/mol, and compounds with a predicted energy value higher than the average energy calculated for the known active compounds (−8.00 kcal/mol) were discarded. On the basis of this criterion, 7,468 compounds were retained, representing almost 52% of the database. Consequently, a second filtering step, using a different methodology, was required in order to identify a reasonable number of potential hits for subsequent testing.
We used MolPrint2D [55] to perform a similarity search on the MayBridge database, which allowed us to re-rank the 7468 hits. We retained 106 compounds that had a predicted free energy of binding lower than −8.00 kcal/mol and a MolPrint2D score higher than 10. We were encouraged that this MolPrint2D cutoff (>10) allowed us to also retain compounds that were only vaguely similar to the known inhibitors. Finally, as a last criterion for selection, we visually inspected each docked complex, removing compounds that did not display interactions with the ALR2 anion and the specificity binding pocket. As a result, 57 candidates were selected and evaluated for their efficacy against ALR2. Twelve out of the 53 soluble compounds were shown to be inhibitors, with IC50 values in the range of 1–100 µM, resulting in a success rate of 22%. Within this set, six new chemotypes were identified that feature novel molecular scaffolds structurally unrelated to the known inhibitors. Analysis of the binding poses calculated for these ligands in complex with ALR2 allowed us to suggest different structural modifications, which provided valuable alternative strategies for ongoing medicinal chemistry optimization.
Conclusions
AutoDock has shown continued success for the application of virtual screening to a variety of targets. Applications studied in our laboratory have ranged from the development of enzyme inhibitors, compounds that stabilize protein dimerization, antitoxins, and nucleic-acid-binding compounds. Through the use of simple and automated scripting methods such as Raccoon, virtual screening with AutoDock is currently available to a wide user community. However, several challenges still face the virtual screening community, most notably, the continued development of scoring methods to improve the ranking of compounds. The AutoDock suite of programs is currently available as open source at http://autodock.scripps.edu.
Expert Opinion
Virtual screening is currently an effective tool for the discovery of compounds for use as leads in drug discovery. Many options are available both for compounds libraries and for computational techniques for evaluating compounds in these libraries. However, the limitations of the scoring functions used in these computations are such that results are not guaranteed, and virtual screening currently requires significant expertise and manual intervention to yield a successful result.
In our hands, AutoDock has been a successful technique for the discovery of drug leads. In most systems that we have attempted, virtual screening has found several compounds that showed binding upon experimental testing. These have included a diverse range of targets, including enzyme active sites, protein dimerization interfaces, and nucleic acids. Looking at the field at large, we see many similar successes using AutoDock and similar computational docking techniques.
The simplified force fields used in current docking techniques are the major limiting factor of current virtual screening efforts. These force fields typically estimate the free energy of binding using a single conformation of the complex, and thus are unable to evaluate the conformational entropy and other contributions to the free energy that are a consequence of the thermodynamics of the entire ensemble of possible complex conformations. Advanced statistical mechanics methods are being developed to address this limitation, unfortunately, they are computationally intensive and thus not suitable for use in virtual screening. We have tested methods for approximating these properties using the information available in a docking simulation, with encouraging results. However, a major advance in scoring functions will be needed to turn virtual screening into a turn-key method for lead discovery.
Acknowledgements
This work is supported by grants P01-GM083658 and R01-GM069832 from the National Institutes of Health. This is manuscript 20616 from the Scripps Research Institute.
References
- 1.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comp. Chem. 1998;19:1639–1662. [Google Scholar]
- 3.Morris GM, Goodsell DS, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J Comput Aided Mol Des. 1996;10:293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 4.Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 5.Beuscher AE, Olson AJ. Iterative docking strategies for virtual ligand screening. In: Stroud RM, Finer-Moore J, editors. Computational and Structural Approaches to Drug Discovery. London: RSC Publishing; 2007. pp. 242–264. [Google Scholar]
- 6.Leach AR, Shoichet BK, Peishoff CE. Prediction of protein-ligand interactions. Docking and scoring: successes and gaps. J. Med. Chem. 2006;49:5851–5855. doi: 10.1021/jm060999m. [DOI] [PubMed] [Google Scholar]
- 7.Coupez B, Lewis RA. Docking and scoring--theoretically easy, practically impossible? Curr. Med. Chem. 2006;13:2995–3003. doi: 10.2174/092986706778521797. [DOI] [PubMed] [Google Scholar]
- 8.Sousa SF, Fernandes PA, Ramos MJ. Protein-ligand docking: current status and future challenges. Proteins. 2006;65:15–26. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- 9.Mohan V, Gibbs AC, Cummings MD, Jaeger EP, DesJarlais RL. Docking: successes and challenges. Curr. Pharm. Des. 2005;11:323–333. doi: 10.2174/1381612053382106. [DOI] [PubMed] [Google Scholar]
- 10.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nature Rev. Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 11.Brooijmans N, Kuntz ID. Molecular recognition and docking algorithms. Annu. Rev. Biophys. Biomol. Struct. 2003;32:335–373. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- 12.Taylor RD, Jewsbury PJ, Essex JW. A review of protein-small molecule docking methods. J. Comp. Aided Mol. Design. 2002;16:151–166. doi: 10.1023/a:1020155510718. [DOI] [PubMed] [Google Scholar]
- 13.Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: and overview of search algorithms and a guide to scoring functions. Proteins: Struct. Funct. Genet. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 14.Kolb P, Ferreira RS, Irwin JJ, Shoichet BK. Docking and chemoinformatic screens for new ligands and targets. Curr Opin Biotechnol. 2009;20:429–436. doi: 10.1016/j.copbio.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolb P, Irwin JJ. Docking screens: right for the right reasons? Curr Top Med Chem. 2009;9:755–770. doi: 10.2174/156802609789207091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koppen H. Virtual screening - what does it give us? Curr Opin Drug Discov Devel. 2009;12:397–407. [PubMed] [Google Scholar]
- 17.Rester U. From virtuality to reality - Virtual screening in lead discovery and lead optimization: a medicinal chemistry perspective. Curr Opin Drug Discov Devel. 2008;11:559–568. [PubMed] [Google Scholar]
- 18.McInnes C. Virtual screening strategies in drug discovery. Curr Opin Chem Biol. 2007;11:494–502. doi: 10.1016/j.cbpa.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 19.Seifert MH, Kraus J, Kramer B. Virtual high-throughput screening of molecular databases. Curr Opin Drug Discov Devel. 2007;10:298–307. [PubMed] [Google Scholar]
- 20.Shoichet BK. Virtual screening of chemical libraries. Nature. 2004;432:862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2006;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 22.Osterberg F, Morris GM, Sanner MF, Olson AJ, Goodsell DS. Automated docking to multiple target structures: incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins: Struct. Funct. Genet. 2001 doi: 10.1002/prot.10028. in press. [DOI] [PubMed] [Google Scholar]
- 23.Olson AJ, Goodsell DS. Automated docking and the search for HIV protease inhibitors. SAR and QSAR in Environ. Res. 1998;8:273–285. doi: 10.1080/10629369808039144. [DOI] [PubMed] [Google Scholar]
- 24.Irwin JJ, Shoichet BK. ZINC--a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irwin JJ. Using ZINC to acquire a virtual screening library. Curr Protoc Bioinformatics. 2008;Chapter 14(Unit 14):16. doi: 10.1002/0471250953.bi1406s22. [DOI] [PubMed] [Google Scholar]
- 26.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunkel M, Fullbeck M, Neumann S, Preissner R. SuperNatural: a searchable database of available natural compounds. Nucleic Acids Res. 2006;34:D678–D683. doi: 10.1093/nar/gkj132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timmers LF, Pauli I, Caceres RA, de Azevedo WF., Jr Drug-binding databases. Curr Drug Targets. 2008;9:1092–1099. doi: 10.2174/138945008786949379. [DOI] [PubMed] [Google Scholar]
- 30.Kirkpatrick P, Ellis C. Chemical space. Nature. 2004;432:823–823. [Google Scholar]
- 31.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 32.Congreve M, Carr R, Murray C, Jhoti H. A 'rule of three' for fragment-based lead discovery? Drug Discov Today. 2003;8:876–877. doi: 10.1016/s1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- 33.Harris R, Olson AJ, Goodsell DS. Automated prediction of ligand-binding sites in proteins. Proteins. 2008;70:1506–1517. doi: 10.1002/prot.21645. [DOI] [PubMed] [Google Scholar]
- 34.Kalliokoski T, Salo HS, Lahtela-Kakkonen M, Poso A. The effect of ligand-based tautomer and protomer prediction on structure-based virtual screening. J Chem Inf Model. 2009;49:2742–2748. doi: 10.1021/ci900364w. [DOI] [PubMed] [Google Scholar]
- 35.ten Brink T, Exner TE. Influence of protonation, tautomeric, and stereoisomeric states on protein-ligand docking results. J Chem Inf Model. 2009;49:1535–1546. doi: 10.1021/ci800420z. [DOI] [PubMed] [Google Scholar]
- 36.Lin JH, Perryman AL, Schames JR, McCammon JA. The relaxed complex method: Accommodating receptor flexibility for drug design with an improved scoring scheme. Biopolymers. 2003;68:47–62. doi: 10.1002/bip.10218. [DOI] [PubMed] [Google Scholar]
- 37.Amaro RE, Baron R, McCammon JA. An improved relaxed complex scheme for receptor flexibility in computer-aided drug design. J Comput Aided Mol Des. 2008;22:693–705. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perryman AL, Lin JH, McCammon JA. HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: possible contributions to drug resistance and a potential new target site for drugs. Protein Sci. 2004;13:1108–1123. doi: 10.1110/ps.03468904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang MW, Lindstrom W, Olson AJ, Belew RK. Analysis of HIV wild-type and mutant structures via in silico docking against diverse ligand libraries. J Chem Inf Model. 2007;47:1258–1262. doi: 10.1021/ci700044s. [DOI] [PubMed] [Google Scholar]
- 40.Amaro RE, Schnaufer A, Interthal H, Hol W, Stuart KD, McCammon JA. Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. Proc Natl Acad Sci U S A. 2008;105:17278–17283. doi: 10.1073/pnas.0805820105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lerner MG, Bowman AL, Carlson HA. Incorporating dynamics in E. coli dihydrofolate reductase enhances structure-based drug discovery. J Chem Inf Model. 2007;47:2358–2365. doi: 10.1021/ci700167n. [DOI] [PubMed] [Google Scholar]
- 42.Sotriffer CA, Dramburg I. "In situ cross-docking" to simultaneously address multiple targets. J Med Chem. 2005;48:3122–3125. doi: 10.1021/jm050075j. [DOI] [PubMed] [Google Scholar]
- 43.Cosconati S, Huey R, Marinella L, Novellino E, Goodsell DS, Olson AJ. Identification of novel beta-secretase inhibitors through inclusion of protein flexibility in virtual screening calculations. FASEB Journal. 2008;22:791–798. [Google Scholar]
- 44.Chang MW, Belew RK, Carroll KS, Olson AJ, Goodsell DS. Empirical entropic contributions in computational docking: evaluation in APS reductase complexes. J Comput Chem. 2008;29:1753–1761. doi: 10.1002/jcc.20936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenfeld RJ, Goodsell DS, Musah RA, Morris GM, Goodin DB, Olson AJ. Automated docking of ligands to an artificial active site: augmenting crystallographic analysis with computer modeling. J Comput Aided Mol Des. 2003;17:525–536. doi: 10.1023/b:jcam.0000004604.87558.02. [DOI] [PubMed] [Google Scholar]
- 46.Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 47.Chen X, Reynolds CH. Performance of similarity measures in 2D fragment-based similarity searching: comparison of structural descriptors and similarity coefficients. J Chem Inf Comput Sci. 2002;42:1407–1414. doi: 10.1021/ci025531g. [DOI] [PubMed] [Google Scholar]
- 48.Li C, Xu L, Wolan DW, Wilson IA, Olson AJ. Virtual screening of human 5-aminoimidazole-4-carboxamide ribonucleotide transformylase against the NCI diversity set by use of AutoDock to identify novel nonfolate inhibitors. J Med Chem. 2004;47:6681–6690. doi: 10.1021/jm049504o. [DOI] [PubMed] [Google Scholar]
- 49.Rogers JP, Beuscher AEt, Flajolet M, McAvoy T, Nairn AC, Olson AJ, Greengard P. Discovery of protein phosphatase 2C inhibitors by virtual screening. J Med Chem. 2006;49:1658–1667. doi: 10.1021/jm051033y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cosconati S, Hong JA, Novellino E, Carroll KS, Goodsell DS, Olson AJ. Structure-based virtual screening and biological evaluation of Mycobacterium tuberculosis adenosine 5'-phosphosulfate reductase inhibitors. J Med Chem. 2008;51:6627–6630. doi: 10.1021/jm800571m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang H, Bower KE, Beuscher AEt, Zhou B, Bobkov AA, Olson AJ, Vogt PK. Stabilizers of the Max homodimer identified in virtual ligand screening inhibit Myc function. Mol Pharmacol. 2009;76:491–502. doi: 10.1124/mol.109.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Utsintong M, Talley TT, Taylor PW, Olson AJ, Vajragupta O. Virtual screening against alpha-cobratoxin. J Biomol Screen. 2009;14:1109–1118. doi: 10.1177/1087057109344617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cosconati S, Marinelli L, Trotta R, Virno A, Mayol L, Novellino E, Olson AJ, Randazzo A. Tandem application of virtual screening and NMR experiments in the discovery of brand new DNA quadruplex groove binders. J Am Chem Soc. 2009;131:16336–16337. doi: 10.1021/ja9063662. [DOI] [PubMed] [Google Scholar]
- 54.Cosconati S, Marinelli L, La Motta C, Sartini S, Da Settimo F, Olson AJ, Novellino E. Pursuing aldose reductase inhibitors through in situ cross-docking and similarity-based virtual screening. J Med Chem. 2009;52:5578–5581. doi: 10.1021/jm901045w. [DOI] [PubMed] [Google Scholar]
- 55.Bender A, Mussa HY, Glen RC, Reiling S. Molecular similarity searching using atom environments, information-based feature selection, and a naive Bayesian classifier. J Chem Inf Comput Sci. 2004;44:170–178. doi: 10.1021/ci034207y. [DOI] [PubMed] [Google Scholar]
- 56.Sanner MF. Python: A Programming Language for Software Integration and Development. J. Mol. Graphics Mod. 1999;17:57–61. [PubMed] [Google Scholar]
- 57.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]