Abstract
In any given cell, thousands of genes are expressed and work in concert to ensure the cell's function, fitness, and survival. Each gene, in turn, must be expressed at the proper time and in the proper amounts to ensure the appropriate functional outcome. The regulation and expression of some genes are highly robust; their expression is controlled by invariable expression programs. For instance, developmental gene expression is extremely similar in a given cell type from one individual to another. The expression of other genes is more variable: Their levels are noisy and are different from cell to cell and from individual to individual. This can be highly beneficial in physiological responses to outside cues and stresses. Recent advances have enabled the analysis of differential gene expression at a systems level. Gene regulatory networks (GRNs) involving interactions between large numbers of genes and their regulators have been mapped onto graphic diagrams that are used to visualize the regulatory relationships. The further characterization of GRNs has already uncovered global principles of gene regulation. Together with synthetic network biology, such studies are starting to provide insights into the transcriptional mechanisms that cause robust versus stochastic gene expression and their relationships to phenotypic robustness and variability. Here, we discuss GRNs and their topological properties in relation to transcriptional and phenotypic outputs in development and organismal physiology.
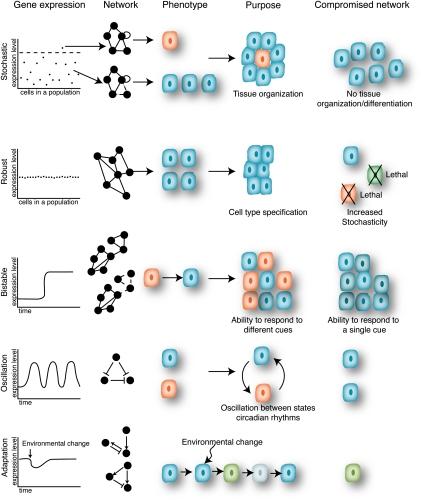
Biological processes can be deterministic and robust, or more stochastic and variable. For example, in development and differentiation, little deviation is tolerated. However, responses to stress can be more stochastic, thereby providing a population of cells or organisms with different outputs to adapt or survive under adverse conditions. Biological robustness and stochasticity can be controlled, at least in part, at the level of differential gene expression. In the last decade or so, the field of systems biology has extensively studied the mechanisms of differential gene expression at the level of gene regulatory networks (GRNs). Technological advances in high-throughput molecular biology have enabled the characterization of large sets of genes and their regulators. The computational modeling and analysis of GRNs, together with the field of synthetic biology, have provided numerous insights into the importance of network architecture and topology in generating differential gene expression and phenotypic outputs. Here, we describe examples of robustness and stochasticity at the organismal or cellular level, as well as at the gene expression level. We discuss the GRN principles and mechanisms that generate these different types of biological outputs (Fig. 1). An overview of frequently used terms is provided in Table 1.
Figure 1.
Robust or stochastic gene expression can generate diverse phenotypes. (Left) Hypothetical gene expression profiles for a population of cells or an individual cell over time (time). Network diagrams are shown to highlight the relationship of gene expression and network dynamics. Cells colored differently reflect different phenotypic fates.
Table 1.
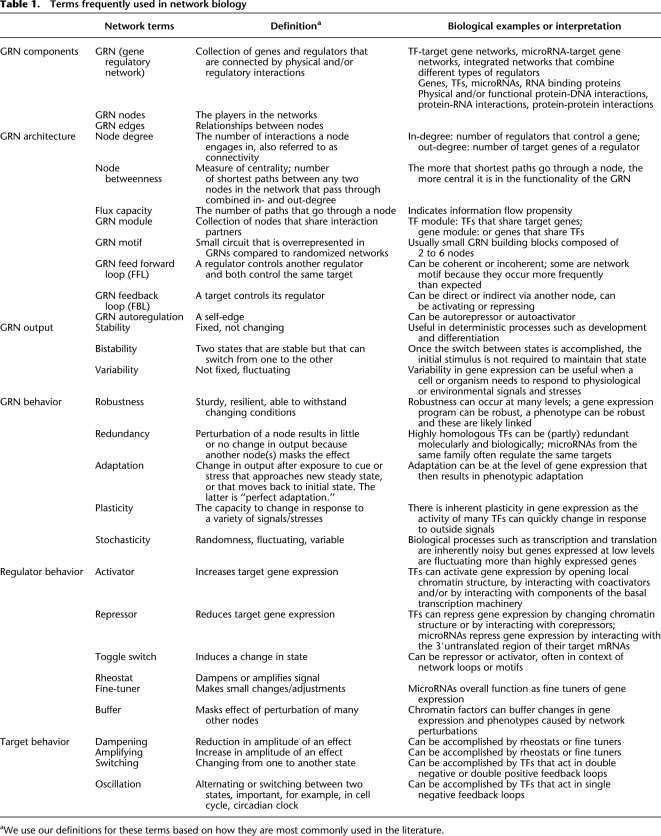
Terms frequently used in network biology
aWe use our definitions for these terms based on how they are most commonly used in the literature.
Differential gene expression
The human genome contains ∼20,000 protein-coding genes. Most of these genes are differentially expressed to generate proteins that are present at specific levels and in specific cells and tissues. These complex patterns of gene expression are generated in response to specific intracellular and extracellular cues and can be controlled at different levels, including transcription, mRNA stability, mRNA translation, and protein stability.
At the level of mRNA synthesis, transcription factors (TFs) control gene expression by directly interacting with cis-regulatory genomic DNA sequences that are usually located in or around their target genes. Changes in the abundance or activity of TFs can result in an increase or decrease in the expression of their downstream targets. Other regulators of gene expression include transcriptional cofactors that can physically interact with TFs, chromatin or components of the basal transcriptional machinery; RNA binding proteins that interact with mRNAs and regulate translation or mRNA stability; and microRNAs that repress mRNA stability by hybridizing to sequences within their mRNA targets. Here, we mainly focus on gene regulation at the transcriptional level.
In all complex organisms, specific transcriptional programs are required for proper developmental patterning and to ensure appropriate responses to changing environmental conditions and stresses. During development, appropriate cell numbers and cell fates together dictate the generation and organization of tissues and organs at the right place and the right time. The expression of developmental genes is often tightly controlled to be within particular levels and to only occur in certain cells irrespective of changes in environmental conditions. For instance, Hox genes are expressed in precise patterns that provide anterio–posterior positional information and segment identity to the developing Drosophila embryo (Lewis 1978; Harding et al. 1985). The specific expression of a subset of Hox genes within a particular embryonic segment determines which body structure that segment will become, such as a leg or an antenna. As a consequence, mis-expression of these genes can result in the formation of an extra pair of wings or the generation of legs in the place of antennae (Casares et al. 1996). The expression of other genes, however, is more variable and can be differentially modulated in response to environmental cues. For instance, yeast cells decrease the expression of RNA metabolism and protein synthesis genes, while increasing the expression of detoxification and protein-folding genes, in response to a broad variety of stressful conditions (Gasch et al. 2000).
Gene regulation is not a linear one-to-one process, but rather occurs in the context of complex networks of interactions between multiple genes and multiple TFs. For instance, the TFs that control the expression of a gene often act together with other TFs. Furthermore, TFs are themselves extensively regulated. The study of GRNs has already provided insights into the systems-level mechanisms of gene regulation that control growth, development, physiology, and stress responses.
Gene regulatory networks
Over the last decade or so, several approaches have been developed that enable the experimental or computational identification of interactions between genes and TFs. These relationships have been mapped onto graphic GRN diagrams that can be interrogated to gain insight into the mechanisms of differential gene expression at a systems level (for review, see Davidson and Levine 2005; Stathopoulos and Levine 2005; Walhout 2006; Long et al. 2008). These GRNs are useful for biologists who seek to understand the regulatory relationships between genes that confer similar phenotypes. These networks can also be used to understand the flow of information in a biological system, to identify circuits that may be used for a specific purpose, and to model changes in gene expression under different conditions.
GRNs are comprised of “nodes,” the genes and their regulators, joined together by “edges,” which represent physical and/or regulatory interactions. Physical interactions between genes and TFs can be delineated using two conceptually and practically different strategies that are highly complementary. The first, TF-centered (protein-to-DNA) methods start with a TF of interest and identify the regions of the genome with which that TF interacts. The most widely used TF-centered methods include chromatin immunoprecipitation (Kim and Ren 2006) and DamID (van Steensel et al. 2001). ChIP has been applied to the majority of yeast TFs, under a variety of experimental conditions (Harbison et al. 2004), as well as to individual regulatory TFs in multicellular organisms (e.g., Odom et al. 2004; Carroll et al. 2005; Cao et al. 2010). The second, gene-centered (DNA-to-protein) methods for the identification of TF–target gene interactions start with a regulatory genomic DNA sequence and identify the repertoire of TFs with which this sequence can interact. One of the few available gene-centered methods is the yeast one-hybrid (Y1H) system, which has been modified for use with large sets of genes and many TFs simultaneously (Deplancke et al. 2004, 2006; Vermeirssen et al. 2007b). The Y1H system has been used to delineate various medium-scale Caenorhabditis elegans GRNs pertaining to tissues, cells, or processes of interest (Deplancke et al. 2006; Vermeirssen et al. 2007a; Martinez et al. 2008; Arda et al. 2010). More recently, the Y1H system has also been used for the first gene-centered Arabidopsis GRN (Brady et al. 2011).
In addition to representing physical interactions, GRN edges can also represent regulatory relationships that can, for instance, be inferred by correlating gene expression profiles between genes and potential regulators. Initially this approach was developed for the yeast Saccharomyces cerevisiae (Segal et al. 2003). More recently, a complex mammalian GRN involved in a pathogen response was delineated using a similar strategy. The resulting relationships were then tested in vivo by RNA interference (RNAi), combined with expression profiling (Amit et al. 2009). While powerful, such methods do not easily identify redundancies in the GRN that may mask relevant relationships, and not all relationships inferred necessarily reflect direct interactions between TFs and their target genes. In addition, these predictive approaches are biased against proteins that are constitutively expressed, but that require post-translational modifications or interactions with cofactors for their activity. Thus, ideally, physical and regulatory relationships need to be combined to obtain the most complete and accurate GRN models.
GRNs can be visualized and analyzed using a variety of computational and mathematical tools (for review, see Babu et al. 2004). Cytoscape is a widely used and easy to implement tool that has been extremely useful for GRN visualization (Shannon et al. 2003). GRNs can be analyzed at different levels (Babu et al. 2004; Arda and Walhout 2010). At the level of overall network architecture and topology, measures include “node degree,” which indicates the connectivity, or number of relationships in which a node engages. There are two types of nodes: Genes and TFs, and therefore GRNs are bipartite. GRNs are also directional as TFs regulate their targets and usually not vice versa. As a consequence, there are two types of degree: The in-degree refers to the number of TFs that bind a gene, and the out-degree refers to the number of genes bound by a TF. While most GRN nodes have a relatively low degree, some are extremely highly connected and are referred to as either “TF hubs” that bind a disproportionately large number of target genes or “gene hubs” that are bound by many TFs (Luscombe et al. 2004; Deplancke et al. 2006; Yu and Gerstein 2006). When the regulation of TF-encoding genes is included in GRNs, these nodes can have both an in-degree and an out-degree. The number of potential information paths that go through such a regulator is the product of the in-degree and out-degree and is referred to by that node's “flux capacity” (Martinez et al. 2008). Finally, the “betweenness” of a node is the number of shortest paths that connect any pair of nodes in the network that pass through that node (Joy et al. 2005). Nodes with high betweenness are centrally located in the network and can connect different network modules.
The overall topology of GRNs has been analyzed in many systems. First, GRNs are not random, as evidenced by the occurrence of TF and gene hubs. Interestingly, however, the out-going degree follows a power-law distribution, whereas the incoming degree best fits an exponential distribution (Luscombe et al. 2004). At present, the significance of this difference is not clear.
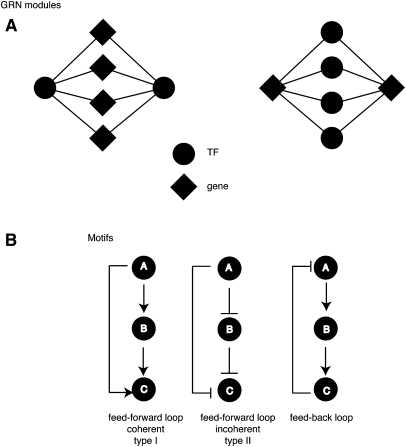
GRN modules are highly interconnected network neighborhoods that point to shared functionality between the nodes involved (Fig. 2). Two types of modules can be identified: “gene modules,” which are defined as sets of genes bound by similar TFs; and “TF modules,” which are sets of TFs that share similar target genes (Vermeirssen et al. 2007a; Arda et al. 2010). A high degree of modularity has been proposed to promote information flow through the network, to generate rapid responses to outside cues (Ravasz et al. 2002; Babu et al. 2004). In Caenorhabditis elegans, the metabolic GRN that must respond to changes in food availability or other stimuli is highly modular (Arda et al. 2010). This GRN contains two modules that are mainly composed of nuclear hormone receptors: TFs that are poised to regulate their targets through interactions with their cognate ligands. Interestingly, most of these nuclear hormone receptors confer a metabolic phenotype when knocked down by RNAi, which indicates that, indeed, they share biological functions, as well as target genes in vivo.
Figure 2.
Common network architectures in GRNs. Examples of modules and motifs are shown. (A) GRN modules. (Left) A TF module; (right) a gene module. (B) GRN motifs. A, B, and C represent three genes that interact in a GRN. FFLs can be divided into two types—coherent and incoherent. In coherent FFLs, the effects of A on C from direct and indirect paths are the same. The type I coherent FFL is the most common (shown here), where A activates B and C, and B activates C. In incoherent FFLs, the effects from A on C are opposite. In this type II incoherent FFL, A represses C, and it activates C by repressing the repressor B. (Right) A feedback loop (FBL) is illustrated in which A activates B, B activates C, and the product of C negatively regulates A.
GRNs can also be analyzed at the level of small circuits or network building blocks that involve only few nodes. These include autoregulatory loops, feed-forward loops (FFLs), and feedback loops (FBLs) (Fig. 2). When such circuits occur more frequently than expected by chance, they are referred to as “network motifs,” which is analogous to the notion of TF binding motifs that are overrepresented in TF target genes (Milo et al. 2002). Because they occur frequently, network motifs likely represent circuits that have provided selective advantages in evolution. Below, we discuss how network topologies and circuits can confer distinct gene expression and phenotypic outputs, such as robustness, stochasticity, and adaptation (Fig. 1).
Robustness
Robustness is the capacity to generate a reproducible trait despite changing conditions. A trait can be a phenotype, as in phenotypic robustness, or a level or pattern of gene expression. Biological systems are overall highly robust because most genes or gene products can be removed without compromising viability. Although minor changes in growth rate have been observed in Saccharomyces cerevisiae for many single gene mutations (Breslow et al. 2008), <20% of the 6000 yeast genes confer lethality when deleted (Giaever 2002; Dixon et al. 2009). Similarly, C. elegans can withstand individual knockdowns of the vast majority of its genes without any obvious effects on viability, reproduction, or longevity (Kamath et al. 2003). Obviously, the ability to disrupt most individual genes with only minor consequences does not mean that they are not important. Rather, there appear to be great levels of redundancy because the effects of gene loss become more obvious in double mutants (Lehner et al. 2006; Byrne et al. 2007; Costanzo et al. 2010). At the level of gene expression, it has been observed that essential genes display lower variability, i.e., are more robust, than other genes, which demonstrates that robustness in gene expression correlates with phenotypic robustness (Batada and Hurst 2007; Lehner 2008).
Stochasticity
Stochasticity indicates variability in phenotype and/or gene expression between genetically identical or highly similar cells that are exposed to the same condition. Stochastic gene expression can be highly advantageous because it can result in phenotypic differences that generate diversity in a population of cells or organisms. Together, different degrees of gene expression enable biological plasticity such that an organism is poised to change or adapt in response to a variety of signals/stresses (Fig. 1). Indeed, genes that are specialized for stress response and energy production display greater variability in expression under stable optimal conditions than other genes (Bar-Even et al. 2006).
There are numerous examples in which stochastic gene expression can be functionally beneficial. For instance, for single-cell organisms, the survival of the population can be ensured under rapidly changing conditions when individuals display very different patterns of gene expression. Indeed, increased gene expression noise has been shown to provide a fitness advantage when cells are exposed to extreme stress (Blake et al. 2006). Stochasticity in gene expression would thus be favored in populations undergoing a broad variety of environmental changes: Greater variability results in greater plasticity, and therefore a higher probability that at least some cells can survive many changes in condition. For instance, in a population of bacteria, a small fraction of cells enters into a slow-growing state called “persistence.” Entry into and exit from this state relies on stochastic expression of the hipA gene. While in the persistence state, bacteria are less sensitive to environmental stresses such as antibiotic treatment and will therefore have a competitive advantage (Rotem et al. 2010). However, when conditions are optimal, these slow-growing cells are less competitive than their fast-growing counterparts. Thus, cells within a bacterial population cycle between two growth states—one that favors growth and reproduction, and one that favors survivability. This type of “bet-hedging” does not depend on the bacteria first sensing stresses in order to induce a response and thus provides a higher probability of survival for the population following sudden environmental changes.
In complex multicellular organisms, similar patterns of stochastic gene expression can be used to cycle between two states. Chang et al. (2008) analyzed SCA1 expression in a population of hematopoietic progenitor cells. For the most part, SCA1 levels in the population followed a normal distribution; however, spontaneously arising outlier cells display either very high or very low levels of SCA1 expression. These outlier cells were isolated and propagated, and the level of SCA1 was measured over time. Populations arising from such outlier cells initially expressed similar levels of SCA1 as the cloned parental cell. However, they displayed a slow resetting to the more broadly distributed SCA1 levels of the original cell population. In embryonic stem (ES) cells, the expression of the pluripotency marker NANOG is similarly noisy, with two peaks in expression levels that define two populations of cells: one that expresses low levels of NANOG and one that expresses high levels of NANOG (Kalmar et al. 2009). These populations differ in their expression of differentiation markers: The population with low levels of NANOG expresses these markers and is as a consequence more susceptible to induced differentiation. For both hematopoietic progenitor cells and ES cells, keeping two populations may guarantee that some cells in the population are poised to differentiate in response to a variety of cues, while a pool of uncommitted cells is maintained as well.
Stochastic processes can also be used to generate noise in gene expression that leads to permanent cellular variation within an individual. For example, in mammals, the expression of olfactory receptors is highly stochastic such that each olfactory sensory neuron expresses only one of the hundreds of olfactory receptors encoded by the genome (Mombaerts 1999). This selection results in the generation of a population of cells in which each cell expresses a different receptor. After exposure to an odor, it is important to have this type of gene expression to enable a sophisticated and appropriate response. This would not be feasible when all cells express the same olfactory receptor, or when one cell expresses all of them. Interestingly, however, this mechanism is not universal: The C. elegans genome also encodes hundreds of olfactory receptors, but its sensory neurons each expresses multiple receptors, and likely in a nonstochastic manner (Bargmann 2006). C. elegans only has a limited number of sensory neurons, and odors are interpreted as attractants or repellents depending on the cell in which these odorant receptors are expressed. Thus, even for similar biological processes, evolutionary differences can result in robustness in one organism and stochasticity in another.
Stochastic gene expression can result in robust phenotypes
Stochasticity and robustness are not mutually exclusive (Fig. 1). For instance, stochastic gene expression is not restricted to phenotypes that are variable. It can also be used to generate robust phenotypes. Developmental processes often rely on the differentiation of a single cell in a population of identical cells to guide the subsequent formation of an organ or tissue. The differentiation of this individual cell is a random and rare event. It is caused by stochastic gene expression that results in levels of expression of one or several genes crossing a threshold level within an individual cell, which promotes differentiation of that cell. Once this cell differentiates, the same process in adjacent cells is blocked. For example, during eye development in Drosophila, a single cell in each ommatidium is selected to become the R8 photoreceptor. Once this cell is selected, it prevents other cells from becoming R8 cells and directs assembly of the rest of the photoreceptor cells within each ommatidium (for review, see Roignant and Treisman 2009).
Stochasticity can also be used as a switch to a more robust state that can subsequently become stabilized by additional mechanisms. One example of this is the reprogramming of somatic cells into induced pluripotency (iPS) cells by the TFs OCT4 (also known as POU5F1), SOX2, KLF4, and MYC (Wernig et al. 2007). Overexpression of these TFs can induce de-differentiation; however, it is a rare event, and after 3–4 wk of treatment with these factors, only 1%–20% of the cells present are iPS cells. Using individually isolated B-cells, Hanna et al. (2009) expressed these factors and followed cell fate over an extended period of time. Cells were separated into individual wells, and after 18 wk, iPS cells were present in 92% of the wells. This demonstrates that following expression of these factors, all B-cells are capable of reprogramming, but illustrates that it is not a frequent event. Rather, the findings point to a stochastic process whereby a rare event triggers a switch that promotes de-differentiation. Once this switch has been triggered, however, the iPS state is stable, and its gene expression program is likely highly robust; e.g., tissue-specific genes are turned off, and pluripotency genes are turned on (Boyer et al. 2005).
The fact that some processes favor stochasticity while others have selected robustness suggests that these are both extensively regulated. We discuss below how robustness and stochasticity can be generated at the level of individual genes and regulators, or at the network level.
Mechanisms of robustness and stochasticity at the node level
Noise can be separated into two types: intrinsic and extrinsic (Swain 2002). Noise that occurs as a result of the inherent probabilistic nature of transcription or translation of a gene is called “intrinsic noise.” In contrast, “extrinsic noise” is transmitted to a gene from elsewhere in the network, for instance, due to fluctuations in the concentrations or activation of the trans-acting regulators of that gene (Swain et al. 2002). Noise is inherent to biological systems, but it can be regulated or reduced in a gene-specific manner to tailor the appropriate degree of variability.
Single-cell analysis in yeast has shown that noise in protein expression is predominantly the result of changes in mRNA expression and stability (Newman et al. 2006). High-plasticity yeast genes have been identified by the application of multiple stresses, followed by expression analysis (Tirosh and Barkai 2008). High-plasticity genes can be defined as genes that change in expression between many different conditions. Such genes are generally noisier in expression even under stable conditions (Bar-Even et al. 2006). Tirosh and Barkai (2008) compared these genes with low-plasticity genes whose expression was unchanged under the different conditions, and identified two main classes of promoters. The promoters of high-plasticity genes tend to exhibit high nucleosome occupancy directly upstream of their transcriptional start site but low nucleosome occupancy more distally. In contrast, low-plasticity genes more frequently display nucleosome free regions at their promoters and well-positioned nucleosomes further upstream. Nucleosomes can decrease the accessibility of TFs to their target DNA sequences. Therefore, competition between TFs and nucleosomes may also be a source of variability (Raser and O'Shea 2004; Choi and Kim 2009). Active promoters tend to have overall decreased nucleosome occupancy. As such, essential genes tend to cluster around open chromatin, which may lead to more robust expression levels (Batada and Hurst 2007; Field et al. 2008). Specific histones may also play a role, as H2A.Z is enriched at low-plasticity gene promoters (Tirosh and Barkai 2008).
Several studies suggest that gene architecture may also be an important determinant of gene expression noise. For instance, genes that are controlled by promoters that possess a TATA box are noisier in their expression (Becksei and Serrano 2000; Blake et al. 2006; Batada and Hurst 2007; Tirosh and Barkai 2008). It is likely that other promoter or gene elements may be involved in the determination of gene expression noise as well. Some of these elements may be more specific to small sets of genes, and others may be more abundant (such as the TATA box).
In addition to mRNA levels, the timing of gene expression can also be stochastic or robust. In Drosophila, many promoters of genes that are required for embryogenesis are preloaded with RNA polymerase II (Pol II), allowing accelerated induction of gene expression (Hendrix et al. 2008; Boettiger and Levine 2009). It has been shown that such preloading can minimize variability between cells associated with transcriptional induction. Genes lacking stalled Pol II exhibited greater variability in their activation times across different cells in the Drosophila presumptive mesoderm and much greater stochasticity in their expression profiles (Boettiger and Levine 2009). The time between activated gene expression in the first and the last nucleus is as little as 2–3 min for genes with stalled Pol II. In contrast, this variation in activation is 15–20 min or more for promoters lacking stalled Pol II (Boettiger and Levine 2009). Polymerase preloading may therefore minimize stochasticity across a tissue to promote proper developmental timing and coordination.
Mechanisms of robustness and stochasticity at the network level
Redundancy
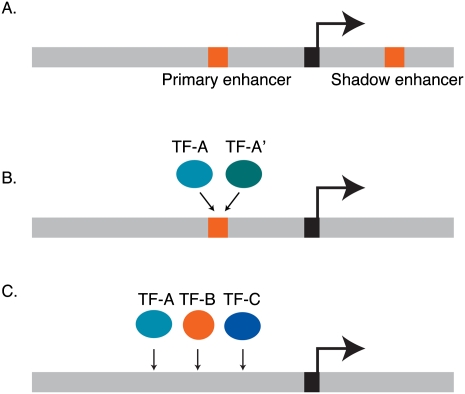
In the last few years, several organizing principles have been discovered that can dictate different types of gene expression outputs at the GRN level. For instance, robustness can be conferred through the redundant wiring of TFs that converge onto individual genes in three different ways. First, “shadow enhancers” have been found in Drosophila that function redundantly with “primary enhancers” but that are often located relatively far from the transcriptional start site (Fig. 3A; Werner et al. 2007; Hong et al. 2008; Frankel et al. 2010; Perry et al. 2010). When cloned in front of a reporter gene, primary and shadow enhancers both confer a highly similar pattern of expression that recapitulates part of the expression of the endogenous gene. A recent example involves the Drosophila shavenbaby (svb [also known as ovo]) gene that is required for the proper formation of hair-like projections called trichomes (Frankel et al. 2010). svb expression is regulated by three enhancers that generate the complete svb expression pattern as well as two shadow enhancers that drive expression in overlapping regions. At the optimal temperature, loss of these distal enhancers results only in minor alterations in the number of trichomes. At higher temperatures, however, loss of these enhancers results in a significant decrease in trichome number. Similarly, in sensitized genetic backgrounds, in which another gene involved is mutated, loss of these enhancers resulted in increased defects in trichome formation. Similar results were obtained for several targets of the Dorsal TF, and with proximal and distal enhancers of the Drosophila snail gene (Hong et al. 2008; Perry et al. 2010). When either the shadow or the primary enhancer was deleted from the snail locus, embryos reared at 22°C developed normally and no discernible differences in expression were observed. At a higher temperature (30°C), however, wild-type embryos were not affected, whereas 20% of the nuclei in the presumptive mesoderm lacked expression in embryos where one of the enhancers was deleted. Together, these data demonstrate that one enhancer is sufficient under optimal conditions, but that redundancy is important under adverse environmental conditions. Enhancer redundancy, therefore, increases robustness and buffers changes that might result from environmental stresses. The mechanism of buffering gene expression and downstream phenotypes has not yet been elucidated. We speculate that adverse conditions may perturb the upstream networks that converge onto the two enhancers and that these perturbations together result in new network states that cannot support full gene expression and, as a result, wild-type phenotypes. This mechanism is likely conserved in other organisms, as similar redundant enhancers have been identified in the mouse Sox10 gene (Werner et al. 2007). Finally, we propose that shadow enhancers will be enriched in essential and other developmental genes compared to stress genes that benefit from higher degrees of stochasticity.
Figure 3.
Redundancy can be conferred by different mechanisms. (A) Genes can be controlled by redundant enhancers: a primary, proximal enhancer and a distal, secondary shadow enhancer. (B) Different TFs from the same family can bind the same cis-regulatory site and control the gene redundantly. (C) Multiple different types of TFs can redundantly control gene expression by together binding to a cis-regulatory module (e.g., enhancer).
The second mechanism that can confer redundancy is when multiple TFs from one TF family can bind the same cis-regulatory DNA element (Fig. 3B). For example, highly similar ETS-type TFs control overlapping sets of targets in human T-cells (Hollenhorst et al. 2007). In C. elegans, FLH-1 and FLH-2 are FLYWCH-type TFs that both bind to a set of microRNA gene promoters. In either flh-1 or flh-2 mutant embryos, little effect on microRNA expression was observed. However, in double mutants, microRNA expression increased dramatically, and there was an increase in larval lethality as well (Ow et al. 2008). Currently, we do not know the extent to which this mechanism of redundancy is used in different organisms, or which types of genes it applies to. Comprehensive GRN mapping studies will shed further light onto the generality of this type of transcriptional redundancy.
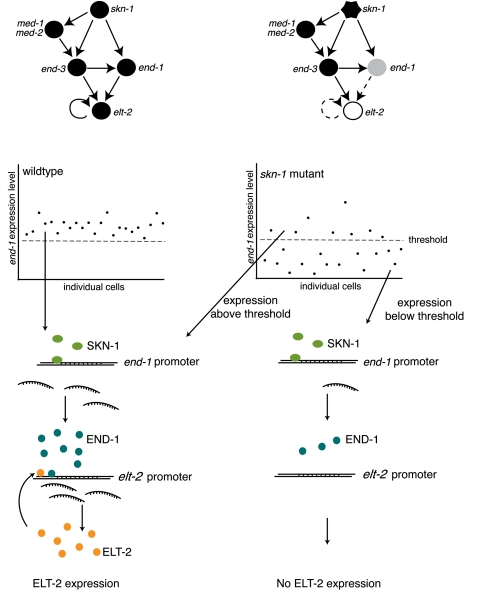
The third mechanism by which redundancy can occur is when TFs from different families interact with a single enhancer or cis-regulatory module that harbors multiple cis-regulatory elements (Fig. 3C). For instance, C. elegans intestinal development is a tightly regulated process that is controlled by a GRN of highly connected TF-encoding genes (Maduro et al. 2007). A key component in this network is SKN-1, a bZIP family TF that regulates the expression of four GATA-type TFs: END-1, END-3, MED-1, and MED-2. The output of the GRN is the activation of the elt-2 gene, which encodes the master regulator of the downstream intestinal gene program (Fig. 4). Interestingly, the expression of these TFs is robust from embryo to embryo, and disregulation of the network can result in partially penetrant lethality because the intestine fails to develop. “Penetrance” refers to the proportion of genetically identical organisms that exhibit a particular phenotype. While some mutations result in a phenotype that is observed in all individuals carrying that mutation, other mutations can result in phenotypes that are only observed in some individuals, despite identical genotypes and environments. For the intestinal GRN, Raj et al. (2010) have proposed that such incomplete penetrance can result from variability in gene expression levels. The investigators found that expression of END-1 becomes highly variable in skn-1 mutants, although the cause of this stochasticity is not fully understood. Interestingly, the percent of embryos that survived was similar to the percent of embryos that express relatively high levels of END-1. Although these data are correlative, they predict that levels of END-1 must exceed a threshold level that results in activation of elt-2 expression, and thus intestinal development and survival. Such threshold effects may be a general mechanism to explain phenotypic differences between genotypically identical cells or organisms.
Figure 4.
Stochasticity in gene expression underlies partial phenotypic penetrance. Summary of data from Raj and colleagues (Raj et al. 2010) explaining partial penetrance of defects in skn-1 mutants. A network controlling intestinal development in C. elegans in wild-type (left) and skn-1 mutants (right). END-1 expression levels must reach a threshold to activate expression of elt-2 and downstream intestinal expression programs.
Recently, it has become apparent that many TF binding events, for instance, detected by ChIP, appear to be nonfunctional because the removal of that TF does not result in a change in the level of the mRNA encoded by the target gene bound (e.g., Li et al. 2008; Cao et al. 2010). In light of the mechanisms that govern redundancy in GRNs that we discussed, it is likely that removal of a single TF would not result in an observable gene expression or biological phenotype. This is in agreement with the observation that most genes and, indeed, also TFs are dispensable for viability and other phenotypes that have been systematically examined. Rather, the removal of multiple TFs would be required to unmask their function in gene expression or a particular biological process. It may well be, however, that loss of individual TFs does result in a gene expression and phenotypic change under adverse conditions, for example, when the organism is exposed to an environmental or physiological stress.
We propose that genes bound by many TFs (high in-degree) could exhibit either high robustness or high plasticity, depending on the (local) network configuration. First, as discussed above, a gene that is bound by a relatively large number of TFs that provide different degrees of redundancy could exhibit more robust expression. This notion is supported by two observations: (1) it has been shown that master regulators of development or differentiation often have a high in-degree (Borneman et al. 2006; Vermeirssen et al. 2007a); and (2) such essential genes generally exhibit less noise than other genes (Batada and Hurst 2007). A high in-degree can also correlate with high plasticity in gene expression (Promislow 2005). This could be a consequence of multiple cis-regulatory elements each of which has its own intrinsic level of noise. In the absence of an outside signal and the resulting activation of one of the interacting TFs, each element could provide a little bit of noise. When many elements occur in a cis-regulatory module, these elements could function additively. However, a robust activation of that gene could ensue when one of the interacting TFs is activated. Further systematic analyses will reveal which of these mechanisms is more pervasive and to which types of genes these apply (see below).
Hierarchy
Analysis of transcriptional networks in yeast revealed a hierarchical organization of regulatory interactions (Jothi et al. 2009). TFs could be organized into three layers: top, core, and bottom. Proteins within a given layer have similar properties. Surprisingly, proteins at the top level are abundant and display high levels of noise when compared to proteins at the bottom level. Low levels of noise in the bottom layer may suggest that additional mechanisms act to buffer noise at this level. Jothi et al. (2009) suggest that this organization may have evolved to minimize transcriptional changes resulting from inadvertent activation of pathways. In addition, they hypothesize that higher levels of noise in the top layer of these hierarchical networks could result in different cells within a population using the same network in different ways and thus be a general strategy to facilitate adaptation to diverse environments.
Noise propagation
The expression level and activity of TFs can affect the variability in expression of their downstream targets. This is because protein–DNA interactions are inherently noisy, probabilistic events: The interaction strength and residence time of a TF with any of its regulatory DNA sites is determined by its concentration, diffusion rate through the nucleus, affinity for each particular DNA sequence, and accessibility to the DNA. When the expression level of a TF is low, the probability of it finding any target sequence is decreased relative to a more highly expressed TF that binds the same DNA sequence. This results in greater variability between cells that respond to the lowly expressed TF. In general, genes that are expressed at low levels display greater variability (Bar-Even et al. 2006). Noise in a network can be transmitted from a TF to its downstream targets (Becksei and Serrano 2000; Pedraza and van Oudenaarden 2005). When some of these targets are themselves TFs, further noise propagation follows. In sum, TFs that are expressed at low levels display greater noise, and this noise is likely transmitted to their target genes.
Within a GRN different nodes can display different levels of noise. In protein–protein interaction networks, for example, highly connected nodes display less expression noise compared to nodes with fewer edges (Lehner 2008). In addition, highly connected nodes are more pleiotropic (i.e., they are involved in multiple biological processes) and more likely to be essential than other nodes, and as stated above, essential nodes are less noisy. Thus, although noise can generate diversity, it is not advantageous for every node in a network.
GRN modules, motifs, and biological outputs
GRN modules can be identified using topological overlap coefficient analysis that determines the sharing of targets between TFs or vice versa (Ravasz et al. 2002; Vermeirssen et al. 2007a). A modular network organization has been postulated to confer several advantages. First, TF modules can provide redundancy in target gene regulation and therefore affect the robustness of the network. Second, the compartmentalization of functions into modules could act to insulate the rest of the network from large changes in expression that occur within a module. Finally, modularity can ensure the coordinated regulation of genes in response to a specific stimulus. Indeed, modularity is often a feature of networks that respond to environmental changes. For instance, bacteria that are subject to greater environmental variation display greater modularity in their metabolic networks than bacteria that are subject to a constant environment (Parter et al. 2007). Similarly, as stated above, a C. elegans metabolic GRN is highly modular and likely responds to different nutritional states (Arda et al. 2010).
Different types of network motifs can generate different gene expression and phenotypic outputs. In GRNs, autoregulation is common, wherein a TF binds to its own promoter and either activates or represses its own expression. Positive autoregulation has been suggested to enhance stochasticity, while negative autoregulation can decrease stochasticity (Alon 2007). Positive autoregulation can induce variability in a population. This is especially true when the initial stimulus generates a weak pulse of TF expression. At low levels of expression, the induced mRNA production is slow because it relies on probabilistic interactions between the TF and its own promoter. Production accelerates gradually as protein levels accumulate (Maeda and Sano 2006). Negative autoregulatory loops function conversely to positive autoregulatory loops. They can be used in situations in which steady-state levels need to be reached quickly. An initial stimulus can induce a strong induction of the promoter resulting in production of the TF. Following the initial stimulus, production levels are decreased as protein levels increase, generating a steady-state level of expression with decreased variability (Becksei and Serrano 2000; Dublanche et al. 2006).
FFLs are one of the most widely occurring network motifs (Milo et al. 2002; Shen-Orr et al. 2002). They are defined as a set of three genes, with two regulators controlling a downstream target and one regulator also affecting the other regulator (Fig. 2). There are a total of eight possible FFLs because regulatory interactions can be either positive or negative, and these can be divided into two types, coherent and incoherent. If we consider three genes—A, B, and C, where C is regulated by both A and B and A regulates B—then both pathways leading from A (direct and indirect) have the same net effect on C in a coherent FFL. In an incoherent FFL, however, the two paths from A would have opposing effects, for example, A activates B and C, but B represses C. FFLs can modulate the timing of gene expression following a stimulation. The most common coherent FFL is the type-1 FFL, wherein A activates B and C, and B activates C (Fig. 2). This circuit can generate delays following either introduction or removal of a stimulus. If A and B are both required for the activation of C (an “AND gate”), the FFL can induce a delay in the activation of C following introduction of a stimulus, but no delay occurs following the removal of a stimulus. A delay can be used to ensure that only a sustained stimulus activates expression of C. This circuit can thus filter out the propagation of noise to C that may occur due to the random, brief activation of A. In contrast, if either activation of A or B is sufficient for the activation of C (an “OR gate”), the FFL can induce a delay following the removal of stimulus but no delay following addition of a stimulus (Alon 2007). The incoherent FFL can be used to generate a transient pulse of expression (Kuttykrishnan et al. 2010). Such a circuit is used in response to glucose in S. cerevisiae: Following stimulation with glucose, an incoherent FFL involving Rgt1 and Mig2 transcriptional repressors generates a brief pulse of expression of the glucose transporter Hxt4. Rgt1 suppresses transcription of both MIG2 and HXT4; Mig2, in turn, also suppresses expression of the HXT4. Interestingly, the same circuit regulates expression of both HXT2 and HXT3, neither of which displays a pulse in gene expression following glucose stimulation. In this instance, the efficiency with which Rgt1 and Mig2 repress expression determines whether or not a pulse in expression will result from this circuit (Kuttykrishnan et al. 2010). This raises an important point, that although certain network motifs are capable of generating a specific behavior, they do not necessarily always do so because they are also subject to input from elsewhere in the network. Examples of all possible FFLs have been identified in a range of organisms. Interestingly, gene expression simulations suggest that the most abundant coherent FFLs display the lowest noise, while the most abundant incoherent FFLs display the highest noise (Ghosh et al. 2005). These observations highlight the importance of FFLs, and likely other network motifs, in the generation and management of gene expression noise.
Surprisingly, FBLs are rare in pure transcriptional networks (Milo et al. 2002; Shen-Orr et al. 2002). By mapping a genome-scale and integrated C. elegans microRNA GRN, however, we found a composite FBL of reciprocal regulation between TFs and microRNAs (Martinez et al. 2008). This type of FBL is a network motif because it is overrepresented in the experimentally mapped network, compared to computationally randomized networks. This suggests that this type of circuitry may provide a selective advantage. Because microRNAs repress target gene expression, there are only two types of TF–microRNA FBLs: one in which the TF activates the microRNA (single-negative FBL) and one in which it represses the microRNA (double-negative FBL). In double-negative FBLs, only one of the two nodes is “on,” and thus such FBLs can function as toggle switches that create bistable states, depending on additional inputs. The activity of single-negative FBLs can result in oscillatory behavior or in stable coexpression of both nodes.
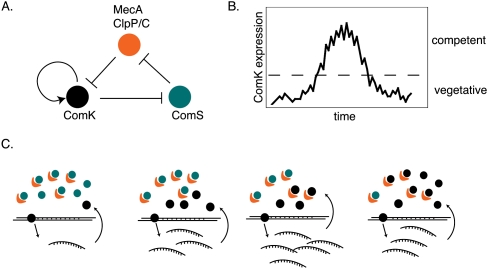
An example of a GRN circuit that uses both autoregulation and positive and negative FBLs is entry into and exit from competence in Bacillus subtilis (Fig. 5). Competence is the ability to take up DNA from the environment and occurs transiently in a small number of cells in the population. It relies on stochastic gene expression of ComK, a TF that is necessary and sufficient to induce competence (Suel et al. 2006). ComK promotes its own expression, but is negatively regulated by a protease complex, CLP–ClpC–MecA. ComK inhibits ComS, which competitively inhibits degradation of ComK by ClpC–MecA. The positive autoregulatory loop of ComK generates stochastic gene expression that, after reaching threshold levels, promotes entry into the competence state. Following the buildup of ComK levels, the negative feedback loop that occurs through ComS and CLP-ClpC-MecA promotes ComK degradation and thus decreases ComK levels and promotes exit from competence. This combination of motifs generates stochastic ComK expression that results in cycling between these two states.
Figure 5.
Entry and exit from competence relies on stochastic gene expression induced by a circuit composed of ComK, ComS, and the protease complex MecA ClpP/C. (A) ComK is necessary and sufficient to induce competence in bacteria. The ComK protein is degraded by the MecA ClpP/C protease complex. ComK negatively regulates the expression of ComS, which, in turn, negatively regulates the protease complex by competing for binding with ComK. (B) The entire circuit cycles over time, resulting in changing levels of ComK, and cycling between competent and vegetative states. (C) The changes that occur over time to network components that result in entry into the competence state.
In the future, individual network motifs that occur in GRNs may allow us to predict the behavior of the target genes with respect to noise generation or noise control. Knowledge of the concentration, expression pattern, and activity of the TFs involved will be of crucial importance. It will also be critical to understand the local network context in which different FFLs and FBLs operate: For instance, when FFLs or FBLs can be affected by many other nodes, they could confer a variety of gene expression outputs, potentially under different conditions.
Adaptation
Adaptation occurs widely in biology and can be defined as adjusting the state of a system following continued exposure to physiological, environmental, or pathological signal (Fig. 1). Either a new state can be acquired, or in the case of perfect adaptation, the system returns precisely to its original state. A clear example of adaptation is the neuronal response to stimuli. This has been extensively studied with respect to odor response. Following an initial stimulus, there is a rapid increase in the frequency of nerve impulses in sensory neurons. The frequency of these impulses then decreases until they reach a plateau level. Although the initial stimulus is still present, the frequency of nerve impulses has returned to a level similar to the unstimulated state (for review, see Kaissling et al. 1987). A recent study by Ma et al. (2009) investigated different three-node network structures that could accommodate biochemical adaptation. They focused on enzymatic networks and found, surprisingly, that only two core topologies support adaptation: one that includes negative FBL with a buffering node, and another that includes an incoherent FFL with a “proportioner node” that is activated in proportion to the most upstream node in the FFL. Their observations suggest that there are clear design principles that enable adaptation in enzymatic networks and that such designs are highly robust because they may provide a selective advantage. Future studies will show whether similar topologies are required for adaptation in GRNs and how such small network circuits behave in the context of larger, system-scale networks.
GRN evolution
Non-coding regions evolve much faster than coding regions, which can result in the loss or gain of TF binding sites. Changes in TF binding can result in alterations of gene expression, which can drive speciation. TF binding sites can differ greatly between species, even in genes and pathways that are highly conserved. For example, CEBPA and HNF4A binding in livers of human, mouse, and dog revealed that binding was shared only 10%–22% of the time between any two of these mammals (Schmidt et al. 2010). Differences in TF binding have also been observed between different individuals of the same species. Analysis of NFKB and Pol II binding in human lymphoblastoid cell lines revealed a significant difference in binding sites between individuals. One-quarter of NFKB binding and 7.5% of Pol II binding differed between individuals (Kasowski et al. 2010). Considering that only a single regulatory TF was examined in this study, the generality of this observation is not yet clear. The extent of variability in binding for essential TFs during development or in other deterministic processes remains an open question. It could be that some TFs functioning throughout the lifetime of the organism are divergent in binding, whereas others are not. In the future, more comprehensive studies, both TF-centered and gene-centered, will reveal the distribution of differential TF binding between and within species. Finally, regulation of a gene by a TF may be conserved between species or individuals, but the physical binding may occur at a different genomic location, either within the same cis-regulatory module, or elsewhere. Indeed, a comparison of similar genomic regions bound by HNF4A between mouse and human revealed that the binding sites for these TFs do not precisely align in two-thirds of the cases examined (Odom et al. 2007). Altogether, these analyses demonstrate the rapid evolution of regulatory regions, and the consequences of this on TF binding.
As GRNs evolve, regulatory interactions are lost and gained. These changes affect not only the edges that are created or lost but also often cause larger changes in network dynamics that affect the expression of additional genes. Landry et al. (2007) measured the effects of nonlethal spontaneous mutations on global gene expression in yeast. Specifically, they measured gene expression in four separate populations that were generated by serially transferring random colonies over 4000 generations. Gene expression that is more sensitive to noise is also more sensitive to mutation. Genes with the greatest changes in expression tended to be underrepresented in key biological processes such as the cell cycle, cell growth, maintenance, and transcription, which suggests that robust and stable gene expression is favored and selected for in these processes (Landry et al. 2007).
Studies comparing a common laboratory strain and a wild isolate of S. cerevisiae revealed a total of 1528 genes, about one-quarter of all yeast genes, that showed significant differences in their expression, and 62 genes differed by greater than eightfold. After mating these two strains, haploid segregants were isolated for gene expression profiling. Analysis of gene expression levels combined with linkage analysis demonstrated that the expression level of most genes is affected by multiple loci. In fact, the investigators estimated that at least five loci affect each gene. Interestingly, a high rate of cis-acting alleles but a relatively small number of trans-acting alleles affected such variation in gene expression (Brem et al. 2002). Similarly, in a study using hybrids of S. cerevisiae and Saccharomyces paradoxus, it was found that under most conditions, differences in gene expression between the two species could be attributed to cis-effects (Tirosh et al. 2009). This indicates that most variation between similar species or strains occurs in regulatory DNA sequences, rather than in protein regulators such as TFs. Interestingly, however, in the presence of glycerol, in which S. cerevisiae grows poorly, the number of differences due to trans-effects was higher (Tirosh et al. 2009). The analysis of gene expression profiles under a number of conditions led to the conclusion that trans-effects are more condition-dependent, whereas cis-effects are more condition-independent. Condition-dependent effects could result from changes in the activity of a sensory TF between the two species, which is then transmitted to its downstream target genes. Mutations in regulatory TFs that respond to particular environmental or physiological conditions are probably less likely to affect the viability of the organism under optimal conditions, whereas mutations in TFs that control genes involved in essential processes are likely less tolerated, and, thus less frequently observed.
In a robust system, it is likely that mutations in cis-regulatory sequences can be tolerated because they can be buffered by the different redundancy mechanisms discussed above. However, it could be that such mutations are unmasked under detrimental environmental conditions or if multiple mutations perturb the same GRN module or motif. The rapid evolution of non-coding DNA sequences is likely more easily tolerated because non-coding mutations can frequently occur without dramatically altering gene function. Most TFs can bind a variety of highly similar DNA sequences (Badis et al. 2009; Grove et al. 2009), and, therefore, a single change in a TF binding site may not result in a complete loss of TF binding. Trichome patterning in different species of Drosophila provides an example of the evolution of regulatory regions, because it differs greatly in Drosophila sechellia compared to Drosophila melanogaster. Body segments that are covered by trichomes in D. melanogaster are not in D. sechellia, and this correlates with the expression of the trichome-regulating gene svb (as discussed above). These differences are the result of individual changes in three enhancers. This suggests that the evolution of svb expression was the result of multiple small changes rather than a more drastic mutation resulting in the elimination of an entire enhancer region (McGregor et al. 2007). Interestingly, shadow enhancers are located relatively far from their target gene, at least in the few cases examined, whereas the primary enhancer is located close to the gene. This organization may permit genetic changes in promoter regions that drive evolution or speciation, while maintaining at least some functionality through the shadow enhancer, or vice versa.
Finally, changes in TF binding sites may change stochasticity in downstream target gene expression. Either for transcriptional activators or repressors, a mutation that results in a lower binding affinity could result in more noise in the expression of the target (see above). Conversely, when a binding site becomes closer to the optimal TF motif, robust binding may be favored, which can result in more robust expression of the downstream target gene. For some target genes, such changes in noise can be beneficial, e.g., stress genes; whereas it would be detrimental to genes involved in essential processes such as development and reproduction.
Future challenges
Knowledge regarding the relationships between the organization of GRNs and stochastic or robust gene expression and phenotypic outputs is only just beginning to emerge. In the future, it will be important to interrogate increasingly comprehensive GRNs, in a broad variety of species and under many different conditions, to gain insights into the differential GRN topologies and circuits that drive specific outputs. In addition, it will be important to integrate different types of networks with GRNs, including protein–protein interaction, microRNA, RNA binding protein, and metabolic and signaling networks.
Variance in binding between different types of TFs in different species or within species has only been studied for a handful of TFs. In the future, more TFs will be examined using various TF-centered methods and preferably under different developmental or physiological conditions. Additionally, gene-centered methods need to be used to shed light on the collection of TFs that contribute to the regulation of homologous or orthologous genes. It is likely that the degree of variance in regulation differs greatly from one TF or gene to another.
Throughout development there is a theme of conserved building blocks that can be combined in different ways to produce different outcomes. Indeed, the same signaling pathways can be used for very different purposes within an organism, but also between different species. For instance, Ras signaling is used in vulval development and germline function in C. elegans and in eye development in the fly, and mutations in the Ras pathway can result in cancer in humans (Han 1992). It is likely that networks are coopted for different functions as well. At the network level, an exciting possibility is that there are “core networks” that are conserved between related species, with additional differences in nodes and edges contributing to species-specific adaptations. Alternatively, some networks may be highly conserved between species, but the levels of expression or noise of individual nodes may be vastly different, resulting in different outcomes in different species using the same overall network connections. It is interesting to speculate that changes in robustness or stochasticity in a network add to this diversity and could essentially generate two very different network dynamics and as a consequence different outputs. The incorporation of different models for TF action, such as Boolean or thermodynamic models, will help to gain further insights into how TF concentration and activity affect GRN states. It will also be essential to incorporate GRN dynamics, for instance, by correlating GRN states, expression outputs, and phenotypes in single cells.
Finally, it will be important not only to study networks that pertain to development or physiology, but also to address the network perturbations that may cause or correlate with disease. It is likely that changes in GRN robustness could play a part in human disease. For instance, when robustness is favored, the introduction of stochasticity can be detrimental. Increased transcriptional noise has been observed in aging cardiomyocytes, leading researchers to hypothesize that cell death resulting from DNA damage in the aging heart may in part be due to an increase in stochasticity in gene expression (Bahar et al. 2006). Many diseases show linkage to multiple loci; others have mutations identified in a number of different genes in different individuals. These findings indicate that the genes that are involved in these diseases are connected in a network and that the disease state is a result of perturbations in that network. Identification of these networks and a better understanding of their dynamics may allow the identification of key nodes or edges that could be targets for therapeutic intervention.
Acknowledgments
We thank Job Dekker, Richelle Sopko, and members of the Walhout lab, especially Efsun Arda and Alex Tamburino, for discussions and critical reading of the manuscript. This work was supported by a CIHR Postdoctoral Fellowship to L.T.M. and by grants R01DK068429 and R01GM082971 to A.J.M.W.
Footnotes
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.097378.109.
References
- Alon U 2007. Network motifs: Theory and experimental approaches. Nat Rev Genet 8: 450–461 [DOI] [PubMed] [Google Scholar]
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. 2009. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326: 257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Walhout AJM 2010. Gene-centered regulatory networks. Brief Funct Genomics 9: 4–12 doi: 10.1093/elp049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Taubert S, Conine C, Tsuda B, Van Gilst MR, Sequerra R, Doucette-Stam L, Yamamoto KR, Walhout AJM 2010. Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans gene regulatory network. Mol Syst Biol 6: 367 doi: 10.1038/msb.2010.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA 2004. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol 14: 283–291 [DOI] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. 2009. Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, et al. 2006. Increased cell-to-cell variation in gene expression in the ageing mouse heart. Nature 441: 1011–1014 [DOI] [PubMed] [Google Scholar]
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, Barkai N 2006. Noise in protein expression scales with natural protein abundance. Nat Genet 38: 636–643 [DOI] [PubMed] [Google Scholar]
- Bargmann CI 2006. Chemosensation in C. elegans. In WormBook (ed. The C. elegans Research Community), pp. 1–29. doi: 10.1895/wormbook.1.123.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- Batada NN, Hurst LD 2007. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat Genet 39: 945–949 [DOI] [PubMed] [Google Scholar]
- Becksei A, Serrano L 2000. Engineering stability in gene networks by autoregulation. Nature 405: 590–593 [DOI] [PubMed] [Google Scholar]
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ 2006. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell 24: 853–865 [DOI] [PubMed] [Google Scholar]
- Boettiger AN, Levine M 2009. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325: 471–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Leigh-Bell JA, Yu H, Bertone P, Gerstein M, Snyder M 2006. Target hub proteins serve as master regulators of development in yeast. Genes Dev 20: 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Zhang L, Megraw M, Martinez NJ, Jiang E, Yi CS, Liu W, Zeng A, Taylor-Teeples M, Kim D, et al. 2011. A stele-enriched gene regulatory network in the Arabidopsis root. Mol Syst Biol 7: 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755 [DOI] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS 2008. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Genome Res 5: 711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne AB, Weirauch MT, Wong V, Koeva M, Dixon SJ, Stuart J, Roy PJ 2007. A global analysis of genetic interactions in Caenorhabditis elegans. J Biol 6: 8 doi: 10.1186/jbiol58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. 2010. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell 18: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao WL, Hestermann EV, Geistlinger TR, et al. 2005. Chomosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Casares F, Calleja M, Sanchez-Herrero E 1996. Functional similarity in appendage specification by the Ultrabithorax and abdominal-A Drosophila HOX genes. EMBO J 15: 3934–3942 [PMC free article] [PubMed] [Google Scholar]
- Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S 2008. Transcriptome-wide noise controls lienage choice in mammalian progenitor cells. Nature 453: 544–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K-Y, Kim Y-J 2009. Intrinsic variability of gene expression encoded in nucleosome positioning sequences. Nat Genet 41: 498–503 [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. 2010. The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E, Levine M 2005. Gene regulatory networks. Proc Natl Acad Sci 102: 4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Dupuy D, Vidal M, Walhout AJM 2004. A Gateway-compatible yeast one-hybrid system. Genome Res 14: 2093–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Mukhopadhyay A, Ao W, Elewa AM, Grove CA, Martinez NJ, Sequerra R, Doucette-Stam L, Reece-Hoyes JS, Hope IA, et al. 2006. A gene-centered C. elegans protein–DNA interaction network. Cell 125: 1193–1205 [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C 2009. Systematic mapping of genetic interaction networks. Annu Rev Genet 43: 601–625 [DOI] [PubMed] [Google Scholar]
- Dublanche Y, Michalodimitrakis K, Kummerer N, Foglierini M, Serrano L 2006. Noise in transcription negative feedback loops: simulation and experimental analysis. Mol Syst Biol 2: 41 doi: 10.1038/msb4100081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E 2008. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 4: e1000216 doi: 10.1371/journal.pcbi.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL 2010. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466: 490–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh B, Karmakar R, Bose I 2005. Noise characteristics of feed forward loops. Phys Biol 2: 36–45 [DOI] [PubMed] [Google Scholar]
- Giaever Ga 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Grove CA, deMasi F, Barrasa MI, Newburger D, Alkema MJ, Bulyk ML, Walhout AJ 2009. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell 138: 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M 1992. Ras proteins in developmental pattern formation in Caenorhabditis elegans and Drosophila. Semin Cancer Biol 3: 219–228 [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R 2009. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462: 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding K, Wedeen C, McGinnis W, Levine M 1985. Spatially regulated expression of homeotic genes in Drosophila. Science 229: 1236–1242 [DOI] [PubMed] [Google Scholar]
- Hendrix DA, Hong JW, Zeitlinger J, Rokhsar D, Levine M 2008. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci 105: 7762–7767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ 2007. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev 21: 1882–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS 2008. Shadow enhancers as a source of evolutionary novelty. Science 321: 1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Balaji S, Wuster A, Grochow JA, Gsponer J, Przytycka TM, Aravind L, Babu MM 2009. Genomic analysis reveals a tight link between transcription factor dynamics and regulatory network architecture. Mol Syst Biol 5: 294 doi: 10.1038/msb.2009.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy MP, Brock A, Ingber DE, Huang S 2005. High-betweenness proteins in the yeast protein interaction network. J Biomed Biotechnol 2: 96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaissling KE, Zack Strausfeld C, Rumbo ER 1987. Adaptation processes in insect olfactory receptors. Mechanisms and behavioral significance. Ann NY Acad Sci 510: 104–112 [DOI] [PubMed] [Google Scholar]
- Kalmar T, Lim C, Hayward P, Munoz-Descalzo S, Nichols J, Garcia-Ojalvo J, Martinez Arias A 2009. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol 7: e1000149 doi: 10.1371/journal.pbio.1000149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak SM, Habegger L, Rozowsky J, Shi M, Urban AE, et al. 2010. Variation in transcription factor binding among humans. Science 328: 232–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Ren B 2006. Genome-wide analysis of protein–DNA interactions. Annu Rev Genomics Hum Genet 7: 81–102 [DOI] [PubMed] [Google Scholar]
- Kuttykrishnan S, Sabina J, Langton LL, Johnston M, Brent MR 2010. A quantitative model of glucose signaling in yeast reveals an incoherent feed forward loop leading to a specific, transient pulse of transcription. Proc Natl Acad Sci 107: 16743–16748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL 2007. Genetic properties influencing the evolvability of gene expression. Science 317: 118–121 [DOI] [PubMed] [Google Scholar]
- Lehner B 2008. Selection to minimise noise in living systems and its implications for the evolution of gene expression. Mol Syst Biol 4: 170 doi: 10.1038/msb.2008.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG 2006. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet 38: 896–903 [DOI] [PubMed] [Google Scholar]
- Lewis EB 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570 [DOI] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6: e27 doi: 10.1371/journal.pbio.0060027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Brady SM, Benfey PN 2008. Systems approaches to identifying gene regulatory networks in plants. Annu Rev Cell Dev Biol 24: 81–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Madan Babu M, Yu H, Snyder M, Teichmann SA, Gerstein M 2004. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 431: 308–312 [DOI] [PubMed] [Google Scholar]
- Ma W, Trusina A, El-Samad H, Lim WA, Tang C 2009. Defining network topologies that can achieve biochemical adaptation. Cell 138: 760–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Broitman-Maduro G, Mengarelli I, Rothman JH 2007. Maternal deployment of the embryonic SKN-1 → MED-1,2 cell specification pathway in C. elegans. Dev Biol 301: 590–601 [DOI] [PubMed] [Google Scholar]
- Maeda YT, Sano M 2006. Regulatory dynamics of synthetic gene networks with positive feedback. J Mol Biol 359: 1107–1124 [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros V, Walhout AJM 2008. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev 22: 2535–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL 2007. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448: 587–590 [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U 2002. Network motifs: Simple building blocks of complex networks. Science 298: 824–827 [DOI] [PubMed] [Google Scholar]
- Mombaerts P 1999. Molecular biology of odorant receptors in vertebrates. Annu Rev Neurosci 1999: 487–509 [DOI] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846 [DOI] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. 2004. Control of pancreas and liver gene expression by HNF transcription factors. Science 303: 1378–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E 2007. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 39: 730–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow MC, Martinez NJ, Olsen P, Silverman S, Barrasa MI, Conradt B, Walhout AJM, Ambros VR 2008. The FLYWCH transcription factors FLH-1, FLH-2 and FLH-3 repress embryonic expression of microRNA genes in C. elegans. Genes Dev 22: 2520–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parter M, Kashtan N, Alon U 2007. Environmental variability and modularity of bacterial metabolic networks. BMC Evol Biol 7: 169 doi: 10.1186/1471-2148-7-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza JM, van Oudenaarden A 2005. Noise propagation in gene networks. Science 307: 1965–1969 [DOI] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M 2010. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol 20: 1562–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promislow D 2005. A regulatory network analysis of phenotypic plasticity in yeast. Am Nat 165: 515–523 [DOI] [PubMed] [Google Scholar]
- Raj A, Rifkin SA, Andersen EC, van Oudenaarden A 2010. Variability in gene expression underlies incomplete penetrance. Nature 463: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK 2004. Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL 2002. Hierarchical organization of modularity in metabolic networks. Science 297: 1551–1555 [DOI] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE 2009. Pattern formation in the Drosophila eye disc. Int J Dev Biol 53: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem E, Loinger A, Ronin I, Levin-Reisman I, Gabay C, Shoresh N, Biham O, Balaban NQ 2010. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc Natl Acad Sci 107: 12541–12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe'er D, Boststein D, Koller D, Friedman N 2003. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet 34: 166–176 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31: 64–68 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M 2005. Genomic regulatory networks and animal development. Dev Cell 9: 449–462 [DOI] [PubMed] [Google Scholar]
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB 2006. An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440: 545–550 [DOI] [PubMed] [Google Scholar]
- Swain PS, Elowitz MB, Siggia ED 2002. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci 99: 12795–12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N 2008. Two strategies for gene regulation by promoter nucleosomes. Genome Res 18: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N 2009. A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324: 659–662 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Henikoff S 2001. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet 27: 304–308 [DOI] [PubMed] [Google Scholar]
- Vermeirssen V, Barrasa MI, Hidalgo C, Babon JAB, Sequerra R, Doucette-Stam L, Barabasi AL, Walhout AJM 2007a. Transcription factor modularity in a gene-centered C. elegans core neuronal protein–DNA interaction network. Genome Res 17: 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V, Deplancke B, Barrasa MI, Reece-Hoyes JS, Arda HE, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Brent M, et al. 2007b. Matrix and Steiner-triple-system smart pooling assays for high-performance transcription regulatory network mapping. Nat Methods 4: 659–664 [DOI] [PubMed] [Google Scholar]
- Walhout AJM 2006. Unraveling transcription regulatory networks by protein–DNA and protein–protein interaction mapping. Genome Res 16: 1445–1454 [DOI] [PubMed] [Google Scholar]
- Werner T, Hammer A, Wahlbuhl M, Bosl MR, Wegner M 2007. Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res 35: 6526–6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R 2007. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448: 318–324 [DOI] [PubMed] [Google Scholar]
- Yu H, Gerstein M 2006. Genomic analysis of the hierarchical structure of regulatory networks. Proc Natl Acad Sci 103: 14724–14731 [DOI] [PMC free article] [PubMed] [Google Scholar]