Abstract
Somatic genome rearrangements are thought to play important roles in cancer development. We optimized a long-span paired-end-tag (PET) sequencing approach using 10-Kb genomic DNA inserts to study human genome structural variations (SVs). The use of a 10-Kb insert size allows the identification of breakpoints within repetitive or homology-containing regions of a few kilobases in size and results in a higher physical coverage compared with small insert libraries with the same sequencing effort. We have applied this approach to comprehensively characterize the SVs of 15 cancer and two noncancer genomes and used a filtering approach to strongly enrich for somatic SVs in the cancer genomes. Our analyses revealed that most inversions, deletions, and insertions are germ-line SVs, whereas tandem duplications, unpaired inversions, interchromosomal translocations, and complex rearrangements are over-represented among somatic rearrangements in cancer genomes. We demonstrate that the quantitative and connective nature of DNA–PET data is precise in delineating the genealogy of complex rearrangement events, we observe signatures that are compatible with breakage-fusion-bridge cycles, and we discover that large duplications are among the initial rearrangements that trigger genome instability for extensive amplification in epithelial cancers.
Genomic alterations are a major characteristic of human cancers. Inherited susceptibility alleles can increase the risk for cancer development and epigenetic changes; somatic mutations including genome rearrangements are believed to drive the development and progression of human malignancies (Sadikovic et al. 2008). Individual tumors frequently exhibit different patterns of somatic mutations, gene copy-number variations (CNV), differential gene expression profiles, and epigenetic modifications (Weir et al. 2007; The Cancer Genome Atlas Research Network 2008). Most of the currently identified genes associated with cancers contribute to oncogenesis as a result of somatic genome rearrangements that result in fusion transcripts or transcriptional deregulation (Largo et al. 2006).
Traditional cytogenetic methods, such as spectral karyotyping (SKY), fluorescence in situ hybridization (FISH), and array comparative genomic hybridization (aCGH), are valuable tools for the analysis of (cancer) genome structural alterations. Although each of the methods bears individual advantages, they all have specific limitations regarding resolution, throughput, and the ability to detect balanced translocations, respectively. Recent advances in DNA sequencing technologies render it possible to systematically identify the majority of genomic rearrangements using genomic DNA–paired-end-tag (DNA–PET) sequencing and mapping strategy (Fullwood et al. 2009). This strategy has been applied using short genomic DNA fragments for human genome analysis (Korbel et al. 2007; Campbell et al. 2008; Wheeler et al. 2008), but has not been fully exploited for the identification of the different types of structural variations (SVs) and their joint architecture, primarily because of cost and the complexity of the analysis.
One of the limitations in the use of short DNA fragments (200–500 bp) for mapping SVs of human cancer genomes as reported in previous studies (Campbell et al. 2008; Stephens et al. 2009) is that such a method is highly dependent on the local complexity of DNA sequence features and requires more sequencing to achieve comparable physical (fragment) coverage. Although short insert libraries have an advantage to identify subkilobase-level SVs, many rearrangements will be missed due to genomic features such as short repetitive sequences, duplicated segments, or segmental transpositions. In theory, large DNA fragments for DNA–PET analysis would have benefits over short fragments in the analysis of complicated DNA sequence features (Fullwood et al. 2009). This is especially pertinent in cancer genomes harboring complex rearrangements and copy-number imbalances. We have performed DNA–PET mapping using fragment sizes of 1, 10, and 20 Kb, and have found that 10 Kb is the optimal fragment size for SV analysis. Here, we report the characteristics of the genomic architectures of two epithelial cancers, i.e., breast and gastric cancers. We comprehensively mapped genome SVs of eight breast cancer samples including five primary breast cancer tumors and three well-established cell lines (MCF-7, T47D, and SKBR3), five gastric cancer samples including four primary gastric cancer tumors, and one cell line (TMK1). These are contrasted to genomes of a colon cancer cell line (HCT116), a chronic myelogenous leukemia (CML) cell line (K562), and two normal individuals (an African and a European). Cross comparison of the cancer and normal genomic maps enabled us to distinguish possible somatic rearrangements from germ-line events and revealed characteristic patterns of SVs that are prominent in breast and gastric cancer genomes. Using the connectivity and quantitative nature of the DNA–PET data, we delineated the genealogy of rearrangement events involved in amplified regions in individual cancer genomes and elucidated potential underlying mechanisms involved in cancer genome instability and aneuploidy.
Results
Genomic DNA–PET sequencing and mapping
The genomic DNAs of 17 human genomes were sheared randomly and gel purified. Fragments ∼10 Kb in length were selected from the gel, except in case of breast tumor 13, where 5-Kb fragments were selected due to limited DNA quality. The DNA fragments of each genome were processed for PET construction and paired-end sequencing analysis (Supplemental Fig. 1). In total, we generated >25.9 Gb of DNA sequence derived from >476 million nonredundant PET sequences from these 17 genomes, and achieved, on average, 81-fold physical (fragment) coverage of each genome (Supplemental Tables 1, 2). Some libraries (n = 5) with larger fragment size and more sequence reads have achieved more than 100-fold physical coverage. The vast majority of PET sequences (89%) were mapped to the reference genome concordantly (concordant PET or cPET) with expected mapping patterns (5′tag → 3′tag) and expected mapping distance (Supplemental Figs. 2, 3; Supplemental Table 3; Supplemental text). The density of the cPETs in any region of the genome was used to reveal chromosomal copy-number variations (Supplemental text; Supplemental Figs. 4–8; Supplemental Tables 4, 5). The rest of the PETs (11%) mapped discordantly (discordant PET or dPET) to the reference genome (wrong paired tag orientation, distance, etc.) (Supplemental Fig. 9). These dPETs provide information about bona fide genomic rearrangements, but include technical noise due to chimeric ligation products during library construction and tag mapping artifacts. Artifactual chimeric ligation products are expected to be randomly scattered over the genome, whereas true rearrangements will be represented by dPET clusters with multiple counts, and we considered nonidentical but overlapping dPETs as likely representing real genomic aberrations.
Detection of rearrangement points and structural variations (SVs)
To distinguish the dPET sequences mapping over rearrangement points from technical noise, we used the PET-mapping overlap scheme (Supplemental Fig. 10; Supplemental Methods) and considered three or more overlapping dPETs as reliable PET mapping across potential rearrangement points (Supplemental text; Supplemental Fig. 11; Supplemental Table 6). A rearrangement point is the junction of two genomic breakpoints. The numbers of rearrangement points identified by dPET clusters with three or more PETs in the 17 genomes ranged from 242 in breast tumor 5 to 1255 in the gastric cancer cell line TMK1 (Supplemental Table 1). The low sequence (base pair) coverage and the short sequence tag length of 25 bp did not allow an efficient assembly of breakpoints. To validate the SVs predicted by dPET clusters, we tested 336 sites passing the ≥3 dPETs criterion by genomic PCR and sequencing, and confirmed 244 (72.6%, Supplemental Tables 7, 8). The remainder either showed no PCR product or gave many bands due to sequence complexity at the junction regions (i.e., short sequence homologies across the genome). By plotting the increasing curve of dPET clusters against the sequencing depth (nonredundant PETs), it is estimated that with 27 million or more nonredundant PETs, we would be able to identify ∼80% of SVs that could be discovered by this technology (Supplemental Fig. 12). We calculated that by using standard short-tag sequencing strategies with 500-bp fragments, we would require 540 million nonredundant PET reads to match this threshold. Most of the 17 genome data sets were either above or close to this mark, except that of three breast tumors (BT1, BT2, BT5), which had only 10 or 5 million nonredundant PET sequences for approximately only 40%–50% of SVs. In our later analyses, we noted that these three tumors showed under-representation of SVs most likely due to lack of comparable coverage.
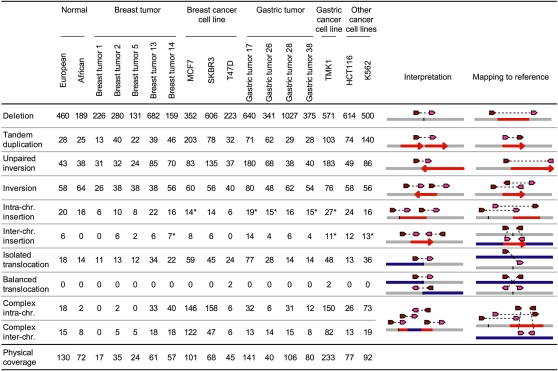
Based on the mapping patterns of dPET clusters that define rearrangement points, various types of SVs can be deduced (Fig. 1; Methods). In addition to isolated SVs, of which architectures can be clearly classified by up to three rearrangement points, we observed a significant number of SVs that were connected by multiple dPET clusters to form complex rearrangement units in which the exact architecture was complicated by overlapping of multiple SV events. Therefore, we classified four or more connected SVs as “complex” (Fig. 1; Supplemental Fig. 13; Supplemental Table 9; Supplemental text). Comparing the dPET cluster sizes of the established SV categories, we found larger cluster sizes for deletions and inversions compared with the other SV categories (Supplemental text; Supplemental Fig. 14).
Figure 1.
Structural variations (SVs) identified by dPET clusters of 15 cancer and two normal genomes. Column “Interpretation” indicates the genomic structure of the sequenced genome deduced from the mapping pattern of the dPET clusters to the human reference sequence (mapping to reference). Dark red arrows represent 5′ anchor regions and pink arrows represent 3′ anchor regions. Gray, blue, and red horizontal lines represent chromosomal segments. Red arrows indicate orientation of chromosomal segments. Asterisks indicate that clusters have been used for more than one insertion.
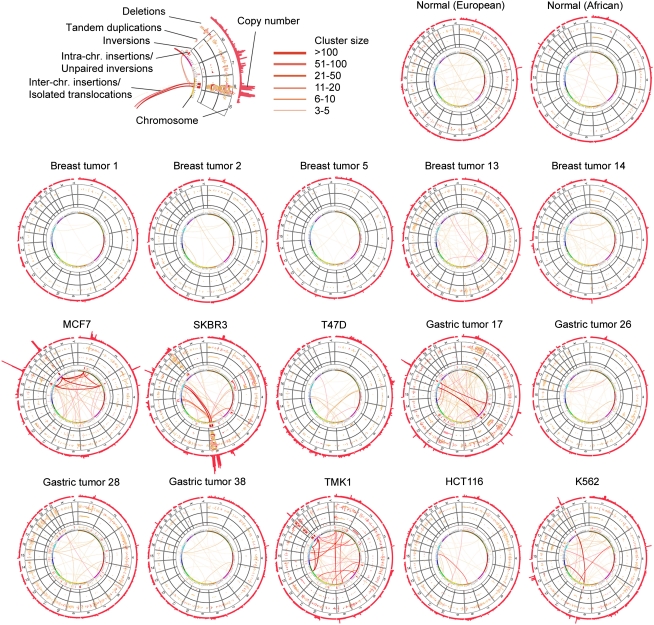
Collectively, the concordant and discordant DNA–PET mapping data constitute the comprehensive SV map for each cancer and normal genome (Fig. 2), displaying the precise genome architecture and quantitative measurements of copy-number variations. For example, the SV map of K562 showed the accurate position of the known BCR–ABL1 translocation between chromosomes 9 and 22 (Groffen et al. 1984; Daley et al. 1990; Heisterkamp et al. 1990) and the previously reported BCAS3–BCAS4 fusion between chromosomes 17 and 20 in MCF-7 (Ruan et al. 2007).
Figure 2.
Karyo-genomic maps of 15 cancer and two normal human genomes. Genomes are arranged in a circular manner with SV categories arranged in concentric layers as indicated on the top, left. Circular plots have been generated using Circos (Krzywinski et al. 2009).
Characteristics of SVs in cancer genomes
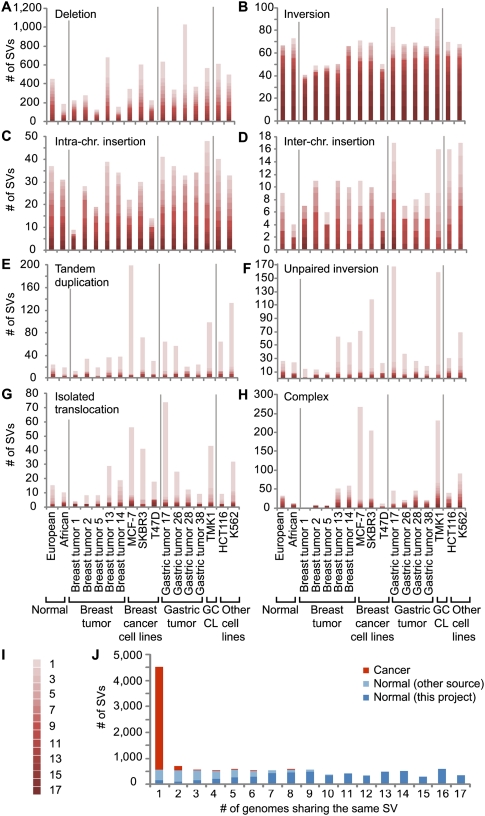
The structural maps of the 17 genomes should include both germ-line and somatic SVs, as well as mapping artifacts and assembly errors in the reference genome sequence. We reasoned that if a particular SV was shared among all 17 genomes, this would most likely represent a rare allele or an assembly error in the reference genome. If an SV was observed in multiple, but not all genomes, it would most possibly represent a germ-line SV that was accumulated in human populations through evolution history. In contrast, if an SV was unique to one particular cancer genome, then it might be considered as derived somatically. Thus, we conducted comparative analysis of all SVs identified by dPET mapping in the 17 genomes, and found 57 different SVs that were common in at least 16 of the 17 genomes (Supplemental Fig. 15; Supplemental Table 10); 1290 SVs that were shared by multiple genomes (two to 15 genomes; Supplemental Table 11), indicating potential “germ-line” origin; 4527 SVs that were unique to single genomes (Supplemental Table 12), of which 4489 SVs were found only in cancer genomes, and which we considered as, most likely, “somatic” events. The median fraction of uniquely observed SVs in the 15 cancer genomes was 26.3% compared with 7.2% in 16 normal genomes (including DNA–PET data of 14 additional normal genomes; P = 4.46 × 10−7; Supplemental Fig. 16), suggesting that a large fraction (1–7.2/26.3 = 72.6%) of unique SVs in cancer genomes was of somatic origin. The PCR validation rate for multiply observed SVs was higher than the rate for uniquely observed SVs (78.4% vs. 69.2%, respectively), indicating a higher false discovery rate for the unique category. Comparing the 17 samples, we found 62 and 96 unique SVs, respectively, in the normal genomes, which could represent novel private germ-line SVs. We also found an average of 115 in breast tumors (range: 43–306), 344 in gastric tumors (range: 73–669), 428 in breast cancer cell lines (range: 104–651), and 584 in the single gastric cancer cell line, TMK1. Although the comparison of the European and African normal samples with European (breast tumors) and East-Asian (gastric tumors) cancer samples is not straightforward, the increase of unique SVs in primary tumors and cell lines can be explained by somatic rearrangements. Some SV classes appear to be more likely germ-line variants than others. Most inversions and intrachromosomal insertions found in the 17 genomes were highly shared among multiple genomes and were significantly represented in the two normal genomes, suggesting that the majority of SVs in these two categories are most likely of “germ-line” origin (Fig. 3). Most of the isolated deletions and interchromosomal insertions could also be considered “germ-line” SVs. In contrast, tandem duplications, isolated translocations, unpaired inversions, and complex rearrangements were over-represented in genome-unique “somatic” SVs (Fig. 3).
Figure 3.
Comparison of SVs across 15 cancer and two normal genomes. (A–H) Frequencies (y-axis) of the indicated SV categories are shown for the individual genomes (x-axis). Cancer groups are separated by vertical gray lines. Degree of recurrent observation of the same SV is indicated in I, where 1 represents the observation in one genome and 17 represents the observation in all 17 genomes. (J) SVs that were observed in the normal individual(s) or which were observed in the cancer genomes, but match those observed in the normal individuals or match by >80% earlier described events (Korbel et al. 2007; Kidd et al. 2008) are indicated in dark blue. SVs that were also observed in the other 14 normal individuals are indicated in light blue. SVs observed only in cancer genomes are indicated in orange. The x-axis represents the number of genomes that share a particular SV, and the y-axis represents the frequency.
As no DNA samples of paired noncancer (normal) tissues were available for the 15 cancer genomes, we used the SVs identified in 12 unrelated normal individuals, two of this study and 10 published previously (Korbel et al. 2007; Kidd et al. 2008), to filter and thereby strongly enrich the set of cancer SVs for somatic events (Supplemental text; Supplemental Fig. 17). Since the aim of this project was to analyze the general characteristics of genome structural changes in breast and gastric cancer, the strong enrichment for somatic events was considered sufficient. Using the common SV filtering approach, we classified the SVs of the 15 cancer genomes that were shared with normal genomes in this study or in previous reports (Korbel et al. 2007; Kidd et al. 2008) as “normal genome SVs” (n = 5105), and the SVs found only in at least one of the 15 cancer genomes but not represented in normal genomes as “cancer genome SVs” (n = 6410; Supplemental Fig. 18). Most cancer SVs (87.3%) were identified only in one genome, whereas normal SVs were mostly shared by multiple genomes (Fig. 3; Supplemental Fig. 19). This suggests that most cancer-specific SVs are likely to be private mutations. We could not identify recurrent rearrangements among our breast and gastric cancer samples, respectively (Supplemental text; Supplemental Tables 13, 14), and observed only regional overlap of potential somatic SVs in MCF-7 and SKBR3 with SVs reported in a recent analysis of breast cancer genomes by paired-end short-fragment sequencing (Stephens et al. 2009; Supplemental Table 15; Supplemental text).
We also investigated whether the segmental size of SVs involved in cancer genomes has some specific characteristics. Interestingly, the sizes of tandem duplications of breast and gastric cancers were clearly larger than those found in normal genomes (median tandem duplication size in normals—22 Kb vs. 100 Kb in breast cancer cell lines [P = 8 × 10−6], 143 Kb in gastric tumors [P = 4.5 × 10−4], and 91 Kb in TMK1 [P = 5 × 10−4]; Supplemental Fig. 20). The size of unpaired inversions was also larger than that of other SV categories. Compared with normal genomes, the size of unpaired inversions was significantly larger in breast-cancer cell lines (P = 0.007). Inversions and insertions did not show significant size differences between cancer and normal.
Characteristics of breakpoints
We investigated the sequence features of germ-line vs. potential somatic breakpoints and observed that significantly more normal SVs (from germ-line DNA) had breakpoint sequence homology than cancer SVs, with striking differences for tandem duplications, unpaired inversions, and complex rearrangements: ∼60% of the normal category had breakpoint homology as compared with ∼20% of the cancer category (Supplemental Fig. 21, P < 10−15). This result suggests that nonhomology-based rearrangements are characteristic for cancer genomes, which is in accordance with the understanding that a significant proportion of normal SVs is mediated by nonallelic homologous recombination (NAHR), whereas the majority of somatic events in rearranged cancer genomes is based on nonhomologous end-joining (NHEJ) (Raphael et al. 2008; Hampton et al. 2009). Further, we investigated the impact of SVs on genes (Supplemental Figs. 22, 23; Supplemental Tables 9, 17; Supplemental text) (a detailed analysis of the genes affected by SVs of the eight breast cancer genomes is provided by Inaki et al. [2011]) and found that breakpoints in normal genomes occurred as frequently in gene deserts as in other regions, whereas cancer breakpoints were significantly under-represented in gene deserts compared with expectation (P < 10−15; Supplemental Fig. 24). Intriguingly, we found that cancer breakpoints were not enriched within gene bodies but within 10 Kb up- and downstream from genes (P < 10−15). Taken together, these data suggest that perturbation of gene regulation may be under positive selection in the evolution of a cancer cell.
Genomic architecture of amplified regions in cancer genomes
A hallmark of cancer genomes is the complex amplification of DNA segments (Hicks et al. 2006; Jönsson et al. 2007; The Cancer Genome Atlas Research Network 2008). This is evident in the 15 structural maps of cancer genomes (Fig. 2). Indeed, amplifications and interchromosomal translocations were observed in both primary tumors and cancer cell lines. The MCF-7 genome has been extensively studied by targeted sequencing analyses of the amplified regions (Volik et al. 2003, 2006; Raphael et al. 2008; Hampton et al. 2009; Supplemental Table 17; Supplemental text) that have revealed complicated sequence structures. However, it is still not clear what events trigger such amplification cascades.
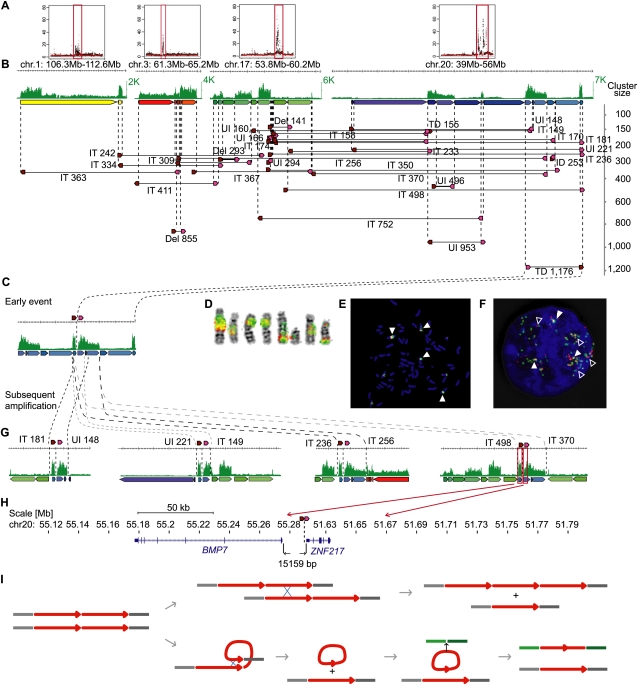
In MCF-7, 26% (268/1047) (Fig. 1) of the rearrangement points were interconnected into only six highly complex units. The largest of these units involved 205 dPET clusters (Supplemental Fig. 13), including mixed types of mapping patterns, with an over-representation of tandem duplications, unpaired inversions, and interchromosomal translocations (Supplemental Table 18), which were tightly associated with the amplified regions on chromosomes 1, 3, 17, and 20. In this complex unit, the SV with the highest dPET cluster count (cluster size, n = 1176) was mapped to a highly amplified region on 20q13, representing a tandem duplication of a large fragment (3.67 Mb) at position 51–55 Mb (Fig. 4). Double-probed DNA–FISH experiments validated this rearrangement, and the extensive FISH signals of the mixed probes in linear position and in multiple chromosomal locations indicated that the junction region of this tandem duplication was further multiplied locally as well as dispatched to other chromosomal locations. Thus, this junction appeared as an epicenter for subordinate dPET clusters that were connected to other parts of the genome, either intra- or interchromosomally. The dPET clusters to the left and right of the initial tandem duplication junction were smaller in size than the initial event, but their sum on each side was comparable to the cluster size of the initial event (Fig. 4; Supplemental text). This suggests that the tandem duplication junction was the origin for the subsequent segmental amplification and dissemination. The dPET connectivity and PET counts together delineate a possible genealogy of rearrangements in the MCF-7 genome. We hypothesize that this 3.67-Mb segment in tandem duplication was the first rearrangement; it then probably created a state of genomic instability and triggered a cascade of subsequent rearrangements that centered around the junction point of the initial tandem duplication, probably by providing the substance for NAHR. Such recombination could take place between sister chromatids to result in further linear amplification of this duplicated segment or intrachromatid generating potential “double minute” constructs that could be further amplified and eventually inserted in other parts of the genome (Fig. 4I).
Figure 4.
Architecture and genealogy of amplifications in MCF-7. (A) Copy-number plots of chromosomes 1, 3, 17, and 20 with amplified regions (red boxes). (B) Concordant tag distributions are shown for amplified genomic regions (top, green track). Genomic segments between predicted breakpoints are indicated by colored arrows (middle) and dPET clusters with cluster sizes greater than 140 are represented by horizontal lines flanked by dark red and pink arrows indicating 5′ and 3′ anchor regions (bottom). Small to large dPET clusters are arranged from top to bottom. All but three dPET clusters were classified as complex. Mapping characteristics are described by: (Del) deletion; (IT) isolated translocation; (UI) unpaired inversion; (TD) tandem duplication. Cluster sizes are given for each cluster. (C) Possible genealogy of amplification. TD1,176 occurred early and subsequent rearrangements have pasted TD1,176 in different genomic contexts (G). (D–F) Double-color FISH using probes flanking TD1,176. Red, chr20:51,920,860–52,096,191; green, chr20:55,137,293–55,311,637. Double signals (filled arrowheads) indicate the fusion of the two loci and single signals indicate the normal genomic distance (open arrowhead). (D) Metaphase chromosomes, (E) metaphase nucleus, and (F) interphase nucleus showing amplification and fusion of breakpoint flanking sequences. (H) BMP7 (left) and ZNF217 (right) are juxtaposed by the TD1,176 rearrangement in a distance of 15,159 bp. (I) Models of local and interchromosomal amplification. Chromosomes are represented by gray and green horizontal lines. Amplified segment is represented by a red arrow. The initial tandem duplication (left) allows local amplification between two sister chromatids or homologous chromosomes (top) or interchromosomal translocation (bottom).
The extensive proliferation of this rearranged structure suggests that some driver element(s) were created to favor the selection of this junction segment during the evolutionary course of this genome. This tandem duplication juxtaposes the BMP7 gene immediately downstream of the ZNF217 gene (Fig. 4H), and this two-gene construct is intact in the minimal core segment that has been amplified most extensively, suggesting that it was advantageous for its extensive amplification. Quantitative reverse transcription PCR (qRT–PCR) of BMP7 and ZNF217 proved that both genes are highly expressed in MCF-7 (Supplemental Fig. 25), suggesting that MCF-7 cells achieve high expression of these two genes through high gene-copy numbers. It is still not entirely understood how this two-gene locus could functionally achieve such superior propagation in MCF-7.
Similarly, the SKBR3 genome also has a few complex units of rearrangements located in highly amplified regions. The largest complex unit consists of 50 rearrangements (Supplemental Fig. 13). The rearrangement with the highest dPET cluster size (n = 624) in this genome is also a large tandem duplication (9.1 Mb) and mapped to chromosome 8 at location 72.8–82 Mb (Supplemental Fig. 26). It is also involved in highly amplified regions and is connected to other rearrangement sites. Based on dPET connectivity and PET counts, we reconstructed the amplified regions that involved at least two levels of subordinate tandem duplications and an interchromosomal translocation that connected chromosome 8 (71.4–82 Mb, 87.2–92.6 Mb, 109.8–129.2 Mb) to chromosome 17 (34.5–37.8 Mb). Similar to the amplicon regions in MCF-7, the SKBR3 data implies that the fusion point created by the tandem duplication occurred early in the genealogy of this breast cancer genome, and that subsequent events have led to the amplification of that fusion junction.
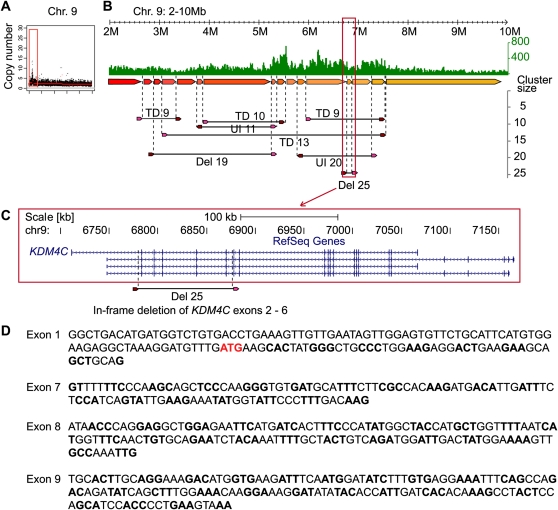
Primary tumor genomes also have extensive amplifications (Fig. 2). For instance, breast tumor 14 displayed a local amplification on chromosome 9p (Fig. 5), where 8 dPET clusters (PET counts >8) were connected to this amplified locus, including four large tandem duplications, two unpaired inversions, and two deletions. The deletion with the largest cluster size excises exons 2–6 of KDM4C (Fig. 5), and the exon 1–7 fusion was validated by RT–PCR. KDM4C (also known as GASC1) has been described as an oncogene in breast cancer (Liu et al. 2009). If translated, this truncated protein would lack the entire JmjN domain, have a partial JmjC domain, and an intact PHD-finger for possible new function.
Figure 5.
The architecture of an amplification in primary breast tumor 14. (A) Concordant tag based copy-number estimate for chromosome 9 indicates an amplification of the distal region of 9p. (B) Concordant tag distribution of chromosome 9 position 2–10 Mb (top, green track). Genomic segments between predicted breakpoints are indicated by colored arrows (middle) and dPET clusters with cluster sizes greater than eight are represented by horizontal lines flanked by dark red and pink arrows (bottom). Abbreviations for mapping characteristics of dPET clusters are described in Figure 4. (C) Genomic structure of KDM4C. Location of amplified deletion (Del25) is indicated by dashed vertical lines. (D) Sequencing result of RT–PCR confirms the in-frame deletion transcript with the more upstream located exon 1.
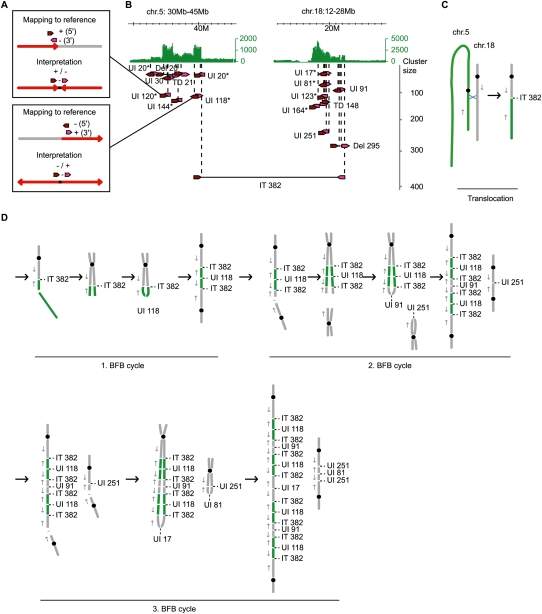
We observed many long-distance unpaired inversions in the breast and gastric cancer genomes, which could indicate the inversion of whole chromosomal arms, large inversions or inverted insertions involved in further rearrangements, or a failure to detect the paired rearrangement point that would classify the event as an inversion. On the other hand, unpaired inversions with a relatively short distance between their breakpoints could occur when a DNA double-strand break results in a truncated chromosome, followed by the replication of the DNA and the joining of the two neighboring ends by a DNA repair mechanism in a head-to-head or tail-to-tail fashion (resulting in a fusion of + and – strands of the sister chromatids) (Fig. 6). Due to the fusion, the two sister chromatids cannot be separated in mitosis and a new break could occur to initiate a new fusion. This mechanism has been described previously as a breakage-fusion-bridge (BFB) cycle (for review, see Tanaka and Yao 2009) and a distance of a few kilobases between head-to-head fusion points has been reported (Lo et al. 2002; Okuno et al. 2004; Bignell et al. 2007). Gastric tumor 17, which had the most rearranged and amplified genome among the four gastric tumor samples (Fig. 2), showed an accumulation of short-distance unpaired inversions in the amplified regions on chromosomes 5, 11, 12, and 18 (Supplemental Fig. 27). This pattern can be explained by BFB cycles that are known to result in amplifications (Tanaka and Yao 2009). The dPET counts implied that a translocation between chromosomes 5 and 18 (cluster size, n = 382) preceded a double-strand break and a subsequent tail-to-tail fusion of chromosome 5 at 39.2 Mb by an unpaired inversion (cluster size, n = 118) (Fig. 6). Further breaks and fusions amplified the chromosomes 5 and 18 segments. A break in a postulated second BFB cycle resulted in two sister chromatid fusions, which showed a larger distance between their breakpoints of 390 and 450 Kb, respectively, and involved a loss of 1.5 Mb. The data imply the propagation of different populations of rearranged chromosomes, which together result in the amplification of the two loci. We observed a larger number of small (<10 Kb) unpaired inversions per chromosome in the gastric cancer samples than in the breast cancer samples (P = 0.00587). This might indicate that BFB cycles are more characteristic for gastric rather than for breast cancer.
Figure 6.
Accumulation of short-span unpaired inversions in amplified regions of gastric tumor 17. (A) PET mapping pattern of short-span unpaired inversions and the interpretation. The mapping of a 5′ anchor (dark red arrow) to the + strand and a 3′ anchor (pink arrow) to the – strand indicates a head-to-head fusion (red arrows) with increasing chromosomal coordinates closer to the breakpoint (top) and a 5′ – strand/3′ + strand mapping indicates a tail-to-tail fusion with decreasing chromosomal coordinates closer to the breakpoint (bottom). UI120 and UI118 in B are examples of head-to-head and tail-to-tail fusions, respectively. (B) Amplifications on chromosomes 5 and 18 of gastric tumor 17 are indicated by concordant tag counts (green). Cancer structural rearrangements with dPET cluster sizes >15 are indicated by dark red and pink arrows for 5′ and 3′ anchors, respectively. Abbreviations and figure structure are described in the legend of Figure 4B. Unpaired inversions with a breakpoint distance <40 Kb are indicated by asterisks. (C) Schematic representation of an isolated translocation between chromosome 5 (green) and 18 (gray). Black circles represent centromeres; blue X represents site of recombination; gray arrows indicate the direction of increasing genomic coordinates. (D) Interpretation of accumulated short unpaired inversions in amplifications by BFB cycles.
Discussion
We have comprehensively characterized SVs of 15 human cancer genomes and two normal human genomes by paired-end-tag sequencing and mapping analysis. The use of a 10-Kb insert size for DNA–PET analysis allows the identification of breakpoints within repetitive or homology-containing regions of a few kilobases in size and results in a higher physical coverage compared with small insert libraries with the same sequencing effort. The latter is based on the fact that a given number of long fragments (long-distance PETs) in a genomic region results in more overlap than the same number of short fragments. The recently reported study by Stephens et al. (2009) on breast cancer genome structures using short DNA fragments may have insufficient physical coverage of the genome to map rearrangements in complicated genomic regions without dramatically increasing the sequencing coverage. Thus, long span DNA paired-end approaches such as outlined here represent a parsimonious and cost-effective approach to comprehensively map structural mutations in cancers.
From the data generated in this study, some characteristic patterns of SVs in cancer genomes emerged. We observed that inversions, insertions, and deletions are more commonly seen in germ-line SVs; whereas somatic rearrangements present in cancer genomes are over-represented in tandem duplications, unpaired inversions, isolated translocations, and in amplified complex regions. Such distinction is likely due to mechanistic differences: SVs with germ-line origins are meiotic recombinants, whereas somatic SVs may use a variety of mechanisms including mitotic DNA repair, transcription-mediated recombination, and generation of double-minute structures (Murnane and Sabatier 2004; Kuttler and Mai 2007; Gu et al. 2008; Lin et al. 2009).
The precise and quantitative connectivity assessment of fusion points in amplified regions by dPET clusters provided an opportunity to delineate the genealogy of amplifications in cancer genomes. In the examples of breast and gastric cancer that we examined, we have gathered evidence to show that large tandem duplications as well as unpaired inversions appear to be early events triggering a subsequent cascade of extensive amplification centered around the junction region. Though it remains a possibility that the tandem duplication may simply function as a “marker” for regional genomic instability, the propagation of the precise tandem duplication through progressive amplification in several cancer genomes suggests that particular tandem duplications have “driver” function. The evolutionary signature of an initiator–amplification cycle is further supported by the observation in the K562 cell line where the BCR-ABL1 balanced translocation between chromosomes 9 and 20 has been subsequently amplified in other parts of the genome (Fig. 2).
It is not clear what mechanisms dictate the initial structure of the early events for amplification and why epithelial cancer genomes favor local duplication, whereas leukemia genomes prefer interchromosomal translocation. The difference at this level may be associated with the spatial state of chromosome conformation in particular cell types and the microenvironment of selective pressure where the primary cells reside. However, the subsequent amplification mechanisms appear to be similar in different cancer types.
Complex rearrangements and extensive amplification are much more abundant in the cancer cell lines than in primary tumors. This is likely due to additional rearrangements that are acquired during in vitro passages of the cell lines. However, detailed analysis of these rearrangements could provide an evolutionary model that “amplifies” the functional importance of any particular rearrangement. To a degree, such a rearrangement map of a highly passaged cell line may represent a steady state of genome fitness for a specific cancer, especially for in vitro conditions.
It is well known that cancers from different lineages such as epithelial and mesenchymal origins harbor very different genetic rearrangements: Balanced translocations are predominantly found in mesenchymal cancers, whereas complex rearrangements are a hallmark of mature epithelial cancers. Our focus on two epithelial cancers, breast and gastric, was an attempt to assess the differences between two distinct epithelial cancers that arise from very different epidemiologic etiologies. We found, within the limitations of sample size, that breast and gastric cancer genomes have some comparable structural characteristics. Both cancers show an enrichment of tandem duplications, unpaired inversions, isolated translocations, and complex rearrangements. In breast and gastric cancer, tandem duplications are larger than other SV categories and have a higher chance of enclosing genes. However, gastric cancer rather than breast cancer shows signatures that are compatible with the breakage-fusion-bridge (BFB) model, which might suggest different mechanisms of genome instability.
The mapping of primary tumor genomes demonstrated in this study validated the feasibility of using large-span paired-end-tag sequencing to characterize clinical samples for genome structural variations. With further optimization for the current prototype of DNA–PET analysis and the continuous drop of sequencing cost, we expect this approach to be sufficiently robust and cost effective to be applied in clinical settings for genetic diagnostics of cancer patients and other genetic disease patients.
Methods
Cell culture
The human cell lines MCF-7, SKBR3, T47D, HCT116, and K562 were obtained from the American Type Culture Collection (ATCC) and TMK1 was kindly provided by Dr Y. Ito (National University of Singapore; Agency for Science, Technology and Research, Singapore). Cell lines were grown under standard culture conditions and harvested at log phase.
Clinical tumor samples
Tissue samples were obtained from five patients who had undergone surgery for breast cancer at the University Hospital Stockholm, Sweden, and from four patients who had undergone surgery for gastric cancer at the National University Hospital of Singapore. All breast tumors were from European patients and belonged to the basal-like subgroup based on microarray expression data. These samples were anonymized prior to sequencing and analysis; therefore, no clinical data are available. The gastric cancer specimens were from four male patients with advanced-stage gastric cancer (TNM stage 3a—gastric tumor 28, stage 3b—gastric tumor 26, stage 4—gastric tumors 17 and 38, respectively) of Chinese (gastric tumors 17, 28, 38) and Malay (gastric tumor 26) ethnicity. Histologically, gastric tumor 26 was a Lauren classification diffuse-type, poorly differentiated signet ring cell carcinoma, while gastric tumors 17 and 38 were Lauren classification mixed-type, poorly differentiated signet ring cell carcinomas, and gastric tumor 28 was a Lauren classification intestinal-type moderately differentiated adenocarcinoma.
Genomic DNA extraction
The genomic DNA of cell lines was extracted by Blood & Cell Culture DNA Kits (Qiagen) and DNA of tumor samples was extracted using AllPrep DNA/RNA Mini Kit (Qiagen) according to the manufacturer's instruction.
DNA–PET library construction, sequencing, and mapping
We prepared and sequenced DNA fragments with the Applied Biosystems SOLiD system using 5–11.5 Kb fragments of hydro-sheared genomic DNA (Supplemental Fig. 2). Paired-end (Applied Biosystems terminology: mate-paired) libraries were constructed as described in Supplemental Figure 1. For gastric tumor 17, LMP CAP adaptors with only a single 5′ phosphorylated end were ligated to the hydro-sheared DNA, thus creating a nick on each strand after circularization of the DNA. Both nicks were translated >50 bp into the circularized genomic DNA fragment by DNA polymerase I, and paired-end tags of >50 bp were released by T7 exonuclease and S1 nuclease. SOLiD sequencing adaptors P1 and P2 were ligated to the library DNA. High-throughput sequencing of the 2 × 25-bp libraries and the 2 × 50-bp gastric tumor 17 library was performed on SOLiD sequencers according to the manufacturer's recommendations (Applied Biosystems). Sequence tags were mapped to the human reference sequence (NCBI Build 36), allowing two color-code mismatches for 25-bp reads and six mismatches for 50-bp reads and paired using the SOLiD System Analysis Pipeline Tool, Corona Lite (Applied Biosystems) (Supplemental text). If sequence tags had multiple mapping locations and one of them was located in the expected distance and orientation to its mate, this location was chosen by a process termed “rescue” (Supplemental text).
PET sequence analysis
PET extraction, classification, clustering of dPETs, identification of SVs and analysis of dPET cluster connectivity by superclustering was performed as described in the Supplemental text.
Cross-genome comparison
Comparison of clusters across different genomes was performed based on an overlap of the 5′ and 3′ anchor regions extended by 10 Kb on both sides. If the 5′ anchor region of a cluster of a second library was overlapping with the 5′ extended anchor region of a cluster of the first library and the same was true for the 3′ anchor regions, the two clusters were grouped together and the 10-Kb extension of the anchor regions were adjusted according to the outermost start and end anchor coordinates. Breakpoint locations of the pooled coordinates were used to compare the identified SVs with SVs in the database of genomic variants (http://projects.tcag.ca/variation/) (Iafrate et al. 2004), paired-end sequencing studies of noncancer individuals (Korbel et al. 2007; Kidd et al. 2008), and paired-end sequencing data of 24 breast cancer genomes (Stephens et al. 2009). The fraction of an SV that overlapped with another event was calculated by the percentage of overlap relative to the larger event. Gene annotations were based on RefSeq Genes downloaded from UCSC (http://genome.ucsc.edu/) (Rhead et al. 2010) on May 14, 2009 using library-specific breakpoints.
Statistical analysis
A two-tailed χ2 test was used to test for differences between the fractions of cancer and normal breakpoints with sequence homologies and to test whether the proportion of normal and cancer breakpoints in gene deserts, genes, and regulatory regions was in accordance with the size proportion of the respective regions relative to the human genome. Mann-Whitney U Test was applied to compare SV size distributions between normal samples and the different cancer categories and to test for differences between the frequency of short unpaired inversions per chromosome of breast and gastric cancer genomes.
Acknowledgments
This work was supported by the Agency for Science Technology and Research (A*STAR), Singapore, and from a grant from the National Cancer Institute (USA) NCI: 5 R33 CA126996-02 (Pair-end-ditag technologies for the complete annotation of fusion genes).
Footnotes
[Supplemental material is available for this article. The sequencing data from this study have been submitted to NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE26954.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.113555.110.
References
- Bignell GR, Santarius T, Pole JC, Butler AP, Perry J, Pleasance E, Greenman C, Menzies A, Taylor S, Edkins S, et al. 2007. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res 17: 1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. 2008. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 40: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley GQ, Van Etten RA, Baltimore D 1990. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science 247: 824–830 [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Wei CL, Liu ET, Ruan Y 2009. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res 19: 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G 1984. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell 36: 93–99 [DOI] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR 2008. Mechanisms for human genomic rearrangements. Pathogenetics 1: 4 doi: 10.1186/1755-8417-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton OA, Den Hollander P, Miller CA, Delgado DA, Li J, Coarfa C, Harris RA, Richards S, Scherer SE, Muzny DM, et al. 2009. A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res 19: 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisterkamp N, Jenster G, ten Hoeve J, Zovich D, Pattengale PK, Groffen J 1990. Acute leukaemia in bcr/abl transgenic mice. Nature 344: 251–253 [DOI] [PubMed] [Google Scholar]
- Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, et al. 2006. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res 16: 1465–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C 2004. Detection of large-scale variation in the human genome. Nat Genet 36: 949–951 [DOI] [PubMed] [Google Scholar]
- Inaki K, Hillmer AM, Ukil L, Yao F, Woo XY, Vardy LA, Zawack KFB, Lee CWH, Ariyaratne PN, Chan YS, et al. 2011. Transcriptional consequences of genomic structural aberrations in breast cancer. Genome Res (this issue). doi 10.1101/gr.113225.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson G, Staaf J, Olsson E, Heidenblad M, Vallon-Christersson J, Osoegawa K, de Jong P, Oredsson S, Ringner M, Höglund M, et al. 2007. High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization. Genes Chromosomes Cancer 46: 543–558 [DOI] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. 2008. Mapping and sequencing of structural variation from eight human genomes. Nature 453: 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA 2009. Circos: An information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuttler F, Mai S 2007. Formation of non-random extrachromosomal elements during development, differentiation and oncogenesis. Semin Cancer Biol 17: 56–64 [DOI] [PubMed] [Google Scholar]
- Largo C, Alvarez S, Saez B, Blesa D, Martin-Subero JI, Gonzalez-Garcia I, Brieva JA, Dopazo J, Siebert R, Calasanz MJ, et al. 2006. Identification of overexpressed genes in frequently gained/amplified chromosome regions in multiple myeloma. Haematologica 91: 184–191 [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. 2009. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 139: 1069–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Bollig-Fischer A, Kreike B, van de Vijver MJ, Abrams J, Ethier SP, Yang ZQ 2009. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene 28: 4491–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AW, Sprung CN, Fouladi B, Pedram M, Sabatier L, Ricoul M, Reynolds GE, Murnane JP 2002. Chromosome instability as a result of double-strand breaks near telomeres in mouse embryonic stem cells. Mol Cell Biol 22: 4836–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane JP, Sabatier L 2004. Chromosome rearrangements resulting from telomere dysfunction and their role in cancer. Bioessays 26: 1164–1174 [DOI] [PubMed] [Google Scholar]
- Okuno Y, Hahn PJ, Gilbert DM 2004. Structure of a palindromic amplicon junction implicates microhomology-mediated end joining as a mechanism of sister chromatid fusion during gene amplification. Nucleic Acids Res 32: 749–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raphael BJ, Volik S, Yu P, Wu C, Huang G, Linardopoulou EV, Trask BJ, Waldman F, Costello J, Pienta KJ, et al. 2008. A sequence-based survey of the complex structural organization of tumor genomes. Genome Biol 9: R59 doi: 10.1186/gb-2008-9-3-r59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2010. The UCSC Genome Browser database: update 2010. Nucleic Acids Res 38: D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Ooi HS, Choo SW, Chiu KP, Zhao XD, Srinivasan KG, Yao F, Choo CY, Liu J, Ariyaratne P, et al. 2007. Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using Paired-End diTags (PETs). Genome Res 17: 828–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadikovic B, Al-Romaih K, Squire JA, Zielenska M 2008. Cause and consequences of genetic and epigenetic alterations in human cancer. Curr Genomics 9: 394–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Yao MC 2009. Palindromic gene amplification–an evolutionarily conserved role for DNA inverted repeats in the genome. Nat Rev Cancer 9: 216–224 [DOI] [PubMed] [Google Scholar]
- Volik S, Zhao S, Chin K, Brebner JH, Herndon DR, Tao Q, Kowbel D, Huang G, Lapuk A, Kuo WL, et al. 2003. End-sequence profiling: sequence-based analysis of aberrant genomes. Proc Natl Acad Sci 100: 7696–7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volik S, Raphael BJ, Huang G, Stratton MR, Bignel G, Murnane J, Brebner JH, Bajsarowicz K, Paris PL, Tao Q, et al. 2006. Decoding the fine-scale structure of a breast cancer genome and transcriptome. Genome Res 16: 394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. 2007. Characterizing the cancer genome in lung adenocarcinoma. Nature 450: 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. 2008. The complete genome of an individual by massively parallel DNA sequencing. Nature 452: 872–876 [DOI] [PubMed] [Google Scholar]