Abstract
Using a long-span, paired-end deep sequencing strategy, we have comprehensively identified cancer genome rearrangements in eight breast cancer genomes. Herein, we show that 40%–54% of these structural genomic rearrangements result in different forms of fusion transcripts and that 44% are potentially translated. We find that single segmental tandem duplication spanning several genes is a major source of the fusion gene transcripts in both cell lines and primary tumors involving adjacent genes placed in the reverse-order position by the duplication event. Certain other structural mutations, however, tend to attenuate gene expression. From these candidate gene fusions, we have found a fusion transcript (RPS6KB1–VMP1) recurrently expressed in ∼30% of breast cancers associated with potential clinical consequences. This gene fusion is caused by tandem duplication on 17q23 and appears to be an indicator of local genomic instability altering the expression of oncogenic components such as MIR21 and RPS6KB1.
Genomic abnormalities in cancer include point mutations, copy number changes, and genomic rearrangements that lead either to transcriptional dysregulation or the generation of fusion gene transcripts, in which two discrete genes are truncated and joined together. The expression and the function of such fusion genes have been extensively studied in hematopoietic cancers and soft tissue tumors (Mitelman et al. 2007; Rabbitts 2009) with the prototype rearrangement being the t(9:22)(q34:q11) translocation in chronic myeloid leukemia (CML) that generates the BCR–ABL1 fusion gene. Recent findings of fusion gene expression in prostate and lung cancers suggest that such fusion transcripts can also be found in solid tumors (Tomlins et al. 2005; Rikova et al. 2007; Soda et al. 2007). The most notable is the TMPRSS2–ERG fusion transcript seen in ∼50% of prostate cancers (Tomlins et al. 2005). These TMPRSS2-ETS family fusions have been shown to enhance invasive activity in prostate cancer, implying a functionality of fusion transcripts in solid tumors (Tomlins et al. 2007, 2008; Helgeson et al. 2008). On the other hand, fusion genes of low frequency but showing transforming activity have also been documented, including the ETV6–NTRK3 fusion found in a rare breast cancer (secretory breast carcinoma) and the EML4–ALK fusion found in a small proportion of non-small-cell lung cancer (6.7%, 5/75) and breast cancer patients (2.4%, 5/209), suggesting that rare mutations may have important driver biological functions in solid tumor development (Tognon et al. 2002; Li et al. 2007; Soda et al. 2007; Lin et al. 2009).
In contrast, gene fusions in breast cancer have not been well studied. To this end, we have pursued the comprehensive identification of cancer genome rearrangements in eight breast cancer genomes (three cell lines and five primary tumors) (Hillmer et al. 2011). Herein, we show transcriptional consequences of these structural genomic rearrangements, especially in generating fusion transcripts.
Results
Transcriptional consequences of structural mutations in breast cancer
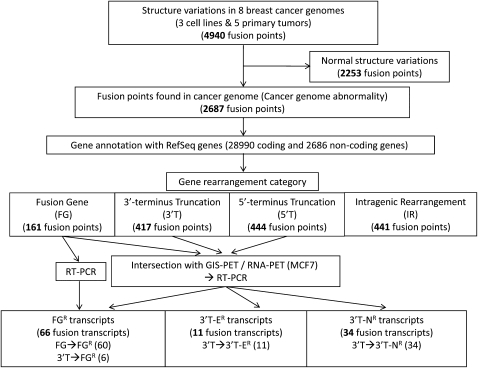
We have described the precise identification of structural abnormalities in cancer cells using a long-span, paired-end-tag sequencing approach called DNA-PET (Hillmer et al. 2011). We now explore the transcriptional consequences of these structural aberrations on a genomic scale, focusing on the results from three breast cancer cell lines and five primary tissues. Using a breast cancer cell line, MCF-7, as a testing platform, we found that gene expression levels as assessed by Affymetrix arrays are significantly correlated with copy numbers as expected: Regions of amplification had higher expression, and regions with putative deletions had lower expression than copy number–neutral regions (Supplemental Fig. 1). Given that deletion and amplification analysis has been studied extensively in breast cancer, we pursued an experimental pipeline with a primary goal of discovering novel cancer-associated transcripts (Fig. 1). Our first analysis revealed 4940 genomic rearrangement points in the three cell lines and five primary breast cancers. Of these, 2253 were identified as normal structural variants, i.e., with potential germline origins. The remaining 2687 fusion points were mapped to the boundaries of 28,990 coding and 2686 noncoding RefSeq genes, of which 1463 fusion points were found to be potential candidates for creating fusion genes. Our analysis does not compare normal–tumor pairs. Therefore, even though these candidate fusions have been screened against known germline SVs (Hillmer et al. 2011), we cannot exclude the possibility that polymorphic germline SVs are included in this candidate list.
Figure 1.
Overview of experimental framework to identify fusion transcripts in breast cancer genomes. (FGR) Transcripts in which exons from two distinct RefSeq genes are fused together in the same direction. (3′T-ER) Fusion transcripts in which the 3′-terminus portion of a given 5′ partner gene is truncated and fused to a non-RefSeq but annotated segment that has evidence for being part of a transcript. (3′T-NR) Any genomic segment that is in the anti-sense strand of any known gene/transcript or in an unannotated region that has no evidence for a transcript in current databases (see Supplemental Glossary).
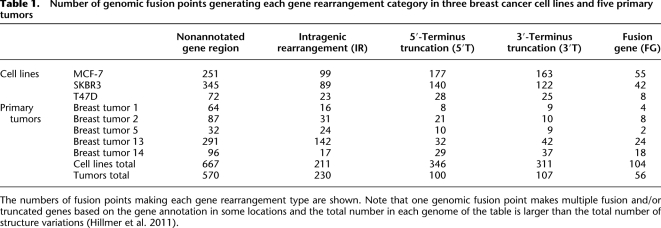
The location of the fusion or rearrangement points and the assessment of the directionality to the gene components allowed us to categorize the impact of genomic rearrangements to gene structures into four categories (Supplemental Glossary; Supplemental Fig. 2A): fusion genes (FG), in which two distinct RefSeq genes are fused together in the same direction; 3′-terminus truncations (3′T), in which the 3′-terminus portion of a given gene is truncated and fused to a segment encoding a non-RefSeq transcript (3′T-E), a nonannotated gene region, or the anti-sense strand of a gene (3′T-N); 5′-terminus truncations (5′T), in which the 5′-terminus is truncated and fused to the nonannotated gene region similarly to 3′T; and intragenic rearrangements (IR) in which the genomic abnormalities (deletion, tandem duplication, inversion, or insertion) are located inside the gene body that result in an internal rearrangement or deletion. As expected, the cell lines harbored ∼2.6 times more structural mutations than primary tumors (546/cell line vs. 213/primary tumor) (Table1). However, the distribution of the category of gene rearrangements differed between the cell lines and the primary tumors. FGs, 3′Ts, and 5′Ts accounted for 46% of the variants in the cell lines versus 24% in the primary tumors, but IRs were proportionately more common in the primary tumors (22% vs. 13% in cell lines) (Supplemental Fig. 2B; Table 1). Even in the IRs, structural mutations associated with exon rearrangements were more frequent in cell lines than primary tumors (Supplemental Table 1; 38% cell lines vs. 24% primary tumors). This may be because of the absence of normal tissue contamination in cell lines making rearrangement detection easier, or because cell lines are under selective pressure to generate potential fusion transcripts through specific types of genomic rearrangements.
Table 1.
Number of genomic fusion points generating each gene rearrangement category in three breast cancer cell lines and five primary tumors
The numbers of fusion points making each gene rearrangement type are shown. Note that one genomic fusion point makes multiple fusion and/or truncated genes based on the gene annotation in some locations and the total number in each genome of the table is larger than the total number of structure variations (Hillmer et al. 2011).
The frequency and heterogeneity of the potential aberrant transcripts made extensive validation a daunting prospect. We therefore pursued a sampling strategy that first examined, in detail, the transcriptional consequences of genomic rearrangements in the MCF-7 cell line. Then, based on the principles uncovered, we expanded the optimized analytical strategy to primary tumors. We had previously pioneered pair-end-tag sequencing of full-length cDNA from the MCF-7 transcriptome (Ruan et al. 2007). This RNA paired-end-tag (RNA-PET) approach captures the sequence of the ends of full-length transcripts with high sequencing depth and is complementary to the DNA-PET approach for identifying unconventional fusion transcripts. To further comprehensively disclose the transcriptome and saturate the discovery of fusion transcripts in MCF-7, we made a new RNA-PET library to increase the coverage to ∼1.8 M full-length transcript equivalents (see Methods). We then sought the intersection between the fusion transcripts detected by RNA-PET data and the DNA-PET library from MCF-7 as a genomic “validation-scan” of the categories of putative aberrant transcripts arising from genomic structural mutations (Supplemental Fig. 2C). This analysis revealed that whereas 35% (19/54) of the predicted FG structural events predicted by DNA-PET data actually intersected with putative fusion transcripts in the RNA-PET libraries, only 24% (40/164) of the 3′T events and 0% of the 5′T events resulted in transcripts. This suggested that the specific structure of the gene rearrangement had consequences in generating certain classes of abnormal transcripts. 5′T events appear to silence genes, whereas other structural rearrangements involving two gene units had a significant probability of generating fusion transcripts. Of the 250 fusion points in MCF-7 within nongenic regions, only one corresponded to a novel gene transcript in the transcriptome libraries arising from a region with no transcript annotation (0.4%). This novel transcript arose from the transcription of genomic sequences in the opposite direction of the FOXA1 gene promoter, but has no open reading frames (ORFs) and is excluded from our further analysis (Supplemental Fig. 2D). The identified transcripts from this exercise and their genomic details are listed in Supplemental Table 2A,C, Supplemental Figure 2E, and Supplemental File 1.
We found that the copy number of genes affected by 98 IRs (rearrangements occurring within gene boundaries) found in MCF-7 were significantly lower than the other categories (data not shown). Furthermore, these genes were significantly enriched in regions reduced to homozygosity as determined by SNP array analysis in MCF-7, suggesting that the allele bearing the IR may be the only allele remaining (WJW Soon and ET Liu, unpubl.). When we assessed the expression levels of genes involved in IR by Affymetrix U133 arrays, we found significantly lower expression levels compared with other categories (Supplemental Fig. 2F). Taken together, these data suggested that the main effect of such IRs was either to attenuate or to silence expression levels of a remaining mutant allele. Alternatively, the IRs may have a tendency to take place in already transcriptionally silent genes.
Because 5′T events would have eliminated the transcriptional start site of the resultant fusion gene, we anticipated that such a rearrangement would not be expressed. Indeed, 5′T rearrangements (N = 177 in MCF-7) do not appear to generate detectable aberrant transcripts (Supplemental Fig. 2C), while the RNA-PET and microarray analyses showed that gene expression from the remaining intact allele was detected at average expression levels (Supplemental Fig. 2F). These data suggest that the main effect of a 5′T event is to silence the affected allele.
With this framework of potential transcriptional consequences for the different classes of genomic rearrangements in breast cancer, we examined the Gene Ontology (GO) of the rearrangement partners based on these genomic features, separating the genes into 5′T + IR that may silence gene expression, FG + 3′T that give rise to aberrant transcripts, and FG alone (Supplemental Table 3). We found that GO terms for biological processes involving cell adhesion and cell signaling are significantly (P < 0.005) enriched in all categories compared with all RefSeq genes, whereas the 5′T + IR class of rearrangements shows significant disruption of gene categories involved in developmental processes, such as nervous system and ectoderm, when compared with other categories (P < 0.005). Taken together, we find that genomic structural mutations in breast cancer appear likely to perturb cell adhesion, cell signaling, and developmental process functions of the involved genes.
Given the interest in discovering novel transcripts in cancer, we decided to focus only on the FG and 3′T rearrangements that putatively generate fusion transcripts (which we also call FGR + 3′TR transcripts) in our extended analysis of other breast cancer cell lines and primary tumors. Because of the difficulty in ascertaining PCR primers for unannotated genomic segments representing the 3′ portion of the putative chimeric transcript, we pursued the validation of the 3′T-ER subset in which the 3′ partner is a known transcript albeit not a RefSeq gene (Methods; Supplemental Glossary) rather than all possible 3′TR transcripts. In this manner, we could predict exon structures in the fused region of 3′T-ER for primer design.
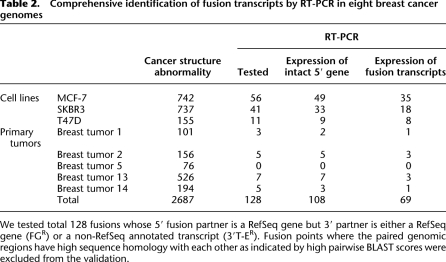
To this end, we sought to validate 128 FG + 3′T-E candidates in the tissues of origin for the presence of fusion transcripts by RT-PCR. Of these, 108 (84%) showed expression of the intact 5′ gene partner, and 69 (54% of the genomic rearrangements or 64% of the expressed 5′ gene rearrangement partners) showed expression of fusion transcripts (Table 2). Fifty-six percent (61/108) of the validated genomic rearrangement points resulted in fusion transcripts in the cell lines as compared to 40% (8/20) in the primary tumors, suggesting that fusion transcript generation is favored in the development of cell lines (P = 2 × 10−4). The increased discovery rate for fusion transcripts from genomic structural variants as compared to the analysis using RNA-PET alone is likely to be due to the negative bias for large transcripts in the RNA-PET protocol. The identified fusion transcripts and their genomic details are listed in Supplemental Table 2A,B and Supplemental File 1.
Table 2.
Comprehensive identification of fusion transcripts by RT-PCR in eight breast cancer genomes
We tested total 128 fusions whose 5′ fusion partner is a RefSeq gene but 3′ partner is either a RefSeq gene (FGR) or a non-RefSeq annotated transcript (3′T-ER). Fusion points where the paired genomic regions have high sequence homology with each other as indicated by high pairwise BLAST scores were excluded from the validation.
Association between tandem duplication and fusion transcripts
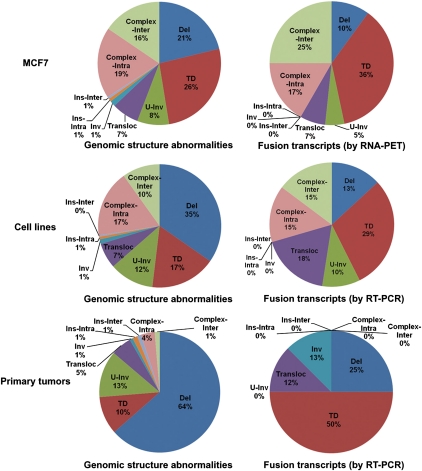
We have noted that single tandem duplications (TDs) in genic regions were common events in both cancer cell lines and primary breast cancers after common variants had been filtered (Hillmer et al. 2011). However, when we examined the SVs that generate validated fusion transcripts, we found that TD was significantly overrepresented among the possible originating structural lesions (Fig. 2; Supplemental Table 4). In all cases, the fusion transcript generated by the TD was a product of a reverse-order variant whereby an originally 5′ gene is now the 3′ contributor to the fusion transcript. Interestingly, the length of these TDs enriched in cancer appeared to be between 50 and 300 kb compared with other SVs (Supplemental Fig. 2G). Since the median size of 28,990 RefSeq coding genes is 23.6 kb, the cancer-associated TDs would be expected to involve the reordering of several genes generating reverse-order fusion transcripts. We confirmed by FISH the presence of the TD as a local amplification event at one TD locus involving the FOXA1 gene that produces a validated novel transcript in MCF-7 (Supplemental Fig. 2D,H). Moreover, expression levels of genes affected by TD are higher than those affected by deletion (Supplemental Fig. 2I,J), with TD breakpoints located frequently within 20 kb of the transcription start site (TSS) (Supplemental Fig. 2K). Taken together, these data suggest an association of TDs with actively expressed genes and with the expression of fusion transcripts.
Figure 2.
Different structure variation types seen in all genomic structure abnormalities and in only those giving rise to fusion transcripts in breast cancer genomes. Fusion transcripts detected by RNA-PET and validated by RT-PCR (top). Fusion transcripts (FGR + 3′T-ER) identified through the RT-PCR screening in three cell lines (middle) and five primary tumors (bottom). (Del) Deletion; (TD) tandem duplication; (U-Inv) unpaired-inversion; (Transloc) isolated translocation; (Inv) inversion; (Ins-Intra) intrachromosomal insertion; (Ins-Inter) interchromosomal insertion; (Complex-Intra and Complex-Inter) intra- and interchromosomal connections in hot spot of genome breakpoints (super cluster size ≥3) (Hillmer et al. 2011).
Translational index and functional architecture of fusion transcripts
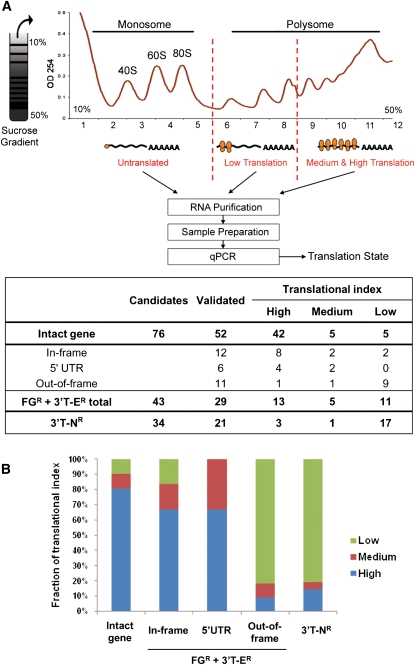
We have validated that the significant fraction of gene rearrangements (FG + 3′T-E) results in fusion transcripts in the entire tumor and cell line set under study. A major question is whether these fusion transcripts are translated or represent noncoding RNAs. We ascertained that individual fusion transcripts could be HA-tagged and shown to be expressed as an intact protein upon transfection into recipient cell lines (data not shown). However, since this does not address whether the chimeric transcript is, indeed, translated within the cell of origin, we asked whether the abnormal fusion mRNAs were bound to cellular polysomes. Determination of the ribosomal loading of an mRNA is an established and reliable method to predict active translation and protein production including cancer cells, in which the number of ribosomes on mRNA has been shown to reflect the translational state of a transcript as examined by Western blot and 35S-methione incorporation (Beilharz and Preiss 2004; Joosten et al. 2004; Tominaga et al. 2005; Provenzani et al. 2006; Sampath et al. 2008; Zhang et al. 2009b; Wang et al. 2010). We purified polysomal fractions by sucrose gradient from extracts of the MCF-7 cell line and analyzed the amount of fusion transcripts in each fraction (Fig. 3A). First, as expected, we observed that 90% of the intact genes of fusion partners were engaged with polysomes (high or intermediate fractions). Intriguingly, when the fusion transcripts were assessed, we found that 44% were bound to either the high or intermediate polysomal content fractions, suggesting that nearly half of all fusion transcripts were likely to be translated in the MCF-7 cell line. We then separated the FGR + 3′T-ER transcripts into three categories based on the intactness of the ORFs of the fusion partners (Supplemental Fig. 3A). Strikingly, in-frame transcripts in which the 3′-gene ORF is in-frame to the 5′ gene ORF, and 5′ UTR transcripts in which the 5′ UTR of the 5′-gene is fused with a recognizable 3′-gene, showed higher translational indexes (83% and 100%, respectively) that are comparable to the cognate intact gene. In contrast, out-of-frame transcripts, in which the 3′-gene ORF is out-of-frame to the 5′-gene ORF, and 3′T-NR transcripts showed much lower indexes (18% and 19%, respectively) (Fig. 3B). This suggests that some types of fusion transcripts (in-frame and 5′ UTR) are translationally enabled, but the others are inactive. The presence of an optimal initiating codon with the Kozak sequence [(A/G)xxATGG], ORF size, and 3′ UTR structure also predicted high polysome content of the RNA (Supplemental Fig. 3B,C; Supplemental Table 5).
Figure 3.
Translational index of fusion transcripts. (A) The figure depicts a typical sucrose gradient fractionation profile demonstrating the separation between translationally active polysomal RNA and the nontranslated monosomal RNA. The number of the ribosomes associated is an indication of its translational potential. The table below shows the numbers of candidates, validated transcripts, and results for the polysomal assay in MCF-7. Candidates include fusion point/splicing variants. ORF structures of each category are explained in Supplemental Figure 3A. The definition of translational index is given in Methods. (B) The fraction of translational index (Low, Medium, High) in each category showing a high translational index for in-frame fusion genes.
Forty percent (44/111) of the identified fusion transcripts (i.e., from FG + 3′T mutations) have at least one predicted protein functional domain (Supplemental Table 6; Supplemental File 2). When the 76 functional domains present in the fusion transcripts were assessed using Pfam and compared with all domains in NCBI, we found a significant enrichment of four domain families: WD40, RhoGEF, DEP, and protein kinase domains (P = 2.0 × 10−6, P = 2.6 × 10−5, P = 4.5 × 10−5, and P = 8.5 × 10−5, respectively) (Supplemental Table 7), all of which have been found in documented oncogenes (Cerione and Zheng 1996; Li and Roberts 2001; Manning et al. 2002; Chen and Hamm 2006).
Potential clinical consequences of transcript rearrangements
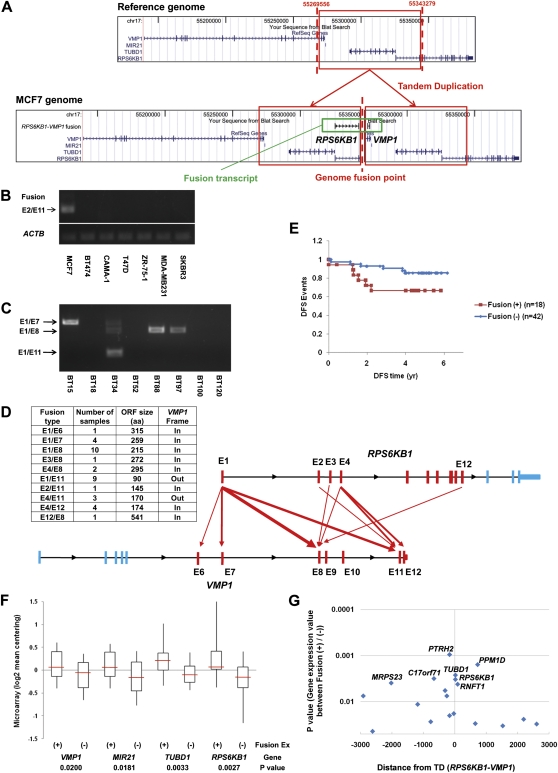
In our analysis, we found a transcript involving adjacent genes on a locus of 17q23 that is known to be amplified in up to 30% of breast cancers (Sinclair et al. 2003; Beroukhim et al. 2010). The DNA-PET data in MCF-7 showed a TD between RPS6KB1 and VMP1 (previously known as TMEM49) genes generating a discernable RPS6KB1–VMP1 fusion transcript (called S6K-fusion hereafter) (Supplemental Fig. 4). The TD juxtaposes the 3′ gene (RPS6KB1) adjacent to the 5′ gene (VMP1) such that the fusion transcript represents a chimera of adjacent genes that are in the incorrect or reverse order (i.e., with RPS6KB1 as the 5′ partner). Our general analysis of TDs suggests that many generate such out-of-order fusion transcripts.
RT-PCR analysis revealed that the major transcript in MCF-7 is the fusion between exon 2 of RPS6KB1 and exon 11 of VMP1. This corresponds to the mapped genome fusion structure found in the long-span DNA-PET library (a fusion between intron 2 of RPS6KB1 and intron 10 of VMP1) (Fig. 4A). However, we did not detect this fusion transcript in the other six breast cancer cell lines tested by RT-PCR (Fig. 4B), and we did not find any TDs at this locus in the other seven breast cancer genomes (Hillmer et al. 2011) and 23 normal DNAs from peripheral blood sources whose genomic structures were analyzed by DNA-PET (data not shown), or in any of the 24 breast cancer genomes sequenced by Stephens et al. (2009). Finally, we extending this analysis to databases of known structural variations (Database of Genomic Variants, DGV) and saw no evidence for this TD. We therefore surmised that such a TD generating a reverse-order fusion transcript is not likely to be a common genomic variant.
Figure 4.
Expression of recurrent fusion gene transcript (RPS6KB1-VMP1) in breast cancer. (A) Structure of the fusion gene transcript created by a tandem duplication (TD) in the 17q23 locus in the MCF-7 genome. The genome DNA rearrangement point detected by DNA-PET data was validated by genomic PCR and sequencing. Expression of the fusion transcript in breast cancer cell lines (B) and in primary tumor samples (C) determined by RT-PCR. (D) Fusion pattern of the transcripts in primary tumor samples showing a range of exons included in the fusions. (E) Correlation of the fusion expression and disease-free survival (DFS) in breast cancer patients. Sixty patients with available information were separated into two groups (18 fusion [+] and 42 fusion [−]) and disease-free survival events were analyzed. Expression of the fusion showed a trend toward correlation (P = 0.06) with poor prognosis of the patients. Correlation of the expression of the fusion with those of genes involved in the TD (F) and with those of neighboring genes (G) within a 3-Mb window from the TD in breast cancer primary tumors. Sixty-eight patient samples with available information were separated into 21 fusion (+) and 47 fusion (−), and the expression of the genes were compared. P-values (<0.05) for the difference of each gene expression between the fusion (+) and (−) samples are shown on each gene location. PTRH2 (P = 0.0010), PPM1D (P = 0.0016), C17orf71 (P = 0.0031), MRPS23 (P = 0.0039), and RNFT1 (P = 0.0042) showed significant correlation as well as TUBD1 and RPS6KB1.
Nonetheless, when we examined the expression of this S6K-fusion transcript in 70 breast primary tumors from Singaporean patients (Ivshina et al. 2006), we found that the fusions are recurrently expressed in 31.4% (22/70 tumors) of the breast cancers (Fig. 4C). Similar to the multiple fusion patterns in TMPRESS-ETS gene family fusions in prostate cancer (Tomlins et al. 2005), we also found multiple fusion points in the abnormal transcripts by RT-PCR and sequencing (total 10 fusion types) (Fig. 4D). The frequent fusion types were E1/E8 (i.e., between exon 1 of RPS6KB1 and exon 8 of VMP1) seen in 10 cases and E1/E11 (nine cases). Of note is that the plurality of the predicted fusion junction at the RPS6KB1 locus (24 fusions use exon 1, while nine fusions use exon 4) is within the two larger introns (intron 1 is the largest and 4 is the second). Similarly, the most common predicted fusion point in the VMP1 gene (14 fusions use exon 8) is also in the largest intron (intron 7). These observations imply random somatic DNA breakage across the gene as the mechanism of TD formation, although the precise DNA breakpoints could not be ascertained because of the unavailability of tumor DNA from these samples.
Among the 70 breast tumor samples, we could obtain limited clinical information for 18 fusion-positive and 42 fusion-negative cases. We could detect a marginally significant difference (P = 0.06, Cox-proportional hazards model) in disease-free survival (DFS) between fusion-positive and fusion-negative patients (Fig. 4E), with no correlation (Fisher's exact test) with other prognostic parameters such as age, tumor size, grade, NPI score, lymph node status, and molecular markers (ER, PgR, and HER2). Thus, S6K-fusion expression may have correlation with poor prognostic outcome of breast cancer patients.
Since the predicted fusion protein from the S6K-fusion transcript does not maintain intact protein domains except one minor species (E12/E8), we speculated that the fusion transcript is a marker for other genomic events at this locus, which may have greater bearing on breast cancer cell growth and biology. Of the neighboring genes in the TD, TUBD1 (an isoform of tubulin, P = 0.0033) and RPS6KB1 (P = 0.0027; whose promoter also drives the fusion transcript) are highly positively correlated with S6K-fusion expression (Fig. 4F). In addition, overexpression of the oncogenic MIR21, which is also inside the TD, also showed positive correlation with the fusion expression (P = 0.0181). Given the small numbers of tumors in our series, it is intriguing that two of the three adjacent genes involved in the TD structure have associations with oncogenesis (MIR21 and RPS6KB1).
Since the TD locus of S6K-fusion is located in the center of an amplicon of 17q22-24 (Zhang et al. 2009a; Beroukhim et al. 2010), we asked if the presence of the S6K-fusion in primary breast cancers can be associated with potential amplification in those tumors (Supplemental Fig. 5). Although we do not have DNA from the tumors from which we assessed fusion gene expression, we tested the possible presence of an amplicon by coexpression of adjacent genes in the 17q23 locus. Using local singular value decomposition analysis of conjoint expression as an indicator for genomic amplification (Zhang et al. 2009a), we found that 41% (28/69) of tumors in our series showed evidence of gene amplification at this locus. Notably those tumors with putative amplification of 17q22-24 are enriched in S6K-fusion (+) tumors (76% [16/21]; P = 8.15 × 10−4). Within 3 Mb upstream and downstream from the TD, we found that expression of 15 genes among the total 49 genes in the region (excluding four genes involved with the TD) is significantly correlated (P < 0.05) with the presence of the S6K-fusion transcript (Fig. 4G). Therefore, when the entire locus is taken into account, the possibility is raised that the S6K-fusion is a marker for genomic instability and/or transcriptional activation at the 17q23 locus. The consequences of this instability include TD and gene amplification events associated with deregulated expression of cancer-associated elements such as MIR21 and RPS6KB1.
When we examined the fusion expression by RT-PCR in five normal breast RNAs from commercial sources (see Methods), however, we detected both E1/E8 and E9/E12 fusions in a single sample (one out of five) (Supplemental Fig. 6). All fusion junctions were confirmed by sequencing. These species occurred at the limits of RT-PCR detection with faint bands detected in a fraction of multiple repeats, but are unlikely to be a PCR artifact since E9/E12 was never isolated before. The result suggested that the fusion transcripts could be detected also in normal breast, albeit at very low levels, which we suspect is a result of trans-splicing (see below).
Discussion
This study comprehensively identified fusion transcripts in five primary tumors and three breast cancer cell lines. Seven of the 43 fusion transcripts we found in MCF-7 have been described in previous studies (Supplemental Table 2A; Bärlund et al. 2002; Hahn et al. 2004; Volik et al. 2006; Bashir et al. 2008; Raphael et al. 2008; Hampton et al. 2009), validating our approach and also indicating the extent of discovery from the clonal depth of this rearrangement-focused sequencing.
The totality of the analysis revealed several distinct findings. First, >50% of the structural rearrangements that intersected two genes generated fusion transcripts when validated by RT-PCR. By intersecting these genomic structural changes with expression of putative fusions as detected by RNA-PET, we found that structural abnormalities involved one gene at the 5′ domain, and either a second gene at the 3′ end or an unannotated region in the 3′ domain are likely to generate an abnormal transcript. However, rearrangements that harbor an unannotated 5′ domain are usually transcriptionally silent. Our polysomal analysis revealed that nearly half of the fusion transcripts are likely to be translated when ascertained in this model cell line. This suggests that a significant proportion of these potentially private structural abnormalities in cancer may generate fusion protein product. Our analysis also showed that single TDs of a large DNA segment are a major source for cancer-associated fusion transcripts. These TDs commonly span several genes, with the resultant fusions having a distinct RNA structure in which the normally downstream gene contributes the 5′ component of the fusion transcript. Moreover, we found that TD breakpoints are enriched within 20 kb of TSS, suggesting that there is an advantage to cancer cells for maintaining the transcriptional regulatory mechanisms of TDs intact. These trends were found not only in cell lines but also in primary tumors. Recently, Stephens et al. (2009) described the deep sequencing of 24 primary breast cancers and also found an enrichment of TDs in these tissues and that many of these TDs also generate fusion transcripts (19 TDs/35 rearrangements making fusion transcripts; 54%) (Supplemental Fig. 7), although none of them are overlapped with identified fusion genes in this study.
Among the eight breast cancer genomes we sequenced, we found no evidence of recurrent genomic rearrangements associated with fusion transcripts, which is consistent with the observations by Stephens et al. (2009). However, when we scanned the expression of the RPS6KB1–VMP1 fusion (S6K-fusion) in 70 primary breast cancer RNA samples in which we had expression array information, we found ∼30% harboring the S6K-fusion RNA. There was, however, considerable heterogeneity in the exons participating in the fusion, suggesting no consistent protein structure correlation. Nevertheless, the expression of the S6K-fusion showed a trend toward association with poor prognosis. The presence of this recurrent fusion transcript was associated with higher expression of adjacent genes, including MIR21 and RPS6KB1, which have known mechanistic association with the cancer phenotype (Bärlund et al. 2000; Sinclair et al. 2003; Iorio et al. 2005; Yan et al. 2008). It is intriguing that the transcriptional start site of the oncogenic MIR21 gene resides within the tenth intron of VMP1 and therefore is an intimate partner in the TD generating the S6K-fusion and that is at the center of a known amplicon in breast cancer. Given the proximity of RPS6KB1, MIR21, and VMP1, it is surprising that evidence of a TD in this specific locus is so commonly linked to overexpression and amplification of this cluster. However, as noted before (Hillmer et al. 2011), TD formation may be involved in the genesis of cancer-associated amplicons. Since the S6K-fusions do not sustain an intact functional protein domain (although some are in-frame and predicted to be translated), we surmise that the fusion transcript may be a complex-genomic “indicator” of genetic instability at the 17q23 locus that leads to gene amplification and/or overexpression of critical oncogenic elements. Thus, in this scenario, this TD is simply a marker that other oncogenic drivers are or will be activated in the evolution of a cancer.
Although we found a recurrent expression of an S6K-fusion gene in our series, the same TD rearrangement was not reported by Stephens et al. (2009) in 24 breast cancer genomes. We believe one reason might be the underrepresentation of 17q23-amplified genomes in the Stephens set. Only one genome (HCC2218 cell line) among 24 genomes showed “amplified” rearrangement on 17q23 that includes a genomic fusion (inverted orientation) between intron 4 of RPS6KB1 and a downstream intergenic region. In contrast, there is evidence that 41% of tumors in our series showed “expression amplicon” footprints in this locus (Zhang et al. 2009a), indicating the likelihood of some genomic amplification in the tumors tested. This difference may be because of differential patient selection by the two groups since all our patients are of Asian descent. Such difference in mutational frequency is well documented in EGFR mutations in Asian lung cancers when compared to Caucasian cases. Since we could not detect the RPS6KB1–VMP1 fusion gene in our preliminary DNA-PET analyses for 23 normal genomes including 10 Chinese individuals, or in the Database of Genomic Variants (DGV), we suspect that the TD-generated fusion gene is at least not likely to be a common germline SV. However, we found S6K-fusions (E1/E8 and E9/E12) in a normal breast RNA among five individuals tested but at very low expression levels. Although it is possible that the RNA from this “normal” breast may indeed have included a small amount of cancerous tissues, an alternative explanation is also possible. It has been reported before that oncogenic fusion transcripts such as BCR–ABL1 could be detected in normal cells of many healthy donors despite normal configuration of the germline DNA (Janz et al. 2003; Hahn et al. 2004; Li et al. 2008). This has been attributed either to structure abnormalities in a small and possibly transient fractional population of normal cells or to trans-splicing. Of note, the JAZF1–SUZ12 (previously known as JJAZ1) fusion transcript detected in normal endometrial stromal cells without any genomic rearrangement is identical to the rearranged fusion gene found in unrelated endometrial stromal tumors (Li et al. 2008). The preponderant evidence was that this fusion transcript was the result of a trans-splicing event in normal tissues but of genomic rearrangements in tumors. The presence of two reverse-order fusions in one normal breast sample despite the lack of any evidence that this TD is present in any other human genomes makes likely that our observation is also the result of trans-splicing. These observations raise the possibility that trans-splicing in normal tissues may mark “fusion-enabled” genes that might be subject to subsequent fixed genomic rearrangements in the carcinogenic process.
Finally, when taken together, our data and that of Stephens et al. (2009) suggest that although the majority of the genomic abnormalities seem to be private mutations, tumor selection and sampling may give a false impression of the rarity of some recurrent rearrangements. That we find multiples of these “rare” fusion transcripts in any cancer suggests that it is the sum of a number of private oncogenic drivers that sustains the cancer phenotype.
Methods
DNA-PET
DNA-PET library construction, sequencing, mapping, discordant PET clustering, and cross-comparison are described precisely elsewhere (Hillmer et al. 2011). In brief, we prepared and sequenced DNA (hydro-sheared 5–11.5 kb of genomic DNA) with the Applied Biosystems SOLiD system. Paired-end libraries were constructed by ligation of EcoP15I CAP adaptors and digested by EcoP15I to release 5′ and 3′ PET, and SOLiD sequencing adaptors P1 and P2 were ligated to the library DNA. High-throughput sequencing of the 2×25 bp was performed on SOLiD sequencers according to the manufacturer's recommendations (Applied Biosystems). Sequence tags were mapped to the human reference sequence (NCBI Build 36), allowing two color code mismatches for 25-bp reads using Corona Lite (Applied Biosystems). Discordant PETs within windows of maximum library size in both directions were clustered, and the number of discordant PETs in the window was represented by the cluster size. Similarly, other discordant PET clusters within windows of maximum library size from the end of each discordant PET cluster in both directions were “super-clustered,” and the number of clusters in the window was represented by the super-cluster size. In cases in which more than three dPET clusters were interconnected, the structure variations were classified as “complex” (intra- and interchromosomal). Comparison of discordant PET clusters across different genomes was performed based on an overlap of the 5′ and 3′ anchor regions extended by 10 kb on both sides. We compared cluster coordinates in 12 genomes (five breast tumors obtained from five patients in Sweden whose clinical information is not available, three breast cancer cell lines [MCF-7, T47D, and SKBR3], two normal controls, and two other cell lines [K562 and HCT116]). Breakpoint locations were also compared with the identified SVs in paired-end sequencing studies of noncancer individuals (Korbel et al. 2007; Kidd et al. 2008), the structure variations were matched to the studies, and our two controls were removed from further validations. We annotated the coordinates of genomic fusion points into RefSeq genes (hg18) and defined gene truncation by each breakpoint. Thus 3′-terminus truncation (3′T) or 5′-terminus truncation (5′T) means that the gene possesses the 5′-terminal or 3′-terminal portion in the fused region, respectively. A combination of 3′T and 5′T in different genes is defined as a fusion gene (FG), where 3′T and 5′T genes indicate the 5′ and 3′ genes of the fusion gene, respectively. On the other hand, 3′T and 5′T in the same gene caused by a deletion or tandem duplication or the combination of a pair of clusters of an inversion or insertion is defined as intragenic rearrangement (IR). A combination of two 3′Ts or 3′T with a nonannotated gene region, including an intergenic region and an anti-sense strand of any genes, is defined as 3′T category, with the combinations for 5′Ts as 5′T category.
Statistics analyses
Statistical analyses in Figure 4F,G and Supplemental Figures 1, 2F,I,J, and 3B,C were done by a Student's t-test. GO analysis was done by PANTHER software as mentioned below. Analysis for the correlation between the fusion gene expression and patient disease-free survival or clinical parameters was done by a Cox-proportional hazards model or Fisher's exact test, respectively. All other analyses were done by binomial distribution probability.
Copy number estimation
Copy number across the genome is predicted by computing the density of all uniquely mapped 5′ and 3′ tags generated from the DNA-PET data across variable-size windows. To access the varying mapping efficiency of the tags across the genome due to repetitive sequences, 25-bp paired-end tags were randomly simulated and mapped across the human genome (∼76 million uniquely mapped tags). The nonoverlapping, variable-size windows were generated by intervals of 300 uniquely mapped simulated tags. Windows that contain satellite repeats, genomic gaps, as well as 100 kb extending from centromeric, heterochromatin, and telomeric regions are removed due to erroneous mapping in these regions. The GC bias of the PET coverage as a result of the DNA preparation step varies for different DNA-PET libraries and can be quantified by the GC content of the concordant-PETs. As copy number variations are not expected to vary with GC content, the average GC content of the DNA fragments of the concordant-PETs and the randomly simulated DNA fragments of similar size were computed, and the relative difference of their quantities for different GC content was used to determine the GC bias. The number of uniquely mapped tags for each DNA-PET in each window was computed, and the tag count was corrected for GC bias based on the GC content of the given window. The copy number of each window was inferred by assuming that the median tag density represents two copies of the genome [copy number = 2 × (corrected tag density)/(median corrected tag density)].
RT-PCR for validation of the fusion transcripts
Total RNA for breast cancer cell lines was purified with RNAeasy (QIAGEN) and reverse-transcribed using Superscript III (Invitrogen) according to the manufacturers' instructions. Total RNA of five primary tumor samples used for DNA-PET analysis was purified with an AllPrep DNA/RNA kit (QIAGEN) and reverse-transcribed using SuperScript VILO (Invitrogen). PCR was done with Pfu Ultra Hotstart (Stratagene), Phusion Flash (Finnzymes), or Hot Star Taq (QIAGEN). PCR products were directly cloned by a StrataClone (Blunt) PCR cloning kit (Stratagene), and purified plasmids were analyzed for sequence. The primers used and the sequence of fusion points are listed in Supplemental File 1. Primers were designed using Primer3 (http://frodo.wi.mit.edu/primer3/) based on putative fusion transcript sequence predicted by DNA-PET information. The primers used in Figure 4B are as follows: 5′-ATAGACCTGGACCAGCCAGA-3′ and 5′-CTCTGAGTTCAACCGCTGCTGG-3′ match to exon 1 of RPS6KB1 and exon 12 of VMP1, respectively; 5′-TCCCTGGAGAAGAGCTACGA-3′ and 5′-AGGAAGGAAGGCTGGAAGAG-3′ for the ACTB gene.
Validation of the difference classes of the structural mutations has varying challenges. The identification of the exonic joins of putative chimeric transcripts when the fusion partners are known genes requires only computational assessment of the exons involved. The other classes of rearrangements such as 3′T-N and 5′T-N are more difficult since, although the genomic breakpoint is identified by DNA-PET, the potentially unknown 3′ or 5′ segment from an untranscribed region of the genome in these truncation–fusion transcripts preclude simple identification of PCR primers to isolate the chimeric transcript. In the case of the MCF-7 validation approach, an RNA-PET library provided the sequence information for all fusion transcripts. Without this library, fusion transcripts involving novel genomic segments would have been difficult to ascertain. For this reason, we pursued a hierarchical strategy in the validation of 3′TR transcripts by first assessing the 3′T-ER putative transcripts where the 3′ fusion partner is an annotated transcript but not a RefSeq gene.
RNA-PET library
An MCF-7 cell was treated with 100 nM of β-estradiol grown in charcoal-stripped serum media for 45 min (IHM101). A control experiment using ethanol as a treatment was performed in parallel to provide a control sample (IHM098). The precise protocol for cloning-free RNA-PET library construction is described elsewhere (Ruan and Ruan 2011). The RNA-PET libraries were generated from poly(A) mRNA samples. Total RNA in good quality was used as the starting material and purified through a MACs poly(T) column to obtain full-length poly(A) mRNAs. Approximately 5 μg of enriched poly(A) mRNA was used for reverse transcription to convert poly(A) mRNA to full-length cDNA. The full-length cDNA obtained was modified and ligated with specific linker sequences, followed by circularization through ligation to generate circular cDNA molecules. The 25-bp tag from each end of the full-length cDNA was extracted by type II enzyme EcoP15I digestion. The resulting PETs were ligated with sequencing adaptors at both ends, amplified by PCR, and further purified as complex templates for paired-end (PE) sequencing using the Illumina platform.
RNA-PET data analysis
The sequenced RNA-PETs are unified in 25/25-bp length from each end of a cDNA. After filtering out redundant and noise tags, the unique PETs were processed by the analysis pipeline. Initially, the orientation of each tag was screened out by the “barcode” built in the sequencing template, then paired into a given orientation-PET. The orientation-determined RNA-PET was mapped onto a reference genome allowing up to two mismatches. The majority of PETs were mapped on the known transcripts, or splice variants. We could map 958,464 concordant PET (82%) and 212,158 discordant PET (18%) in IHM098, while 2,250,056 (92%) and 193,370 (8%), respectively, in IHM101. Discordant PETs were further categorized as follows: (1) mapped to different chromosomes; (2) mapped to the same chromosome, different strand; (3) mapped to the same chromosome, same strand, but in incorrect order (5′Tag is downstream and 3′Tag is upstream); (4) mapped to the same chromosome, the same strand, in correct order but different genes.
Intersection of DNA-PET and GIS-PET/RNA-PET in MCF-7
We searched DNA-PET clusters whose orientations fit to corresponding RNA-PETs within 500-kb windows. Gene annotation was done in 10-kb windows (5′-to-3′ direction) of RNA-PETs. In Supplemental Figure 2C, numbers of validated fusion transcripts by RT-PCR are shown in the table (bottom), while predicted fusion transcripts by the analysis are given in the parentheses. To identify further FG and 3′T-derived transcripts in MCF-7, we intersected DNA-PET data with GIS-PET and RNA-PET data. GIS-PET data sets for MCF-7 were described previously (Ng et al. 2006; Ruan et al. 2007). We compared 5′ fused gene symbols of FG and 3′T predicted in the DNA-PET data and those of the transcriptome data. For the matched genes, we searched 3′ GIS-PETs/RNA-PETs whose orientations fit to corresponding 3′ DNA-PET in location and direction within 1 Mb.
Protein functional domain analysis
Based on the fusion points that were identified by RT-PCR and sequencing, we predicted full-length fusion coding sequence and determined that the 3′-gene ORF is in-frame or out-of-frame to the 5′-gene ORF. Then we searched protein functional domains in the fusion structure. Protein functional domain units were defined by Pfam (http://pfam.sanger.ac.uk/) and SMART (http://smart.embl-heidelberg.de/). In the case of out-of-frame fusions, we supposed a bicistronic transcript and/or a de novo translation start site for the domains of the 3′ gene.
GO analysis
Gene Ontology analysis was done using PANTHER software (http://www.pantherdb.org/). We compared the fusion partner gene data with all RefSeq gene data that were used in the gene annotation step.
Polysomal assay
MCF-7 cells were subjected to polysome fractionation via sucrose gradient centrifugation to determine the translational index of the fusion transcripts (Tominaga et al. 2005). Figure 3A depicts a typical sucrose gradient fractionation profile demonstrating the separation between translationally active polysomal RNA and the untranslated monosomal RNA. MCF-7 cells were incubated in 100 μg/mL cycloheximide (Sigma) for 10 min followed by harvesting with trypsin. All buffers for harvesting contained 100 μg/mL cycloheximide. Twenty million cells were resuspended in RSB buffer (10 mM Tris-HCl at pH 7.4, 10 mM NaCl, 15 mM MgCl, 2 U/μL RNase inhibitor, 100 μg/mL cycloheximide, and 100 μg/mL heparin) followed by lysis with 1.2% Triton X-100 and 1.2% sodium deoxycholate for 10 min on ice. Cell lysates were centrifuged at 12,000g for 10 min at 4°C to pellet nuclei, and the supernatant was diluted with 1 volume of dilution buffer (25 mM Tris-HCl at pH 7.4, 25 mM NaCl, 25 mM MgCl, 0.05% Triton X-100, and 500 μg/mL heparin). One milliliter of the polysome extract was then loaded onto 11 mL of linear 10%–50% (w/v) sucrose gradients made using BioComp gradient master. Following centrifugation in an SW41 Ti Rotor (Beckman) for 2 h at 36,000 rpm at 8°C, gradients were fractionated into 12 1-mL fractions using the BioComp piston gradient fractionator attached to an EM-1 UV Monitor (Bio-Rad). All fractions were collected by continuous monitoring of the A254, which indicated the positions of the ribosomal subunits and the polysomes. Fractions were then incubated for 30 min at 42°C with 1% SDS and 12 μL of Proteinase K (10 mg/mL; Invitrogen). RNA was extracted by phenol chloroform followed by RNeasy column purification (QIAGEN) and used to make cDNA using Superscript III Reverse Transcriptase (Invitrogen).
Real-time PCR was performed with SYBR Green PCR master mix (ABI) to assess enrichment in certain fractions. Primers were designed to uniquely identify the fusion transcripts as well as uniquely identify the intact transcripts of the 5′ and 3′ fusion partner genes. Duplicate samples and negative controls for each were included to ensure accuracy.
In order to find the percentage of specific transcripts in each fraction of the polysome gradient, we calculated the relative amount of RNA (RNA units) normalized by total units of a given gene and showed a percentage of each group composed by several fractions (Fig. 3A) among total RNA units. Group 1 represents fractions 1 to 5 containing mRNP particles, ribosomal subunits, and transcripts with monosomes indicative of no translation. Group 2 represents fractions 6 to 8 containing transcripts with two to four attached ribosomes. Transcripts could occur in these fractions due to inefficient translation or short transcript length. Group 3 consists of fractions 9 to 11 containing transcripts with five or more ribosomes. This group is defined as a highly translated group. Group 4 contains fraction 12 containing transcripts that sediment rapidly due to an excessive number of attached ribosomes or due to their association to other larger cellular bodies. If the total percentage of RNA units was >60% in any one group, then we define the following translation index: >60% in group 1 = NONE, >60% in group 2 = LOW, >60% in group 3 = HIGH. However, if the total percentage of RNA units did not clearly peak in a single group but spread across groups 2 and 3 (e.g., 40% in group 2 and 40% in group 3), then we defined it as MEDIUM. In our study, we did not find any transcripts spread across groups 1 and 2.
Microarray gene expression data for MCF-7
A control datum (DMSO 0 h, n = 3) of a comprehensive estrogen treatment time-course microarray experiment (Fullwood et al. 2009) was used as basal gene expression data for MCF-7. After 3 d of serum starvation, RNA was extracted and the labeled probes were hybridized to microarrays (HG-U133 Plus). In case of multiple probes for a given gene, the average data of microarray log2 intensity were used, except for using the highest value in the whole genome analysis in relation to copy number (Supplemental Fig. 1).
Fluorescence in situ hybridization (FISH)
Nuclei preparation
MCF-7 nuclei were harvested by treating cells with 0.75 M KCl for 20 min at 37°C. Then, after a few fixations, nuclei were dropped on slides for FISH.
Fosmid probe preparation
DNA was labeled by nick translation in the presence of biotin-16-dUTP using the Nick translation system (Invitrogen). In the presence of 1 μg/μL Cot1 DNA, a DNA cosmid clone was resuspended at a concentration of 5 ng/μL in hybridization buffer (2SSC, 10% dextran sulfate, 1× PBS, 50% formamide).
FISH
Prior to hybridization, MCF-7 nuclei slides were treated with 0.01% pepsin for 5 min at 37°C followed by 1× PBS rinse, 1% formaldehyde 10-min treatment, 1× PBS rinse (5 min), and dehydratation through ethanol series (70%, 80%, and 100%). A denaturated probe was applied to these pretreated slides and codenaturated for 5 min at 75°C and hybridized overnight at 37°C. Two post-hybridization washes were performed at 45°C in 2SSC/50% formamide for 7 min each followed by two washes in 2SSC for 7 min each at 45°C. After blocking, the slides were revealed with avidin-conjugated fluorescein isothiocyanate (FITC; Vector Laboratories). After washing, slides were mounted with Vectashield and observed under epifluorescence microscope. Image analysis was done using Metasystem software.
Breast tumor clinical samples (Singapore cohort)
Total RNA for 70 clinical breast tumor samples of the National University Hospital (Singapore) cohort was described previously (Ivshina et al. 2006). Normal breast control RNAs were purchased from Stratagene (NB41, NB52, and NB71), Ambion (NB82), and Clontech (NB27). For RT-PCR detection of the fusion transcripts, 200 ng of total RNA was amplified using the SuperScript III RNA amplification kit (Invitrogen) according to the manufacturer's protocol. Amplified RNA (700 ng) was used for cDNA synthesis with random primer and SuperScript III (Invitrogen). The quality of cDNA was checked by housekeeping genes (ACTB and GUSB). PCR was done by Hot Star Taq (QIAGEN) using these primers: 5′-AGACAGGGAAGCTGAGGACA-3′ and 5′-CATTTCGCTTTTGTGGTGAA-3′ match to exon 1 of RPS6KB1 and exon 11 of VMP1, respectively (Fig. 4C); 5′-AACAGGAGCAAATACTGGGA-3′ and 5′-TCAAACATCCAGGACAACCA-3′ match to exon 4 of RPS6KB1 and exon 12 of VMP1, respectively. PCR products were cloned and sequenced as above. Microarray analysis and data processing for clinical outcomes were done as described previously (Ivshina et al. 2006).
Acknowledgments
We thank Q.Y. Pang, L.C. Tan, L.P. Yap, S.L. Theng, C.T. Lee, K. Thomas, B.H. Eng, L.J. Lee, S.B. Mohamed Asrafali, Y.W. Chee, H.L. Lee, P.Y. Lee, S.S. Rahman, and Y. B. Mohamed for technical support. This work is supported by grants from the Agency for Science Technology and Research (ASTAR), Singapore and from U.S. NIH (NCI: 5 R33 CA126996, “Pair-End-Editing Technologies for the Complete Annotation of Fusion Genes”).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.113225.110.
References
- Bärlund M, Forozan F, Kononen J, Bubendorf L, Chen Y, Bittner ML, Torhorst J, Haas P, Bucher C, Sauter G, et al. 2000. Detecting activation of ribosomal protein S6 kinase by complementary DNA and tissue microarray analysis. J Natl Cancer Inst 92: 1252–1259 [DOI] [PubMed] [Google Scholar]
- Bärlund M, Monni O, Weaver JD, Kauraniemi P, Sauter G, Heiskanen M, Kallioniemi OP, Kallioniemi A 2002. Cloning of BCAS3 (17q23) and BCAS4 (20q13) genes that undergo amplification, overexpression, and fusion in breast cancer. Genes Chromosomes Cancer 35: 311–317 [DOI] [PubMed] [Google Scholar]
- Bashir A, Volik S, Collins C, Bafna V, Raphael BJ 2008. Evaluation of paired-end sequencing strategies for detection of genome rearrangements in cancer. PLoS Comput Biol 4: e1000051 doi: 10.1371/journal.pcbi.1000051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz TH, Preiss T 2004. Translational profiling: The genome-wide measure of the nascent proteome. Brief Funct Genomic Proteomic 3: 103–111 [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerione RA, Zheng Y 1996. The Dbl family of oncogenes. Curr Opin Cell Biol 8: 216–222 [DOI] [PubMed] [Google Scholar]
- Chen S, Hamm HE 2006. DEP domains: more than just membrane anchors. Dev Cell 11: 436–438 [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. 2009. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462: 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y, Bera TK, Gehlhaus K, Kirsch IR, Pastan IH, Lee B 2004. Finding fusion genes resulting from chromosome rearrangement by analyzing the expressed sequence databases. Proc Natl Acad Sci 101: 13257–13261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton OA, Den Hollander P, Miller CA, Delgado DA, Li J, Coarfa C, Harris RA, Richards S, Scherer SE, Muzny DM, et al. 2009. A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res 19: 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeson BE, Tomlins SA, Shah N, Laxman B, Cao Q, Prensner JR, Cao X, Singla N, Montie JE, Varambally S, et al. 2008. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res 68: 73–80 [DOI] [PubMed] [Google Scholar]
- Hillmer AM, Yao F, Inaki K, Lee WH, Ariyaratne PN, Teo ASM, Woo XY, Zhang Z, Zhao H, Ukil L, et al. 2011. Comprehensive long-span paired-end-tag mapping reveals characteristic patterns of structural variations in epithelial cancer genomes. Genome Res (this issue). doi: 10.1101/gr.113555.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65: 7065–7070 [DOI] [PubMed] [Google Scholar]
- Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, Lindahl T, Pawitan Y, Hall P, Nordgren H, et al. 2006. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res 66: 10292–10301 [DOI] [PubMed] [Google Scholar]
- Janz S, Potter M, Rabkin CS 2003. Lymphoma- and leukemia-associated chromosomal translocations in healthy individuals. Genes Chromosomes Cancer 36: 211–223 [DOI] [PubMed] [Google Scholar]
- Joosten M, Blázquez-Domingo M, Lindeboom F, Boulmé F, Van Hoven-Beijen A, Habermann B, Löwenberg B, Beug H, Müllner EW, Delwel R, et al. 2004. Translational control of putative protooncogene Nm23-M2 by cytokines via phosphoinositide 3-kinase signaling. J Biol Chem 279: 38169–38176 [DOI] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. 2008. Mapping and sequencing of structural variation from eight human genomes. Nature 453: 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Roberts R 2001. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci 58: 2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Tognon CE, Godinho FJ, Yasaitis L, Hock H, Herschkowitz JI, Lannon CL, Cho E, Kim SJ, Bronson RT, et al. 2007. ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell 12: 542–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang J, Mor G, Sklar J 2008. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science 321: 1357–1361 [DOI] [PubMed] [Google Scholar]
- Lin E, Li L, Guan Y, Soriano R, Rivers CS, Mohan S, Pandita A, Tang J, Modrusan Z 2009. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res 7: 1466–1476 [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S 2002. The protein kinase complement of the human genome. Science 298: 1912–1934 [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F 2007. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 7: 233–245 [DOI] [PubMed] [Google Scholar]
- Ng P, Tan JJ, Ooi HS, Lee YL, Chiu KP, Fullwood MJ, Srinivasan KG, Perbost C, Du L, Sung WK, et al. 2006. Multiplex sequencing of paired-end ditags (MS-PET): a strategy for the ultra-high-throughput analysis of transcriptomes and genomes. Nucleic Acids Res 34: e84 doi: 10.1093/nar/gkl444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzani A, Fronza R, Loreni F, Pascale A, Amadio M, Quattrone A 2006. Global alterations in mRNA polysomal recruitment in a cell model of colorectal cancer progression to metastasis. Carcinogenesis 27: 1323–1333 [DOI] [PubMed] [Google Scholar]
- Rabbitts TH 2009. Commonality but diversity in cancer gene fusions. Cell 137: 391–395 [DOI] [PubMed] [Google Scholar]
- Raphael BJ, Volik S, Yu P, Wu C, Huang G, Linardopoulou EV, Trask BJ, Waldman F, Costello J, Pienta KJ, et al. 2008. A sequence-based survey of the complex structural organization of tumor genomes. Genome Biol 9: R59 doi: 10.1186/gb-2008-9-3-r59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. 2007. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131: 1190–1203 [DOI] [PubMed] [Google Scholar]
- Ruan X, Ruan Y 2011. Genome wide full-length transcript analysis using 5′ and 3′ paired-end-tag next generation sequencing (RNA-PET). In Transcriptional regulation: Methods and protocols. Methods in molecular biology (ed. Vancura Ales). Humana Press, Totowa, NJ: (in press). [DOI] [PubMed] [Google Scholar]
- Ruan Y, Ooi HS, Choo SW, Chiu KP, Zhao XD, Srinivasan KG, Yao F, Choo CY, Liu J, Ariyaratne P, et al. 2007. Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using Paired-End diTags (PETs). Genome Res 17: 828–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE 2008. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2: 448–460 [DOI] [PubMed] [Google Scholar]
- Sinclair CS, Rowley M, Naderi A, Couch FJ 2003. The 17q23 amplicon and breast cancer. Breast Cancer Res Treat 78: 313–322 [DOI] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. 2007. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448: 561–566 [DOI] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, Becker L, Carneiro F, MacPherson N, Horsman D, et al. 2002. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell 2: 367–376 [DOI] [PubMed] [Google Scholar]
- Tominaga Y, Tamgüney T, Kolesnichenko M, Bilanges B, Stokoe D 2005. Translational deregulation in PDK-1−/− embryonic stem cells. Mol Cell Biol 25: 8465–8475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. 2005. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310: 644–648 [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. 2007. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 448: 595–599 [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, et al. 2008. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia 10: 177–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volik S, Raphael BJ, Huang G, Stratton MR, Bignel G, Murnane J, Brebner JH, Bajsarowicz K, Paris PL, Tao Q, et al. 2006. Decoding the fine-scale structure of a breast cancer genome and transcriptome. Genome Res 16: 394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Vardy LA, Tan CP, Loo JM, Guo K, Li J, Lim SG, Zhou J, Chng WJ, Ng SB, et al. 2010. PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase. Cancer Cell 18: 52–62 [DOI] [PubMed] [Google Scholar]
- Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY 2008. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14: 2348–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, Han J, George J, Wong C, Ramnarayanan K, Phua TY, et al. 2009a. RCP is a human breast cancer-promoting gene with Ras-activating function. J Clin Invest 119: 2171–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE 2009b. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 113: 2526–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]