Abstract
The spatial organization of genes in the interphase nucleus plays an important role in establishment and regulation of gene expression. Contradicting results have been reported to date, with little consensus about the dynamics of nuclear organization and the features of the contact loci. In this study, we investigated the properties and dynamics of genomic loci that are in contact with glucocorticoid receptor (GR)–responsive loci. We took a systematic approach, combining genome-wide interaction profiling by the chromosome conformation capture on chip (4C) technology with expression, protein occupancy, and chromatin accessibility profiles. This approach allowed a comprehensive analysis of how distinct features of the linear genome are organized in the three-dimensional nuclear space in the context of rapid gene regulation. We found that the transcriptional response to GR occurs without dramatic nuclear reorganization. Moreover, contrary to the view of transcription-driven organization, even genes with opposite transcriptional responses colocalize. Regions contacting GR-regulated genes are not particularly enriched for GR-regulated loci or for any functional group of genes, suggesting that these subnuclear environments are not organized to respond to a specific factor. The contact regions are, however, highly enriched for DNase I–hypersensitive sites that comprehensively mark cell-type–specific regulatory sites. These findings indicate that the nucleus is pre-organized in a conformation allowing rapid transcriptional reprogramming, and this organization is significantly correlated with cell-type–specific chromatin sites accessible to regulatory factors. Numerous open chromatin loci may be arranged in nuclear domains that are poised to respond to diverse signals in general and to permit efficient gene regulation.
The spatial organization of the genome in the interphase nucleus has nonrandom features, yet its role in gene regulation remains elusive (de Laat and Grosveld 2007). The nuclear periphery is mostly gene-poor and transcriptionally inactive (Guelen et al. 2008), while the inner nuclear space may include various environments for distinct transcriptional activities (Fraser and Bickmore 2007). Although genes can adopt different positions in the nucleus in relation to changes in their expression status during cellular differentiation (Kosak et al. 2002; Chambeyron and Bickmore 2004), the extent of reorganization in response to rapid transcriptional modulation is controversial. Some groups argue that nuclear architecture plays a dynamic and gene-specific role in gene regulation (Apostolou and Thanos 2008; Hu et al. 2008; Sandhu et al. 2009; Schoenfelder et al. 2009), while others have contradicting results (Simonis et al. 2006; Kocanova et al. 2010). The discrepancy may have arisen from the limited number of genomic sites under study. Studies relying primarily on a few sites do not reveal details about the genomic environments that the genes leave and enter, and therefore questions remain whether the large-scale alterations in nuclear architecture are a global response to transcriptional reprogramming and, moreover, what the features of the spatial environment of these rapidly responsive loci are.
In this study, we took a systematic approach that integrates genome-wide profiles of spatial interactions, gene expression patterns, transcription factor occupancy, and chromatin accessibility. To identify genome reorganizational events associated with rapid transcriptional reprogramming, we characterized genomic profiles before and after activation of glucocorticoid receptor (GR) by dexamethasone (Dex). Hormone-activated GR alters the transcriptional state of several hundred genes, producing complex expression kinetics both for induced and for repressed genes (John et al. 2009). GR, being the sole receptor for Dex in the cells, allowed us to study the specific action of a single transcription factor, as opposed to inducers affecting complex, multifactorial systems. We report that the rapid transcriptional reprogramming by GR occurs in a pre-organized nuclear architecture. Unexpectedly, we found that gene repression as well as gene activation by GR can happen in the same location in the nucleus. These preexisting subnuclear environments contained a surprisingly large number of GR-binding sites, even though GR is not present in the nucleus before hormone treatment. Importantly, the contact loci are highly enriched for DNase I–hypersensitive sites (DHS), representing cell-type–specific regulatory sites. These results suggest that DNA-interacting proteins may potentially play an important role in shaping the nuclear architecture.
Results
GR-inducible genes are engaged in multiple inter- and intrachromosomal interactions
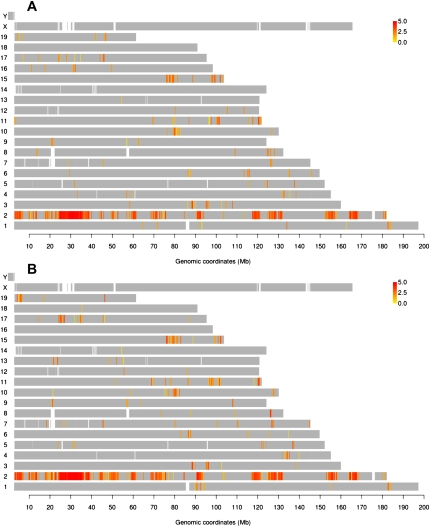
Using chromosome conformation capture on chip (4C), which quantitatively captures genome-wide interactions with a locus of interest (Simonis et al. 2006), we characterized genomic loci in close physical proximity to lipocalin 2 (Lcn2), which is highly induced by GR in murine mammary adenocarcinoma cells (Hakim et al. 2009). We used tailored NimbleGen microarrays that contain a single probe near every unique HindIII restriction enzyme recognition site in the entire mouse genome, providing an average resolution of 7 kb. Statistical analysis was applied to the average log2 (4C/input DNA) values of two repeats, to assign a p score to every probe, based on its signal and that of neighboring probes within a 100-kb window (see Methods). Clusters of contiguous positive probes, potentially representing genomic contact loci, occurred on almost all chromosomes before Dex treatment (Fig. 1A). The most prominent positive-probe cluster was readily noticeable at the bait locus (Chr2: 32.2 Mb), as observed in other 4C data (Simonis et al. 2006). Regardless of cell types and baits, the contact frequency within the bait-proximal region is distinctively higher than the contact frequency with more distant loci and loci on other chromosomes. Moreover, the near-maximal probe signals from this region do not provide any discriminating profiles of interaction frequency (Supplemental Fig. S1). Therefore, we excluded this 10-Mb region centered on the bait from all computational analyses regarding enrichment for a given genomic feature.
Figure 1.
Lcn2 is engaged with genomic loci in cis and in trans across the mouse genome before and after induction by Dex. Genomic map of Lcn2 contacts before (A) and 1 h after induction by Dex (B). Probes with P-values above the threshold (p score = −log10 p > 4: 0.3% FDR) are labeled in a yellow-to-red color scale according to the intensity of the probe signal (log2 4C/genomic DNA). (Gray) Probes with P-values below the threshold.
Nuclear organization dynamics during transcriptional reprogramming
After GR activation by hormone (Dex), hundreds of genes are either repressed or activated with various complex kinetics (John et al. 2009). To examine the dynamics of nuclear organization during active transcriptional reprogramming, we probed spatial environments of Lcn2 1 h after hormone treatment (Fig. 1B). Increased numbers of positive probes were detected from Dex-treated samples, but the overall interaction profiles, as seen in the genome plots, remained largely similar to those before Dex, ruling out gross chromosomal reorganization (Fig. 1). To further distinguish the dynamic components, we assessed the differences between Lcn2-contacting regions before and after hormone treatment in more detail. For better delineation of genomic regions with changes in interaction frequency, we retrieved the probes whose p score was altered by more than 4.5 (0.4% FDR) after Dex treatment, and plotted their 4C log ratio (+Dex/−Dex) values (Supplemental Fig. S2A). While most of these regions show increased frequency of contact with Lcn2 after Dex, the magnitude of these changes was modest overall. Strengthened contacts after Dex treatment were also evident by the fact that the number of robust contact regions with more than 10 contiguous positive probes increased from 101 to 121 during Dex activation (Supplemental Tables S1, S2). A close examination showed that many of the newly appearing contacts arose from regions with a preexisting 4C signal (below threshold) prior to hormone treatment (Supplemental Fig. S3). Moreover, the median size of the contact loci grew from 47 kb to 56 kb (Supplemental Fig. S2B). These data support the conclusion that the predominant hormone-induced changes for Lcn2-contacting loci can be attributed to an increased frequency of preexisting interactions.
As our findings are in contrast to a report showing a dramatic nuclear reorganization of selected estrogen receptor (ER)–responsive loci post–hormone treatment (Hu et al. 2008), we decided to look for such a phenomenon in the regions with the most pronounced changes in contact frequency. We retrieved all the probes that had their p score increased (n = 636 of 384,942, or 0.17%) or decreased (n = 79, or 0.02%) by more than 5 after hormone induction. Only a small fraction of these probes were within 10 kb of any known genes, and none were near GR-regulated genes (data not shown). This argues against the possibility that GR-regulated loci preferentially come into close proximity in response to hormone.
We also considered whether the most pronounced reorganization takes place before the examined time point (1 h after Dex). Because transcriptional response to GR activation is rapid, with new transcripts appearing within 15 min after Dex, we profiled the Lcn2 contacts 10 and 30 min after hormone induction. Still, there were no major changes in the nuclear architecture around Lcn2 (Supplemental Fig. S4).
4C profiles reflect quantitative interaction frequencies from DNA FISH
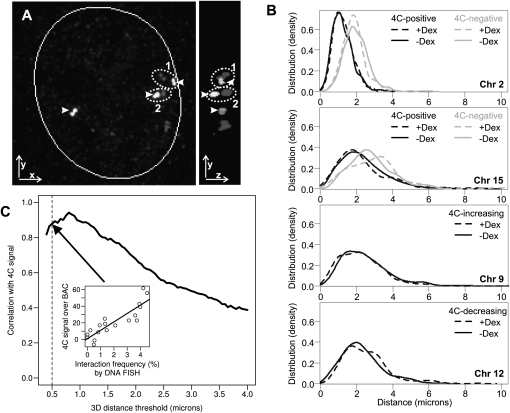
For independent evaluation of 4C data, three-dimensional (3D) DNA fluorescence in situ hybridization (3D DNA FISH) was applied to Lcn2 and nine loci. These regions were selected to encompass various Lcn2 contact characteristics (positive, negative, cis, trans, and dramatic changes in contact frequency after Dex) (Supplemental Table S3). To prevent bias in the analysis, high z-resolution 3D images were analyzed with a custom-designed computational approach. This algorithm automatically identifies positions for Lcn2 and a partner locus as the center of the FISH signal and calculates the 3D distance between them within each nucleus (Fig. 2A) (see Methods). We found a remarkable concordance between the interaction probability from Lcn2 4C and the distribution of 3D distances to Lcn2, both intra- and interchromosomal loci (Fig. 2B, upper two plots; Supplemental Table S3). In addition, Dex-mediated increases or decreases in contact probability measured by 4C were recapitulated in DNA FISH for all examined loci (Fig. 2B, lower two plots; Supplemental Table S3). The redistribution of FISH signals in response to hormone activation was modest even for loci showing the most dramatic changes in contact frequency by 4C, confirming the lack of large-scale nuclear reorganization in response to transcriptional reprogramming by GR (Fig. 2B, lower two plots).
Figure 2.
High correlation between 4C and 3D DNA FISH. (A) Automated image analysis computer algorithm for measuring the three-dimensional distance between the centers of two DNA FISH signals. (Left panel) Example of a nucleus from imaging DNA FISH as presented by 2D xy projection. The Lcn2 locus on chromosome 2 (arrowhead) and a locus on chromosome 8 in the nucleus. From this view, pair 2 is the closest one. (Right panel) yz view of the nucleus showing that the upper pair of FISH foci (1) is closer than pair 2 in 3D. (B) Distribution of 3D distances between Lcn2 and other loci as determined by DNA FISH, before (solid line) and after (dashed line) induction by Dex. (Top panel) Distances between Lcn2 (at Chr2: 32 Mb) and a locus scored positive (at Chr2: 164 Mb, black) or a locus called negative (at Chr2: 141 Mb, gray) by 4C for intrachromosomal interaction. (Second from top) Distances between Lcn2 and a locus scored positive (Chr15: 102 Mb, black) or a locus called negative (Chr15: 20 Mb, gray) by 4C for interchromosomal interaction. (Bottom two panels) Distance distributions for loci that had an increase (Chr9: 108 Mb) or a decrease (Chr12: 105 Mb) in 4C signals with respect to Lcn2. To estimate the distribution density, 200–1000 cells were examined by the automated algorithm for each probe and condition (see Supplemental Table S3; BACs [A, C], [H, I], E, and G correspond to the four panels, from top to bottom). (C) Robust correlation between the 4C contact probability and interaction frequency by 3D DNA FISH over different FISH distance thresholds. Interaction frequency (percentage of cells containing Lcn2 contact) was determined by calculating the proportion of nuclei that had the two FISH signals within a set distance threshold (center-to-center in 3D). The distance threshold was varied from 0.4 to 4 μm, and the corresponding correlation coefficient between the integrated 4C signal over BAC and interaction frequency by DNA FISH was calculated for each threshold on all the experiments (see Supplemental Table S3). (Inset) An example correlation for a distance threshold at 0.5 μm.
Finally, we assessed the correlation between 4C signal and the interaction frequency by 3D FISH, two independent estimates of contact probability. Since the FISH interaction frequency is commonly determined as the percentage of cells with two FISH signals within a preset distance threshold, we calculated the frequency over a range of thresholds. We found a high correlation between 4C and 3D DNA FISH that is quite robustly maintained over different distance thresholds of up to 1 μm or so (Fig. 2C). The slightly lower correlation coefficients for the extremely small thresholds only reflect the fact that statistical estimates for low-frequency events are more prone to sampling error. This is not surprising given the relatively small sample size (200–1000 cells) of DNA FISH measurements. These findings indicate that the comprehensive profiles in Lcn2 4C data accurately reflect the interaction frequency with other genomic loci regardless of particular distance thresholds used.
Genes from diverse ontological groups and with opposite transcriptional responses to GR are colocalized
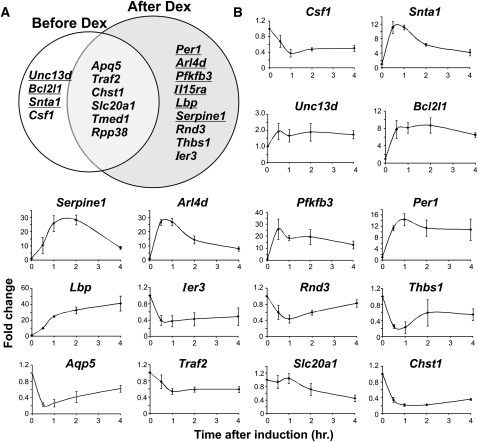
To discern organizational features of the genome, we next explored properties of loci colocalized in the physical neighborhood of Lcn2. Current models suggest that functionally related genes, regulated by common factors, colocalize in specialized nuclear microenvironments to facilitate their coordinated regulation (Sexton et al. 2007; Schoenfelder et al. 2009). We therefore asked whether the Lcn2 contact regions comprise genes with similar transcriptional responses to GR. First, we investigated the correlation between pre-hormone genome configuration and transcriptional regulation by GR. Numerous GR-regulated genes were found near Lcn2-interacting loci prior to Dex treatment. Surprisingly, the effects of hormone on the expression of these genes are remarkably diverse, even showing cases of opposite responses to GR regulation (Fig. 3). Because our expression profiles were obtained using Affymetrix microarrays, we evaluated the expression level of nascent transcripts by RT-qPCR, to exclude mRNA stability as a contributing factor. Transcriptional induction or repression by GR was confirmed for most genes (Fig. 3B). In summary, the transcriptional response of the genes positioned in this spatial environment is markedly heterogeneous.
Figure 3.
Genes with opposite transcriptional responses to GR colocalize with Lcn2. (A) GR induced (underlined) and repressed (not underlined) genes from Affymetrix exon expression array analysis, located within Lcn2 contact regions before and after 1 h of hormone treatment. First, all genes in the Lcn2 contact regions were obtained by retrieving genes whose TSS is located within 10 kb of 4C positive probes (excluding the bait region). Genes were considered GR-responsive if their expression changed by more than twofold after Dex treatment. (B) Time-course analysis by qRT-PCR of nascent transcript levels of GR-regulated genes located within Lcn2 contact regions. Samples were collected before Dex treatment and 0.5, 1, 2, and 4 h after Dex induction. Results shown are the average of three independent experiments, with SD presented as error bars.
Equally complex were the hormone-induced changes in contacts between Lcn2 and other GR-regulated genes. Although hormone treatment results in a 50% increase in the number of GR-regulated genes proximal to Lcn2 (10 to 15 genes, identified by Affymetrix exon microarrays), neither induced genes nor repressed loci had coherent changes in contact probability (Fig. 3A). In particular, both induced genes and repressed genes were found within the four loci that lost Lcn2 contact after Dex treatment, as well as within the nine that gained contact. Overall, the proportion of induced versus repressed genes near Lcn2 remained roughly the same and similar to that found genome-wide (42% of all GR-regulated genes are induced, 58% are repressed). Dex-induced loss of contact with Lcn2 for some GR-regulated genes was particularly unexpected in light of the overall hormone-dependent increase in interaction frequency. These findings argue against hormone-induced colocalization of similarly regulated genes within the Lcn2 subnuclear environment after GR activation. We also note that no dominant ontological group of genes is over-represented among colocalized genes at Lcn2. In spite of the well-documented action of GR in repressing numerous inflammatory genes while inducing important anti-inflammatory genes, the genes in proximity to Lcn2, either before or after Dex, do not share a common functional theme. Taken together, our data indicate a lack of a simple relationship between spatial organization and coregulation of genes, but instead suggest that the Lcn2 neighborhood can support diverse gene-reprogramming events.
Contact loci are most significantly enriched for DNase I–hypersensitive sites
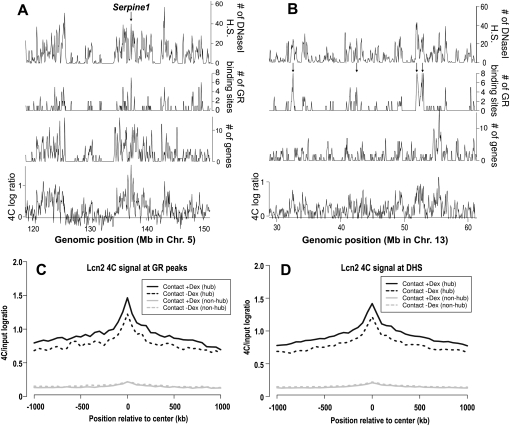
Since transcriptional response to GR did not appear to be a major theme in the spatial environment of Lcn2, we investigated other features of the contact loci. In view of previous reports on colocalization of gene-rich and highly expressed loci (Simonis et al. 2006; Brown et al. 2008; Lieberman-Aiden et al. 2009), we asked whether Lcn2, resident in a gene-dense region, preferentially interacts with other gene-rich regions. A significant fraction of Lcn2-contacting regions harbor genes (transcription start sites [TSS] within 10 kb of 4C-positive probes), encompassing 515 and 742 genes pre-hormone and post-hormone, respectively (both p < 0.01). In fact, this is the reason why we observed as many GR-regulated genes in the Lcn2 microenvironment as mentioned above (10 before Dex and 15 after Dex). These genes comprise ∼2% of colocalized genes in each condition, which is completely in line with the genome-wide estimate of 2.6% for genes regulated by GR (John et al. 2011). Furthermore, continuous running average profiles of 4C data and gene density have a genome-wide correlation coefficient of 0.29 and 0.33 (p < 0.01 for both) before and after hormone treatment, respectively (Fig. 4A,B; Supplemental Fig. S5; Supplemental Table S5). We also note a significant but slightly lower correlation between Lcn2 contact probability and expression profiles (0.23 and 0.24 for before and after Dex, respectively) (Supplemental Table S5). Moreover, the average expression level of genes in Lcn2-interacting loci is nearly identical to that from other gene-rich regions not in contact with Lcn2. These data suggest that the gene density was a more relevant correlative feature for Lcn2 4C than expression pattern or presence of GR-regulated genes.
Figure 4.
Lcn2 contact regions tend to be gene-dense or in close proximity to numerous GR-binding sites and DHS. (A,B) Profiles of Lcn2 4C signal 1 h after Dex treatment (average log2 ratios/100 kb), gene density (number of TSS/100 kb), GR-binding density (number of binding sites/100 kb), and the density of DHS after Dex (number of DHS/100 kb). (Arrow in A) The location of a GR-regulated gene, Serpine1. (Arrows in B) Point to contact regions in gene-poor regions that have numerous clustered GR-binding and DHS sites, showing that the enrichment of GR-binding and DHS sites can be observed independently of gene density in the contact loci. The genomic position in mm8 coordinates is indicated on the horizontal axis. Additional examples are provided in Supplemental Figure S5. (C,D) Average Lcn2 contact probabilities increase at GR-binding sites and DHS after Dex induction. Mean 4C signal (solid, after Dex; dashed, before Dex) centered at genome-wide GR binding sites (C) or DHS (D). Sites that are within 10 kb of Lcn2 contact loci (pooled from loci before and after Dex) were under the “hub” category (black), and those located at least 200 kb from Lcn2 or Arrdc2 contacts were included in the “non-hub” category (gray).
We next examined genome-wide binding profiles of GR, a specific factor that directly regulates Lcn2. We found a significant enrichment for GR-binding sites in Lcn2-interacting loci, with 454 GR ChIP-seq peaks located within 4C-positive regions (within 10 kb of positive probes). This is roughly twice as many binding events as would be expected from these contact loci based on the total number of binding sites (8373) identified in our ChIP-seq data. The enrichment for GR binding is also noteworthy, given that it is the only genomic profile that is completely independent of gene density in these cells (Fig. 4B; Supplemental Fig. S5; John et al. 2011). In fact, these Lcn2-colocalized GR-binding sites are mostly distal from known promoters, with only 11% within 2.5 kb of the nearest TSS. Moreover, GR binding near the most dramatically increased Lcn2 interactions after Dex treatment (Supplemental Fig. S2A) occurs even less at promoter-proximal sites (7% within 2.5 kb of TSS). These are similar to the genome-wide proportion of promoter-proximal GR ChIP peaks found in this cell line (John et al. 2011). Notably, the enrichment for GR-binding sites is apparently not mediated by Dex activation of GR, as a similar extent of enrichment was observed in Lcn2 contacts before hormone treatment. This implies that the Lcn2 subnuclear environment is pre-configured to allow numerous GR-binding events, even when GR is in the cytoplasm before hormone stimulation.
Given that GR activation is one of a repertoire of signals the cells can receive and the Lcn2 contacts harbor genes from diverse functional groups, we asked whether the Lcn2 neighborhood may be enriched for binding sites of the various factors that mediate different cellular responses. We addressed this question by integrating the genome-wide DNase I-seq data for these cells (John et al. 2008, 2011). DNase I–hypersensitive sites (DHS) represent accessible chromatin regions, serving as a proxy for the regulatory sites that are functional in a given cell type. The genome-wide correlation of the DHS profile to the Lcn2 4C profile was by far the highest (0.39 and 0.44 before and after Dex, respectively) among all genomic features that we examined. The Lcn2 neighborhood was, indeed, highly enriched for DHS sites beyond the level expected from their gene density, with a majority of 4C-positive probes located near DHS (Fig. 4A,B; Supplemental Fig. S5; Supplemental Table S5). Together, Lcn2 contact loci cover several thousand DHSs (4505 and 6940 before and after Dex treatment, respectively, out of ∼100,000 total DHSs in each condition). Similar to GR-binding sites near contact loci, the DHSs found in Lcn2-interacting regions are largely distal from genes (70% of DHS at least 2.5 kb from TSS) (Fig. 4B; Supplemental Fig. S5). Together with the fact that the majority of GR-binding sites are associated with DHS that exist before hormone treatment (John et al. 2011), these results explain how GR-binding sites are enriched in the neighborhood of Lcn2 via these putative distal regulatory elements in the contact regions before and after hormone. In summary, the most prominent feature of the contact loci is the presence of regulatory sites whose enrichment far exceeds those of gene density and expression.
To gain further insight into the proteins that are potentially associated with these DHSs, we performed motif discovery analysis on the top 1500 sites with the strongest DNase I hypersensitivity in this spatial environment before and after hormone induction. In both states, we found significant enrichment for recognition motifs of Pitx2, Pax-6, Sp1, and CTCF (Supplemental Fig. S6). Notably, CTCF has been implicated in multiple regulatory functions, including transcriptional activation, repression, and insulation (Phillips and Corces 2009).
To single out the Lcn2 contact probabilities specifically at GR-binding sites and DHS, we analyzed the 4C signal centered at the GR ChIP peaks or DHSs. We found that the average Lcn2 contact probability increases in response to GR activation, not only at the binding/DHS sites but also over a broad surrounding domain (up to 1 Mb) (Fig. 4C,D, black curve). This hormone-induced change is absent for sites that do not belong in this environment, for which the Lcn2 contact probability remained clearly in a far lower range (Fig. 4C,D, gray curves). Such a distinctive dichotomy is consistent with a recent report on the organization of the genome into two large-scale compartments (Lieberman-Aiden et al. 2009).
There are multiple subnuclear environments with similar organizational features
To investigate whether the characteristics of genome organization near Lcn2 are unique to the locus, we analyzed by 4C the spatial environment of two other loci. We selected arrestin domain containing 2 (Arrdc2) for another GR-induced gene, and annexin A5 (Anxa5) for a gene that is not responsive to GR but resides in a gene- and DHS-rich locus, much like Lcn2 and Arrdc2. Both loci did not interact with Lcn2 based on the Lcn2 4C data. Indeed, we confirmed that the baits Lcn2, Arrdc2, and Anxa5 manifest distinct contacts across the genome, with little overlap in interacting regions (Supplemental Fig. S7), allowing us to expand our genomic coverage. These new spatial environments did not exhibit a global reorganization in response to Dex treatment (Supplemental Figs. S8, S9), and the contact profiles were more strongly correlated with DHS than with gene density, expression, or GR binding (Supplemental Table S5).
These findings were not uniquely associated with our mammary cell line because very similar nuclear dynamics and organization features of the Lcn2 contacts were also observed in hepatocytes (Hepa1C1C7 cell line) (Supplemental Fig. S10; Supplemental Table S5). GR function is well documented in liver, and Lcn2 is induced by GR in hepatocytes (Tronche et al. 2004; Engblom et al. 2007; Hakim et al. 2009).
With all the baits and cell types that were examined, virtually all the organizational properties found for Lcn2 are recapitulated in the other 4C data, including the hormone-induced dynamics, association with gene-dense regions, lack of correlation with expression or gene regulation by GR, no dominant functional group of genes within colocalized loci, and significant enrichment for distal GR-binding sites and DHS (Supplemental Figs. S7–S10; Supplemental Table S5). We therefore conclude that GR reprogramming of target loci can occur in physically independent subnuclear environments with highly similar spatial and dynamic characteristics.
Discussion
In this study, we investigated the underlying principles of genome organization in the context of the rapid and highly specific transcriptional reprogramming. Our genome-wide analysis demonstrated that although the transcriptional response is dramatic, GR target genes show modest dynamic changes in their subnuclear environment. Hormone activation of GR mostly leads to increases of interaction frequency with existing contact partners. These findings are in contrast to the observation that liganded estrogen receptors rapidly induce specific interchromosomal contacts between target genes (Hu et al. 2008) but in agreement with other reports showing that activation of these genes does not affect their positioning (Kocanova et al. 2010). The discrepancy could arise from the different culture conditions or source cells (Belmont 2010). The moderate changes in the spatial environments of our GR-regulated loci are in agreement with a previous finding that chromatin has constrained mobility, limited to a range of ∼0.5 μm, in mammalian cells (Chubb et al. 2002). The observation that individual genes have limited capacity to search for specific contacts also bears well with the fact that each locus is physically linked to other neighboring loci whose own preferred interactions (Simonis et al. 2006) may hinder dramatic movements. It is therefore likely that the genomic environment of a given gene in the nuclear space depends not only on its own characteristics but also on the properties of its flanking sequences and, by extrapolation, of those of the entire chromosome (de Laat and Grosveld 2007; Misteli 2007). Further examination of global chromatin organization will be required to resolve to what extent nuclear hormone-responsive genes are dominant over other genomic sequences in determining subnuclear environments.
The GR-responsive genes in the same physical environments show unexpectedly diverse and even opposite transcriptional responses to hormone. The choice of GR, a rapid transcriptional regulator that can efficiently repress as well as activate genes, allowed us to uncover the existence of disparate regulatory activities within the same subnuclear environment. Diverse transcriptional regulation by GR apparently does not require reorganization of target genes into specialized environments for induction and those for repression, for example. Our data do not exclude the existence of environments dedicated for a specific transcriptional regulation (Sandhu et al. 2009; Schoenfelder et al. 2009). It is possible that specialized environments may coexist with other transcriptionally active environments in the nucleus or may be more pronounced in certain types of cells (Hakim et al. 2010). When characterizing subnuclear environments, it would also be useful to assess whether the enrichment of functionally related genes is above and beyond that expected from the association of gene-rich regions.
Regardless of the extent of hormone-induced changes, our comprehensive analysis reveals a significant spatial clustering of genes and regulatory elements around GR-regulated genes. Hundreds of genes and thousands of DHSs are found in proximity of a GR target gene in resting cells, and the interaction network expands further after hormone treatment. We also discovered DNase I hypersensitivity to be the most salient genomic feature of loci interacting with Lcn2, Arrdc2, and Anxa5. Because chromatin accessibility is cell-type–specific, the correlation of 4C with DHS is more relevant than with gene density as potential determinants of cell-specific genome architecture. Some of these DHS sites may be occupied by architectural proteins, whereas others may contain sites for transient binding of chromatin factors, both of which may support and influence the 3D organization. Interestingly, the CTCF motif is enriched at DHS sites within the Lcn2 contact regions as well as recognition motifs for other transcription factors and coregulators. CTCF has been implicated in nuclear organization through association with cohesin (Wendt et al. 2008; Hadjur et al. 2009).
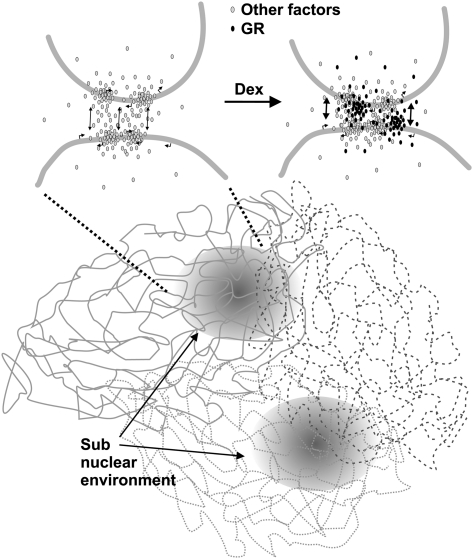
Instead of inducing large-scale chromosomal rearrangements, we find that a transcription factor uses pre-organized subnuclear environments, enriched with distal regulatory DNA elements and specific response elements. Because these environments are also gene-rich, they provide numerous targets for gene regulation by the transcription factor (Schoenfelder et al. 2009). The high concentration of GR recognition sites and DHSs in the microenvironments of GR-regulated genes already prior to hormone activation facilitates a rapid local accumulation of freely diffusing GR molecules and other cofactors upon stimulation (Fig. 5). This may be a fundamental and prevalent spatial feature of the mammalian genome. Numerous other transcription factors and regulators also bind to thousands of genomic sites far away from the genes that they regulate (Nielsen et al. 2008; Yu et al. 2009). These abundant binding sites may help maintain the nuclear architecture that allows rapid preprogrammed responses to diverse signal-responsive factors. Therefore, the remarkable clustering of distal regulatory elements may not simply be a secondary feature, but perhaps a major driving force behind the cell-type–specific 3D configuration of the genome. Altogether, our results suggest that the genome serves as a physical platform to enable efficient transcriptional regulation by promoting high local concentrations of freely diffusing protein factors in distinct spatial environments.
Figure 5.
Model for the structure of subnuclear environments enriched for GR and other distal regulatory elements and active gene loci. Chromosomes of a mammalian cell are organized into territories in an interphase nucleus. Within the chromosome territories, regions enriched for regulatory sites and active genes are nonrandomly organized in localized environments. These DHS-rich spatial domains are likely to provide binding sites for transcription factors and chromatin-remodeling proteins (gray), further reinforcing the organization and maintaining a high local concentration of these proteins. Although cross-linking protocols immobilize interacting regions, the local interactions are presumed to be highly dynamic. Hormone-activated GR (black) may increase the frequency of these dynamic contacts in preexisting spatial environments.
Methods
Cell culture
Mouse mammary epithelial adenocarcinoma (3134) and Hepa1C1C7 cells were maintained as described (Hakim et al. 2009). Cells were transferred to 10% charcoal-dextran-treated, heat-inactivated fetal bovine serum for 24 h before hormone treatment.
Chromosome conformation capture on chip (4C)
4C assay was performed as previously described (Simonis et al. 2006, 2007) with minor modifications.
Cultured mouse 3134 epithelial breast carcinoma or Hepa1C1C7 cells were fixed with 2% formaldehyde for 10 min, after 1 h treatment with 100 nM Dex or vehicle (EtOH). The initial steps in the 4C procedure giving rise to HindIII 3C ligated molecules were performed as described (Hakim et al. 2009). Cross-linked chromatin was digested overnight with excess of HindIII enzyme, and then DNA ends were ligated under dilute conditions that favor junctions between cross-linked DNA fragments. The Csp6I four-base restriction enzyme (Roche) was used to create the 4C circular DNA molecules.
Primers were used to PCR-amplify 4C DNA: Lcn2_4CF, 5′-GGGAGGTTAGTCACAGGGA-3′; Lcn2_4CR, 5′-AGGGAGGATGACTTAAGGAG-3′; Arrdc2_4CF, 5′-CAAATGCCAAAAGGAAGGAA-3′; Arrdc2_4CR, 5′-TCAGTCAACATGCATCTCAGAA-3′; Anxa5_4CF, 5′-CACCAGTTAACTAGCTTGCTTGC-3′; and Anxa5_4CR, 5′-TGAACAGGTCAGTCACATTGG-3′.
4C amplified DNA was hybridized to custom-made microarrays (Roche Nimblegen) that contained probes (60-mers) within 100 bp of HindIII sites across the mouse genome (mm8) excluding repetitive DNA elements. 4C DNA was labeled and hybridized on the array together with alternatively labeled genomic DNA cut with HindIII and Csp6I as a control, following the Nimblegen ChIP-chip protocol. For each experiment, two independently processed samples were used. For Lcn2 in the Hepa1C1C7 cell line and Lcn2 in the 3134 cell line, 10 min and 30 min after Dex treatment, and for Anxa5, the two independent samples were combined after PCR amplification and labeled on one microarray.
Real-time quantitative RT-PCR
RNA extraction and real-time quantitative RT-PCR were performed as previously described (Hakim et al. 2009). The forward and reverse primers flanking an intron–exon junction were designed to amplify the nascent transcript (Supplemental Table S4). Each experiment was performed in three independent repeats. Error bars represent SD of the biological repeats.
DNA FISH
For 3D FISH, we followed Meaburn and Misteli (2008) with minor modifications using the mouse BAC clones listed in Supplemental Table S6. Nuclei and probes were denatured together for 5 min at 85°C and left to hybridize overnight at 37°C in a humidified chamber. BACs hybridization specificity was confirmed by verifying the colocalization of two overlapping BACs labeled with biotin or digoxigenin and by metaphase spreads together with RP23-61N22 BAC for the Lcn2 locus.
Microscopy
Cells were imaged with an IX70 microscope (Olympus) controlled by a Deltavision System (Applied Precision) with SoftWoRx 3.5.1 (Applied Precision) and fitted with a charge-coupled device camera (CoolSnap; Photometrics) using a 60 × 1.4 oil objective lens (Olympus) and 29 0.3-μm optical steps covering the entire nucleus. Images were deconvolved by Applied Precision Softworks 1.0 software.
Automated image analysis
To quantitate distances between FISH signals, an automated image analysis computer algorithm was developed that determines the center-to-center distances in 3D. Nuclei and DNA FISH loci were automatically identified by a series of custom algorithms that were developed and executed using Matlab technical computing software and the Matlab Image Processing toolbox (The Mathworks, Inc.). All automatically defined regions of interest (ROIs) within the images were confirmed by manual inspection of the automated data. The resulting morphometric information from each nucleus was automatically stored in a Matlab database file, which could be accessed by the custom algorithms. An algorithm first used a combination of intensity-based and edge-detection-based computational methods to identify the nuclear ROI (nucROI) in the DAPI fluorescent channel images. Briefly, this was accomplished by a marker-based watershed segmentation method, which used simple fixed ratio intensity-based thresholding to define the nuclear markers and the first derivative of the Dapi image to define the nuclear edges. A separate algorithm used the nucROI positional information and simple fixed ratio intensity-based thresholding of the FISH fluorescent channel images to identify the ROIs containing the DNA FISH loci (FISH ROI). A third algorithm assigned the nuclear position of each FISH locus, based on the x–y–z location of the center of the brightest 3 × 3 FISH fluorescent channel pixel that was located within the FISH ROI. For each nucleus, 3D distances (distance = √[(x2 − x1)2 + (y2 − y1)2 + (z2 − z1)2]) were calculated between all possible pairs of FISH loci for the two probes being evaluated. The closest distance between the two probes in each nucleus was output to the database for further statistical analysis.
Statistical analysis for significant interactions from 4C data
Quantile-normalized 4C/input log ratios from duplicate chips were averaged for each condition. Each probe tiled in the NimbleGen chip was assessed by a chi-squared test using a running window of size 100 kb centered at the probe. The procedure generated a P-value for each probe, based on the number of neighboring probes (within the 100-kb window) whose 4C log ratio values were in the top 10% of all log ratio values from the chip. Probes were considered positive if the p score (= −log10 p) is greater than 4, which corresponded to FDR of ∼0.3%. Probe p scores before and after Dex treatment were considered significantly different if their absolute difference was greater than 4.5 (0.4% FDR). Contiguous probe clusters were obtained by merging positive 4C probes within 100 kb. Interacting probe clusters were considered robust if they contained more than 10 4C probes. All the microarray data have been deposited in the NCBI GEO database under accession number GSE27115 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27115).
FDR was empirically determined from the data as follows: Randomized 4C data were generated for each condition by permuting the association between genomic positions and log ratios of all NimbleGen probes. The chi-squared test on a window running through the genome was performed as above. The number of positive probes in the random data divided by the number of positive probes in the actual 4C data was taken as the FDR. Similarly, FDR for significant p score differences between ±Dex was calculated by dividing the number of probes with absolute p score differences greater than 4.5 by that observed from 4C. The analysis was implemented by using the R package ChIP-chip (available as ACME at http://www.bioconductor.org).
Integrative analyses of 4C, gene expression microarray, GR ChIP-seq, and DNase I-seq data
The expression data set was obtained by hybridization of samples from the 3134 cells either untreated or treated with Dex for 2 h onto Affymetrix Mouse Exon 1.0 ST arrays (John et al. 2011). GR ChIP-seq and DNase I-seq data were generated using the Illumina massively parallel sequencing platform for the 3134 cells treated with Dex for 1 h as described in John et al. (2011).
An enrichment analysis was applied to the set of 4C positive probes against each genomic data set. The 10-Mb bait region was excluded from all enrichment analyses. The number of positive 4C probes that had a target event (occurrence of TSS, TSS for expressed genes, TSS for GR-regulated expression, or center of GR ChIP peaks or DHS hotspots) within 10 kb was counted, and the corresponding targets were retrieved (genes, expressed genes, regulated genes, or GR-binding sites, DHS). Based on the Affymetrix exon array data, genes were considered to have detectable expression if the probe intensity was greater than 90 percentile of the microarray, and were scored as GR-regulated if the Dex-dependent fold change was greater than two. To assess statistical significance, random sampling of the same number of NimbleGen probes was performed (n = 100 or 1000), which provided a P-value for encountering the observed number of target-proximal probes by chance.
In addition, genome-wide correlation between 4C and each of the other profiles was calculated. Because enrichment analyses involve setting thresholds for various quantities (chi-squared test P-values for positive 4C probes, expression log ratios to define GR-regulated genes, minimum base-pair distances to define proximity, etc.), it is possible that subtle but systematic relationships may be missed in such an approach. To maintain the quantitative aspect of each genomic data set despite their different genomic scales, we obtained a fixed resolution profile of each datum by computing either the average value (for 4C and expression) or the total number of occurrences (TSS for gene density; GR ChIP peaks for GR binding; hotspots for DHS) in a window of size 100 kb, sliding along the genome at 50-kb increments. The resulting profiles were used for Figure 4 and Supplemental Figure S5, and for calculating the Pearson correlation coefficient over the genome excluding the 10-Mb bait region (Supplemental Table S5).
Motif discovery analysis was performed using the algorithm MEME 4.4.0 (Bailey and Elkan 1994) implemented at the NIH supercomputing cluster Biowulf. One thousand five hundred input regions were submitted to MEME, and output files were examined to retrieve significantly enriched motifs. Each motif was queried against the database TRANSFAC using Tomtom to identify matching motifs for known transcription factors.
Acknowledgments
We acknowledge A. Indrawan for technical help; T. Karpova for help with microscopy; Y. Dalal and K. Gardner for helpful discussions; D. Meerzaman and R.L. Walker for help with the microarray scanner; S. Baek for submitting motif analysis jobs to the NIH supercomputing cluster; and K. Meaburn for help with DNA FISH. Fluorescence imaging was performed at the National Cancer Institute Fluorescence Imaging Facility. This work was supported in part by the Intramural Research Program of the National Institute of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
[Supplemental material is available for this article. The microarray data from this study have been submitted to Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE27115.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.111153.110.
References
- Apostolou E, Thanos D 2008. Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell 134: 85–96 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Belmont AS 2010. Estrogen fueled, nuclear kiss: Did it move for you? Nucleus 1: 440–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, et al. 2008. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol 182: 1083–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev 18: 1119–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA 2002. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol 12: 439–445 [DOI] [PubMed] [Google Scholar]
- de Laat W, Grosveld F 2007. Inter-chromosomal gene regulation in the mammalian cell nucleus. Curr Opin Genet Dev 17: 456–464 [DOI] [PubMed] [Google Scholar]
- Engblom D, Kornfeld JW, Schwake L, Tronche F, Reimann A, Beug H, Hennighausen L, Moriggl R, Schutz G 2007. Direct glucocorticoid receptor–Stat5 interaction in hepatocytes controls body size and maturation-related gene expression. Genes Dev 21: 1157–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P, Bickmore W 2007. Nuclear organization of the genome and the potential for gene regulation. Nature 447: 413–417 [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, John S, Ling JQ, Biddie SC, Hoffman AR, Hager GL 2009. Glucocorticoid receptor activation of the Ciz1-Lcn2 locus by long range interactions. J Biol Chem 284: 6048–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, Sung MH, Hager GL 2010. 3D shortcuts to gene regulation. Curr Opin Cell Biol 22: 305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, et al. 2008. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci 105: 19199–19204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, et al. 2008. Interaction of the glucocorticoid receptor with the global chromatin landscape. Mol Cell 29: 611–624 [DOI] [PubMed] [Google Scholar]
- John S, Johnson TA, Sung MH, Koch-Paiz CA, Davis SR, Walker R, Meltzer P, Hager GL 2009. Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology 150: 1766–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43: 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocanova S, Kerr EA, Rafique S, Boyle S, Katz E, Caze-Subra S, Bickmore WA, Bystricky K 2010. Activation of estrogen-responsive genes does not require their nuclear co-localization. PLoS Genet 6: e1000922 doi: 10.1371/journal.pgen.1000922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296: 158–162 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T 2008. Locus-specific and activity-independent gene repositioning during early tumorigenesis. J Cell Biol 180: 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T 2007. Beyond the sequence: Cellular organization of genome function. Cell 128: 787–800 [DOI] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, et al. 2008. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev 22: 2953–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG 2009. CTCF: Master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS, Shi C, Sjolinder M, Zhao Z, Gondor A, Liu L, Tiwari VK, Guibert S, Emilsson L, Imreh MP, et al. 2009. Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development. Genes Dev 23: 2598–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. 2009. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Schober H, Fraser P, Gasser SM 2007. Gene regulation through nuclear organization. Nat Struct Mol Biol 14: 1049–1055 [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38: 1348–1354 [DOI] [PubMed] [Google Scholar]
- Simonis M, Kooren J, de Laat W 2007. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods 4: 895–901 [DOI] [PubMed] [Google Scholar]
- Tronche F, Opherk C, Moriggl R, Kellendonk C, Reimann A, Schwake L, Reichardt HM, Stangl K, Gau D, Hoeflich A, et al. 2004. Glucocorticoid receptor function in hepatocytes is essential to promote postnatal body growth. Genes Dev 18: 492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. 2009. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell 36: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]