Abstract
Core promoters mediate transcription initiation by the integration of diverse regulatory signals encoded in the proximal promoter and enhancers. It has been suggested that genes under simple regulation may have low-complexity permissive promoters. For these genes, the core promoter may serve as the principal regulatory element; however, the mechanism by which this occurs is unclear. We report here a periodic poly-thymine motif, which we term T-blocks, enriched in occurrences within core promoter forward strands in Caenorhabditis elegans. An increasing number of T-blocks on either strand is associated with increasing nucleosome eviction. Strikingly, only forward strand T-blocks are correlated with expression levels, whereby genes with ≥6 T-blocks have fivefold higher expression levels than genes with ≤3 T-blocks. We further demonstrate that differences in T-block numbers between strains predictably affect expression levels of orthologs. Highly expressed genes and genes in operons tend to have a large number of T-blocks, as well as the previously characterized SL1 motif involved in trans-splicing. The presence of T-blocks thus correlates with low nucleosome occupancy and the precision of a trans-splicing motif, suggesting its role at both the DNA and RNA levels. Collectively, our results suggest that core promoters may tune gene expression levels through the occurrences of T-blocks, independently of the spatio-temporal regulation mediated by the proximal promoter.
The genome contains information encoded according to both a genetic code for translation and a separate code for its regulation. Recent research has provided strong evidence that in metazoans much of the morphological differences among species can be accounted for by changes in gene regulation rather than by changes to the protein-coding regions (Davidson 2006; Carroll 2008). The genetic code is well-characterized, allowing for the conceptual translation of a gene given only its DNA sequence. However, the regulatory code is far less understood. For example, transcription factors (TFs) bind highly redundant motifs of variable lengths, distributed in a relaxed manner across a wide promoter region, in multiple numbers and tolerance for different polarities (Stark et al. 2007; Meireles-Filho and Stark 2009; Schmidt et al. 2010). Juxtaposed between the proximal promoter and the coding region is the core promoter, unique in its role as the gateway to a gene's transcription (Juven-Gershon et al. 2008), containing both spatially restricted motifs as well as flexibility in the occurrences of its composing motifs.
Previous work has highlighted the important role of the core promoter as an integrator of a complex array of signals toward the assembly of the RNA polymerase (Butler and Kadonaga 2001). The core promoter is usually defined as the −50 to +50 base pair (bp) region with respect to the start of transcription. Yet, despite its short length, the core promoter harbors a number of conserved elements including the TATA box, Initiator, downstream promoter element (DPE), TFIIB recognition element (BRE), motif ten element (MTE), and downstream core element (DCE) (Smale and Baltimore 1989; Burke and Kadonaga 1996; Lagrange et al. 1998; Ohler et al. 2002). These elements are involved in the recruitment of additional auxiliary factors (general transcription factors [GTFs] and TBP-associated factors) mediating RNA polymerase assembly during transcription initiation and thus determining the actual composition of the RNA polymerase complex (Hoey et al. 1990; Burke and Kadonaga 1996; Lagrange et al. 1998; Chalkley and Verrijzer 1999). The action of many TFs relies upon their direct or mediated interactions with the RNA polymerase complex (Butler and Kadonaga 2001). Thus, the set of TFs distributed along the proximal promoter and enhancer—regulating gene expression along spatial and temporal axes—might be dependent upon the composition of the core promoter. Indeed, a design of a “super core promoter” including four conserved core promoter motifs (TATA box, MTE, DPE, and Initiator) exhibited significantly elevated expression levels, both in vitro and in vivo (Juven-Gershon et al. 2006). However, natural core promoters do not generally have a characteristic structure but rather exhibit a surprisingly wide variation in terms of their motif composition (Juven-Gershon and Kadonaga 2009).
While the core promoter is typically thought of as a regulatory integrator, recent work has suggested that, for certain classes of genes, the core promoter may constitute the principal regulatory element (Tirosh and Barkai 2008; Cairns 2009; Juven-Gershon and Kadonaga 2009). In yeast, a significant fraction of genes can be classified as having one of two contrasting promoter architectures (Tirosh and Barkai 2008). The open promoters are free of nucleosomes and relatively independent of TF regulation. Such simple promoters are enriched with poly(dA:dT) motifs, yet depleted with the TATA box. Alternatively, covered promoters are tightly bound by nucleosomes and exhibit the opposite properties to open promoters (Tirosh and Barkai 2008; Cairns 2009). The distinction between these two groups essentially relies upon the sequence composition of the core promoters (Cairns 2009). Open genes might therefore be more heavily dependent upon the core promoter because of their loose association with TFs. However, the mechanism by which the core promoter may be precisely tuned to encode expression levels is unclear.
To study this topic we selected the nematode Caenorhabditis elegans, which provides an efficient system, given its compact well-characterized genome and a wide range of available whole-genome experimental data. C. elegans is possibly the best understood metazoan, and consequently, the mechanism by which its transcription is regulated stands as a formidable challenge. C. elegans also contains post-transcriptional regulatory mechanisms that impinge upon the sequence of the core promoter. Most C. elegans mRNA transcripts undergo trans-splicing, where the 5′ UTR is replaced by a short splice leader, donated by a 100 nucleotide noncoding nuclear RNA, called SL1 (Blumenthal 2005). Trans-splicing is similar to cis-splicing and depends upon similar components (Blumenthal 2005). Moreover, trans-splicing relies upon a DNA sequence motif that is nearly identical to the 3′ splice sites of introns (Graber et al. 2007). Two additional elements found in tight association with trans-splice sites—the U-rich element and Kozak sequence—are implicated in this process (Graber et al. 2007). Trans-splicing is also present in genes organized into operons, which in C. elegans amounts to 15% of all coding genes. Most operon genes have housekeeping functions and are associated with basal cellular processes such as transcription, translation, and respiration (Blumenthal and Gleason 2003; Blumenthal 2005). mRNA transcribed from internal operon genes is trans-spliced by a second RNA, called SL2 (Blumenthal 2005).
In the present work, we report a motif found in the core promoter sequence that is composed of periodic strand-specific thymine repeats with a phase of ∼10 bp. We propose that occurrences of this repeat are shaped by selective pressures to tune gene expression levels through the modulation of nucleosome occupancy, transcription stability, and translation efficiency. According to this view, core promoter T-blocks tune gene expression levels by evicting nucleosomes and mediating post-transcriptional regulation in C. elegans.
Results
Motif composition of the Caenorhabditis core promoter
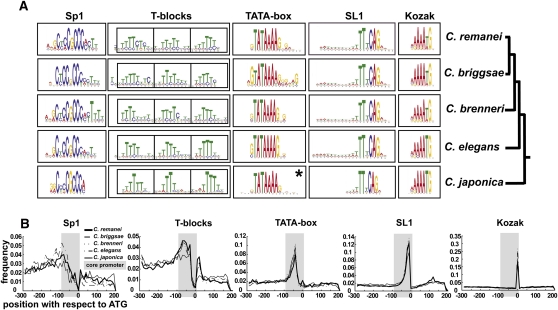
To study the role of the core promoter in gene regulation, we first sought to identify sequence motifs. A global motif search in C. elegans core promoters using MEME (Bailey and Elkan 1994) yielded five dominant motifs (Fig. 1A). Two have been previously associated with 5′ UTR sequences (Conrad et al. 1991; Graber et al. 2007): the SL1 (TTnCAG) motif and the Kozak sequence (AAAATG), comprising ∼60% and ∼37% (Supplemental Table S1) of C. elegans genes, respectively. The canonical SL1 motif is very similar in sequence to the 3′ splice signal of introns (Conrad et al. 1991). Consistently, SL1 possesses a 5′ 10–15-bp tail enriched with T/A nucleotides. The Kozak sequence contains the start codon and is nearly identical to its counterpart in Drosophila ([CA]AA[CA]ATG) (Cavener 1987). Further, the TATA box and SP1 motifs have been previously described in promoters (Goldberg 1979; Xi et al. 2007). The TATA box (GTATA[TA][TA]AG) of C. elegans is highly similar to that of Drosophila (Ohler et al. 2002). The TATA box has a wide phylogenetic distribution and is mainly found in the core promoters of stress-response (Basehoar et al. 2004; Huisinga and Pugh 2004), noisily expressed (Blake et al. 2006), and evolvable genes (Tirosh et al. 2009). Consistent with other works, we find that the TATA box is present in ∼6% of the genes (Supplemental Table S1). The SP1 motif has been previously characterized in promoters of vertebrates where it regulates expression in TATA-less promoters (Huber et al. 1998). The fifth conserved element identified by MEME is a periodic occurrence of three to five thymines (Fig. 1A). We named a single group of these neighboring thymines a “T-block.” T-blocks are widely distributed in the genome; ∼42% of C. elegans genes contain the motif (Supplemental Table S1).
Figure 1.
Motif composition of the Caenorhabditis core promoter. (A) Five conserved motifs in each of the five examined Caenorhabditis species are shown as sequence logos. (*) In contrast to all other motifs which were found in the initial search, the Caenorhabditis japonica TATA box motif was detected only in sequences whose orthologs contained the “TATA” motif. (B) Distribution of motif occurrences along the sequence. The gray box in each plot corresponds to the core promoter. The C. japonica SL1 motif was normalized to the length of the other species.
The five motifs identified in C. elegans are well-conserved in four additional nematode genomes (Fig. 1A). The frequency of genes containing each motif is also generally conserved (Supplemental Table S1). To characterize the conservation of the location of the five motifs across species, we compared the distributions of locations of each motif in each genome (Fig. 1B). The distributions are remarkably similar, demonstrating that the five motifs are constrained to the same relative positions along the core promoter.
The T-blocks motif has a periodicity of ∼10 bp
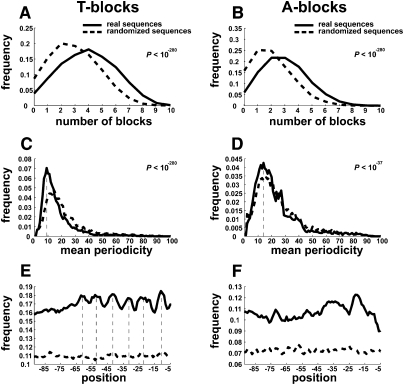
The detected T-blocks motif shown in Figure 1A contains three T-blocks; however, individual promoters may have more. To summarize the number of T-blocks per gene, we developed the following simple procedure. Parsing the promoter sequence into 10 nonoverlapping 10-bp windows, the number of T-blocks is equal to the number of windows with at least three consecutive thymines. We found a wide distribution in the number of T-blocks per gene in C. elegans, with a mode of four (Fig. 2A). Over 5000 genes have six or more T-blocks. Random sequences of the same length and nucleotide composition exhibit a significantly smaller number of T-blocks (Fig. 2A; P < 10−280). Since only the forward strand was examined, the appearance of T-blocks and not (also) A-blocks suggests a strand bias. Supplemental Table S4 indicates the instances of genes with each number of T-blocks and A-blocks in the core promoter. While we found that there are more A-blocks than expected (Fig. 2B), they are depleted in the total number of blocks with respect to T-blocks (P < 10−280).
Figure 2.
Occurrences and periodicity of the T-blocks motif in core promoters. (A,B) Distributions of T/A-blocks per gene, respectively. A block of a given nucleotide is defined as three or more occurrences of that nucleotide. The overall number of blocks is computed as the number of nonoverlapping 10-bp windows containing the blocks (see Methods). (C,D). Distributions of mean periodicity for all genes. For each gene, the mean distance between T-blocks was calculated (see Methods). The random plot corresponds to the mean periodicity in randomly permutated sequences. The observed plot for T-blocks (left) indicates a periodicity with a mode of nine, while A-blocks show a periodicity of 15 (vertical dashed lines). (E,F). Locations of T-blocks and A-blocks along the core promoter. For each location, the fraction comprising T/A-blocks is shown. Random sequences are computed as in C and D. A periodicity of 10 bp is found in the −60 to −10 region (vertical dashed lines). Indicated P-values relate to differences between the distributions.
Since DNA (in its B-form) contains 10.6 bp per turn, the ∼10-bp period of the T-blocks motif (Fig. 1A) indicates a role relating to DNA structure as opposed to specific TF-binding. Two additional analyses support ∼10-bp periodicity. First, for each gene, we computed the average number of nucleotides separating the starts of T-blocks. The distribution of such mean periodicities shows a mode of nine nucleotides, while the mode for the random sequences is 15 (dashed lines in Figure 2C). On the contrary, A-blocks show a similar mean periodicity to randomized sequences, suggesting an absence of ∼10-bp periodicity for A-blocks (Fig. 2D). Second, we analyzed the locations of T-blocks along the core promoter. We found that T-blocks overall are separated by ∼10 bp, while no periodicity was observed for random sequences (Fig. 2E). Further, no periodicity was observed for A-blocks (Fig. 2F). This analysis also suggests that T-blocks occupy fairly fixed positions along the promoter. We conclude from these analyses that T-blocks are enriched in their occurrence in promoter sequences and exhibit a periodicity of ∼10 bp. We note, however, that while patterns are evident at the genome level, for each particular gene there is a wide variation in occurrences and periodicities.
The number of T-blocks and A-blocks correlates with nucleosome eviction
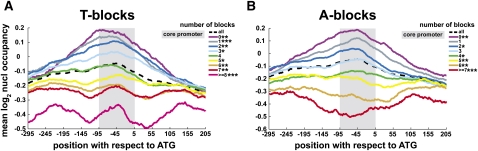
The periodicity of the T-blocks motif led us to hypothesize a role for T-blocks in nucleosome occupancy. Recent studies demonstrated that the DNA sequence plays an important role in nucleosome positioning (Rhodes 1979; Prunell 1982; Struhl 1985; Koch and Thiele 1999; Anderson and Widom 2001; Segal et al. 2006; Kaplan et al. 2009). Long stretches of adenine nucleotides known as A-tracks, for instance, have the lowest nucleosome occupancy in vitro (Anderson and Widom 2001), while AA/TT/TA dinucleotides occurring with a periodicity of ∼10 bp are enriched in nucleosome occupancy (Segal et al. 2006; Kaplan et al. 2009). Furthermore, the start and end of C. elegans genes are enriched with nucleosome occupancy (Gu and Fire 2010). To examine nucleosome positioning within C. elegans core promoters, we invoked a published data set (Valouev et al. 2008) to determine genome-wide nucleosome occupancies. For each set of genes with a particular number of T-blocks, we calculated the mean nucleosome occupancy (Fig. 3A). We found a negative correlation between the number of T-blocks and nucleosome occupancy in the core promoter (Fig. 3A). For example, genes with six T-blocks are significantly depleted of nucleosomes relative to the set of all genes (P < 10−19). We also observed a gradual correlation of nucleosome occupancy with an increasing number of T-blocks, whereby genes with two or less T-blocks are enriched in nucleosome occupancy relative to the genome average, while an increasing number of T-blocks have, on average, less nucleosome occupancy. This same correlation is also observed for A-blocks, suggesting strand-independence (Fig. 3B). Further, genes with both many T-blocks and many A-blocks display the lowest nucleosome occupancy, while those with both few T-blocks and few A-blocks are the most heavily occupied (Supplemental Fig. S1). However, we did not find a significant difference between the nucleosome coverage of genes with many T-blocks and few A-blocks and vice versa (Supplemental Fig. S1). Thus, we conclude that both T-blocks and A-blocks in the core promoter have a similar effect on nucleosome eviction.
Figure 3.
The number of T-blocks and A-blocks correlates with nucleosome eviction. (A) For each set of genes with a given number of T-blocks, the mean log2 nucleosome occupancy is shown for each base pair in the −296 to +205 region with respect to the ATG. The mean nucleosome occupancy for all genes in this region is indicated by the dashed line. Significance of differences between the distribution of particular T-block counts and the genome average are indicated as stars in the legend (*, P < 10−5; **, P < 10−10; ***, P < 10−20). (B) The same as A for A-blocks. The normalized nucleosome occupancy data was obtained from Valouev et al. (2008).
The number of core promoter T-blocks—but not the number of A-blocks—correlates with gene expression levels
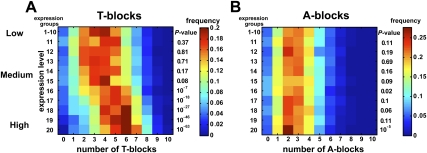
We reasoned that the level of nucleosome occupancy would correlate with gene expression levels, since the initiation of transcription requires the eviction of the nucleosomes (Fuda et al. 2009). To test this, we invoked gene expression data from the four-cell stage of C. elegans embryos (Yanai and Hunter 2009). We defined 20 equally populated gene groups according to mRNA transcript levels. The first 10 groups were collapsed to one, representing nonexpressed to lowly expressed genes. For each group of genes, we computed the distribution of the number of T-blocks in their core promoters. We found a significant correlation between the expression levels and the number of T-blocks (Fig. 4A). While genes with low and medium expression levels have a relatively low number of T-blocks (generally, no more than four), the distribution gradually shifts to higher numbers of T-blocks with higher expression levels. Genes with ≥6 T-blocks have fivefold higher expression levels than genes with ≤3 T-blocks. Surprisingly, this correlation is absent for A-blocks (Fig. 4B), despite the similar effect of A-blocks on nucleosome eviction (Fig. 3B). A similar correlation was also observed in an independent expression data set using mRNA from adult (Supplemental Fig. S2A,B) and L1 (first larvae) worms (Supplemental Fig. S2C,D).
Figure 4.
The number of T-blocks, but not the number of A-blocks, correlates with gene expression levels. (A) For each set of genes with a given level of expression, the distribution of the number of T-blocks is shown as a heat map. Twenty equally populated groups of genes were defined based upon their gene expression levels. Most genes have a low level of expression, and thus the ten groups with the lowest expression levels were merged into one, “1” group. The remaining ten are shown in the figures as groups 11 through 20. The distribution of T-blocks of each category was compared with that of the “1” group and the P-value (Kolmogrov-Smirnov test) is indicated to the right. (B) Same as (A) for A-blocks.
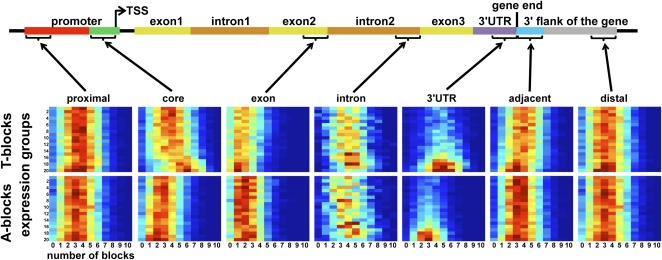
We asked if the correlation between the number of T-blocks and expression levels is unique to the core promoter. We did not find a similar correlation in the proximal promoter, exons, introns, 3′ UTR, and adjacent and distal gene ends (Fig. 5; Supplemental Fig. S3). The absence of correlation is most evident in the proximal promoter and in adjacent and distal 3′ flanking sequences, suggesting that T-blocks exert their effect only in the core promoter. Consistent with previous works, we observed an overall increase in stretches of T's and A's in the intergenic regions and introns as compared with coding sequences (VanWye et al. 1991; Widom 1996; Fukushima et al. 2002; Cohanim et al. 2006; Fire et al. 2006; Kumar et al. 2006; Moreno-Herrero et al. 2006). Interestingly, the 3′ UTR is also rich in T-blocks relative to A-blocks (Mangone et al. 2010), although in an expression-level independent manner (Fig. 5). Since 3′ UTR sequences < 100 bp were excluded from this analysis, the cloud in Figure 5 reflects the observation that low-expression genes contain shorter 3′ UTRs (see also Supplemental Fig. S4). This is also consistent with the recent result that 3′ UTR lengths decrease with developmental time (Mangone et al. 2010).
Figure 5.
The correlation between T-block occurrences and expression level is unique to the core promoter. For each of seven gene regions, including the proximal promoter, end of second exon and intron, 3′ UTR, and the 3′ adjacent and 3′ distal end regions, the T-block and A-block frequency is shown for different expression groups as in Figure 4. The 3′ UTR and core promoter are T-rich, while exons are A-rich. For each region, 100 bp were selected as the core promoter. Ensembl annotation was used to identify the second exon and intron, ignoring alternatively spliced sequences.
Differences in T-block occurrences between orthologous core promoters correlate with expression level changes
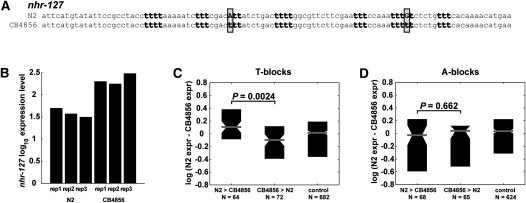
The correlation between T-blocks and expression levels may be indirect due to an effect of other properties such as gene neighborhood and gene function. For example, different sets of genes may have both more T-blocks and higher expression due to their particular function. We thus sought to investigate how changes in T-block numbers influence the expression level of particular genes. We examined gene expression data from embryos of two C. elegans strains (N2 and CB4856), together with single nucleotide polymorphisms (SNPs) between these strains (see Methods). For example, the core promoter of nhr-127 in CB4856 has two SNPs relative to the N2 promoter, both affecting T-blocks (Fig. 6A). The higher T-block count in nhr-127 in CB4856 correlates with the significantly higher gene expression level relative to its N2 counterpart (Fig. 6B).
Figure 6.
Differences in T-block occurrences between orthologous core promoters correlate with expression differences. (A) The nhr-127 gene has seven T-blocks in the strain CB4856, while its ortholog in the N2 strain has one less T-block. (B) The nhr-127 gene is more highly expressed in CB4856 than in N2. (C) Ratio of expression differences for three groups of genes: (1) Genes with more T-blocks in N2; (2) genes with more T-blocks in CB4856; and (3) genes with core promoter SNPs, yet the same number of T-blocks in both strains. More T-blocks correlate with higher expression. (D) Same format as C for A-blocks. A correlation with expression change was not detected.
To examine this globally, we compiled two groups of genes. The first contained genes with a higher T-block count in N2, and the second contained genes with a higher T-block count in CB4856. The analysis was limited to genes with relatively large differences in T-block numbers (see Methods) and at least four T-blocks in either strain. For each group, we calculated the ratio of N2 to CB4856 expression levels between orthologs. We found that for the 64 genes with a higher T-block count in the N2 strain, expression was higher in N2 (Fig. 6C). Similarly, in the 72 genes with more T-blocks in CB4856, expression was generally higher in that strain (Fig. 6C). The expression ratios between these two groups are significantly different (P = 0.0024). As a control, we examined genes that contain core promoter SNPs that do not change the T-block count, and the distribution of their expression ratios lies between those of the two groups (Fig. 6C). Further, combining the groups together, we also found a significant difference between those and the control category (P = 0.0014). Interestingly, this trend was not apparent for SNPs affecting instances of A-blocks (Fig. 6D; P = 0.6331). Thus, in spite of the 98.8% genetic identity between the two C. elegans strains (Wicks et al. 2001), more core promoter T-blocks across orthologs are associated with higher gene expression levels.
T-blocks form a supra-motif with the SL1 motif
T-blocks may occur independently of other motifs or in combination as a supra-motif. We found that genes with a significant match to the MEME T-blocks motif (Fig. 1A) tend to also possess the SL1 motif (P < 10−16; hypergeometric distribution; Supplemental Table S2). Also, the sets of genes containing T-blocks and SL1 are strongly depleted for occurrences of the TATA box (P < 10−32 and P < 10−56 for SL1 and T-blocks, respectively; hypergeometric distribution; Supplemental Table S2). These results were also observed in each of the other Caenorhabditis species (Supplemental Table S2). One shortcoming of this analysis is the use of the MEME motifs which limits T-blocks to three blocks and is based upon an arbitrary P-value threshold. To overcome these constraints, we further examined the dependency of the T-blocks and SL1 motifs by comparing the distribution of T-blocks across equally populated groups of genes with different matches to the SL1 motif, based upon the MEME P-value. We term the latter measure the “fuzziness” of the occurrence of the SL1 motif, similar to a previous formulation (Bilu and Barkai 2005). We found a pronounced correlation between the number of T-blocks and the fuzziness of the SL1 sequence (Fig. 7A). Genes with the best matches to the SL1 MEME motif in their core promoters also have the highest number of T-blocks, whereas genes most likely to lack the SL1 motif (most fuzzy) exhibit the lowest number of T-blocks (Fig. 7A). The same analysis for A-blocks did not reveal such a correlation (Fig. 7E). We thus conclude that T-blocks and SL1 form a supra-motif.
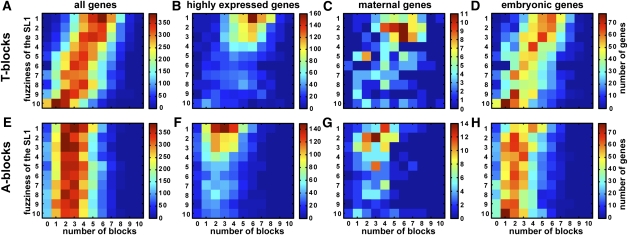
Figure 7.
Highly expressed genes have both a preponderance of T-blocks and a precise SL1 motif. In each heat map, genes are mapped onto a plane of the number of T-blocks (and A-blocks) and the fuzziness of the SL1 motif. SL1 fuzziness is determined by dividing the set into 10 equally populated groups according to the P-value of the best match of the SL1 motif to the gene's core promoter (see Methods). The heat maps correspond to the number of T-blocks (top) and A-blocks (bottom) for all genes (A,E), highly expressed genes (B,F), maternally expressed genes (C,G), and embryonically expressed genes (D,H). The latter three groups are mutually exclusive by definition (see Methods).
The SL1 MEME motif (Fig. 1A) contains a long tail composed of thymines and adenines which may contribute to the number of computed T-blocks for genes with well-matching SL1 motifs. To test whether this tail can alone account for this correlation, we repeated the analysis on a set of core promoter sequences truncated in their 3′ ends by 40 bp. As the correlation was still evident (Supplemental Fig. S5), we conclude that the thymine/adenine-rich 5′ tail of the SL1 motif cannot explain the increase in the number of T-blocks in groups of genes with precise SL1 motifs (Supplemental Fig. S5).
Highly expressed genes have a preponderance of T-blocks and a precise SL1 motif
To gain insight into the use of different core promoters throughout the genome, we next asked which gene ontology (GO) terms are enriched in genes with either the T-blocks–SL1 supra-motif or the TATA box. We found a significant association of SL1–T-blocks genes with growth, reproduction, and development, accounting for seven of the 15 GO terms with the highest level of enrichment (Supplemental Table S3). This enrichment likely reflects a strong correlation between highly expressed genes and these essential functions. The TATA box motif, in contrast, was constrained to stress-related genes and genes involved in nucleosome assembly (six GO terms from 15) (Supplemental Table S3). Consistently, an association of the TATA box with stress-related genes in Saccharomyces cerevisiae has been reported (Basehoar et al. 2004).
We next asked whether genes with the T-blocks–SL1 supra-motif show a bias toward particular patterns of expression. We defined three mutually exclusive expression groups: maternal, embryonic, and genes with high expression throughout embryonic development (see Methods). For embryonic genes, we found roughly the same distribution of genes along the T-blocks and SL1 fuzziness axes as for the total set of genes (Fig. 7D). However, highly expressed genes are distinguished with high numbers of T-blocks and a precise SL1 motif in their core promoters (Fig. 7B). Maternal genes also possess high numbers of T-blocks and well-conserved SL1 motifs similar to highly expressed genes, though with a slight shift toward less precise SL1 motifs and fewer T-blocks (Fig. 7C). Again, a similar analysis repeated for A-blocks did not reveal a correlation with SL1 fuzziness among highly expressed, maternal, and embryonic genes (Fig. 7F,G,H).
Operon genes are enriched with T-blocks
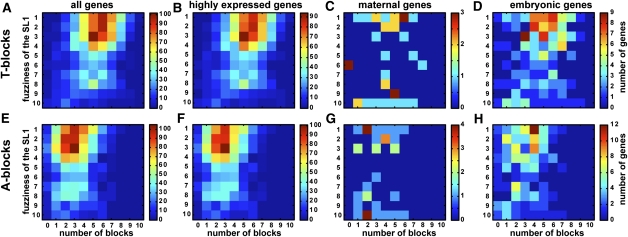
Operons are estimated to account for ∼15% of C. elegans genes, which tend to be highly expressed (Blumenthal 2005). The SL1 motif, as well as the similar SL2 motif, is involved in the processing of the polycistronic transcript (Blumenthal 2005). Given our findings that highly expressed genes should have both multiple T-blocks and a precise SL1 motif, we reasoned that, in addition to the splicing motifs (SL1 and SL2), operon genes as a group should be enriched with T-blocks. We found that operon genes have a median of 5.08 T-blocks, significantly higher than the median for nonoperon genes (3.95; P < 10−118). Operon genes may be classified according to their order within the operon, and we find that both the first operon genes and the internal operon genes have a similar enrichment for T-blocks (medians of 5.08 and 4.97, respectively). The overwhelming majority of operon genes are highly expressed (90%) and contain a clearly shifted distribution of T-blocks, though not A-blocks (Fig. 8A,E). To test whether the correlation between T-blocks and expression levels is not driven by misannotated operon genes, we repeated the analysis on a restricted set of genes not annotated as operon genes and with at least 1-kb intergenic region on either end. The correlation remained after this restriction (Supplemental Fig. S6). This analysis also excludes the possibility that T-blocks appear only in core promoters shared by close gene neighbors in reverse orientation. We conclude that highly expressed nonoperon genes and operon genes share a similar core promoter architecture involving the presence of the SL1 motif and a large number of T-blocks.
Figure 8.
Operon genes have a high number of T-blocks, a precise SL1 motif and are highly expressed. (A–H) The heat maps are in the same format as in Figure 7.
Preponderance of thymines in metazoan core promoters
Many core promoter motifs are highly conserved throughout eukaryotes, leading to the question of the generality of core promoter T-blocks beyond the Caenorhabditis genus (Fig. 1). We found that thymines are enriched relative to adenines in metazoan genomes in the core promoter forward strands of many model species (Supplemental Fig. S7A). Furthermore, T-blocks are more frequent than A-blocks in all examined species with the exception of Drosophila melanogaster (Supplemental Fig. S7B). It will be interesting to further characterize the metazoan-wide evolution of this promoter element and to study its effect on regulation in other systems.
Discussion
We report here the existence of a simple motif, which we term T-blocks, that occurs with a periodicity of ∼10 bp in a dominant fraction of C. elegans core promoters. T-blocks can be regarded as a composite motif occurring in a wide range of numbers of individual blocks. The presence of an increasing number of T-blocks specifically in the core promoter is correlated with a graded paucity of nucleosomes and a higher level of expression. Our results suggest that T-blocks are an important factor in the regulation of C. elegans gene expression levels. In this discussion, we consider the genomic and transcriptomic constraints that may shape the structure, function, and evolution of the core promoter T-blocks. What constraints impinge on this DNA region? What evolutionary forces underlie the periodic structure? What mechanism is responsible for the graded correlation with expression level? Why do different genes have different numbers of T-blocks?
The 100 bp preceding the start of translation in C. elegans are under strong constraints since these simultaneously represent both the 5′ UTR and the core promoter and consequently contain their associated regulatory motifs. This apparently stems from the general shortness of the C. elegans 5′ UTR, as evidenced by the following observations. The regulatory elements we find in this region are known functional elements of the 5′ UTR: the SL1 motif and the Kozak sequence (Fig. 1). Of 15,537 well-evidenced genes, ∼82% (12,702) contain 5′ UTRs of length ≤ 60 bases (Supplemental Fig. S8). Since the core promoter is defined as the 100-bp region flanking the start of transcription, in cases of short 5′ UTR's, these will be in (nearly) complete overlap with the downstream part of the core promoter. Previous work has also shown that in C. elegans the 5′ UTR is generally short (Blumenthal and Steward 1997; Rhoads et al. 2006). The nucleosome occupancy of the region preceding the start of translation is also heavily bound (Fig. 3), implicating the nearby location of the transcription start site (TSS) (Gu and Fire 2010). Furthermore, the distance between the TATA box motif and the start codon is generally 50 bases, consistent with a typically short 5′ UTR (Fig. 1B). Such an overlap between a short 5′ UTR and the downstream part of the core promoter imposes constraints to combine the functions of the initiation and later processing of transcription.
As the gateway to transcription, the core promoter should have the ability to evict nucleosomes and enable access to RNA polymerase II and GTFs (Fuda et al. 2009; Juven-Gershon and Kadonaga 2009). Recent work has identified two classes of promoters with respect to their nucleosome occupancy: open and covered promoters (Tirosh and Barkai 2008). Among the different attributes associated with each group is the presence or absence of a poly(dA:dT) track, a motif previously associated with areas of the lowest nucleosome occupancy in vitro (Rhodes 1979; Prunell 1982; Struhl 1985; Koch and Thiele 1999; Anderson and Widom 2001; Segal et al. 2006; Kaplan et al. 2009). Our results indicated that T-blocks are correlated with an intrinsic ability to also cause nucleosome eviction. Moreover, similar to yeast, where poly(dA:dT) tracks (often referred to as A-tracks) are associated with essential genes (Struhl 1985), in C. elegans, high numbers of T-blocks are found in core promoters of housekeeping genes. These observations suggest that T-blocks function as the C. elegans analog of the poly(dA:dT) track. However, while poly(dA:dT) tracks do not show a strand preference, we find a significant asymmetry in the frequency of T-blocks across the two strands of the core promoter. Despite this asymmetry, T-blocks located on the reverse strand are correlated with similar patterns of nucleosome eviction as those on the forward strand. Another difference between the poly(dA:dT) tracks and T-blocks are their lengths. While poly(dA:dT) tracks can span 15–30 bp (Struhl 1985), T-blocks are distributed as multiple blocks of ≥3 uninterrupted thymines. The periodicity we observe places the T-blocks on the same groove of the DNA helix, while higher numbers of blocks correlate with higher nucleosome eviction. This observation is somewhat unexpected in light of some previous works (Satchwell et al. 1986; Hsieh and Griffith 1988; Costanzo et al. 1990; Collings et al. 2010) showing the preferential binding of nucleosomes to short periodic A/T stretches that produce DNA bending. Our findings that nucleosome occupancy is anti-correlated with A/T-block-rich regions may thus be interpreted in light of reports of nucleosome absences in DNA with long stretches of A's and T's (Rhodes 1979; Prunell 1982; Struhl 1985; Koch and Thiele 1999; Segal and Widom 2009), whereby periodic stretches of A/Ts can be seen as imperfect A/T stretches. The resolution of this paradox requires additional research and may involve the distinctive effect that A/T-blocks could have on nucleosomes comprised of differentially modified histones, which may not have been queried in previous studies. Most previous work assaying DNA-nucleosome interaction invoked nucleosomes in general without distinguishing histone modifications. However, core promoters of actively expressed genes are occupied by nucleosomes that are differentially modified relative to untranscribed genes (Li et al. 2007). Thus, histone modifications might have an effect on the nucleosome binding capacity of the T-blocks in the core promoter.
The dependence upon T-blocks as opposed to A-blocks is likely due to the involvement of a uracil-rich motif for trans-splicing. Trans-splicing is important for the expression of most C. elegans genes. Previous work has shown that trans-splicing efficiency depends upon the presence of a uracil-rich (U-rich) element located upstream to the SL1/SL2 elements (Graber et al. 2007). Since the 5′ UTR overlaps with the core promoter, T-blocks apparently function as these U-rich elements. Indeed, we found a high enrichment in co-occurrence of the SL1 and T-blocks motifs (Fig. 7A,E). Moreover, an increasing number of T-blocks correlates with an increase in the precision of the SL1 element to the canonical form. Thus, the constraint of the U-rich element for the efficient trans-splicing may be the cause of asymmetry in the distribution of T- and A-blocks within C. elegans core promoters. While both T-blocks and A-blocks can cause similar eviction of the nucleosomes, only a U-rich, and not an A-rich, region is required for the trans-splicing. Further, a combination of both A-blocks and T-blocks would also lead to self-complementarity which would interfere with trans-splicing, leading to selection against A-blocks. There is also a mutational bias against uninterrupted T-stretches (Denver et al. 2004), favoring organization of thymines in blocks located on the same side of the double helix. The inter T-blocks regions might also act as sites for the location of downstream regulatory elements of the core promoter. Thus, T-blocks may serve a dual function in the context of the constraints acting on this sequence.
How might the number of T-blocks in a core promoter control the expression level of the downstream gene? The initiation of transcription may be regarded as a competition between nucleosomes that repress the gene, on the one hand, and a preinitiation complex accessing the core promoter, on the other hand. Increasing the number of T-blocks might differentially tune in favor of the preinitiation complex, leading to higher rates of transcription initiation. Our observed correlation between the gradual increase in the number of T-blocks and the levels of gene expression may thus be explained by the fact that more T-blocks would lead to higher rates of nucleosome turn-over, clearing the promoter for transcription. Transcribed T-blocks would then serve as the U-rich element, increasing efficiency of trans-splicing (Graber et al. 2007). Thus, changes to a single motif may simultaneously affect transcription, transcript stability, and translational efficiency of a particular gene's expression. As predicted, genes whose number of T-blocks is reduced by a SNP generally have lower expression levels with respect to orthologs in another C. elegans strain (Fig. 6C).
The dual function of T-blocks may provide a simple mechanism for tuning gene expression levels. Since many essential C. elegans genes are characterized by constitutive expression patterns, such tuning would alleviate the need for transcription factor regulation. Under such a model, few, if any, TFs are required, and the core promoter is the dominant locus of regulation. The proximal promoter may thus be responsible for the spatial and temporal regulation of expression, while the core promoter modulates expression levels. In particular, we propose that expression of housekeeping genes will depend mainly upon the composition of the core promoter and internal gene architecture, rather than on the elements of proximal promoters or enhancers. Consistent with this supposition, it has been recently suggested that housekeeping genes such as those encoded by operons and expressed in the germline have low-complexity, permissive promoters, thus enabling their encoding as operons (Reinke and Cutter 2009).
Our model for the tuning of expression levels using T-blocks makes several important and testable predictions. The construction of a highly expressed transgene should be enabled by a promoter containing multiple T-blocks in combination with a trans-splicing motif. Such a promoter is predicted to drive constitutive expression of a reporter gene in a tuneable fashion across various tissues of C. elegans. In contrast, genes that depend upon precise regulation, such as developmental regulators, should be depleted in T-blocks, and this notion might provide a proxy for their identification. Finally, our results predict that T-blocks and their function will generally occur in organisms with similar selective pressures for short 5′ UTRs and trans-splicing.
Methods
Definition of the core promoter and sequence motif analysis
We defined the core promoter sequence as the 95 to +5 region with respect to the start codon. Sequences were retrieved from genomic sequences using the WormBase 204 annotation release. The motif identification program MEME (Bailey and Elkan 1994) was used to identify overrepresented elements in a set of input sequences, assuming that each sequence contains at most one motif occurrence and using the standard MEME background model. The set of sequences was limited to 4031 orthologs with one copy in each of the five Caenorhabditis species (1-1-1-1-1 orthologs) and not associated with operons. The PSSMs generated by MEME for each motif were used in a MAST-search (Bailey and Gribskov 1998) of the entire set of core promoters. Parameters specified were: “-norc -comp -mt 0.5 -best -seqp -hit_list”, scoring only the forward strand, calculating the P-value based upon a random model using the letter frequencies of the query sequences, and reporting the best motif with P < 0.5. The threshold P-value for each motif was set to 0.005 after correcting for multiple testing using the FDR method (Benjamini and Hochberg 1995). The P-values and the start positions of the motifs in the core promoters were determined using MAST.
T/A-block count, distribution, and periodicity
A block of a given nucleotide was defined as at least three consecutive occurrences of that nucleotide. The number of blocks was computed as the number of nonoverlapping 10-bp windows containing the blocks. We examined all ten frames and selected the one leading to the highest number of blocks. Mean periodicity shown in Figure 2C,D is calculated as the mean of the distances between the first nucleotides of blocks in each core promoter sequence. Random sequences were generated by permutating the observed sequences. To compute block location (shown in Figure 2E,F), the fraction of genes associated with blocks for each base pair of the sequence was determined.
Statistics
Distributions were compared using the Kolmagorov-Smirnov test.
Nucleosome occupancy in the core promoters
Adjusted log2 nucleosome coverage data was obtained from Valouev et al. (2008). For each set of genes with a given number of blocks, the mean nucleosome occupancy was calculated for each base pair in the -296 to +205 region with respect to the start codon. Negative and positive nucleosome occupancy indicates depletion and enrichment, respectively, of nucleosome coverage relative to the average coverage of the genome (Valouev et al. 2008).
Gene expression level analyses
Gene expression data for four-cell stage C. elegans embryos was obtained from a previously published data set (Yanai and Hunter 2009). Genes were assigned to one of 20 equally sized bins of increasing expression levels. For the analysis shown in Figure 4, the 10 groups with the lowest expression levels were combined into a “1” group. The distribution of T/A-blocks in different gene groups was compared with the “1” group using the Kolmagorov-Smirnov test. Sequences corresponding to different regions of C. elegans genes were retrieved from BioMart (Ensembl genes 58, release WS210) (Haider et al. 2009). The core promoter sequence was defined here as the 100 bp upstream to the start of transcription. The proximal promoter corresponds to the −500 to −401 bp. The immediate 3′ UTR flank and distal flank correspond to the 1–100 bp and 401–500 bp, respectively, following the end of the 3′ UTR. The exon and intron sequences correspond to the last 100 bp of each sequence. The 3′UTR corresponds to the last 100 bp of the 3′ UTR sequence. Different expression groups may have a different number of genes in a particular gene region since sequences shorter than 100 bp are filtered out.
Differences in T-blocks number and expression levels between two C. elegans strains
Expression data from the four-cell stage embryos of N2 (Bristol) and CB4856 (Hawaiian) strains of C. elegans was obtained from a previously published data set (Yanai and Hunter 2009). The SNP data for these strains was retrieved from WormBase Release 195 and newly identified SNPs detected in this study (see below). First, we detected genes with SNPs in their core promoters (N = 2591). Next, we selected only those orthologs whose T-blocks number was affected as a result of the SNPs (N = 697). To increase the sensitivity of estimation of T-blocks number change, we counted the mean number of T-blocks for each of the ten windows. We selected only those orthologs with a ≥0.6 T-block difference between strains and with gene pairs with ≥4 T-blocks in at least one strain (N = 136). Expression of each gene pair was represented as the log2 of the ratio between the expression of N2 gene and that of its CB4856 ortholog. Thus, positive values indicate expression higher in the N2 gene, while negative values mean higher expression in the CB4856 ortholog. The same analysis was repeated for A-blocks.
Combinatorial analysis
Enrichments and depletions of genes with pairs of the MEME-defined SL1, T-blocks, and TATA box motifs within the core promoter sequences of each Caenorhabditis species were examined by hypergeometric analysis. The threshold P-value for each motif was set to 0.005 after correcting for multiple testing using FDR.
Gene ontology analysis
Genes with both T-blocks and SL1 motifs (MEME-defined; P < 0.005; FDR-corrected) were examined for enrichment in gene ontology (GO) biological process terms. Only 15 most overrepresented terms were considered. The analysis was repeated for TATA box (MEME-defined; P < 0.005; FDR-corrected).
SL1-fuzziness
Genes were assigned to one of 10 equally-sized groups depending on the P-value of the SL1 motif in their core promoter. Group 1 has the smallest P-values and can be considered to have the most precise SL1, while genes in group 10 completely lack this element. In between, genes may be considered to have different levels of “fuzziness” of the SL1.
Expression pattern analysis
Developmental expression data from four-cell, 28-cell, 55-cell, 95-cell, and 190-cell stage embryos of C. elegans was obtained from the previously defined data set (Yanai and Hunter 2009). For each stage, all genes were assigned into one of two groups according to the level of their expression: highly expressed genes, and genes with middle and low expression levels according to a 3.75 threshold. Next, we defined three alternative categories of genes. Highly expressed genes contain a high level of expression across all five developmental stages. Maternal genes have high expression only at the four-cell stage. Finally, embryonically expressed genes have high expression in any stage other than the four-cell stage.
Operons
Operon genes were defined according to WormBase. Strict nonoperon genes were defined as genes not defined as operons in WormBase and, additionally, also separated from the neighboring gene by at least 1 kb.
Genomic sequencing
Strain CB4856 was obtained from the Caenorhabditis Genetics Center, and maintained at 20°C using standard culture methods (Brenner 1974). Genomic DNA was extracted by proteinase K digestion followed by two rounds of phenol-chloroform extraction, with an intermediate step of RNase A digestion in TE. Genomic DNA libraries were built using Illumina's standard paired-end protocol and 36 × 2 bp were sequenced on the Illumina Genome Analyzer IIx following the manufacturer's recommendations. The number of detected paired-end reads amounted to 25,443,382 with an 8.2× coverage over the sequenced N2 genome. SNPs were detected by requiring at least five instances of the sequenced variant with no heterogeneity. 78,126 SNPs were detected (Supplemental Table S5).
Acknowledgments
We thank Asher Cutter of the University of Toronto and Michal Levin, David Silver, Benjamin Podbilewicz, and Yael Mandel-Gutfreund of the Technion–Israel Institute of Technology for critical readings of and suggestions on this manuscript. This work was supported by Israel Science Foundation grant 1500/09 and the Lorry I. Lokey Interdisciplinary Center for Life Science and Engineering.
Footnotes
[Supplemental material is available for this article. The sequence data from this study have been submitted to the SRA database (http://www.ncbi.nlm.nih.gov/sra) under accession no. SRA028423.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.113381.110.
References
- Anderson JD, Widom J 2001. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol Cell Biol 21: 3830–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bailey TL, Gribskov M 1998. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics 14: 48–54 [DOI] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Bilu Y, Barkai N 2005. The design of transcription-factor binding sites is affected by combinatorial regulation. Genome Biol 6: R103 doi: 10.1186/gb-2005-6-12-r103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ 2006. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell 24: 853–865 [DOI] [PubMed] [Google Scholar]
- Blumenthal T. 2005. Trans-splicing and operons. In WormBook (ed. The C. elegans Research Community), pp. 1–9. doi: 10.1895/wormbook.1.11.1, http://www.wormbook.org. [DOI] [PubMed]
- Blumenthal T, Gleason KS 2003. Caenorhabditis elegans operons: Form and function. Nat Rev Genet 4: 110–118 [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Steward K 1997. RNA processing and gene structure. In C. Elegans II (ed. Riddle D.L. et al. ), pp. 117–145 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT 1996. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev 10: 711–724 [DOI] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT 2001. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev 15: 2515–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198 [DOI] [PubMed] [Google Scholar]
- Carroll SB 2008. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- Cavener DR 1987. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res 15: 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkley GE, Verrijzer CP 1999. DNA binding site selection by RNA polymerase II TAFs: A TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J 18: 4835–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohanim AB, Kashi Y, Trifonov EN 2006. Three sequence rules for chromatin. J Biomol Struct Dyn 23: 559–566 [DOI] [PubMed] [Google Scholar]
- Collings CK, Fernandez AG, Pitschka CG, Hawkins TB, Anderson JN 2010. Oligonucleotide sequence motifs as nucleosome positioning signals. PLoS ONE 5: e10933 doi: 10.1371/journal.pone.0010933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Thomas J, Spieth J, Blumenthal T 1991. Insertion of part of an intron into the 5′ untranslated region of a Caenorhabditis elegans gene converts it into a trans-spliced gene. Mol Cell Biol 11: 1921–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo G, Di Mauro E, Salina G, Negri R 1990. Attraction, phasing and neighbour effects of histone octamers on curved DNA. J Mol Biol 216: 363–374 [DOI] [PubMed] [Google Scholar]
- Davidson EH 2006. The regulatory genome: Gene regulatory networks in development and evolution. Academic Press, Burlington, MA [Google Scholar]
- Denver DR, Morris K, Kewalramani A, Harris KE, Chow A, Estes S, Lynch M, Thomas WK 2004. Abundance, distribution, and mutation rates of homopolymeric nucleotide runs in the genome of Caenorhabditis elegans. J Mol Evol 58: 584–595 [DOI] [PubMed] [Google Scholar]
- Fire A, Alcazar R, Tan F 2006. Unusual DNA structures associated with germline genetic activity in Caenorhabditis elegans. Genetics 173: 1259–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461: 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A, Ikemura T, Kinouchi M, Oshima T, Kudo Y, Mori H, Kanaya S 2002. Periodicity in prokaryotic and eukaryotic genomes identified by power spectrum analysis. Gene 300: 203–211 [DOI] [PubMed] [Google Scholar]
- Goldberg ML 1979. “Sequence analysis of Drosophila histone genes.” PhD thesis, Stanford University, Stanford, CA [Google Scholar]
- Graber JH, Salisbury J, Hutchins LN, Blumenthal T 2007. C. elegans sequences that control trans-splicing and operon pre-mRNA processing. RNA 13: 1409–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SG, Fire A 2010. Partitioning the C. elegans genome by nucleosome modification, occupancy, and positioning. Chromosoma 119: 73–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S, Ballester B, Smedley D, Zhang J, Rice P, Kasprzyk A 2009. BioMart Central Portal–unified access to biological data. Nucleic Acids Res 37 (Suppl 2): W23–W27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T, Dynlacht BD, Peterson MG, Pugh BF, Tjian R 1990. Isolation and characterization of the Drosophila gene encoding the TATA box binding protein, TFIID. Cell 61: 1179–1186 [DOI] [PubMed] [Google Scholar]
- Hsieh CH, Griffith JD 1988. The terminus of SV40 DNA replication and transcription contains a sharp sequence-directed curve. Cell 52: 535–544 [DOI] [PubMed] [Google Scholar]
- Huber R, Schlessinger D, Pilia G 1998. Multiple Sp1 sites efficiently drive transcription of the TATA-less promoter of the human glypican 3 (GPC3) gene. Gene 214: 35–44 [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell 13: 573–585 [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga JT 2009. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Cheng S, Kadonaga JT 2006. Rational design of a super core promoter that enhances gene expression. Nat Methods 3: 917–922 [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT 2008. The RNA polymerase II core promoter—the gateway to transcription. Curr Opin Cell Biol 20: 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KA, Thiele DJ 1999. Functional analysis of a homopolymeric (dA-dT) element that provides nucleosomal access to yeast and mammalian transcription factors. J Biol Chem 274: 23752–23760 [DOI] [PubMed] [Google Scholar]
- Kumar L, Futschik M, Herzel H 2006. DNA motifs and sequence periodicities. In Silico Biol 6: 71–78 [PubMed] [Google Scholar]
- Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH 1998. New core promoter element in RNA polymerase II-dependent transcription: Sequence-specific DNA binding by transcription factor IIB. Genes Dev 12: 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. 2010. The landscape of C. elegans 3′ UTRs. Science 329: 432–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles-Filho AC, Stark A 2009. Comparative genomics of gene regulation-conservation and divergence of cis-regulatory information. Curr Opin Genet Dev 19: 565–570 [DOI] [PubMed] [Google Scholar]
- Moreno-Herrero F, Seidel R, Johnson SM, Fire A, Dekker NH 2006. Structural analysis of hyperperiodic DNA from Caenorhabditis elegans. Nucleic Acids Res 34: 3057–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Liao GC, Niemann H, Rubin GM 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol 3: research0087.1–0087.12 doi: 10.1186/gb-2002-3-12-research0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A 1982. Nucleosome reconstitution on plasmid-inserted poly(dA). poly(dT). EMBO J 1: 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Cutter AD 2009. Germline expression influences operon organization in the Caenorhabditis elegans genome. Genetics 181: 1219–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads RE, Dinkova TD, Korneeva NL 2006. Mechanism and regulation of translation in C. elegans. In WormBook (ed. The C. elegans Research Community), pp. 1–18 doi: 10.1895/wormbook.1.11.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- Rhodes D 1979. Nucleosome cores reconstituted from poly (dA-dT) and the octamer of histones. Nucleic Acids Res 6: 1805–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell SC, Drew HR, Travers AA 1986. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol 191: 659–675 [DOI] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J 2009. Poly(dA:dT) tracts: Major determinants of nucleosome organization. Curr Opin Struct Biol 19: 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J 2006. A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST, Baltimore D 1989. The “initiator” as a transcription control element. Cell 57: 103–113 [DOI] [PubMed] [Google Scholar]
- Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al. 2007. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K 1985. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci 82: 8419–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N 2008. Two strategies for gene regulation by promoter nucleosomes. Genome Res 18: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N, Verstrepen KJ 2009. Promoter architecture and the evolvability of gene expression. J Biol 8: 95 doi: 10.1186/jbiol204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, et al. 2008. A high-resolution nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res 18: 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanWye JD, Bronson EC, Anderson JN 1991. Species-specific patterns of DNA bending and sequence. Nucleic Acids Res 19: 5253–5261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet 28: 160–164 [DOI] [PubMed] [Google Scholar]
- Widom J 1996. Short-range order in two eukaryotic genomes: Relation to chromosome structure. J Mol Biol 259: 579–588 [DOI] [PubMed] [Google Scholar]
- Xi H, Yu Y, Fu Y, Foley J, Halees A, Weng Z 2007. Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1. Genome Res 17: 798–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Hunter CP 2009. Comparison of diverse developmental transcriptomes reveals that coexpression of gene neighbors is not evolutionarily conserved. Genome Res 19: 2214–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]