Abstract
The structural complexity of nucleosomes underlies their functional versatility. Here we report a new type of complexity—nucleosome fragility, manifested as high sensitivity to micrococcal nuclease, in contrast to the common presumption that nucleosomes are similar in resistance to MNase digestion. Using differential MNase digestion of chromatin and high-throughput sequencing, we have identified a special group of nucleosomes termed “fragile nucleosomes” throughout the yeast genome, nearly 1000 of which were at previously determined “nucleosome-free” loci. Nucleosome fragility is broadly implicated in multiple chromatin processes, including transcription, translocation, and replication, in correspondence to specific physiological states of cells. In the environmental-stress-response genes, the presence of fragile nucleosomes prior to the occurrence of environmental changes suggests that nucleosome fragility poises genes for swift up-regulation in response to the environmental changes. We propose that nucleosome fragility underscores distinct functional statuses of the chromatin and provides a new dimension for portraying the landscape of genome organization.
Eukaryotic chromosomal DNA is organized into an array of nucleosomes (Luger et al. 1997), providing the molecular environment for a variety of biological processes (Workman and Kingston 1998). It is well established that the structural complexity of nucleosomes—the incorporation of distinct types of histone isoforms and the post-translational modifications of histones—profoundly modulates the functional states of the chromatin. Thus, extensive knowledge of genome organization at the nucleosome level is crucial for elucidating the molecular mechanisms of all chromatin-related processes. Using the high-throughput tiling microarray and the next-generation massively parallel sequencing technologies, genome-wide nucleosome positions were determined, generating comprehensive nucleosome maps in multiple model organisms (Yuan et al. 2005; Albert et al. 2007; Lee et al. 2007; Mavrich et al. 2008a,b). These studies have led to important insights into chromatin organization and its functional implications.
A hallmark property of nucleosomes is that a 147-bp core DNA segment wraps tightly around the histone octamer core and, therefore, is sterically occluded (Luger et al. 1997) and protected against cleavage by nucleases (e.g., micrococcal nuclease [MNase]); whereas the linker DNA connecting adjacent nucleosomes is highly susceptible to MNase cleavage. As a result, MNase digestion reduces the chromatin length incrementally by an integral number of nucleosomes, eventually resulting in mononucleosomes that are relatively stable. This property has long been recognized (Noll and Kornberg 1977) and used broadly in studies of chromatin and nucleosomes, including the whole-genome nucleosome mapping studies (Yuan et al. 2005; Albert et al. 2007; Lee et al. 2007; Mavrich et al. 2008a,b).
Despite the common presumption that nucleosomes are similar in their resistance to MNase digestion, it is possible that the level of resistance may vary quantitatively among individual nucleosomes in a genome due to their specific histone composition or conformational states. In vertebrates, nucleosomes simultaneously containing the histone isoforms H2A.Z and H3.3 are unstable in solution after release from chromatin and appear sensitive to MNase (Jin and Felsenfeld 2007; Jin et al. 2009). Alternatively, nucleosomes may undergo rapid spontaneous conformational changes, with a portion of the core DNA transiently unwrapped from the histone octamer (Li et al. 2005). If such a transient open conformation is stabilized (e.g., via interaction with specific proteins), it may render a nucleosome less resistant to MNase. Indeed, a recent study has illustrated the existence of such nucleosomes that are less resistant to MNase treatment, in particular, at the promoter regions in the yeast genome (Weiner et al. 2010). Here, we report a systematic analysis of such variations in the resistance to MNase throughout the budding yeast genome and their broad implications in chromatin-related processes. In the group of environmental stress-response (ESR) genes (Gasch et al. 2000), high sensitivity of nucleosomes to MNase (referred to as nucleosome fragility) precedes the impending environmental changes. It may poise the genes for prompt up-regulation.
Results
Fragile nucleosomes are localized throughout the yeast genome in a locus- and physiological state-specific manner
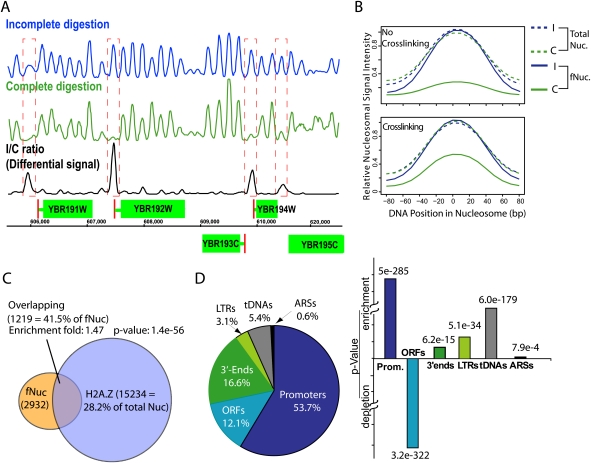
We sought to systematically explore the possible variations in resistance to MNase among all nucleosomes in the genome of the budding yeast Saccharomyces cerevisiae. We reasoned that, experimentally, the nucleosomes providing limited protection to the core DNA may initially be indistinguishable from the more stable ones during MNase digestion, but would become distinctively diminished or eliminated following prolonged MNase treatment. We define nucleosomes that are less resistant to MNase digestion as “fragile nucleosomes.” Accordingly, the following procedure was implemented to identify the fragile nucleosomes. Yeast chromatin was subjected to MNase digestion without formaldehyde cross-linking. Mononucleosomal DNA was recovered at two time points during the digestion: an Incomplete Digestion (I) sample at an early time point with only a minor portion of the chromatin (∼10%) reduced to mononucleosomes, and a Complete Digestion (C) sample recovered at a later time point with nearly all the chromatin reduced to mononucleosomes (Supplemental Fig. S1A; Supplemental material, Section 1). The two mononucleosomal DNA samples were sequenced using the Illumina Genome Analyzer II platform to deduce the positions and the relative abundance of all the nucleosomes, except for those occupying duplicated sequence DNA. We generated two nucleosome maps: I and C, for incomplete and complete digestion, respectively (Supplemental material, Section 2). Fragile nucleosomes were defined by their higher abundance in the I map relative to that in the C map (Fig. 1A).
Figure 1.
Fragile nucleosomes are localized to the functional regions of the chromatin. (A) Nucleosome occupancy of a region in chromosome II. Plots of MNase-protected DNA sequences illustrate the relative abundance (peak heights) of nucleosomes at particular positions (DNA coordinates, x- axis). (Green boxes) ORFs; (red lines) TSSs. Incomplete/complete differential nucleosome occupancies (fragile nucleosome occupancies; black) are peaks that are missing or significantly reduced in the complete digestion map (green) compared with the incomplete digestion map (blue). (B) Nucleosome fragility is partially suppressed by chemical cross-linking. Panels show the average nucleosomal signal intensity of all nucleosomes (Total Nuc) or the identified 3341 fragile nucleosomes with or without chromatin cross-linking of the chromatin prior to MNase digestion in the incomplete digestion (I, blue) and the complete digestion (C, green) maps, respectively. (C) Partial overlapping between fragile nucleosomes and H2A.Z-nucleosomes (Albert et al. 2007). Enrichment of fragile nucleosomes in H2A.Z nucleosomes is as labeled. (D) Fragile nucleosomes are enriched in chromatin functional regions. (Left) Distributions of fragile nucleosomes in the functional regions. (Right) P-values of fragile nucleosome enrichment or depletion at the labeled functional regions.
To test whether nucleosome fragility reflects distinct functional states at specific loci in the genome, we characterized cells under two physiological conditions: optimal growth and heat treatment, each with two biological replicates. Nucleosome profiles were remarkably different between physiological conditions but were very similar between biological replicates (Supplemental Fig. S1B). Globally, these nucleosome maps are highly consistent with the published maps (Supplemental Fig. S2). Using the nucleosome abundance ratio (I/C), we quantified the nucleosome fragility throughout the genome. Fragile nucleosomes represent a significant portion of the total nucleosomes, ∼5.4% of total (3341 of 63,018) in the optimally growing cells and 4.5% in heat-treated cells (Supplemental material, Section 2). We conclude that the fragile nucleosome distribution in the genome is not random but, rather, is affected by the physiological conditions in a specific and reproducible manner.
Nucleosome fragility correlates with multiple biochemical traits
To determine whether fragile nucleosomes are genuine nucleosomes or other types of DNA/protein complexes, we performed a histone H3-specific affinity purification on chromatin after an incomplete digestion, followed by purification and high-throughput sequencing of the mononucleosomal DNA (Supplemental material, Section 1). More than 93% of the fragile nucleosomes, in comparison to 98% of the total nucleosomes, were recovered by H3-specific affinity purification, demonstrating that the majority of the fragile nucleosomes are histone-based DNA/protein particles.
We then tested whether the histone core instability in solution could contribute to the observed nucleosome fragility. In vertebrates, nucleosome particles containing both the H2A.Z and H3.3 histone isoforms are highly unstable in solution due to histone octamer core dissociation and, as a result, exhibit high sensitivity to MNase. Chemical cross-linking suppresses the high sensitivity to MNase in the vertebrate H2A.Z/H3.3 nucleosomes efficiently (Jin and Felsenfeld 2007; Jin et al. 2009). We therefore tested the effect of covalent cross-linking on nucleosome fragility observed in yeast (Supplemental material, Section 1). Fragile nucleosomes identified under the non-cross-linking condition by definition had a high I/C ratio (5.0 on average) (Fig. 1B). Cross-linking with formaldehyde prior to MNase digestion, however, led to a significant reduction in the average I/C ratio to 2.2 in the same group of nucleosomes, although the ratio was still higher than 1—the average I/C ratio of total nucleosomes with or without cross-linking. Examination of individual fragile nucleosomes revealed that while many of the fragile nucleosomes no longer exhibited significant fragility after cross-linking, others retained high fragility (I/C > 3) and appeared insensitive to the effects of covalent cross-linking (Supplemental Fig. S3). These results suggest that instability in histone/DNA interactions or histone cores, similar to that seen in vertebrate H2A.Z/H3.3-containing nucleosomes, partially contributes to nucleosome fragility. Importantly, budding yeast has only one type of histone H3, lacking the diversity in histone H3 isoforms seen in vertebrates. On the other hand, Yeast H2A.Z does display a significant correlation with fragile nucleosomes (Fig. 1C). However, this correlation is not sufficient to account for the fragility of the majority of fragile nucleosomes (Fig. 1C; Supplemental Fig. S4). Therefore, biochemical mechanisms other than H3.3/H2A.Z combinations may underlie the histone core instability or weak histone/DNA interaction in yeast.
To assess the possible contribution of DNA sequence to nucleosome fragility, we compared the fragile and nonfragile nucleosomes in terms of their agreement with the in vitro–assembled nucleosome positions throughout the yeast genome, which is determined by the underlying DNA sequence in the absence of trans factors (Kaplan et al. 2009). We found that fragile nucleosomes matched less well to the in vitro–assembled nucleosome positions in comparison to canonical nucleosomes, suggesting that the specific DNA sequences that are less favorable for nucleosome assembly may contribute to nucleosome fragility and furthermore, in vivo, trans factors may facilitate the positioning of the fragile nucleosomes (Supplemental Fig. S5A).
Fragile nucleosomes are implicated in a multitude of chromatin-related processes
We observed that fragile nucleosomes are highly enriched in specific functional regions on chromatin including the promoters and the 3′ ends (but not the coding sequences) of specific RNA Pol II genes, tRNA genes, long terminal repeats (LTRs), and replication origins (ARSs) (Fig. 1D; Supplemental Table S1). This finding suggests that fragile nucleosomes are involved in a variety of chromatin-related processes, such as RNA Pol II transcription, RNA Pol III transcription (Supplemental Fig. S5B,C; Supplemental material, Section 3), DNA transposition, and replication.
We further characterized the fragile nucleosomes in the promoters of the RNA Pol II genes in detail. Chromatin organization at the promoter is important for transcription regulation, via modulating the accessibility to the cis-elements in the promoter (Field et al. 2008; Tirosh and Barkai 2008; Cairns 2009). Specifically, one analysis of nucleosome positioning genome-wide in yeast has led to the finding of two typical promoter structures, the OPN (occupied proximal nucleosome) and the DPN (depleted proximal nucleosome) (Tirosh and Barkai 2008). The OPN type, characterized by high nucleosome occupancy at the region proximal (<100 bp) to the transcription start site (TSS), is associated with high transcription plasticity, sensitive to chromatin remodeling and high noise in gene expression. Conversely, the DPN type, characterized by nucleosome depletion at the TSS proximal region and high occupancy at the distal region (hence exhibiting a prominent “nucleosome-free region”—NFR—immediate upstream to TSS), is associated with the opposite features in transcription regulation. We found that among 1283 promoters (TSS-400 ∼ TSS + 0) that carry a fragile nucleosome, the OPN-type (494 in total) was significantly enriched (180; null = 110; enrichment P-value: 3.7 × 10−14). In contrast, the DPN-type (544 in total) was noticeably depleted (103; null = 121; depletion P-value: 0.0267). These results are consistent with the notion that nucleosome fragility may manifest distinct biochemical mechanisms underlying the dynamicity of nucleosome positioning implicated in modulating the accessibility of the cis-elements in the OPN-type promoters, which ultimately contributes to the high variability in gene expression (Tirosh and Barkai 2008).
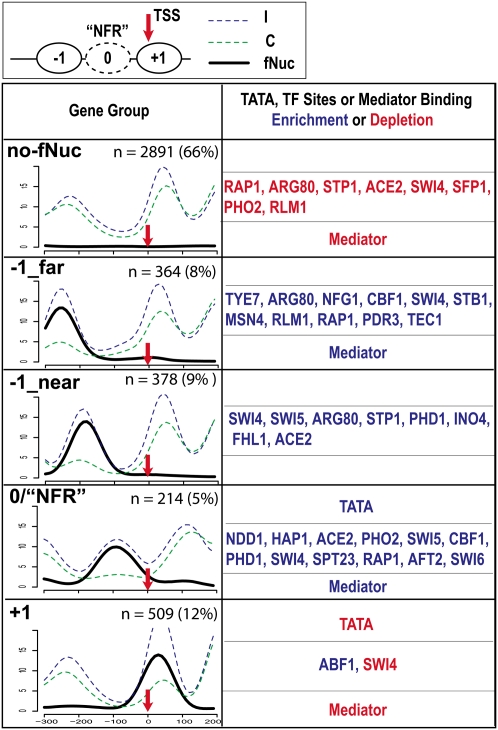
Within the promoter, the nucleosome positioning relative to TSS varies in individual genes. To investigate this feature systematically, we binned all the 4556 RNA Pol II genes with an experimentally defined transcription start site (TSS) (Nagalakshmi et al. 2008) into five groups based on the properties of the nucleosomes in their promoters (Fig. 2; Supplemental material, Section 4). One group (labeled as no-fNuc–no fragile nucleosome), comprising 66% of the genes, contained no fragile nucleosomes and displayed the canonical chromatin architectural pattern at the promoter as previously reported (Yuan et al. 2005; Albert et al. 2007; Lee et al. 2007). Each of the other groups depicted a novel type of chromatin organization at the promoter, with a fragile nucleosome localized at a specific position relative to the TSS, as indicated by the labels, respectively (Fig. 2). This observation suggests the involvement of fragile nucleosomes in the regulation of transcription initiation and the assembly of the transcription machinery on chromatin. Furthermore, each group displays specific correlations—enrichment as well as depletion—in various biological traits (Supplemental material, Section 4), including the presence of a TATA-box or the consensus sites of a multitude of transcription factors and the binding of the mediator, a general transcription coactivator mediating the interaction between RNA Pol II and the gene-specific regulatory factors (Fig. 2; Bjorklund and Gustafsson 2005; Casamassimi and Napoli 2007). No significant difference in the average transcription rate or the abundance of transcripts was observed among the groups of genes. These results suggest that nucleosome fragility is involved in transcription initiation in about one-third of the genes under the optimal growth conditions. Notably, similar to the finding of a recent study (Weiner et al. 2010), the “0/NFR” Group genes (Fig. 2), more than 200 in total, contain a fragile nucleosome at a crucial site, previously characterized as NFR (nucleosome-free-region), immediate upstream of the TSS (Supplemental Fig. S6; Supplemental material, Section 5; Lee et al. 2007; Mavrich et al. 2008a). Group “0/NFR” genes therefore may adopt a distinct mode of regulating transcription initiation: The cis-elements upstream of the TSS may not be readily accessible but, rather, require chromatin remodeling for accession that involves nucleosome fragility.
Figure 2.
Classification of RNAP II genes into five groups by K-means clustering based on fragile nucleosome occupancy in promoters. (Upper) A diagram illustrating the canonical nucleosomal organization at the promoter. TSS (red arrow): transcription start site; ovals labeled with +1 and −1 are nucleosomes on or upstream to TSS. Oval in dashed line (0) is the fragile nucleosome at the NFR. The “Gene Group” column depicts the overlay of nucleosome distribution of all the genes in each group. Group names, based on the fragile nucleosome position, are in bold at upper left corners. (x-axis) The nucleotide positions, with TSS = 0. (y-axis) The normalized number of sequencing counts. (Blue) Incomplete digestion; (green) complete digestion; (black) differential signals representing fragile nucleosome; (n) numbers of genes in each group; the percentage indicates the portion of total characterized genes. Enrichment and depletion of TATA, TF sites, and mediator binding are marked by blue and red, respectively. The P-value cutoff for TF binding site enrichment is 1.0 × 10−10.
Nucleosome fragility may poise stress-response genes for prompt activation
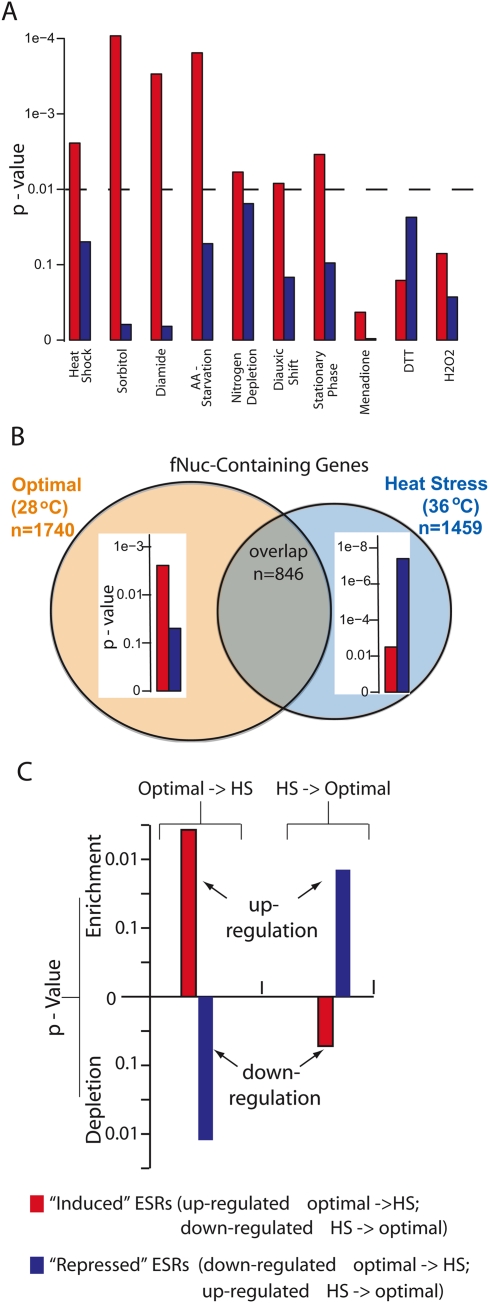
We further explored the functional implications of nucleosome fragility in transcription regulation. We examined whether genes containing fragile nucleosomes in their promoters are enriched for any gene sets that carry out a specific physiological function (GO terms) or exhibit distinct features in transcription activity and/or regulation. To this end, we found that the fragile nucleosome-containing genes in optimally growing cells are enriched in a subset of the Environmental-Stress-Response (ESR) genes that are induced, but not the subset that are repressed, under various environmental stresses (Fig. 3A; Gasch et al. 2000). Transcription of these genes is up-regulated promptly, some as quickly as a few minutes following stress induction. These genes are involved in various metabolic and cellular processes, contributing to cell survival under adverse conditions (Gasch et al. 2000; Causton et al. 2001). Consistently, binding sites of Msn4, a transcription factor that is a key regulator of ESR, are also enriched in the fragile nucleosome-containing promoters (Fig. 2). We propose that the presence of fragile nucleosomes is characteristic of genes that need to be rapidly activated in response to stimuli.
Figure 3.
Fragile nucleosomes in the promoters of the ESR gene correlate with transcription up-regulation and are inclined to be evicted upon environment changes. (A) Bar graphs depict the P-values of the enrichment of subsets of the “induced” ESR genes that are up-regulated (red) or the “repressed” ESR genes that are down-regulated (blue) in response to specific environmental stresses in fragile nucleosome-containing genes. (Dashed line) The P-value cutoff at 0.01. (B) The spectra of fragile nucleosome-containing genes identified under optimal growth (28°C; light brown) or heat stress (36°C; light blue) are significantly different. Inserted bar diagrams illustrate the P-values of enrichment of the “induced” or the “repressed” subgroups of ESR genes (red and blue) in corresponding fragile nucleosome-gene groups. Identifications of ESR subgroups and their transcription changes with temperature shifts are obtained from Gasch et al. (2000). (C) Enrichment of fragile nucleosomes that are converted to nucleosome-free regions in combinations of ESR subgroup plus environment change that lead to transcription up-regulation. The y-axis represents the log-scale P-values calculated by comparing the number of fNuc to NFR conversions in the “induced” or “repressed” ESR genes with that in all genes. Positive and negative P-values indicate enrichment and depletion, respectively.
To further test this hypothesis, we identified the fragile nucleosome-containing genes in cells under heat stress. The spectrum of fragile nucleosome-containing genes in heat-treated cells was significantly different from that in the optimally growing cells (Fig. 3B). Moreover, reversing the environmental change was shown to have the opposite effects on the expression of distinct subgroups of ESR genes: The “induced ESR” genes are up-regulated when the stresses are applied and down-regulated when returned to optimal growth; whereas the “repressed ESR” genes are down-regulated under stress and up-regulated when the stress is relieved (Gasch et al. 2000). Remarkably, we observed that nucleosome fragility is consistently associated with the combinations of gene subgroup and environmental change that lead to transcription activation: Fragile nucleosome-containing genes in optimal growth conditions (brown circle) were enriched for the “induced ESR” genes that are up-regulated in response to heat stress (red column): and fragile nucleosome-containing genes found in heat-stressed cells (light blue circle) were enriched for the “repressed ESR” genes (dark blue column) that are up-regulated when shifted back to the optimal temperature (Fig. 3B). Thus, genes with high nucleosome fragility in their promoters are consistently susceptible for up-regulation in response to environmental changes, regardless of the directions of the changes. Importantly, the presence of fragile nucleosomes in the promoters precedes the induction of environmental changes, suggesting that yeast cells in anticipation of the commonly encountered changes poise the ESR genes for rapid up-regulation, using nucleosome fragility as part of the mechanism.
We further sought to investigate the fate of the fragile nucleosome in the promoter following transcription up-regulation. We reasoned that the fragile nucleosome may either be evicted, converting the corresponding site into NFR; or, it may be stabilized and become a canonical nucleosome; and lastly, it may retain the high fragility. We assessed all three possibilities between optimal growth and heat-stress conditions for each fragile nucleosome within the promoters of ESR genes. Although all three possible fates were observed, we found that the fNuc → NFR conversion was consistently enriched in the combinations of gene subgroup and environmental change that lead to transcription activation: The “induced” ESRs switched from optimal growth to heat stress; and the “repressed” ESRs switched from heat stress to optimal growth (Fig. 3C). This result indicates that nucleosome fragility may represent a mode of chromatin remodeling that primes the nucleosome to be evicted upon stimulation, leading to an “open” promoter for active transcription.
Furthermore, previous studies on chromatin remodeling have shown that heat shock factor 1 (Hsf1), a master regulator of transcription in response to heat stress, preloads onto some of the target promoters prior to the stress and that it may further recruit chromatin remodeling factor, SWI/SNF, leading to nucleosome eviction in a quick response to heat stress (Hahn et al. 2004; Shivaswamy and Iyer 2008). Interestingly, we have observed a noticeable overlap between fNuc-containing promoters (Total: 1740) and Hsf1 targets (Total: 167; overlap: 67; null: 50; enrichment P-value: 0.00243).
Discussion
Taking the approach of differential MNase digestion of the chromatin combined with deep sequencing, we have identified nucleosomes that are distinctively sensitive to MNase throughout the yeast genome, at the resolution of a single nucleosome. Compared to a published complete nucleosome map (Lee et al. 2007), close to 1/3 (961 of 3341 in total) of these fragile nucleosomes identified in the optimally growing cells are at the sites previously characterized as “nucleosome-free.” Consistently, a recent nucleosome mapping study, using differential MNase digestions of yeast chromatin, has identified nucleosomes highly sensitive to MNase treatment, enriched at the promoter and 3′ ends of the genes (Supplemental Fig. S7; Weiner et al. 2010). Our study further provides biochemical evidence that the fragile nucleosomes are genuine nucleosome particles, instead of other protein–DNA complexes that happen to encompass a similar length of DNA. Our work expands the observation of nucleosome fragility by refined categorization of genes based on fNuc positioning in the promoter (Fig. 2), as well as identification of fNucs on a multitude of other functional regions (see below). Thus, our comprehensive annotation of the fragile nucleosomes fills in the gaps in the existing nucleosome maps, many at regions with demonstrated functional importance (e.g., the “0/NFR” gene group) (Fig. 2). Interestingly, we also detected nucleosomes exhibiting unusually high “resistance” to MNase, namely, the abundance of these nucleosome in the C map was significantly higher than in the I map (Supplemental material, Section 7), presumably due to reduced accessibility to these regions. Whether these “resistant” nucleosomes bear any physiological significance awaits future investigation.
We have demonstrated that fragile nucleosomes, depending on their specific locations in the genome, correlate with multiple biochemical traits, indicating that nucleosome fragility may be caused independently by a multitude of (rather than a single) biochemical properties of the nucleosomes. Our results suggest that in some of the nucleosomes, the fragility may be due to the instability of histone core particles that can be rescued by chemical cross-linking prior to MNase treatment, reminiscent of (but not identical in biochemical nature to) the H2.Z/H3.3-containing nucleosomes in vertebrate cells (Jin and Felsenfeld 2007; Jin et al. 2009). This result is also concordant with a recent finding of a group of unstable nucleosomes defined by differential intensity signals in the cross-linked versus the non-cross-linked nucleosome maps, which correlates with transcript factor binding sites (Goh et al. 2010). In other cases in which the fragility is insensitive to cross-linking, we speculate that the particular nucleosome conformation allows MNase to access the particular nucleosomes more easily. Such high accessibility may be caused by DNA rigidity, or, by certain yet-to-be determined biochemical traits such as specific histone modifications or the binding of distinct nonhistone proteins that may stabilize an open conformation of the nucleosome (Li et al. 2005). Together, these results suggest the potential utility of nucleosome fragility as a novel index for the structural complexity and the functional status of chromatin.
It is noteworthy that because chromatin was commonly digested extensively in previous studies, fragile nucleosomes tend to be under-represented or even missing in the previous characterizations of various chromatin properties (e.g., the incorporation of histone isoforms and various types of histone modifications, or, chromatin dynamics at single-nucleosome resolution). This may diminish the detection of possible correlations between nucleosome fragility and other chromatin features at single-nucleosome resolution. Therefore, although we succeeded in detecting some of the correlations (e.g., H2A.Z nucleosomes and OPN-type chromatin organization in the promoters), we consider the apparent lack of correlation with a specific chromatin property not reliable at this stage. More accurate assessment on the relative contribution of distinct biochemical properties to the nucleosome fragility would need refined mapping of chromatin features using MNase digestion conditions that preserve fragile nucleosomes. Such concern, however, does not apply to the genome-wide data sets without using MNase digestion (such as the ESR genes).
The fact that the fragile nucleosomes are highly enriched at various functional regions in the genome including the promoters of protein-coding genes, the tRNA genes, and replication origins, as well as LTRs, strongly suggests that nucleosome fragility is broadly implicated in many important chromatin-related processes. Together with the notion that a large number of fragile nucleosomes are at the loci previously thought to be “nucleosome-free,” our finding should broadly impact the studies of chromatin by revealing a new level of complexity in nucleosome organization and help amend the inaccuracy in delineating the molecular mechanisms of multiple biological processes.
The profound implication of nucleosome fragility in transcription regulation is attested by the presence of fragile nucleosomes in the promoters of nearly one-third of total protein-coding genes. On the other hand, nucleosome fragility is not limited to its correlation with any single group of genes categorized based on either their transcription profile or their cellular functions, underscoring the complexity of this phenomenon overall. Here, we detect the significant association between nucleosome fragility in the promoter of ESR genes and the up-regulation of these genes upon change in growth conditions (optimal growth or heat stress). Hopefully, with future refined mapping of chromatin features under the experimental conditions that preserve fragile nucleosomes, more gene sets using nucleosome fragility for their corresponding biological processes would be discerned. In the case of ESR genes, we presented further evidence that fragile nucleosomes in the promoter tend to be evicted upon gene activation, suggesting that a possible mechanism in which fragile nucleosomes may be in the state primed to be evicted so as to provide prompt access to the underlying cis-elements for gene activation. The observed enrichment of HSF1 target genes among the fNuc-containing population may indicate that HSF1-dependent chromatin remodeling may be one of the possible mechanisms.
In summary, our findings demonstrate that variations in resistance to MNase broadly exist in nucleosomes throughout the yeast genome, revealing a new facet of nucleosome complexity. Delineating the connections between nucleosome fragility and the implicated processes should enrich our understanding of how chromatin organization is modulated to suit the needs of a variety of biological activities, such as coping with frequent environmental fluctuations.
Methods
Standard procedures were used for molecular biology experiments (yeast nuclei preparation, chemical cross-linking, and anti-H3 affinity purification).
Differential MNase digestion was conducted at 36°C, with 100 units/mL MNase. For the incomplete digestion sample, mononucleosomal DNA was recovered from the time point at which mononucleosomal DNA accounts for ∼10% of the total DNA (estimated from band intensity of agarose electrophoresis), whereas at the complete digestion time point, mononucleosomal DNA was the only discrete band. Purified DNA was subjected to Illumina massively parallel sequencing following the procedures by the manufacturer. A detailed description of these procedures is given in the Supplemental material, Section 1.
Established procedures were used to process the sequencing data and map the nucleosomes to the S. cerevisiae reference genome (UCSC Oct 2003) by ELAND, and the raw profiles were denoised using wavelength smoothing.
To call the fragile nucleosomes, sequencing results of two biological replicates were first merged to compensate for the possible experimental variations between the biological replicates. Using the criteria of I/C ratio > 3.0, p < 1.0 × 10−7, 5.4% of total nucleosomes (3341 of 62,018) were identified as fNucs in the optimal growing cells, whereas 4.5% were found in heat-treated cells.
Established statistical tests were applied to determine the correlations between nucleosome fragility to genome-wide traits at single-nucleosome level, or, the correlations between genes containing a fragile nucleosome in the promoters and the gene sets displaying various biological characteristics.
Detailed descriptions of these procedures are given in Supplemental material, Section 2: “Sequencing data processing and fragile nucleosomes calling”; Supplemental material, Section 3: “Correlations between nucleosome fragility and other genome-wide traits”; Supplemental material, Section 4: “K-mean clustering with respect to transcription start sites (TSS)”; Supplemental material, Section 5: “Analysis of the ‘0/NFR’ group promoters”; Supplemental material, Section 6: “Analysis of fragile nucleosome change upon the environment changes between normal growth and heat stress conditions”; and Supplemental material, Section 7: “Identification of resistant nucleosomes.”
Acknowledgments
We thank Craig Kaplan for helpful discussions; X. Shirley Liu and Yong Zhnag for the NPS program; and Sharon Y.R. Dent, Gad Shaulsky, Philip J. Hastings, and Gregory Ira for a critical reading of the manuscript. This work was supported in part by NIH grant GM068676 (to X.H.), NIH grant U01DA025956, DOD grant PC094421, and 973 project 2010CB944900 of China (to W.L).
Footnotes
[Supplemental material is available for this article. The data from this study have been submitted to the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE26412.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.117101.110.
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Bjorklund S, Gustafsson CM 2005. The yeast Mediator complex and its regulation. Trends Biochem Sci 30: 240–244 [DOI] [PubMed] [Google Scholar]
- Cairns BR 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198 [DOI] [PubMed] [Google Scholar]
- Casamassimi A, Napoli C 2007. Mediator complexes and eukaryotic transcription regulation: An overview. Biochimie 89: 1439–1446 [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA 2001. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E 2008. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 4: e1000216 doi: 10.1371/journal.pcbi.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WS, Orlov Y, Li J, Clarke ND 2010. Blurring of high-resolution data shows that the effect of intrinsic nucleosome occupancy on transcription factor binding is mostly regional, not local. PLoS Comput Biol 6: e1000649 doi: 10.1371/journal.pcbi.1000649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn JS, Hu Z, Thiele DJ, Iyer VR 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol 24: 5249–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G 2007. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev 21: 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G 2009. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Li G, Levitus M, Bustamante C, Widom J 2005. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol 12: 46–53 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF 2008a. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. 2008b. Nucleosome organization in the Drosophila genome. Nature 453: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M, Kornberg RD 1977. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol 109: 393–404 [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, Iyer VR 2008. Stress-dependent dynamics of global chromatin remodeling in yeast: Dual role for SWI/SNF in the heat shock stress response. Mol Cell Biol 28: 2221–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N 2008. Two strategies for gene regulation by promoter nucleosomes. Genome Res 18: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N 2010. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20: 90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman JL, Kingston RE 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem 67: 545–579 [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]