Abstract
Studying the genetic regulation of expression variation is a key method to dissect complex phenotypic traits. To examine the genetic architecture of regulatory variation in Arabidopsis thaliana, we performed genome-wide association (GWA) mapping of gene expression in an F1 hybrid diversity panel. At a genome-wide false discovery rate (FDR) of 0.2, an associated single nucleotide polymorphism (SNP) explains >38% of trait variation. In comparison with SNPs that are distant from the genes to which they were associated, locally associated SNPs are preferentially found in regions with extended linkage disequilibrium (LD) and have distinct population frequencies of the derived alleles (where Arabidopsis lyrata has the ancestral allele), suggesting that different selective forces are acting. Locally associated SNPs tend to have additive inheritance, whereas distantly associated SNPs are primarily dominant. In contrast to results from mapping of expression quantitative trait loci (eQTL) in linkage studies, we observe extensive allelic heterogeneity for local regulatory loci in our diversity panel. By association mapping of allele-specific expression (ASE), we detect a significant enrichment for cis-acting variation in local regulatory variation. In addition to gene expression variation, association mapping of splicing variation reveals both local and distant genetic regulation for intron and exon level traits. Finally, we identify candidate genes for 59 diverse phenotypic traits that were mapped to eQTL.
Genetic mapping of gene expression levels (Jansen and Nap 2001) has been used to dissect the genetic architecture of expression regulatory variation in a number of systems (Brem et al. 2002; Schadt et al. 2003; Yvert et al. 2003; Monks et al. 2004; Morley et al. 2004; Cheung et al. 2005; DeCook et al. 2006; Keurentjes et al. 2007; West et al. 2007; Huang et al. 2009; Swanson-Wagner et al. 2009) and has aided in the identification of phenotypic and disease quantitative trait loci (QTL) (Bystrykh et al. 2005; Chesler et al. 2005; Hubner et al. 2005). These studies demonstrate the importance of genetic factors regulating gene expression variation and suggest that polygenic control is common. Cis-acting polymorphisms are located in gene regulatory elements that affect the transcript abundance of the linked allele of the target gene. Trans-acting polymorphisms are located elsewhere in the genome and affect the transcript abundance of both alleles of the target gene (Rockman and Kruglyak 2006). Traditional linkage and association mapping studies can distinguish local and distant expression quantitative trait loci (eQTL), while the cis and trans regulatory mechanisms must be directly tested by allele-specific expression (Doss et al. 2005; Ronald et al. 2005).
The observance of hybrid vigor in commercial agriculture has sparked interest in the inheritance of gene expression in hybrids, to identify an explanatory mechanism. Although most gene expression is inherited additively, studies in several organisms have identified a substantial fraction of genes with non-additive, or dominant, inheritance patterns (Gibson et al. 2004; Auger et al. 2005; Vuylsteke et al. 2005; Cui et al. 2006; Swanson-Wagner et al. 2006; Stupar et al. 2007; Zhang et al. 2008). Whether non-additive regulatory variation is due to common genetic variation segregating in a population remains unclear.
Alternative splicing serves to increase the transcript repertoire and has been identified as an important regulatory mechanism in development (Macknight et al. 1997; Calarco et al. 2009). Population genetic variation in alternative splicing has been identified in humans and is correlated with local polymorphism (Kwan et al. 2008; Montgomery et al. 2010; Pickrell et al. 2010). Plants differ from other higher eukaryotes in that intron retention seems to be a common form of alternative splicing (Wang and Brendel 2006; McGuire et al. 2008). A recent deep mRNA sequencing study found alternative splice forms for >40% of all intron-containing genes, many of which appeared under stress conditions (Filichkin et al. 2009). Alternatively spliced introns were enriched for premature termination codons, suggesting that alternative splicing may play an important role in regulating transcript abundance through nonsense-mediated decay.
In Arabidopsis, eQTL mapping has been reported using recombinant inbred lines (RILs) (DeCook et al. 2006; Keurentjes et al. 2007; West et al. 2007). Linkage mapping in RILs can potentially map local regulatory variation segregating between parental lines, but resolution is limited by recombination. Association mapping in distantly related individuals tests for common genetic variation controlling expression variation while providing higher mapping resolution through ancestral recombination. The combination of near-saturating coverage of SNP markers and the rapid decay in their linkage disequilibrium (LD) has facilitated the discovery of genetic factors for diverse phenotypes in Arabidopsis thaliana (Atwell et al. 2010; Baxter et al. 2010; Li et al. 2010). We used this powerful genetic resource to map gene expression variation in a diverse panel of F1 hybrids. Our study suggests different selective forces shaping the population genetics of local and distant eQTL and highlights a level of complexity in local regulatory variation.
Results
Genome-wide association mapping of gene expression
We mapped 21,803 gene expression traits against 142,048 SNPs, which have minor allele frequency >0.1 in our sample of 57 hybrid lines. This results in a genome scan of 839-bp average resolution. We analyzed associations at several false discovery rate (FDR) thresholds (Supplemental Table 1), but will focus on associations with FDR < 0.2 for a balanced discussion on local and distant regulatory variation. At FDR < 0.2, a total of 1838 gene expression traits were mapped to 6190 SNPs (Table 1). A small fraction of genes were mapped to a large number of SNPs due to long LD blocks (Supplemental Fig. 1A). An associated SNP explains 38.1%–92.6% of the corresponding gene expression variation, with a median effect being 42.3%. To test whether there is any directional bias of new mutations, we compared the ancestral and derived SNP alleles at 3179 associated SNPs using Arabidopsis lyrata as an outgroup. The derived alleles at 1773 SNPs up-regulate 772 genes, whereas derived alleles at 1406 SNPs down-regulate 680 genes, revealing little if any directional bias of new mutations.
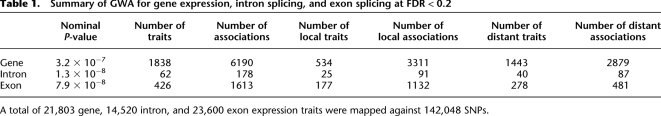
Table 1.
Summary of GWA for gene expression, intron splicing, and exon splicing at FDR < 0.2
A total of 21,803 gene, 14,520 intron, and 23,600 exon expression traits were mapped against 142,048 SNPs.
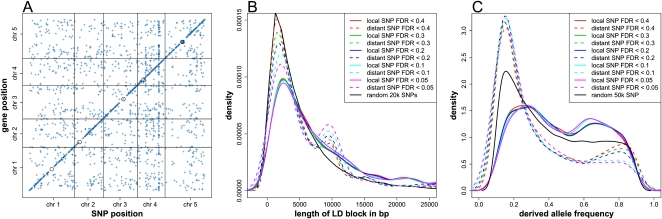
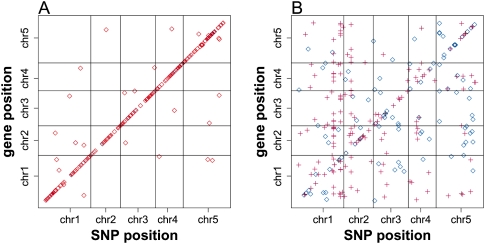
Associated SNPs are enriched at the location of the mapped genes, shown as the diagonal line in Figure 1A. The proportion of associated SNPs peaks around the physical position of the gene and drops down to background level ∼25 kb from the gene (Supplemental Fig. 1B). Therefore, we defined locally associated SNPs as SNPs located within the range from −25 kb relative to the gene transcription start to +25 kb relative to the gene transcription stop. Based on this definition, we found 3311 local associations for 534 genes and 2879 distant associations for 1443 genes (Table 1). Consistent with previous studies (Keurentjes et al. 2007; West et al. 2007), local associations tend to have a larger effect (r2) than distant associations (Supplemental Fig. 1C). As such, the proportion of local associations increases with a more stringent detection threshold (Supplemental Table 1). This implies that the genetic architecture of local regulatory variation is relatively simple, meaning a large proportion of the trait variation is explained by a single additive SNP.
Figure 1.
GWA for gene expression. (A) The middle position of the mapped genes was plotted against the chromosome position of the associated SNPs detected at FDR < 0.2. The intensity of the points indicates the effect (r2) of the corresponding association. (Black circles) Centromeres. (B) The length distribution of LD blocks surrounding locally (solid lines) and distantly (dashed lines) associated SNPs, detected at different FDR thresholds. LD block length was defined as the distance between the first two flanking SNPs, with which focal SNP r2 < 0.1. (Black line) Randomly sampled 20,000 SNPs without local or distant association. The small peak between 8 and 12 kb for the distant associations included 470 associations, 126 of which were with the largest trans hot spot located at 14,423,393 bp on chromosome 4. (C) The frequency distribution of the derived alleles of locally (solid line) and distantly (dashed line) associated SNPs, detected at different FDR thresholds. (Black line) Randomly sampled 50,000 SNPs without local or distant association.
Several SNPs were associated with multiple expression traits. These are the so-called master regulatory loci, or trans hot spots. Consistent with an eQTL study using RILs in A. thaliana (West et al. 2007), trans hot spots tend to regulate genes coordinately either up or down (Supplemental Fig. 1D). Identification of the causal variations tagged by these trans hot spots is not straightforward and requires experimental validation. For this reason, we do not discuss the potential regulatory mechanisms for these trans hot spots but provide the Gene Ontology (GO) analysis for their target genes (Supplemental Table 2).
Population structure if severe could be a confounding factor in association studies. In Arabidopsis, population structure is due to different relatedness among samples (Platt et al. 2010a). The confounding effect also depends on the traits investigated (Atwell et al. 2010). Our F1 lines were generated from a subset of a diversity panel selected to be equally unrelated (Li et al. 2010). We did not detect any discrete subpopulation across these study samples (Supplemental Fig. 1E). The observed P-value distribution indicates an inflation of significant associations for a small fraction of gene expression traits (Supplemental Fig. 1F), suggesting that there may still be some confounding due to population structure for these traits. This minor fraction of traits, however, was not biased toward local or distant regulatory variation (Supplemental Fig. 1F).
Distinct evolutionary histories of local and distant regulatory loci
As shown in Supplemental Table 1, 53.5% of associations with FDR < 0.2 were local, regulating 29.1% of mapped genes; whereas 46.5% of associations were distant, regulating 78.5% of mapped genes. Thus, on average, there are more locally associated SNPs than distantly associated SNPs per target gene. On one hand, a distant regulatory locus could control multiple expression traits, as seen in trans hot spots. On the other hand, LD could be different between local and distant loci, so that a local regulatory locus contains more SNP associations than a distant one. To test the latter, we compared LD surrounding locally and distantly associated SNPs. Locally associated SNPs tend to have a higher level of long-range, weak LD (r2 > 0.1) (Fig. 1B). The short-range, strong LD (r2 > 0.8) is similar between locally and distantly associated SNPs (Supplemental Fig. 2A). This indicates that locally associated SNPs are more likely to be located in long LD blocks and may have experienced different evolutionary histories from distantly associated SNPs. In line with this observation, the population sample frequencies of the derived allele for locally associated SNPs are distributed rather uniformly, whereas those for distantly associated SNPs are highly skewed toward low frequency (Fig. 1C). Interestingly, there was a small peak of extended LD (8–12 kb) for distant associations (Fig. 1B). The 275 genes mapped to these associations were significantly enriched in the GO categories “plant hypersensitive response” (adjusted P < 9.9 × 10−4) and “system acquired resistance” (adjusted P < 4.3 × 10−2), two distinct plant defense responses to pathogens (Ryals et al. 1996). This peak of extended LD was mostly contributed by associations with the largest trans hot spot, located at 14,423,393 bp on chromosome 4 (Supplemental Fig. 2B), suggesting a selective sweep on this master regulatory locus.
Allelic heterogeneity of local regulatory variation
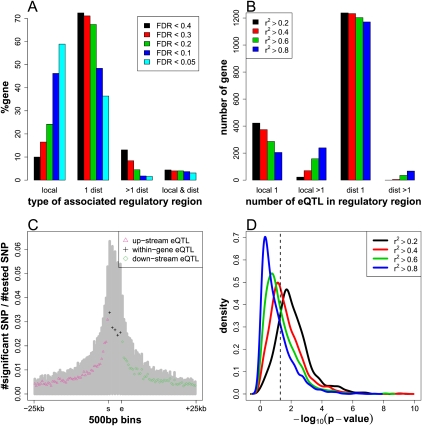
Multiple associations within a regulatory locus, if not in strong LD, suggest allelic heterogeneity, with multiple distinct alleles within the regulatory locus affecting a gene expression trait. For a mapped gene, locally associated SNPs by definition fall within a single locus, whereas distantly associated SNPs could be located in a single locus or across multiple unlinked loci. To delineate regulatory loci, for each gene, we grouped associated SNPs into regions such that within a region all associations were <100 kb apart. Applying a wider 100-kb cutoff is to account for long-range LD in some local regulatory loci. Regions within which the most significant SNP association is ±25 kb from the mapped gene were considered local. We then estimated the number of alleles within each of these regulatory regions by clustering the associated SNPs based on LD (Methods). For most of the mapped genes, associations were grouped into a single distant (67.0%) or local (23.2%) region (Fig. 2A). Only these regions were further compared. The associated SNPs after clustering by LD were called eQTL. When SNPs were clustered at r2 > 0.8, multiple eQTL (≥2) were detected for 53.8% of local regions, while only for 5.4% of distant regions (Fig. 2B). Multiple local eQTL were consistently identified from r2 > 0.8 through r2 > 0.2 (Fig. 2B). This result suggests that many local regulatory regions contain multiple haplotypes.
Figure 2.
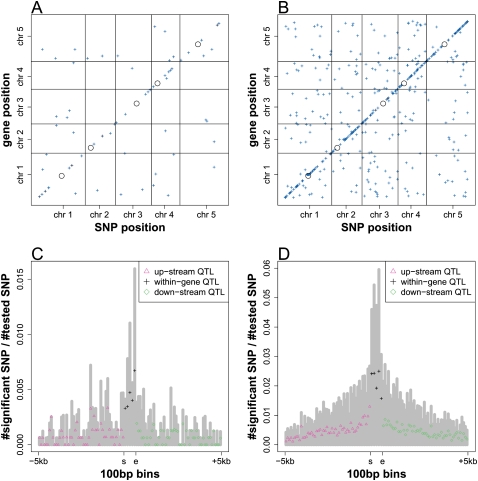
Allelic heterogeneity of local regulatory variation. (A) Associated SNPs detected at different FDR thresholds from GWA of gene expression were grouped to local and distant regulatory regions. At FDR < 0.2, for example, 1238 genes (67.4%) were associated with a single distant region (1 dist), 82 genes (4.5%) were associated with multiple distant regions (>1 dist), 444 genes (24.2%) were only locally associated (local), and an additional 74 genes (4.0%) were associated with both local and distant regions (local & dist). (B) For associations detected at FDR < 0.2 from GWA of gene expression, the number of genes that have single eQTL within local regulatory region (local 1), multiple eQTL within local regulatory region (local >1), single eQTL within distant regulatory region (dist 1), and multiple eQTL within distant regulatory region (dist >1). Associated SNPs within a regulatory region were clustered at different LD thresholds. (C) Distribution of eQTL from local scan. Proportion of eQTL (the number of eQTL after clustering by r2 > 0.8/[the number of SNPs tested]) was plotted along 500-bp bins for 25 kb upstream and 25 kb downstream. Within genes, the positions were binned to five positional quantiles. (Gray bars) The proportion of associated SNPs before LD clustering. (s) Transcriptional start site; (e) transcriptional stop site. (D) The P-value distribution of the F-tests for model comparison. Associated SNPs were clustered at different LD thresholds. (Dashed line) P-value of 0.05.
The above observation led us to a refined scan for local associations, for which we mapped expression traits only against SNPs within ±25 kb of a target gene, to avoid the multiple testing inherit in full-genome analysis. As in genome-wide association (GWA), we analyzed associations at several detection thresholds (Supplemental Table 3). At FDR < 0.05, we detected 21,203 associations for 3365 genes (Table 2). An associated SNP explains 18.7%–92.6% of expression variation, with a median effect of 24.9%. Enrichment for associated SNPs was highest within the gene and the immediately upstream and downstream regions. This trend remains after clustering of the associated SNPs by LD (Fig. 2C). The 5′ portion of the gene and the 500-bp upstream region exhibit the highest enrichment for eQTL, suggesting the importance of these regions in genetic regulatory variation. For eQTL located in upstream and downstream regions, their effect (r2) exhibits a modest negative correlation with their distance to the gene (Supplemental Fig. 3A).
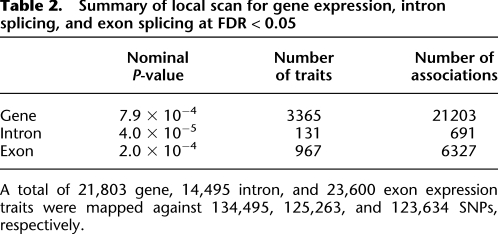
Table 2.
Summary of local scan for gene expression, intron splicing, and exon splicing at FDR < 0.05
A total of 21,803 gene, 14,495 intron, and 23,600 exon expression traits were mapped against 134,495, 125,263, and 123,634 SNPs, respectively.
As in GWA, we observed a high proportion of the mapped genes associated with multiple local eQTL (Supplemental Fig. 3B). The lack of LD among the multiple local eQTL suggests they are functionally distinct. To further address the independence of multiple local eQTL, for each gene mapped to two or more local eQTL, we compared a single SNP (null) model that includes only the local eQTL of the largest marginal effect, with an alternative model that includes all local eQTL in an additive mode. For a substantial proportion of these genes, the improvement of the model fit is significant (Fig. 2D). These results argue that allelic heterogeneity of local regulatory variation could be common in Arabidopsis. This is distinct from the observation in linkage studies, where trait variation is explained by two segregating alleles at a single marker due to wide mapping resolution. It should be noted, however, that in some cases, a single untyped causal polymorphism at relatively rare allele frequency could cause multiple associations at neutral SNPs that are in LD with the latent SNP (Dickson et al. 2010; Platt et al. 2010b).
Association mapping of allele-specific expression
While physically linked to a gene, local regulatory polymorphisms can either act in cis or in trans. True cis-acting polymorphism causes differential transcript abundance between the two alleles of the target gene, or allele-specific expression (ASE). We mapped ASE traits (Supplemental Fig. 4A; Methods) across 18,813 genes, which contain a total of 55,401 transcribed SNPs, against 134,983 local SNPs. At q-value < 0.2, we detected 17,660 associations for 2478 genes (Table 3). An example of cis-acting, locally associated SNP is shown in Figure 3. We then examined how often local regulatory variation is due to cis variation. Among the 3365 genes significant at FDR < 0.05 from local scan of expression level variation, 2877 were also tested for ASE association. We detected cis variation for 727 genes (25.3%) at q < 0.2. This is 1.9-fold enrichment over independence (χ2-test; P < 6.2 × 10−94). The fold enrichment increases with more stringent detection threshold (Table 3). A slightly higher level of allelic heterogeneity was observed in the local regulatory regions of the genes overlapping between expression association and ASE association (Supplemental Fig. 3B).
Table 3.
Summary of local scan for ASE
A total of 18,813 genes, containing 55,401 transcribed SNPs, were tested against 134,983 SNPs.
aThe q-value thresholds for ASE association.
bThe number of genes that were significant for ASE association and which were among the 3,365 genes significant at FDR < 0.05 in local scan of expression level.
cThe fold enrichment for cis regulated genes among locally regulated genes.
dP-value of χ2-test for the overlap between genes with local regulatory variation and genes with cis regulatory variation.
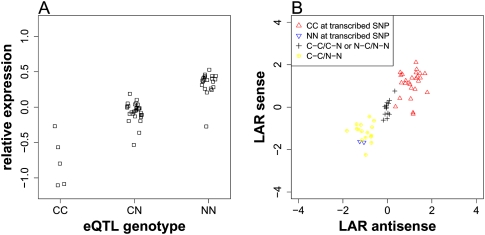
Figure 3.
An example of cis-acting, locally associated SNP. (A) The relative gene expression levels were plotted against the genotypes at the associated SNP, located at 3,041,799 bp on chromosome 1. (C) Col allele; (N) non-Col allele. (B) The log allele ratios (LARs) of the sense strain were plotted against LARs of antisense strain for the transcribed SNP, located at 3,041,022 bp on chromosome 1. An explanation of legends follows: (CC at transcribed SNP) lines for which transcribed SNP is homozygous Col allele; (NN at transcribed SNP) lines for which transcribed SNP is homozygous non-Col allele; (C-C/C-N or N-C/N-N) lines for which transcribed SNP is heterozygous but the regulatory SNP is homozygous; (C-C/N-N) lines for which transcribed SNP is heterozygous and the regulatory SNP is in phase with transcribed SNP. In this case, the non-Col allele at the regulatory SNP up-regulates gene expression.
The 25.3% coverage of locally regulated genes by cis regulated genes is likely an underestimation. The discrepancy could be caused by several factors. First, only samples heterozygous for the transcribed SNPs have ASE traits that could be mapped, which reduced the average sample size from 57 to 21 lines. Many SNPs with rare minor allele frequencies cannot be tested for aseQTL in this reduced sample size. Second, the measurement of ASE (log allele ratio) and gene expression level (log probe intensity) appears to have different detection sensitivities depending on gene expression level (Supplemental Fig. 4B). It is also possible, however, that a fraction of local regulatory variation may not act in cis. To explore this, we focused on the genes highly significant (FDR < 0.001) in expression level association. Among them we further selected genes for which ASE trait can be measured by very discriminative transcribed SNPs (Methods) and can be mapped with at least 28 lines. Among 99 genes selected based on these criteria, only 54.5% were mapped to cis variation at q < 0.2. Close examination identified a fraction of potentially trans-acting local SNPs, an example of which is shown in Supplemental Figure 4C.
Genetic inheritance of gene expression
The additive or dominant inheritance of a QTL affects the trait heritability and fixation rate in a population. To identify the most likely mode of inheritance for expression regulatory variation, we tested the fit of three genetic models—additive, Col allele-dominant, and Col allele-recessive—for gene expression traits, where “Col” refers to the reference accession Columbia. We tested 21,803 expression traits with 56,819 genome-wide SNPs, which have at least six lines for each of the three SNP genotype classes. At FDR < 0.2, we detected 1482 associations, among which 994 were additive and 488 were dominant, for 426 genes (Supplemental Table 4). Additive associations were predominantly found at the location of the mapped genes (Fig. 4A) and tend to have a relatively larger effect (r2) than dominant associations (Supplemental Fig. 5A). This is consistent with the expected additive inheritance for cis regulatory variation, when a heterozygote has the mid-parent expression level. Trans variation may act dominantly if their presence can activate a pathway. To quantify this, we calculated the proportion of additive associations falling in local regulatory loci as well as dominant associations in distant loci. For each mapped gene, we again grouped the associated SNPs into regulatory regions (Supplemental Fig. 5B). In summary, the 426 genes were mapped to 209 local and 253 distance regulatory regions. As much as 86.6% of additive associations falls in local regulatory regions. In contrast, distantly associated regions tend to regulate their target genes dominantly. As such, 61.7% of dominant associations were located in distant regulatory regions (Supplemental Table 4).
Figure 4.
Additive (A) and dominant (B) associations detected at FDR < 0.2 in GWA for genetic inheritance of gene expression. The middle position of the associated genes was plotted against the chromosome position of the associated SNPs. The intensity of the points indicates the effect (r2) of the corresponding association. In B, cross points represent Col allele dominant, while diamond points represent non-Col allele dominant.
The fraction of dominant associations falling in a local regulatory region is interesting (Fig. 4B), as they could be local polymorphisms acting in trans through genetic feedback (Gjuvsland et al. 2010) or transvection as discovered in Drosophila (Lewis 1954). Probe binding, however, could be nonlinear to target concentration in a microarray experiment, which causes the measured expression level of the heterozygous genotype to deviate from the mid-parent level. Heterozygous genotypes may express a target gene at the level of the higher homozygous genotype class, showing positive dominance, or expression could be repressed in the heterozygote showing negative dominance. We found that local dominant associations tend to show partial positive dominance, whereas distant dominant associations are relatively enriched in complete negative dominance (Supplemental Fig. 5C). A clear separation of partial dominance from nonlinear probe binding is challenging; thus, confirmation of local dominant regulation may require independent experimental investigation.
We found that distant dominant eQTL are more likely to regulate multiple expression traits. As such, 7.1% of distant dominant eQTL control two or more expression traits, while this is true for 1.5% of local additive eQTL. Several distant dominant eQTL are clearly regulatory hot spots (Fig. 4B). They are distinct from the additive hot spots detected in GWA using only the additive model (Supplemental Table 2). Nevertheless, these dominant hot spots are similar to additive hot spots in that they regulate gene expression directionally (Supplemental Fig. 5D).
Genetic regulation of splicing variation
Gene expression traits summarize transcript abundance across commonly expressed exons. To examine the genetic regulation for splicing variation, we performed GWA separately for 14,520 intron and 23,600 exon level traits, against 142,048 SNPs (Methods). In comparison with gene expression variation, genetic regulation of intron (Fig. 5A) and exon (Fig. 5B) splicing variation is relatively limited. At FDR < 0.2, we detected 178 associations for 62 intron level traits and 1613 associations for 426 exon level traits (Table 1). Both local and distant associations were detected; as much as 64.5% of associated introns and 65.3% of associated exons were mapped to distant SNPs (Supplemental Table 1). Similar to gene expression traits, local associations have a larger effect (r2) than distant associations for splicing traits (Supplemental Fig. 6).
Figure 5.
Genetic regulation of splicing variation. The middle position of the mapped introns (A) and exons (B) was plotted against the chromosome position of the associated SNPs, detected in GWA at FDR < 0.2. The intensity of the points indicates the effect (r2) of the corresponding association. (Black circles) Centromeres. The distribution of intron (C) and exon (D) splicing QTL from local scan. Proportion of splicing QTL (the number of splicing QTL after clustering by r2 > 0.8/[the number of SNPs tested]) was plotted along 100-bp bins for 5 kb upstream and 5 kb downstream from the mapped introns or exons. Within introns or exons, the positions were binned to five positional quantiles. (Gray bars) The proportion of associated SNPs before LD clustering. (s) Start position of intron or exon; (e) end position of intron or exon.
For a refined local scan, we mapped the intron and exon level traits against SNPs located within ±25 kb of the corresponding gene. At FDR < 0.05, we detected 691 associations for 131 introns and 6327 associations for 967 exons (Table 2). Associated SNPs were highly enriched within the intron (Fig. 5C) and exon (Fig. 5D) itself. The entire exon as well as the middle and 3′ end of the intron appear to be in strong short-range LD, suggesting that these could be relatively conserved regions. While the 3′ end of introns is the most abundant site for splicing QTL (Fig. 5C), the 3′ end of exons is relatively depleted for splicing QTL (Fig. 5D).
eQTL underlying phenotypic QTL
eQTL aid in the identification of causal genes for phenotypic QTL. To identify candidate genes underlying our set of 107 phenotypic traits (Atwell et al. 2010), we examined the SNPs that were detected in phenotypic association and that were linked with eQTL (Methods). The regulatory regions from our GWA eQTL mapping were compared across genes. Regions at close physical positions (<2 kb) were combined. We then searched for phenotypic associations in LD (r2 > 0.6) with eQTL within these regulatory regions. In summary, 59 phenotypic traits were mapped to eQTL within local (Supplemental Table 5) and/or distant (Supplemental Table 6) regulatory regions. The majority of eQTL control a single phenotypic trait or several highly related phenotypic traits. An eQTL located at 1,275,971 bp on chromosome 4 was associated with several phenotypes including flowering time, trichome density, rosette erectness, and germination in the dark (Supplemental Table 5). This eQTL locally regulates expression variation of AT4G02920, which has an unknown function.
The leaf yellowing phenotype was mapped to two local and two distant regulatory regions, among which a local region on chromosome 3 from 18,316,567 bp to 18,356,919 bp overlaps a distant region from 18,353,289 bp to 18,357,216 bp (Supplemental Tables 5, 6). The overlapped region contains a trans hot spot at 18,356,216 bp. This trans hot spot locally regulates the expression of AT3G49510, a gene encoding an F-box family protein, and distantly regulates a set of genes that are significantly enriched in the “chlorophyll biosynthetic process” (Supplemental Table 2) in our leaf samples. The up-regulation of AT3G49510 expression by the non Col-allele of this trans hot spot likely causes down-regulation of chlorophyll biosynthesis, leading to leaf yellowing.
Discussion
The fate of a newly emerged mutation, as determined by genetic drift and natural selection, can affect linked sites. A genome scan can reveal distinct patterns in population allele frequency distribution among sites with different evolutionary histories. Although recent studies have investigated eQTL in various human populations (Stranger et al. 2007; Duan et al. 2008; Montgomery et al. 2010; Pickrell et al. 2010), the population allele frequency distribution of eQTL has not been characterized (Gibson and Weir 2005). In Arabidopsis, we found that the population frequencies of derived SNP alleles of eQTL follow a bimodal distribution. The bimodal distribution is distinct between locally and distantly associated SNPs. For distantly associated SNPs, the bimodal distribution is highly skewed, with a large mode at very low allele frequency and a small mode of very high allele frequency. The bimodal distribution for locally associated SNPs is centered at moderately low and moderately high allele frequency. This suggests distinct selective forces acting on local and distant regulatory variation. Trans-acting polymorphisms could have pleiotropic effects, which may present a larger phenotypic target for selection, whereas cis-acting polymorphisms affect the expression of a single locus and thus could be neutral (Alonso and Wilkins 2005; Wray 2007). Such differences predict abundant distantly associated SNPs with low derived allele frequency, as we found in this study. Interestingly, locally associated SNPs have more common derived allele frequencies than background SNPs without either local or distant association. One explanation is that they are older polymorphisms located in chromosome regions with low recombination, as suggested by the level of long-range LD surrounding locally associated SNPs. In A. thaliana, selection could be particularly stringent against trans-acting variation, while relaxed for cis-acting variation. This is because A. thaliana reproduces largely through self-fertilization, where strongly deleterious mutations are quickly eliminated, but weakly deleterious mutations are purged rather inefficiently due to low effective recombination. Selective sweeps could fix a small proportion of trans polymorphisms that provide an adaptive advantage, such as that trans polymorphism tagged by the largest trans hot spot, which genetically regulates plant defense responses.
Cis-acting regulatory variation is thought to play a major role in gene expression differentiation, within and between species (Denver et al. 2005; Landry et al. 2007; Wray 2007; Wilson et al. 2008; Wittkopp et al. 2008). Traditional linkage studies identify local regulatory variation at a broadly defined locus (Keurentjes et al. 2007; West et al. 2007). In a mapping population derived from two parental lines, such as RIL and F2 lines, segregation of intralocus polymorphisms is rare due to limited recombination. This situation may change when mapping in diverse accessions, where historical recombination has broken down linkage at various degrees across the genome and new mutations accumulate. Allelic heterogeneity has been demonstrated for cis regulatory variation for specific genes (Horan et al. 2003; Tao et al. 2006; Babbitt et al. 2009), and our study suggests that this could be common in A. thaliana. Local regulatory loci tend to have a lower level of recombination and are also more diverse than other chromosome regions. Such patterns may be intertwined with the evolution of gene expression variation within these regions (Tung et al. 2009; Zhang and Borevitz 2009). Association mapping of ASE traits is expected to reveal true cis regulatory variation (Pastinen and Hudson 2004). Trait variance, however, could be high when many rare regulatory alleles are present, as suggested by a recent deep sequencing of human RNA samples (Montgomery et al. 2010). Here, variation in abundance between two transcript alleles is the composite effect of multiple cis regulatory polymorphisms linked to these transcript alleles. Association of an ASE trait with allelic haplotypes could be more appropriate to identify these complex cases of cis regulatory variation.
We found only a small number of introns whose level is genetically controlled by QTL, and a substantial proportion of intron splicing variation mapped to distant SNPs. This is different from a previous study between two parental accessions, wherein cis-acting variation was detected for >25% of the analyzed introns, while trans-acting variation was minor (Zhang and Borevitz 2009). Difference in developmental stage and genetic background between the samples used in these studies could contribute to this. However, it is also possible that cis variation of intron splicing is due to multiple rare variants missed by GWA but detected between two pure lines.
In summary, this study reveals the genetic architecture of regulatory variation among a set of diverse hybrid lines and indicates networks of genes underlying phenotypic traits.
Methods
Plant material and growth conditions
We randomly selected 111 accessions from a diversity panel (Li et al. 2010). These lines were randomly crossed resulting in 57 F1 lines. See Supplemental Table 7 for a list of parental accessions. Seeds from the cross were cold stratified in water for 5 d then sown in 36 cell flats in Promix 1:1 Metro:C2 soil. Plants were grown with 18 h of fluorescent light at 21°C.
RNA isolation and microarray hybridization
A single leaf (the fifth or sixth true leaf) was collected for each F1 line, 3 wk post-germination, resulting in 57 samples with single replication. Total RNA preparation, cDNA synthesis, and labeling were described previously (Zhang and Borevitz 2009). Twenty micrograms of labeled product was hybridized to the AtSNPTILE1 microarray (Affymetrix) using a standard washing/staining protocol for gene expression arrays at the University of Chicago Functional Genomics Facility.
Genotyping
Genotypes for each parent were called from genomic DNA hybridization data generated in a previous study (Li et al. 2010), using a modified version of Corrected Robust Linear Model with Maximum Likelihood Distance (Carvalho et al. 2007) assuming homozygosity. For eQTL mapping, SNPs for which >30% samples have posterior probability of genotype call <0.90 were removed. The remaining genotype calls with posterior probability <0.85 were imputed (Roberts et al. 2007). For ASE mapping, SNPs for which >30% samples have posterior probability of genotype call <0.95 were removed. The remaining genotype calls with posterior probability <0.90 were imputed. F1 genotypes were based on the combination of parental genotypes.
Array preprocessing
Raw intensities from CEL files were log-transformed, background-corrected, and normalized as previously described (Borevitz et al. 2003). Previous studies have found a significant proportion of local eQTL that were due to sequence hybridization polymorphisms within probes (Doss et al. 2005; Huang et al. 2009). We addressed this by removing from further analysis the probes containing Single Feature Polymorphisms (SFPs) (Borevitz et al. 2003). SFPs were detected between the reference accession Columbia and the parental lines, using genomic DNA hybridization data generated previously (Li et al. 2010). The detection threshold was defined so that it corresponds to permutation-based FDR < 0.2 between Columbia and Vancouver accessions using five replicates (Zhang and Borevitz 2009). Only genes interrogated by three or more major probes were considered for expression mapping. Major probes are probes interrogating transcribed sequences that occur in >50% of expression clones of the corresponding gene (Zhang et al. 2008). To minimize the impact of probe hybridization variation, for each probe, the mean value across lines was subtracted from the probe value. For each expression phenotype, the corrected probe values were averaged to return a single expression value for each line. The gene expression values and genotypes for 57 F1 lines are provided in the Supplemental material.
Association mapping
Mapping of 21,803 gene, 14,520 intron, and 23,600 exon expression traits was performed using an additive model, with genotype coded as 0, 1, and 2 for homozygous reference allele, heterozygous, and homozygous nonreference allele, respectively. To estimate FDR, the expression phenotypes were permuted five times, and the model P-values were recorded. FDR was calculated as the (average number of significant tests in permuted data)/(number of significant tests in real data). Permutation orders for the five permutations were kept the same for gene, intron, and exon analysis. For exon analysis, exons were selected that contain two or more major probes and that were interrogated by <20% of probes of their corresponding genes (Zhang et al. 2008). Mean log gene expression values were subtracted from mean log exon expression values to correct gene level expression variation.
For analysis of genetic inheritance of expression association, three models—coded as 0, 1, 2 for additive; 0, 1, 1 for Col allele recessive; and 0, 0, 1 for Col allele dominant—were applied separately for each trait–SNP pair. The maximum F-statistic across three models was recorded for each trait–SNP pair. The same procedure was applied across five permutations to estimate FDR.
Genes and chromosome positions were based on TAIR 9 annotation. All mapping and modeling were carried out in R using lsfit.
Clustering of associated SNPs by LD
For each mapped gene expression trait, the associated SNPs were ranked by the effect size (r2). The SNP with the largest effect was selected as the focal SNP. LD r2 between the focal SNP and all of the remaining SNPs was calculated. SNPs that have r2 exceeding the r2 threshold were removed. The procedure repeats until all associated SNPs were clustered.
Local mapping of ASE traits
Our unique array platform contains ∼1.0 million SNP probes interrogating ∼250,000 SNPs as well as ∼1.6 million tiling probes with an average of 35-bp resolution (Zhang and Borevitz 2009). Only genes containing heterozygous SNPs within the transcribed region were analyzed. For these genes, log allele intensity ratios (LARs) of Col allele over non-Col allele at the transcribed SNPs were mapped against local SNPs (±25 kb of the tested gene). Here we denoted C and N to represent Col and non-Col allele, respectively. Three phase groups were defined based on the allelic combination of the transcribed SNP and the SNP tested for association (regulatory SNP). These three phase groups were C-C/N-N (regulatory SNP and transcribed SNP in phase), C-C/C-N or N-C/N-N (no regulatory variation), and N-C/C-N (regulatory SNP and transcribed SNP out-of-phase). An additive model was applied on these three phase groups coded as 0, 1, and 2, respectively. Thus, the regression coefficients represent the effect of a non-Col allele over a Col allele at the regulatory SNP. Only tests that contained six or more samples, with the sum of two smaller phase groups three or more samples and representing ≥10% of all samples, were analyzed. Association tests were divided into 31 groups based on the sample size. Within each sample size group, a d-statistic was calculated for each association, d = coefficient/(standard deviation + s0), where s0 represents the median of standard deviations across all tests within the sample size group. Ten permutations were performed within the sample size group to obtain q-values (Storey and Tibshirani 2003). The final significant list pooled across sample size groups was based on q-value. It should be noted that due to the insufficient breakdown of LD in a sample size of 57, the majority of ASE traits could only be tested for two phase groups, which usually included the group homozygous at the regulatory SNP. In selection of very discriminative transcribed SNPs to explore trans-acting local variation, homozygous genotypes at the transcribed SNPs formed two clusters, C1 and C2, based on the LARs, each cluster with five or more lines. Discriminative transcribed SNPs were defined as those where |medianC1 − medianC2| ≥ 3 × (SDC1 + SDC2).
Overlap with phenotypic associations
Phenotypic traits (Atwell et al. 2010) were mapped against SNPs with MAF > 0.1, using the Wilcoxon test for quantitative traits or a Fisher's exact test for categorical traits. Significant associations were detected at P < 1 × 10−5 for disease resistance, ion concentration, and general developmental traits, whereas at P < 1 × 10−7 for flowering time traits, which are extensively confounded by population structure (Atwell et al. 2010). Regulatory regions from GWA of gene expression were obtained. For regions containing only one associated SNP, the region was redefined as from −1 kb to +1 kb relative to the associated SNP. Regulatory regions were then compared across mapped genes. Regions <2 kb apart were combined as one. Within each regulatory region, the expression associations and phenotypic associations were clustered by LD at r2 > 0.6.
Acknowledgments
We thank the reviewers for their great comments on this manuscript. We thank Dr. Alexander Platt for helpful discussion. X.Z., A.J.C., and J.O.B. were supported by National Institutes of Health Grant R01GM073822 to J.O.B.
Footnotes
[Supplemental material is available for this article. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE23912.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.115337.110.
References
- Alonso CR, Wilkins AS 2005. Opinion: the molecular elements that underlie developmental evolution. Nat Rev Genet 6: 709–715 [DOI] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger DL, Gray AD, Ream TS, Kato A, Coe EH Jr, Birchler JA 2005. Nonadditive gene expression in diploid and triploid hybrids of maize. Genetics 169: 389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbitt CC, Silverman JS, Haygood R, Reininga JM, Rockman MV, Wray GA 2009. Multiple functional variants in cis modulate PDYN expression. Mol Biol Evol 27: 465–479 [DOI] [PubMed] [Google Scholar]
- Baxter I, Brazelton JN, Yu D, Huang YS, Lahner B, Yakubova E, Li Y, Bergelson J, Borevitz JO, Nordborg M, et al. 2010. A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet 6: e1001193 doi: 10.1371/journal.pgen.1001193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Liang D, Plouffe D, Chang HS, Zhu T, Weigel D, Berry CC, Winzeler E, Chory J 2003. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res 13: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755 [DOI] [PubMed] [Google Scholar]
- Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, Wiltshire T, Su AI, Vellenga E, Wang J, Manly KF, et al. 2005. Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics.’ Nat Genet 37: 225–232 [DOI] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O'Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, et al. 2009. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell 138: 898–910 [DOI] [PubMed] [Google Scholar]
- Carvalho B, Bengtsson H, Speed TP, Irizarry RA 2007. Exploration, normalization, and genotype calls of high-density oligonucleotide SNP array data. Biostatistics 8: 485–499 [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, et al. 2005. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet 37: 233–242 [DOI] [PubMed] [Google Scholar]
- Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT 2005. Mapping determinants of human gene expression by regional and genome-wide association. Nature 437: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Affourtit J, Shockley KR, Woo Y, Churchill GA 2006. Inheritance patterns of transcript levels in F1 hybrid mice. Genetics 174: 627–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCook R, Lall S, Nettleton D, Howell SH 2006. Genetic regulation of gene expression during shoot development in Arabidopsis. Genetics 172: 1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, Thomas WK 2005. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet 37: 544–548 [DOI] [PubMed] [Google Scholar]
- Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB 2010. Rare variants create synthetic genome-wide associations. PLoS Biol 8: e1000294 doi: 10.1371/journal.pbio.1000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss S, Schadt EE, Drake TA, Lusis AJ 2005. Cis-acting expression quantitative trait loci in mice. Genome Res 15: 681–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, Huang RS, Zhang W, Bleibel WK, Roe CA, Clark TA, Chen TX, Schweitzer AC, Blume JE, Cox NJ, et al. 2008. Genetic architecture of transcript-level variation in humans. Am J Hum Genet 82: 1101–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC 2009. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G, Weir B 2005. The quantitative genetics of transcription. Trends Genet 21: 616–623 [DOI] [PubMed] [Google Scholar]
- Gibson G, Riley-Berger R, Harshman L, Kopp A, Vacha S, Nuzhdin S, Wayne M 2004. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics 167: 1791–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjuvsland AB, Plahte E, Adnoy T, Omholt SW 2010. Allele interaction—single locus genetics meets regulatory biology. PLoS ONE 5: e9379 doi: 10.1371/journal.pone.0009379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan M, Millar DS, Hedderich J, Lewis G, Newsway V, Mo N, Fryklund L, Procter AM, Krawczak M, Cooper DN 2003. Human growth hormone 1 (GH1) gene expression: Complex haplotype-dependent influence of polymorphic variation in the proximal promoter and locus control region. Hum Mutat 21: 408–423 [DOI] [PubMed] [Google Scholar]
- Huang GJ, Shifman S, Valdar W, Johannesson M, Yalcin B, Taylor MS, Taylor JM, Mott R, Flint J 2009. High resolution mapping of expression QTLs in heterogeneous stock mice in multiple tissues. Genome Res 19: 1133–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, et al. 2005. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet 37: 243–253 [DOI] [PubMed] [Google Scholar]
- Jansen RC, Nap JP 2001. Genetical genomics: the added value from segregation. Trends Genet 17: 388–391 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, Terpstra IR, Garcia JM, van den Ackerveken G, Snoek LB, Peeters AJ, Vreugdenhil D, Koornneef M, Jansen RC 2007. Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc Natl Acad Sci 104: 1708–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P, Hudson TJ, Sladek R, Majewski J 2008. Genome-wide analysis of transcript isoform variation in humans. Nat Genet 40: 225–231 [DOI] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL 2007. Genetic properties influencing the evolvability of gene expression. Science 317: 118–121 [DOI] [PubMed] [Google Scholar]
- Lewis EB 1954. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am Nat 88: 225–239 [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO 2010. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci 107: 21199–21204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, et al. 1997. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89: 737–745 [DOI] [PubMed] [Google Scholar]
- McGuire AM, Pearson MD, Neafsey DE, Galagan JE 2008. Cross-kingdom patterns of alternative splicing and splice recognition. Genome Biol 9: R50 doi: 10.1186/gb-2008-9-3-r50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks SA, Leonardson A, Zhu H, Cundiff P, Pietrusiak P, Edwards S, Phillips JW, Sachs A, Schadt EE 2004. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet 75: 1094–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET 2010. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464: 773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430: 743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastinen T, Hudson TJ 2004. Cis-acting regulatory variation in the human genome. Science 306: 647–650 [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK 2010. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464: 768–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Horton M, Huang YS, Li Y, Anastasio AE, Mulyati NW, Agren J, Bossdorf O, Byers D, Donohue K, et al. 2010a. The scale of population structure in Arabidopsis thaliana. PLoS Genet 6: e1000843 doi: 10.1371/journal.pgen.1000843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Vilhjalmsson BJ, Nordborg M 2010b. Conditions under which genome-wide association studies will be positively misleading. Genetics 186: 1045–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, McMillan L, Wang W, Parker J, Rusyn I, Threadgill D 2007. Inferring missing genotypes in large SNP panels using fast nearest-neighbor searches over sliding windows. Bioinformatics 23: i401–i407 [DOI] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L 2006. Genetics of global gene expression. Nat Rev Genet 7: 862–872 [DOI] [PubMed] [Google Scholar]
- Ronald J, Brem RB, Whittle J, Kruglyak L 2005. Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet 1: e25 doi: 10.1371/journal.pgen.0010025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD 1996. Systemic acquired resistance. Plant Cell 8: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et al. 2007. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315: 848–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Hermanson PJ, Springer NM 2007. Nonadditive expression and parent-of-origin effects identified by microarray and allele-specific expression profiling of maize endosperm. Plant Physiol 145: 411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS 2006. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci 103: 6805–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, DeCook R, Jia Y, Bancroft T, Ji T, Zhao X, Nettleton D, Schnable PS 2009. Paternal dominance of trans-eQTL influences gene expression patterns in maize hybrids. Science 326: 1118–1120 [DOI] [PubMed] [Google Scholar]
- Tao H, Cox DR, Frazer KA 2006. Allele-specific KRT1 expression is a complex trait. PLoS Genet 2: e93 doi: 10.1371/journal.pgen.0020093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J, Fedrigo O, Haygood R, Mukherjee S, Wray GA 2009. Genomic features that predict allelic imbalance in humans suggest patterns of constraint on gene expression variation. Mol Biol Evol 26: 2047–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuylsteke M, van Eeuwijk F, Van Hummelen P, Kuiper M, Zabeau M 2005. Genetic analysis of variation in gene expression in Arabidopsis thaliana. Genetics 171: 1267–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BB, Brendel V 2006. Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci 103: 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MA, Kim K, Kliebenstein DJ, van Leeuwen H, Michelmore RW, Doerge RW, St Clair DA 2007. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175: 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VL, Fisher EM, Tavare S, Odom DT 2008. Species-specific transcription in mice carrying human chromosome 21. Science 322: 434–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG 2008. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet 40: 346–350 [DOI] [PubMed] [Google Scholar]
- Wray GA 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8: 206–216 [DOI] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L 2003. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet 35: 57–64 [DOI] [PubMed] [Google Scholar]
- Zhang X, Borevitz JO 2009. Global analysis of allele-specific expression in Arabidopsis thaliana. Genetics 182: 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Byrnes JK, Gal TS, Li WH, Borevitz JO 2008. Whole genome transcriptome polymorphisms in Arabidopsis thaliana. Genome Biol 9: R165 doi: 10.1186/gb-2008-9-11-r165 [DOI] [PMC free article] [PubMed] [Google Scholar]