Abstract
Tandem 3′ UTRs produced by alternative polyadenylation (APA) play an important role in gene expression by impacting mRNA stability, translation, and translocation in cells. Several studies have investigated APA site switching in various physiological states; nevertheless, they only focused on either the genes with two known APA sites or several candidate genes. Here, we developed a strategy to study APA sites in a genome-wide fashion with second-generation sequencing technology which could not only identify new polyadenylation sites but also analyze the APA site switching of all genes, especially those with more than two APA sites. We used this strategy to explore the profiling of APA sites in two human breast cancer cell lines, MCF7 and MB231, and one cultured mammary epithelial cell line, MCF10A. More than half of the identified polyadenylation sites are not included in human poly(A) databases. While MCF7 showed shortening 3′ UTRs, more genes in MB231 switched to distal poly(A) sites. Several gene ontology (GO) terms and pathways were enriched in the list of genes with switched APA sites, including cell cycle, apoptosis, and metabolism. These results suggest a more complex regulation of APA sites in cancer cells than previously thought. In short, our novel unbiased method can be a powerful approach to cost-effectively investigate the complex mechanism of 3′ UTR switching in a genome-wide fashion among various physiological processes and diseases.
The complexity of the eukaryotic transcriptome is greatly expanded by alternative splicing of pre-mRNAs, the use of alternative transcription start sites, the use of alternative polyadenylation (APA) sites, and other processes (Nilsen and Graveley 2010). It has been shown that more than half of human and mouse genes have multiple poly(A) sites (Tian et al. 2005; Lutz 2008), which could generate multiple mRNA isoforms from a single gene. APA sites may impact the protein sequences if the cleavage sites are located in the intron or internal exons. Another kind of APA site (herein called tandem APA sites), located in the last exon, results in tandem 3′ UTRs with variable lengths (Tian et al. 2005). Tandem 3′ UTRs play an important role in regulating the gene expression network because they may lead to the loss of regulatory elements, especially microRNA binding sites (Sandberg et al. 2008) in the 3′ UTR. Recent studies have shown that the cells in various physiological states, such as activated T lymphocytes (Sandberg et al. 2008) and neurons (Flavell et al. 2008), tumor cells (Mayr and Bartel 2009), and embryonic cells (Ji et al. 2009; Mangone et al. 2010), are inclined to use the shorter 3′ UTR, benefiting from the increased stability due to the loss of microRNA binding sites. These studies (Sandberg et al. 2008; Ji et al. 2009) were based on the prior knowledge of APAs inferred from the EST database or only focused on some candidate genes (Mayr and Bartel 2009). Moreover, only genes with two known APA sites could be investigated, and some of these genes may harbor other unknown APA sites, which may severely reduce the power of the conventional methods used in these studies.
Recently, second-generation sequencing technology has largely improved our understanding of the prevalence of alternative splicing (Castle et al. 2008; Kwan et al. 2008; Pan et al. 2008; Sultan et al. 2008; Wang et al. 2008) and alternative transcription start sites (Ni et al. 2010). Transcriptome sequencing can identify not only alternative splicing events but also APA sites (Pickrell et al. 2010). However, transcriptome sequencing is a very expensive but ineffective method for the study of APA sites because only a very small number of the reads are tagged with poly(A) tails.
To date, there is still no comprehensive method to analyze APA site switching, including genes with multiple APA sites, in a genome-wide fashion. Here, we describe a novel strategy of sequencing APA sites (SAPAS) with second-generation sequencing technology and a bioinformatic pipeline to analyze the sequencing data. By directly sequencing the 3′ ends of mRNA, our method has the potential to complete genome-wide profiling of APA sites and to identify new poly(A) sites. To apply this new method, we conducted Illumina GA IIx sequencing to conduct genome-wide profiling of APA sites of two human breast cancer lines (MCF7 and MB231) and one cultured human mammary epithelial normal cell line (MCF10A). MCF7 expresses the estrogen receptor (ER) and is an estrogen-sensitive breast cancer line, but MB231 is estrogen-independent and highly invasive. MCF10A is a nontumorigenic epithelial cell line and is frequently used as a normal control in breast cancer research. The proliferation rate of MCF10A is comparable to that of the cancer cell lines and is higher than that of other nontumorigenic epithelial cell lines, such as HMEC (Ramljak et al. 2005) and 184A1 (Bhaskaran et al. 2009). We found a completely opposite pattern of tandem 3′ UTR length between MCF7 and MB231, suggesting a more complex regulation of APA sites in cancer than previously thought. The global matched enrichment of gene ontology terms and pathways in the genes with switched tandem 3′ UTRs indicate the importance of APA site regulation in cancer transformation. Furthermore, we also found some motifs that may contribute to switching.
Results
Deep sequencing of 3′ ends of mRNA
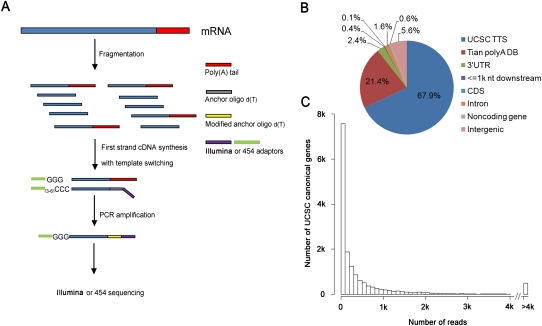
Briefly, in this SAPAS method (Fig. 1A), the total RNA is first fragmented by heating, and then a template switch reverse transcription (RT) reaction is carried out to generate first strand cDNA. The RT reaction is optimized to improve the anchoring effect, which can reduce the proportion of cDNA with long poly(A) tails. After PCR with modified oligo d(T) tagged with sequencing primers, size-selection of fragments of 200–300 bp is performed. The fragments can be sequenced from the 5′ end with 454 Life Sciences (Roche) pyrosequencing technology or from the 3′ end with Illumina GA IIx.
Figure 1.
SAPAS strategy. (A) Experiment outline. (B) Genomic locations of reads that were uniquely mapped to the nuclear genome after internal priming filtering. (C) Histogram of the number of reads for UCSC canonical genes.
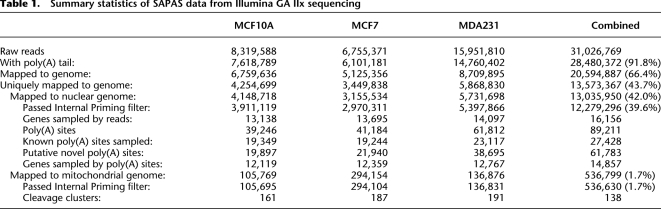
We used the SAPAS strategy to profile the APA sites of two human breast cancer lines and one cultured human mammary epithelial cell line. In total, we obtained 31 million raw reads with lengths of 75 bp from Illumina sequencing; a statistical summary of the data is shown in Table 1. A process of poly(A) site identification was employed (see details in Methods). The modified anchor oligo d(T) was detected in about 28.5 million (91.8%) reads, of which about 13 million reads uniquely mapped to the human nucleus genome (hg19). After filtering the reads with internal priming, we obtained 12.3 million reads that could be used directly to infer transcript cleavage sites.
Table 1.
Summary statistics of SAPAS data from Illumina GA IIx sequencing
The majority of the filtered reads (89.3%) from our experiments mapped to known poly(A) sites listed in the UCSC transcripts ends database and Tian's database (Tian et al. 2005), and an additional 2.4% and 0.5% reads mapped to the 3′ UTR and 1 kb downstream from the UCSC canonical genes, respectively (Fig. 1B). Additionally, 16,156 UCSC canonical genes were sequenced by at least one read, representing 60% of all canonical genes. These results suggest that our SAPAS method can effectively extract the 3′ ends of mRNAs.
Importantly, as a special RNA-seq method, the SAPAS method could also be used to measure the gene expression level in a genome-wide fashion. Since it only sequences the 3′ ends of mRNAs, we do not need to consider the problem of the mapped length of genes. The distribution of the number of reads is shown in Figure 1C. We compared our results for MCF7 and MB231 with previous microarray data (Li et al. 2009). The Spearman correlations were 0.77 and 0.80 for the two cell lines, respectively (Supplemental Fig. 1A,B). Furthermore, for the MCF7 cell line, our result is also highly correlated to previous RNA-seq data (Wang et al. 2008) (R = 0.73) (Supplemental Fig. 1C). Pairwise comparisons of gene expression among the three cell lines were done. It is very interesting that the cell cycle–related GO categories are enriched in the genes with differential expression levels (Supplemental Tables 1–3).
We also sequenced the 3′ UTRs of MCF10A and MCF7 with the SAPAS method using 454 pyrosequencing technology (Supplemental Table 4). However, the data generated via 454 was limited compared with that obtained via Illumina technology; therefore, we only focused on the Illumina GA IIx data for the following analysis.
Characterization of poly(A) sites
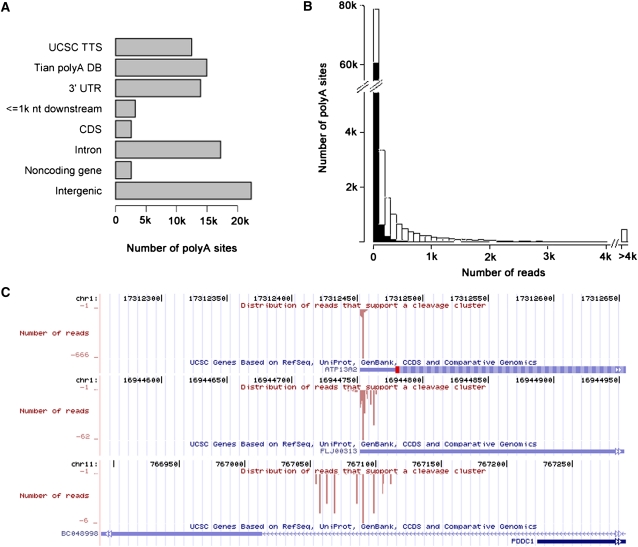
Due to the heterogeneity of the cleavage sites at poly(A) sites, we performed a modified snowball-like clustering (Tian et al. 2005) and took the cleavage clusters with more than one read as poly(A) sites. In total, we identified 89,211 poly(A) sites from the three cell lines, and we found only 30.7% of them in the UCSC and Tian's databases (Fig. 2A), suggesting that the SAPAS method could identify the new poly(A) sites much more effectively, even though the expression levels of these new sites are lower than those of the known sites (Fig. 2B). We also noticed that 5776 genes had more than one tandem APA site, among which 3635 genes harbored more than two tandem APA sites. For the 3443 genes that had two tandem APA sites in Tian's database, we found new tandem APA sites in 1248 (36%) of them. We did 3′ RACE on seven novel APA sites, and all of them were confirmed. One-quarter of the poly(A) sites are located in intergenic regions; this result is not unexpected because transcripts were already observed in the intergenic region (Bertone et al. 2004). Consistently, most of these poly(A) sites were located within 5 kb downstream from the RNA-seq data (Supplemental Table 5), which suggested that these poly(A) sites were real.
Figure 2.
Poly(A) site characteristics. (A) Genomic locations of poly(A) sites. (B) Distribution of poly(A) site expression levels. (C) Examples of type A, B, and C poly(A) sites, which are strong peak, weak peak, and no peak sites, respectively.
The width of the cleavage clusters shows a positive skewed distribution (Supplemental Fig. 2). Most of cleavage clusters have narrow read distributions, whereas some of them have relatively wide and uniform read distributions. To investigate effects of the polyadenylation signals on this difference, we focused on the poly(A) sites with more than 30 reads. For this purpose, we first classified the poly(A) sites into three types: (A) a strong peak cleavage cluster with more than half of the reads falling within 3 nt, (B) a weak peak cleavage cluster with more than one-third of the reads falling within 3 nt, and (C) other cleavage clusters (Fig. 2C). In total, we found 16,127, 2475, and 276 poly(A) sites of type A, B, and C, respectively.
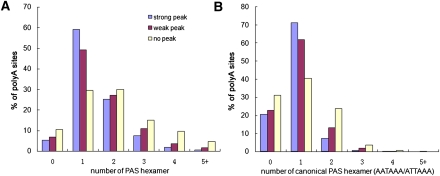
Previously, twelve variant polyadenylation signals including the hexamer AAUAAA were identified, and their efficiencies were determined (Beaudoing et al. 2000). Here, we analyzed the distribution difference of these polyadenylation signals on poly(A) sites corresponding to the three types. As shown in Supplemental Figure 3, the hexamers AAUAAA and AUUAAA were more common than the others, especially for the type A sites, which is consistent with the results of previous studies (Beaudoing et al. 2000; Tian et al. 2005). The polyadenylation signals of the type C sites have a flatter position distribution than the other types, suggesting a higher heterogeneity of type C sites. The distributions of the number of polyadenylation signals are shown in Figure 3. The average numbers of polyadenylation signals of the three types were 1.432, 1.610, and 2.006, respectively, and the differences were statistically significant, according to bootstrap tests. Furthermore, it should be noted that the number of signals for type C sites was severely underestimated because of their heterogeneity and broader read distribution.
Figure 3.
Histogram of the signal number of poly(A) sites. Motifs were searched in the upstream 50 nt of the cleavage cluster range combined with the range itself. (A) Canonical AATAAA and ATTAAA sequences and the other 10 variants were all considered. (B) Only the canonical signal AATAAA and ATTAAA were considered.
Differential usage of poly(A) sites among the three cell lines
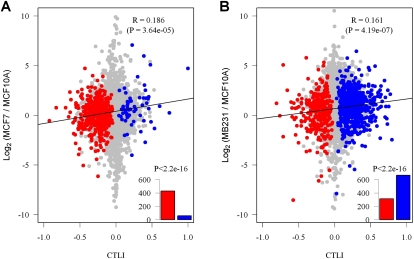
Several previous studies discovered that shortened 3′ UTRs are associated with elevated cell proliferation rates (Sandberg et al. 2008; Ji et al. 2009) or transformation (Mayr and Bartel 2009). Here, we compared the tandem 3′ UTR length of the two cancer cell lines MCF7 and MB231 to that of the normal cell line MCF10A. Because some genes have more than two APA sites and the tandem 3′ UTR length is a quantitative variable, we performed the test of linear trend alternative to independence (Agresti 2002) instead of a normal chi-squared (χ2) test. We denoted the normal cell line MCF10A as 1 and the cancer cell lines as 2. As a result, a positive Pearson correlation r indicates that the cancer cell line harbors longer tandem 3′ UTRs than the normal cell line, and a negative Pearson correlation r indicates that the cancer cell line harbors shorter tandem 3′ UTRs than the normal cell line. Here, we defined this Pearson correlation r as the cancer tandem 3′ UTR length index (CTLI). We found 489 genes [false discovery rate (FDR) = 0.01] with a significant difference in the tandem 3′ UTR length between MCF7 and MCF10A, and the CTLIs of 88% (P < 2.2×10−16 with a binomial test) of these genes were negative (Fig. 4). The switching to shorter 3′ UTRs in MCF7 is consistent with the hypothesis that transformed cells or highly proliferative cells tend to use shortened UTRs. However, among the 977 genes (FDR = 0.01) with a difference in the 3′ UTR length between MB231 and MCF10A, opposite trends of switching were found (only 32% of the genes had negative CTLIs, P < 2.2×10−16 with the binomial test) (Fig. 4). We also compared the 3′ UTR length between MCF7 and MB231. The result showed that, among 1262 genes (FDR = 0.01) with significantly different 3′ UTR lengths, 1122 genes switched to longer 3′ UTRs in MB231. The details of the genes with significant differences among the three cell lines are shown in Supplemental Tables 6–8.
Figure 4.
APA site switching and gene expression levels of normal and cancer cell lines. Cancer tandem 3′ UTR length index (CTLI) is plotted against the logarithm of the expression level ratios between the cancer ([A] MCF7 and [B] MB231) and normal (MCF10A) cell lines. The x-axis denotes CTLI; a larger positive value indicates that longer tandem UTRs are prone to be used in the cancer samples. Genes with significant switching to longer (blue) or shorter (red) tandem UTRs in cancer samples (FDR = 0.01; see Methods) are colored. The y-axis denotes the logarithm of the expression level of genes from the cancer sample relative to the normal sample.
The trend of 3′ UTR switching in the MB231 cell line was so disparate that we were very careful to validate the results. First, we performed quantitative RT-PCR to validate our SAPAS method. Eight genes of MCF7 and MB231 with extreme 3′ UTR length differences relative to MCF10A were chosen. The PCR of one gene failed. Among the remaining seven genes, six of them were confirmed (Supplemental Figs. 4–5). Second, we compared our SAPAS results with Mayr and Bartel's results on MCF7 and MB231 (Mayr and Bartel 2009). They used Northern blotting to measure the expression of different isoforms of six genes with various APA sites. As for the MCF7 and MB231 cells, they successfully quantified three and five genes, respectively (Supplemental Table 9). All of the genes had shorter 3′ UTRs relative to the MCF10A cell line, except for the RAB10 gene in MB231. Our results covered three of these five genes in both MCF7 and MB231. Among the six pairwise comparisons of our results with Mayr and Bartel's, only two genes, DICER1 and RAB10, in MB231 showed a different trend. DICER1, which had a shorter 3′ UTR in MB231 in Mayr and Bartel's data, was not different between MB231 and MCF7, according to our data. The cause of this conflicting result may be that only the shortest and longest isoforms were used in Mayr and Bartel's results, and no intermediate isoforms were considered. When we only took the shortest and longest isoforms into consideration, the result of the shorter 3′ UTR of DICER1 in MB231 appeared. We performed qRT-PCR on these two genes, and the results revealed that both DICER1 and RAB10 were prone to use distal APA sites (Supplemental Fig. 5). In the end, we compared the CTLI values of the genes between the Illumina GA IIx and 454 data sets. A positive correlation was observed (R = 0.243), though it was weak (Supplemental Fig. 6).
One of the obvious effects of 3′ UTR switching may be the gain or loss of miRNA binding sites, which may impact the stability of mRNA and translation. Flavell and coworkers (Flavell et al. 2008) found shorter 3′ UTRs in MEF2-activated genes in neurons, suggesting a correlation between the 3′ UTR length and the gene expression level. For the genes with switched APA sites according to our data, we did not observe a negative correlation between the 3′ UTR length and gene expression (Fig. 4). This result is consistent with those of a previous study on the expression of miRNAs and their target genes (Wu et al. 2009). Another possible consequence of the APA site switching may be changes in protein production, which is of particular interest to be investigated further.
Functional annotation analysis of the genes with switched APA sites
To further understand the biological significance of such switching of APA sites, we conducted a functional annotation of the above genes with DAVID Bioinformatics Resources (Supplemental Tables 10, 11; Huang et al. 2009). In the list of genes with longer 3′ UTRs in the MB231 cell line, 30 genes are associated with apoptosis or programmed cell death, leading to the significant enrichment of cell death–related GO terms. More interestingly, the genes involved in the caspase pathway were found to be enriched (P = 0.08; Supplemental Fig. 7), with four downstream genes (Caspase 6, DFFA [ICAD], DFFB [CAD], and PARP1) switched to distal APA sites in MB231. The lengthening 3′ UTRs of these genes might contribute to the escape of apoptosis of the MB231 cell line.
Cyclin D1 (Rosenwald et al. 2003; Mayr and Bartel 2009) and cyclin D2 (Mayr and Bartel 2009), two members of cyclin-CDK complexes that control passing through the cell cycle, were identified as having shortened 3′ UTRs in cancer cells. Here, we also observed the enrichment of mitotic cell cycle–related GO terms in the list of genes with switched APA sites in both the MCF7 and MB231 cell lines (Supplemental Tables 10–11). Cyclin D1 and CDK6 in MCF7 and cyclin D1 and cyclin A2 in MB231 are switched to shorter 3′ UTRs. Moreover, the CDK inhibitor CDKN2C (p18) gene is prone to use the distant APA sites in MB231 cells. The APA site switching of these genes might promote the cell cycling of the two cancer cell lines. However, the ANAPC5 and ANAPC13 genes encoding subunits of APC, an ubiquitin ligase, are, respectively, switched to shorter and longer 3′ UTRs in MB231 cells. Additionally, the CDC25B and CDC25C genes, which activate cyclin-CDK (Karlsson-Rosenthal and Millar 2006), are prone to use the longer 3′ UTR in MB231 cells. These data indicate the complexity of APA site switching in cell transformation.
One of the characteristics of the cancer cells is their preference for aerobic glycolysis instead of the mitochondrial tricarboxylic acid (TCA) cycle, which is usually called the Warburg effect (Warburg 1956). Vander Heiden and coworkers (Vander Heiden et al. 2009) proposed that the Warburg effect promotes “the uptake and incorporation of nutrients into the biomass (e.g., nucleotides, amino acids, and lipids).” Here, we also found that metabolic-related GO terms were enriched in the genes with shortened 3′ UTRs in the MCF7 cell line. The most interesting finding is the GO terms related to the regulation of glucose import and transport. The switching to proximal APA sites in these genes may be related to the Warburg effect in cancer cells.
Another noted pathway is the antigen processing and presentation pathway; seven genes from this pathway were found to be switched to shorter 3′ UTRs in the MCF7 cell line (P = 0.007; Supplemental Fig. 8). The down-regulated expression of HLA class I genes has been widely reported in many kinds of cancer cells or tissues. This down-regulation could help the cancer cell to escape elimination by cytotoxic T cells, though complete loss of HLA class I molecules could also induce NK cell lysis (See reviews in Aptsiauri et al. [2007a]; Aptsiauri et al. [2007b]). Moreover, the up-regulation of HLA-G (a nonclassical class I gene) has also been reported, and this up-regulation may contribute to the escape from NK cell lysis (Rouas-Freiss et al. 2003). Our results indicate that 3′ UTR switching in cancer cells may contribute to the regulation of the expression of HLA class I genes, which may then result in escape from immune surveillance.
Discussion
Here, we developed a SAPAS strategy to study the APA sites with second-generation sequencing technology. Our method has several advantages over previous methods. It can not only detect APA switching between samples but also identify new poly(A) sites in a genome-wide fashion, while microarray techniques can only analyze known sites. In addition, we can use this new method to quantify the expression levels of mRNAs with various 3′ UTRs. By simultaneously measuring the expression and position of APA sites, our method can efficiently analyze genes with multiple APA sites. Exon array (Sandberg et al. 2008) and transcriptome sequencing (Wang et al. 2008) can only study genes with two known APA sites, and complex statistical tools must be implemented to analyze these data. Although the reads with poly(A) from transcriptome sequencing can also be used to identify novel APA sites (Pickrell et al. 2010), the cost is much higher. Northern blotting has traditionally been used to identify APA switching (Chuvpilo et al. 1999; Mayr and Bartel 2009), but it can only take the shortest and longest isoforms into consideration (Mayr and Bartel 2009), leading to some bias (for example, the DICER1 gene between MB231 and MCF10A). While we were preparing our manuscript, another group published a different 3′ end-capturing method based on 454 sequencing technology (Mangone et al. 2010). They used the 4-bp recognition enzyme DpnII to release the 3′ end of the cDNA, which is also used in the SAGE method. However, the SAGE method has been found to have some bias because of incomplete digestion and enzyme site location (Zaretzki et al. 2010). Torres et al. (2008) also showed the lack of short (less than ∼80 bp) and long (more than ∼300–400 bp) fragments with enzyme digestion compared with nebulization. Thus, our new SAPAS method may be more powerful in a genome-wide study of APA sites, including genome annotation and the regulation of APA site switching in association with many biological processes and diseases.
We also first implemented the test of linear trend alternative to independence in the analysis of UTR length difference between samples. Considering both the proportion of reads in APA sites and their positions, this method is more conservative than the Fisher exact test or χ2 test. For example, assume there are three sites in a gene, and the lengths of 3′ UTRs are 300, 320, and 800. If the numbers of reads are 20, 50, and 30 for sample A, and 50, 20, and 30 for sample B, it is obvious that the 3′ UTR length of this gene between the two samples is not significantly different. However, the P-value from the χ2 test is 2.067 × 10−6, while it is not significant in our linear model (P = 0.85). If we only consider the first and last APA sites in performing the Fisher exact test or χ2 test, the power would be dramatically reduced due to the loss of information of the middle sites.
The global functional enrichment indicates the importance of 3′ UTR switching in cancer cell transformation and proliferation. The different trends of APA site switching between MCF7 and MB231 cells compared to MCF10A cells suggest that cancer cells may not have a simple trend of shortening 3′ tandem UTRs. Although both of the cancer cell lines showed shortened 3′ UTRs in some cell cycle–related genes, different enrichments of GO terms and pathways were found in the APA-switched genes. The differences could be explained by shifting balance or adaptive landscape theory (Wright 1988). Under the adaptive landscape theory, transformation of a normal cell to a cancer cell is an evolutionary, dynamic process driven by some driver-mutations under various environmental selection forces. The cells may transit from one state of gene expression to another on the adaptive landscape by some gene mutation or by responding to changes in its microenvironment (Demicheli and Coradini 2010). The mutations and the changes in microenvironment may only destabilize the present state of gene expression, and natural selection will drive the cells to another adaptive state. MCF7, which is estrogen-sensitive, and MB231, which is estrogen-independent and highly invasive, were isolated from different individuals. These observations indicate that the two cell lines may be at different states on the adaptive landscape. Therefore, it is not surprising that the two cell lines showed different profiles of APA sites and enriched GO terms.
Methods
Cell cultures
Two breast cancer cell lines, MCF7 and MB231, were cultured in Dulbecco's modified Eagle's medium (DMEM), and a human normal mammary epithelial cell line (MCF10A) was cultured in monolayers in DMEM/F12.
SAPAS library preparation
Total RNA was extracted from cells using QIAGEN RNeasy Mini kits, and ∼10 μg total RNA was randomly fragmented by heating (Cloonan et al. 2008). A template switch reverse transcription (RT) reaction was carried out to generate first strand cDNA with SuperScript II from Invitrogen using an anchored oligo d(T) primer and a 5′ template switching adaptor. The 5′ ends of the primers were tagged with 454 or Illumina adaptors. PCR was then performed to amplify the cDNA and to introduce mutations in the poly(A), and the number of cycles was determined to ensure that the ds cDNA remained in the exponential phase of amplification. After PCR with sequencing primers, size-selection of fragments of 200–300 bp with PAGE gel-excision was performed. The final pooled fragments were sequenced from the 5′ end with 454 or from the 3′ end with Illumina GA IIx. All of the primers are listed in Supplemental Table 13.
3′ RACE and qRT-PCR
Five novel APA sites with relative higher expression levels were chosen, and 3′ RACE was done. The PCR products were sequenced with ABI3730. Five genes with extreme 3′ UTR length differences between MCF10A and the two cancer cell lines were chosen. The poly(A) sites of these genes were divided into two supersites (the proximal and distal sites), and the region upstream of the supersites was targeted for qRT-PCR. All of the primers are listed in Supplemental Table 14.
Data analysis
Filtering and trimming of the reads was performed with Perl scripts, and the trimmed reads were mapped to the human genome (hg19) with Bowtie (Langmead et al. 2009). Cleavage sites were clustered into poly(A) sites as described previously (Tian et al. 2005). 3′ UTR switching for each gene between the cancer cell lines and the normal cell line was detected by a test of linear trend alternative to independence (Agresti 2002). The false discovery rate (FDR) of BH was estimated with R software. Functional analysis of the genes with switched 3′ UTRs was performed by DAVID Bioinformatics Resources (Huang et al. 2009).
Acknowledgments
We are grateful to Dr. Erwei Song and Dr. Qiang Liu for the cell lines. This work was supported by Fundamental Research Funds for the Central Universities (to Y.F.), the National Natural Science Foundation of China (No. 30801012) (to Y.F.), the key project of the National Natural Science Foundation of China (No. 30730089) (to A.X.), and the National Basic Research Program of China (973 Program) (No. 2011CB946101 and 2007CB815800) (to A.X.).
Footnotes
[Supplemental material is available for this article. The sequencing data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA023826.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.115295.110.
References
- Agresti A 2002. Categorical data analysis. Wiley-Interscience, New York [Google Scholar]
- Aptsiauri N, Cabrera T, Garcia-Lora A, Lopez-Nevot MA, Ruiz-Cabello F, Garrido F 2007a. MHC class I antigens and immune surveillance in transformed cells. Int Rev Cytol 256: 139–189 [DOI] [PubMed] [Google Scholar]
- Aptsiauri N, Cabrera T, Mendez R, Garcia-Lora A, Ruiz-Cabello F, Garrido F 2007b. Role of altered expression of HLA class I molecules in cancer progression. Adv Exp Med Biol 601: 123–131 [DOI] [PubMed] [Google Scholar]
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res 10: 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Bhaskaran N, Lin KW, Gautier A, Woksepp H, Hellman U, Souchelnytskyi S 2009. Comparative proteome profiling of MCF10A and 184A1 human breast epithelial cells emphasized involvement of CDK4 and cyclin D3 in cell proliferation. Proteomics Clin Appl 3: 68–77 [DOI] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM 2008. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet 40: 1416–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuvpilo S, Zimmer M, Kerstan A, Glockner J, Avots A, Escher C, Fischer C, Inashkina I, Jankevics E, Berberich-Siebelt F, et al. 1999. Alternative polyadenylation events contribute to the induction of NF-ATc in effector T cells. Immunity 10: 261–269 [DOI] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. 2008. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5: 613–619 [DOI] [PubMed] [Google Scholar]
- Demicheli R, Coradini D 2010. Gene regulatory networks: A new conceptual framework to analyse breast cancer behaviour. Ann Oncol. doi: 10.1093/annonc/mdq546 [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME 2008. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson-Rosenthal C, Millar JB 2006. Cdc25: Mechanisms of checkpoint inhibition and recovery. Trends Cell Biol 16: 285–292 [DOI] [PubMed] [Google Scholar]
- Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P, Hudson TJ, Sladek R, Majewski J 2008. Genome-wide analysis of transcript isoform variation in humans. Nat Genet 40: 225–231 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gao F, Li N, Li S, Yin G, Tian G, Jia S, Wang K, Zhang X, Yang H, et al. 2009. An improved method for genome wide DNA methylation profiling correlated to transcription and genomic instability in two breast cancer cell lines. BMC Genomics 10: 223 doi: 10.1186/1471-2164-10-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS 2008. Alternative polyadenylation: A twist on mRNA 3′ end formation. ACS Chem Biol 3: 609–617 [DOI] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T, Corcoran DL, Rach EA, Song S, Spana EP, Gao Y, Ohler U, Zhu J 2010. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat Methods 7: 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK 2010. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464: 768–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramljak D, Romanczyk LJ, Metheny-Barlow LJ, Thompson N, Knezevic V, Galperin M, Ramesh A, Dickson RB 2005. Pentameric procyanidin from Theobroma cacao selectively inhibits growth of human breast cancer cells. Mol Cancer Ther 4: 537–546 [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink HK, Smeland EB, et al. 2003. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell 3: 185–197 [DOI] [PubMed] [Google Scholar]
- Rouas-Freiss N, Moreau P, Menier C, Carosella ED 2003. HLA-G in cancer: A way to turn off the immune system. Semin Cancer Biol 13: 325–336 [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. 2008. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321: 956–960 [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres TT, Metta M, Ottenwalder B, Schlotterer C 2008. Gene expression profiling by massively parallel sequencing. Genome Res 18: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O 1956. On the origin of cancer cells. Science 123: 309–314 [DOI] [PubMed] [Google Scholar]
- Wright S 1988. Surfaces of selective value revisited. Am Nat 131: 115–123 [Google Scholar]
- Wu CI, Shen Y, Tang T 2009. Evolution under canalization and the dual roles of microRNAs: A hypothesis. Genome Res 19: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaretzki RL, Gilchrist MA, Briggs WM, Armagan A 2010. Bias correction and Bayesian analysis of aggregate counts in SAGE libraries. BMC Bioinformatics 11: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]