Abstract
Massively parallel sequencing is a useful approach for characterizing T-cell receptor diversity. However, immune receptors are extraordinarily difficult sequencing targets because any given receptor variant may be present in very low abundance and may differ legitimately by only a single nucleotide. We show that the sensitivity of sequence-based repertoire profiling is limited by both sequencing depth and sequencing accuracy. At two timepoints, 1 wk apart, we isolated bulk PBMC plus naïve (CD45RA+/CD45RO−) and memory (CD45RA−/CD45RO+) T-cell subsets from a healthy donor. From T-cell receptor beta chain (TCRB) mRNA we constructed and sequenced multiple libraries to obtain a total of 1.7 billion paired sequence reads. The sequencing error rate was determined empirically and used to inform a high stringency data filtering procedure. The error filtered data yielded 1,061,522 distinct TCRB nucleotide sequences from this subject which establishes a new, directly measured, lower limit on individual T-cell repertoire size and provides a useful reference set of sequences for repertoire analysis. TCRB nucleotide sequences obtained from two additional donors were compared to those from the first donor and revealed limited sharing (up to 1.1%) of nucleotide sequences among donors, but substantially higher sharing (up to 14.2%) of inferred amino acid sequences. For each donor, shared amino acid sequences were encoded by a much larger diversity of nucleotide sequences than were unshared amino acid sequences. We also observed a highly statistically significant association between numbers of shared sequences and shared HLA class I alleles.
T lymphocytes are key mediators of adaptive immunity that recognize heterologous cells expressing foreign or mutated proteins. Recognition is mediated by the interaction of cell surface molecules, whereby a heterodimeric T-cell receptor (TCR) on the surface of a T lymphocyte will bind to a protein degradation product from the heterologous cell that is presented at the surface of that cell by the major histocompatibility complex (MHC). To generate a repertoire of structurally variant TCRs capable of recognizing diverse peptide–MHC (pMHC) complexes, the locus encoding the receptor undergoes somatic recombination among the Variable (V), Diversity (D), and Joining (J) gene segments, plus the addition/subtraction of nontemplated bases at recombination junctions (Davis and Bjorkman 1988; Bassing et al. 2002). The process is directly analogous to the generation of antibody diversity by somatic VDJ recombination of the B-cell receptor locus. Like antibody diversity, the potential for TCR diversity is nearly infinite, but actual diversity in a biological repertoire is restricted by deletion of over- and under-reactive cells during thymic maturation and is molded continuously by the clonal expansion of antigen responsive cells in the periphery (Nikolich-Zugich et al. 2004; Harty and Badovinac 2008).
By allelic exclusion, a T cell typically expresses only a single TCRB variant (Khor and Sleckman 2002), making beta-chain sequence variation a useful measure of T-cell repertoire diversity. The vast majority of TCRB variation is within the CDR3 (Complementarity Determining Region 3), which encompasses the VDJ recombination junctions and encodes the portion of the TCR that directly contacts pMHC (Davis and Bjorkman 1988). We use the sequence of the CDR3 plus the identity of the flanking V and J gene segments to uniquely classify TCRB variants.
Sequence diversity in both T-cell and B-cell immune repertoires has been surveyed previously (Boyd et al. 2009; Freeman et al. 2009; Robins et al. 2009; Klarenbeek et al. 2010; Wang et al. 2010) but not exhaustively sequenced. Here, we analyze TCR beta-chain sequences from peripheral blood from a single healthy individual, and we compare this immune repertoire to survey sequence from two other healthy individuals. We find that by exhaustive sequencing and careful mitigation of sequencing error, it is possible to saturate the diversity within a single sequencing library and within a sample of peripheral blood. However, determining the true size of an immune repertoire by exhaustive sequencing is intractable because a repertoire can only be subsampled, and by the nature of “next-generation” sequencing technologies where sequencing errors are incurred at a constant rate, it is not possible to distinguish very rare sequences from sequencing errors. Still, immune repertoire analysis by massively parallel sequencing offers tremendous utility, where distinct clonotypes can be readily identified and tracked, diversity can be profiled, and differences among individuals or subsets of sorted cells can be readily distinguished. All data from the present study have been made available as a community resource to facilitate future comparative studies of immune repertoires.
Results
With informed consent, we isolated PBMCs (peripheral blood mononuclear cells) from 20 mL of peripheral blood samples. These samples were obtained at two timepoints, 1 wk apart, from unrelated Caucasian donors (age 29–33 yr) with no self-declared immune-related disorder. Total RNA was isolated from ficoll-purified PBMCs and reverse transcribed using a 3′ primer specific for the two conserved TCR beta-chain C genes. A 5′ priming site was added to cDNA molecules during reverse transcription by template switching (Peters et al. 1999). The TCRB sequence was then amplified by PCR and directionally sheared to remove uninformative V gene nucleotides, leaving the distal part of the V gene, the informative CDR3 sequence, and the J segment (TRBJ) intact. This procedure shortened templates to ∼130–180 bp, a length appropriate for Illumina library construction and paired-end sequencing, and allowed double-strand coverage of the critical CDR3 region.
Recognition and mitigation of sequence error is essential for accurate repertoire enumeration
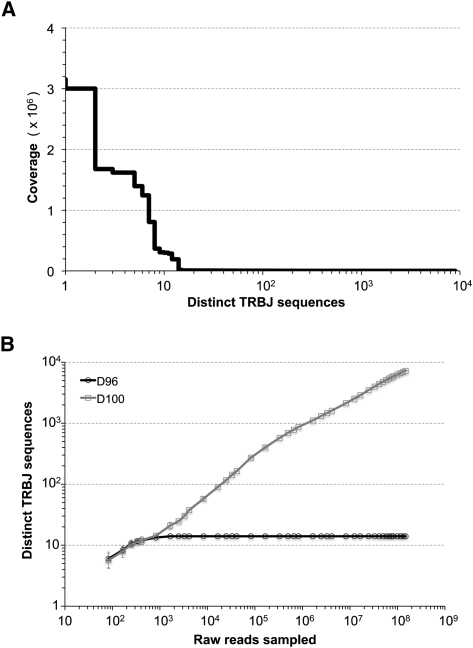
Immune receptors are extraordinarily difficult sequencing targets because any given receptor variant may be present in very low abundance and may differ legitimately from other receptor variants by only a single nucleotide. The first PBMC sample from the first donor, a healthy 29-yr-old male, contained ∼12 million αβ T-cells. By using standard procedures, we constructed an Illumina sequencing library from the shortened 5′-RACE products from this first donor and ran six lanes of Illumina GAIIx sequence to obtain 142.1 million pairs of reads. To assess sequence accuracy, we aligned raw reads to the known TCR J gene segments, of which there are 13 annotated in the human genome (Flicek et al. 2010). From aligned raw single pass reads, we observed 9.4 errors per kilobase, but when we added the requirements of (1) double-strand coverage, (2) minimum quality score (Ewing and Green 1998) of Q30, and (3) no high-quality discrepancy between strands at any position, the error rate fell to 2.2 errors per kilobase. However, when we realigned the quality-filtered data to the 13 reference J gene segments, this residual sequence error still generated thousands of distinct J genes (Fig. 1A). The presence of a huge excess of distinct sequences that are artifacts is clearly problematic when the goal is to enumerate the number of real distinct sequences in a sample. We found that even more aggressive quality filtering (i.e., a higher Q value threshold) was not helpful and that redundancy was the only useful metric for distinguishing real from erroneous sequences. We observed that the 13 J segments were represented by 96% of the data, with 1.1 ± 0.9 (mean ± SD) million–fold coverage, and the remaining 4% of the data represented all of the thousands of artifactual J sequences, with 78 ± 448 (mean ± SD)-fold coverage. Thus, by restricting the data set to those distinct sequences that are represented by 96% of the data, all real sequences were retained and all erroneous sequences were removed (Fig. 1B). We call this a D96 cutoff. A D50 cutoff, for example, would restrict the data set to fewer but higher copy number sequences, which together would account for 50% of the data and could not be artifacts because the same sequence errors could not, by chance, have been incurred at the same position so many times. A D100 data set would be unrestricted but highly error prone because it would contain many distinct sequences at low copy number, where sequences were grouped together because of the chance occurrence of the same sequencing error at the same position not because they represent the same T-cell clone. The corollary of this feature of the data is that it is not possible to distinguish between sequences from very rare T cells and sequencing errors.
Figure 1.
Distinct TCRB sequences. (A) Sequence coverage of distinct TRBJ sequences observed upon alignment of Illumina reads quality filtered to >99.9% predicted accuracy (Q30). There was substantial coverage, up to several million-fold, of the 13 human TRBJ segments, but thousands of artifactual distinct TRBJ sequences were also observed, arising from residual sequencing error. (B) Restricting the data set to D96 effectively retains all real TRBJ sequences and excludes all of the artifactual TRBJ sequences observed when no coverage restriction is applied (D100).
Having determined, empirically, that D96 is an appropriate cutoff for eliminating artifactual sequences, we applied the D96 restriction to CDR3, the highly variable region of TCRB that comprises the VDJ recombination junctions and for which no reference sequence exists. The error model and threshold derived from analysis of J sequence will be even more stringent when applied to CDR3 because CDR3 is located in the middle of the 5′-RACE amplicon, where double-stranded coverage and consensus quality are highest. We used the standard definition of CDR3 as the region between the last conserved cysteine of the V gene and the first conserved phenylalanine of the J gene in the conserved motif FGXG, and we defined a distinct TCRB sequence as having a precise VDJ rearrangement, effectively captured by the CDR3 sequence signature, flanking V and J genes, and deleted 3′V and 5′J bases. The D gene segment is too short to reliably annotate and is contained within CDR3 variation.
TCRB diversity is greater than that captured by a single library or a single blood sample
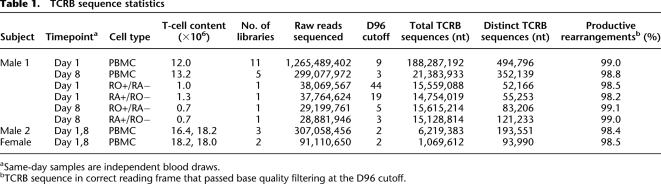
Quality filtering and D96 restriction of 142.1 million raw reads pairs obtained from the first library yielded 181,258 distinct TCRB sequences. Saturation analysis predicted that even deeper sequencing would not produce very many new sequences (Supplemental Fig. S1); however when we obtained an additional lane of Illumina data from a second library that had been constructed from the same blood sample, we observed, unexpectedly, that 74.8% of quality filtered TCRB sequences from this second library were novel. Library exhaustion is a recognized but under-reported phenomenon of massively parallel sequencing, and at least for the present application, our data clearly show that a single library may not adequately capture the diversity of a biological sample. Recognizing this limitation, we constructed 10 additional libraries, consuming all of the starting PBMC RNA from the first blood sample from this donor. We sequenced one to two lanes from each of the new libraries to obtain 632.7 million pairs of raw reads (Table 1). This is a large data set equivalent to about 50-fold coverage of the entire human genome. The data were quality filtered and D96 restricted as described above, yielding 494,796 distinct TCRB sequences, each with ninefold or higher coverage (Table 1).
Table 1.
TCRB sequence statistics
aSame-day samples are independent blood draws.
bTCRB sequence in correct reading frame that passed base quality filtering at the D96 cutoff.
We also sequenced five libraries constructed from a second 20 mL blood sample from the same subject 1 wk later (day 8), obtaining 149.5 million raw read pairs and 352,139 distinct TCRB sequences (Table 1). We performed saturation analyses to determine how much of the diversity, within each blood sample, had been captured. By rarefaction analysis (random resampling of increasingly larger subsets of the data), there is an appearance of saturation sequencing of both blood samples (Fig. 2A) but a higher total number of distinct sequences obtained for blood draw 1, which was sequenced much deeper than blood draw 2. It is important to note, however, that rarefaction curves are not informative regarding total abundance and that by random resampling any data set will show a trend toward saturation. Therefore, in addition to rarefaction, we used accumulation analysis, whereby we plotted the number of distinct sequences found in each new library sequenced against the total number of distinct sequences from the blood sample as a whole. This approach, which takes into account the limitation of library exhaustion, illustrates that nearly all of the diversity present in the first blood sample has been captured (Fig. 2B). Interestingly, 33.5% of total sequences from the second blood sample were observed in the first, but after collapse into distinct sequences, there was only 12.8% overlap (Fig. 2C). This illustrates that independent blood samples contain many of the same abundant clonotypes but, due to the stochastic nature of sampling, fewer rare clonotypes. Further, it illustrates that a 20 mL blood sample captures only a portion of the diversity present within an individual's peripheral blood repertoire.
Figure 2.
TCRB diversity in peripheral blood samples. (A) Rarefaction curves plotted using all data from all libraries from blood draw 1 (round symbols) and blood draw 2, taken 1 wk later (square symbols) from donor 1. Random resampling was done in triplicate, and error bars are contained within symbols. Both curves plateau, suggesting that more sequencing of either sample would not be expected to produce many new sequences. However, this tendency toward leveling is a property of rarefaction curves, and the plateau is not informative regarding absolute abundance. Hence, rarefaction curves must be interpreted with caution. (B) Accumulation analysis provides a more meaningful measure of saturation. Here we show the number of new distinct sequences found in each library (x-axis) against the total number of distinct sequences from the blood sample as a whole. The TCRB diversity within blood draw 1 from donor 1 appears to have been captured, since analyzing additional libraries would not be expected to yield many new TCRB sequences. In contrast to the rarefaction curve present in the previous panel, library-based accumulation shows that the diversity of blood draw 2 is similar to that of blood draw 1 but has not yet been fully captured. (C) Despite saturation of blood draw 1, sequences found in blood draw 2 only partially overlap, indicating that there is considerable un-sampled diversity within the peripheral blood TCRB repertoire of this individual.
Diversity and plasticity of CD45RA+/RO− and CD45RA−/RO+ T-cell subsets
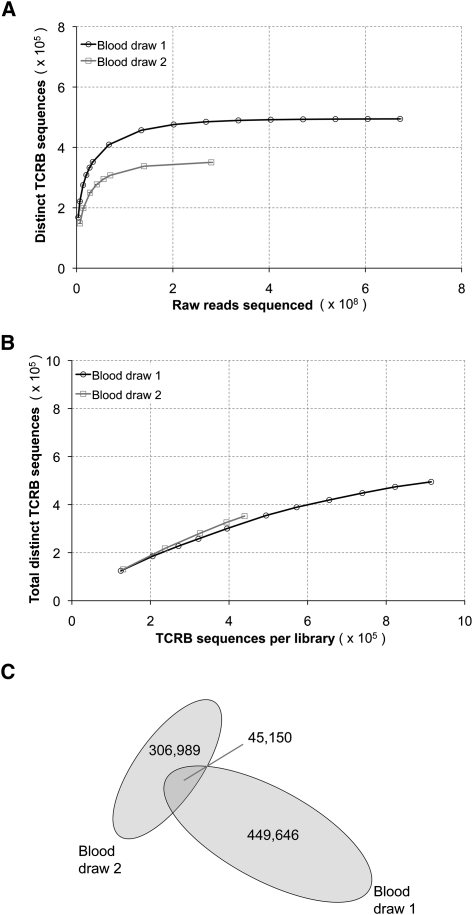
To compare diversity between naïve and memory T-cell subsets, we analyzed a separate 20 mL sample of peripheral blood from the same healthy 29-yr-old donor, taken on day 1. From this sample, we FACS sorted 1.3 million CD3+/CD45RA+/CD45RO− cells (naïve) and 1.0 million CD3+/CD45RA−/CD45RO+ cells (memory) (Supplemental Fig. S2). TCRB mRNA was amplified by 5′-RACE, Illumina sequenced, and quality filtered as described above to yield 55,253 and 52,166 distinct TCRB sequences, respectively (Table 1). We see very similar clonal diversity within these two T-cell subsets (Fig. 3A), with only slightly higher clonality detected within the memory subset. Surprisingly, we found that only a portion of sequences from the naïve and memory subsets matched sequences obtained from the deep analysis of unsorted cells from this donor (Fig. 3B). This is likely due to the fact that the cells are from an independent blood draw, and therefore, sequence signatures would be expected to overlap only partially. Very few (<1% for naïve and <3% for memory) of the new sequences found within the sorted subsets matched anything that had been removed by D96 restriction from the deep sequencing of PBMC-derived amplicons, so it is not the case that there was overly aggressive filtering of the deeply sequenced set.
Figure 3.
TCRB sequence diversity in the naïve and memory compartments of a deeply sequenced individual. (A) Frequency distributions of CD45RA+/CD45RO− (naïve) and CD45RA−/CD45RO+ (memory) T-cell subsets isolated by FACS from PBMCs from a separate blood sample from donor 1, taken on day 1. The similarity of the two curves reflects the presence of high diversity within both subsets, although this is slightly greater for the naïve subset, as evident from the extended tail of the distribution. There are, in both cases, a small number of extreme copy clonotypes and a relative clonotype abundance varying over four orders of magnitude. (B) There is more overlap with the deeply sequenced sorted cells for the memory versus the naïve subset, but even the overlap of the memory subset is modest. This is consistent with a large total repertoire size that can be only partially captured in a given blood draw. (C) Of the sequences that were shared between sorted naïve and memory cells on day 1, there was a preferential transition to the memory subset on day 8.
We also observed 540 TCRB sequences, most having a high copy number, which were present in both populations of sorted cells (Supplemental Fig. S3). The observation of dual marker cells is consistent with previous reports (Deans et al. 1989; Johannisson and Festin 1995, Wang et al. 2010). Because sorted cells were of high purity (Supplemental Fig. S2), it is unlikely that this mixed phenotype is an artifact of sorting. Rather, these cells may represent a population of acutely expanded effectors where the expression of CD45RA+ versus CD45RO+ is in transition. To address this question, we sorted cells from an additional sample of peripheral blood from this donor, taken on day 8, into CD3+/CD45RA+/CD45RO− and CD3+/CD45RA−/CD45RO+ subsets, and sequenced and analyzed as above (Table 1). Of the 540 TCRB sequences that were shared between the memory and naïve compartment on day 1, 453 were re-identified on day 8. Of these, 224 were still found in both populations of sorted cells. However, of those that were found at day 8 in only one cell population or the other, 29 were in the CD45RA+ population and 200 were in the CD45RO+ population. This illustrates plasticity of a T-cell subset, as defined by surface marker expression, and a preferred directionality for transition (Fig. 3C).
A modest proportion of sequences are shared among individuals, and these show a signature of antigen selection
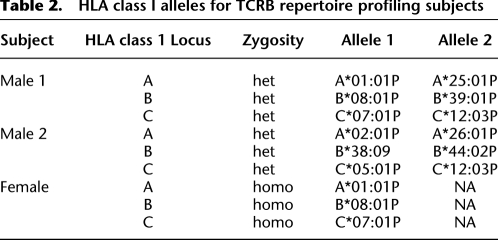
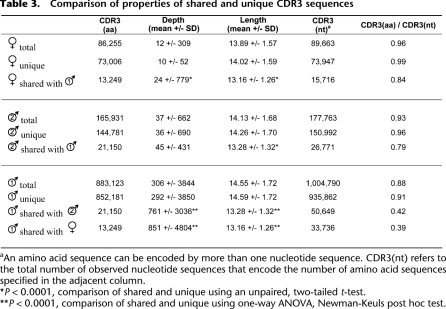
To compare repertoire diversity among the individuals, we obtained, with informed consent, peripheral blood samples (20 mL) from two additional healthy, unrelated donors. One of the new donors (male, 33 yr) had no HLA class I match to the first donor, and the other new donor (female, 33 yr) was matched to the first donor at all three HLA class I loci (Table 2). Two blood samples, taken 1 wk apart, were obtained from each donor, and these contained between 16.4 million and 18.2 million αβ T-cells. PBMCs were purified, and TCRB sequencing was performed as described above to yield 193,551 and 93,990 distinct TCRB nucleotide sequences from each donor, respectively (Table 1). Due to the lower depth of sequencing, data from each of the two samples from the same donor were pooled. We determined the number of TCRB sequences from each of the two additional donors that were observed in any of the sample from the first donor. Doing the comparisons in this manner controls for the different sequencing depths achieved. Each repertoire was found to be mostly unique at the nucleotide level. The female donor shared 1.1% of her sampled TCRB sequences with male donor 1, and male donor 2 shared 0.7% of his sampled TCRB sequences with male donor 1. Next, we translated CDR3 sequences in silico and repeated this analysis, with markedly different results (Table 3). At the amino acid level, the female donor shared 14.2% of her sampled sequences with male donor 1, and male donor 2 shared 11.7% of his sequences with male donor 1. This is consistent with the notion that the response to antigen led to retention of certain preferred amino acid sequences that, due to degeneracy of the genetic code, were specified by a larger diversity of nucleotide sequences. The different nucleotide sequences would be expected to be contributed by different cells, such that in some cases a given T-cell specificity may represent responses from multiple, independent precursor cells. We then segregated sequences into those that were shared versus unique for a given donor. When the ratios of amino acid versus nucleotide sequences in these two groups were compared, the shared sequences have a much stronger signature of selection (Table 3). We observe a strong association between the sharing of HLA class 1 alleles and the proportion of shared TCRB sequences (P < 1 × 10−6, χ2 test for two proportions), and we note that compared with unique sequences, shared sequences have shorter mean CDR3 length and higher mean abundance (P < 1 × 10−4, unpaired t-test or one-way ANOVA with Newman-Keuls post hoc test) (Table 3). Finally, in contrast to the predominant individuality of sequence specificity within repertoires, we observed striking similarity in the pairing frequency between the specific V and J gene segments among individuals (Supplemental Fig. S4). Pairwise comparisons between donors of V and J usage produced Pearson correlation coefficients of 0.87 ± 0.02 (mean ± SD) and 0.79 ± 0.12 (mean ± SD), respectively.
Table 2.
HLA class I alleles for TCRB repertoire profiling subjects
Table 3.
Comparison of properties of shared and unique CDR3 sequences
aAn amino acid sequence can be encoded by more than one nucleotide sequence. CDR3(nt) refers to the total number of observed nucleotide sequences that encode the number of amino acid sequences specified in the adjacent column.
*P < 0.0001, comparison of shared and unique using an unpaired, two-tailed t-test.
**P < 0.0001, comparison of shared and unique using one-way ANOVA, Newman-Keuls post hoc test.
Discussion
Here we report the deepest sequencing of any immune repertoire to date, capturing a total of 1,061,522 TCRB sequences from a single individual. This value places a new lower boundary on T-cell peripheral repertoire size that is consistent with the long-standing estimate of approximately 1 million distinct TCRB sequences in peripheral blood (Arstila et al. 1999), which was derived by amplification and exhaustive sequencing of a small subset of specific V-J recombinants. However, the lower bound figure we report here is definitive because it is directly measured. We see only partial overlap between the two, independent, deeply sequenced blood samples from an individual (Fig. 2C), so it is clear that total peripheral blood repertoire size is higher still. In principle, it should be possible to estimate total repertoire size based on the observed overlap; however, the usual statistical methods, such as those used by ecologists to estimate species richness, are not well suited to this problem. They are confounded by the extreme heterogeneity in the abundance of different sequences observed within a library, the variation in sampling depth (i.e., number of sequence reads) among libraries, and the difficulty in distinguishing very rare sequences from sequencing errors. Thus, an accurate estimation of total repertoire size awaits the development of new statistical methods that can account for these issues.
For sequence-based repertoire profiling, the recognition and mitigation of sequencing error is extremely important. We show that even aggressive error filtering approaches that would be considered adequate for most applications are ineffective here, where there is very intensive sequencing of a short but highly variable target. The situation can be improved by using sequence redundancy as a metric for higher stringency error filtering. In our approach, co-amplified and sequenced J gene segments provide a useful empirical measure of error rate that can be used to inform quality filtering. We find that with appropriate error filtering, it is possible to exhaustively sequence a library and, by interrogating numerous libraries, exhaustively sequence a blood sample. Unethically intensive sampling would be necessary to exhaustively sequence a human immune repertoire.
In addition to measuring total TCRB diversity, we compared diversity within memory (CD45RA−/CD45RO+) and naïve (CD45RA+/CD45RO−) subsets. Following exposure to antigen, naïve cells proliferate rapidly and differentiate into effector cells. The splicing of the signaling molecule CD45 is altered so that the CD45RA+ isoform is replaced by the CD45RO+ isoform typical of effector and memory cells. While we were initially surprised to see comparable diversity in these two subsets, this observation is consistent with, and confirms, a recent report that both the CD45RA+ naïve subset and the CD45RO+ memory subset contain many unexpanded clones (Klarenbeek et al. 2010). The presence of abundant naïve (CD45RA+) clonotypes is puzzling as it is unclear why they would be expanded, if not in response to antigen exposure. It has been suggested, however, that these abundant CD45RA+ clonotypes may originate from proliferating cells that have not yet shifted to CD45RO+ expression or that there may exist populations of effector cells that have reverted to expression of CD45RA+ (Akondy et al. 2009). In our data from sorted cells, most TCRB sequences at day 1 are unique to one population, but there were hundreds of examples of an identical TCRB sequence being found in both the CD45RA+ and CD45RO+ sorted populations. At day 8, many more of these were found only in the CD45RO+ subset than were found only in the CD45RA+ subset (Fig. 3C). These results are consistent with a preferential transition to a memory phenotype, but interestingly, this is not absolute, since some cells did change to expressing CD45RA+ only.
By comparing TCRB nucleotide sequences sampled from two additional subjects to the deeply sequenced repertoire from the first subject, we see that sharing among subjects is minimal and that at the nucleotide level individual repertoires are largely distinct. The observation of any sequence sharing at all is remarkable, however, since the space of theoretically possible TCRB sequences is vast (Davis and Bjorkman 1988) and shared sequences would not be expected by chance. The observation of shared sequences has been reported recently (Robins et al. 2010), where it was noted that sharing was highly elevated compared to what would be expected by chance, when comparing to a model of total theoretical receptor diversity. Here, we confirm sequence sharing, and we show that sharing is much more extensive at the amino acid than at the nucleotide level. This is consistent with selective pressure on TCRs, whereby a TCR amino acid sequence with suitable antigen binding characteristics may be encoded, in different cells, by different nucleotide sequences and selected independently.
One well-established mechanism of antigen selection is positive selection for HLA binding affinity during thymic T-cell maturation, and in our study, we see a clear association between proportions of shared TCR sequences and shared HLA class I alleles. Such an association has not been reported previously, and although ours is a preliminary observation based on only three subjects, it is highly statistically significant (P < 1 × 10−6). It suggests an influence of HLA type on T-cell repertoire features that deserves further scrutiny.
Methods
5′-RACE and preparation of 5′-RACE products for Illumina sequencing
Samples of peripheral blood (∼20 mL each) were obtained by venipuncture. For each of the three donors, samples were obtained on two separate days, 1 wk apart. PBMCs were isolated immediately from each sample by Ficoll-Paque (GE Healthcare) gradient centrifugation. For male donor 1, an additional, independent blood sample was taken on each day for the purpose of sorting the CD45RA+ and CD45RO+ subsets. To estimate numbers of T-cells in each sample, bulk PBMCs were stained with FITC-conjugated anti-human TCRαβ (clone T10B9.1A-31) and PE-Cy5-conjugated anti-human CD3 (clone UCHT-1; both from BD Biosciences) and were analyzed on a FACS Calibur, collecting a total of 50,000 events. For library construction, cells were centrifuged at 400g and resuspended in 2.4 mL buffer RLT (Qiagen), and 600 μL aliquots were passed through a 27-gauge needle. Each aliquot was processed using an RNeasy Plus Mini column (Qiagen) according to the manufacturer's specifications. The eluates were pooled and the concentration determined using a NanoDrop ND-8000 spectrophotometer. First-strand cDNA was synthesized using a published TRBC primer (5′>CACGTGGTCGGGGWAGAAGC<3′) (Ozawa et al. 2008). A target-switching oligo (Peters et al. 1999) (5′>AAGCAGTGGTAACAACGCAGAGTACGCGGG<3′) was added to provide a 5′ template for RACE. First-strand synthesis reaction conditions were as follows: 333 ng RNA, oligonucleotides 1 μM each, 2 mM DTT, 1 mM each dNTP, 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 6 mM MgCl2, 40 U of RNaseOUT (Invitrogen), and 200 U SMARTScribe Reverse Transcriptase (Clontech) in a 20 μL volume. Extension was for 90 min at 42°C followed by inactivation for 15 min at 70°C. PCR was performed using Phusion Hot-Start DNA Polymerase (Finnzymes) and 8.0 μL of first-strand reactions with long and short universal primers (5′>CTAATACGACTCACTATAGGGCAAGCAGTGGTAACAACGCAGAGT<3′ and 5′>CTAATACGACTCACTATAGGGC<3′) and a nested TRBC primer, (5′>TCTCTGCTTCTGATGGCTCAAAC<3′). Reaction conditions were as follows: 16 U enzyme, 1× Phusion HF amplification buffer, 3% DMSO, long universal primer at 0.1 μM, short universal and nested primers at 0.5 μM each, and 0.2 mM each dNTP in an 800 μL volume. Each reaction was split into 50 μL aliquots. For cycling, a 30-sec denaturation at 98°C was followed by 26 cycles of 10 sec at 98°C, 10 sec at 55°C, and 20 sec at 72°C, plus a final extension for 5 min at 72°C. The reaction was purified using two QIAquick columns (Qiagen), and the eluates were pooled and loaded on a 1.5% Tris-acetate low melting temperature agarose gel (Seaplaque GTG, Mandel). The gel segment corresponding to a product size of 500–625 bp was excised and melted at 65°C in 1/10 volume of 3 M NaOAc, digested at 42°C with 1000U beta-Agarase (New England Biolabs) per milliliter for 2 h, and then purified by phenol extraction and ethanol precipitation. PCR was performed on a fraction of the first-round reaction with a nested universal primer (5′>ACGACTCACTATAGGGCAAGCAG<3′) and an equimolar combination of two biotinylated primers (5′>biotin/ACACTTAATTAACGGGTGGGAACACCTTGTTCAGGT<3′) and (5′>biotin/ACACTTAATTAACGGGTGGGAACACGTTTTTCAGGT<3′), which contain 5′ PacI sites and are specific for TRBC1 and TRBC2, respectively. Reaction conditions were as follows: 800 ng purified fragment, 4 U Phusion Hot-Start DNA Polymerase (Finnzymes) 1× Phusion HF amplification buffer, 3% DMSO, 0.5 μM oligonucleotides, and 0.2 mM each dNTP in a 400 μL volume. A 30-sec denaturation at 98°C was followed by eight cycles of 30 sec at 98°C and 20 sec at 72°C, plus a final extension for 5 min at 72°C. The nested PCR reaction was purified using two QIAquick columns. Three micrograms was then sheared using the Covaris S1 (Applied Biosystems). Reaction conditions were as follows: 100 μL reaction volume, with a concentration of 100 ng/μL and 18 cycles with a duty cycle of 10%, intensity of 5, and cycles per burst at 200 for 30 sec. Biotinylated fragments were then purified using 100 μL of Dynabeads M-270 Streptavidin (Invitrogen) prepared according to the manufacturer's specifications. Washed and bound biotinylated fragments were then cleaved with PacI (reaction conditions; 1× NEB buffer 1, 100 μg/mL BSA, 50 U PacI [NEB] in a 300 mL volume for 2 h at 37°C followed by 20 min at 70°C) and ethanol precipitated.
The sample was loaded on a 8% polyacrylamide gel, and the fraction from 125–175 bp was excised, purified, and blunted. Reaction conditions were 1× NEB blunting buffer, 100 μM dNTPs, 1 μL blunting enzyme mix (NEB E1201S) in a 25 μL volume, for 30 min at 21°C. The product was purified by phenol/chloroform extraction and ethanol precipitation prior to A-tailing. Reaction conditions were as follows: 5 U Klenow fragment (3′→5′ exo–; NEB), 1× reaction buffer, 200 μM dATP in a 50 μL volume, 30 min at 37°C. The product was purified by phenol/chloroform extraction and ethanol precipitation in preparation for ligation to Illumina PE adapters. Reaction conditions were as follows: 1× NEB T4 DNA ligase buffer, 1200 U T4 DNA ligase (NEB), 1 μL PE adapters in a 30 μL volume, 15 min at 21°C. The product was purified using a QIAquick column (Qiagen) and eluted in a volume of 30 μL. Ten microliters was then amplified by PCR using Illumina primers 1.0 and 2.0 (Reaction conditions; 1 U Phusion Hot-Start DNA Polymerase, 1× Phusion HF amplification buffer, 3% DMSO, 0.3 μM oligonucleotides, and 0.2 mM each dNTP in a 25 μL volume). A 2-min denaturation at 98°C was followed by 10 cycles of 10 sec at 98°C, 30 sec at 65°C, and 30 sec at 72°C, plus a final extension of 5 min at 72°C. The PCR product was purified using a MinElute column (Qiagen) with a final volume of 13 μL and further purified from a 8% polyacrylamide gel.
We chose to sequence mRNA, not the rearranged genomic locus, because VDJ rearrangement leaves residual and potentially interfering priming sites. For transcript sequencing, 5′-RACE is the method of choice because it mitigates the risk of PCR bias that could be incurred if amplification relied instead on using multiplexed V- and J-segment specific primers.
Illumina sequencing and analysis
Illumina libraries were sequenced (100- to 150-bp paired-end reads) using an Illumina GAIIx analyzer. Most data was of read length 114 bp, and sequencing to 150 bp did not increase substantially the yield of TCRB sequences post quality filtering. A microassembler was developed to join overlapping paired-end reads from each sequencing template. Briefly, the assembler uses Exonerate (Slater and Birney 2005) to perform gapless alignments between any two mate pairs and joins the reads into sequence contigs, provided the reads align on opposite strands, facing inwards. Each alignment is scrutinized at run-time to resolve base conflicts, whenever applicable, and a consensus base sequence and quality score was generated for each newly formed contig. Agreeing bases on opposite strands were given a consensus score that corresponds to the sum of individual Q scores. Disagreeing bases were assigned an N at that position and a score of zero, unless the base call on one strand was 99.9% accurate or higher (≥Q30) and the discrepant base on the other strand was <99% accurate (<Q20). In the latter case, the most accurate base was called. We found that 68.3% of all raw sequence pairs assemble and that attrition of pairs that do not assemble is due to many factors, including mixed clusters on the flow cell, sequence errors, and templates too long for mate pairs to overlap. Annotation of the aligned paired ends contigs was done as previously described (Freeman et al. 2009). Briefly, assembled pairs aligning to the 3′ end of Ensembl TRBV gene predictions (Flicek et al. 2010) were retained and searched for the presence of 18 consecutive TRBJ segment bases. For any TRBJ segment, any 18-letter word from base positions 1–25 characterized uniquely that segment and allowed the identification of the precise TRBJ segment boundary as well as the number of TRBJ bases deleted. The TRBV segment boundaries and exact number of deleted TRBV bases were inferred by tracing back the alignments in the contig under scrutiny. Distinct TCRB sequences were identified as having unambiguous V and J segment annotation paired with a unique CDR3-encoded nucleotide sequence in a specific VDJ rearrangement that accounts for bases added and deleted at the junction. Only CDR3-encoded bases bearing no ambiguous (N) bases and having a base accuracy of 99.9% or higher at each base position were considered further. CDR3 is defined as the region between the last conserved cysteine of the V gene and the first conserved phenylalanine of the J gene in the conserved motif FGXG.
Real error rates were determined by analysis of TRBJ segments, which do not rearrange and for which reference sequences are known. This is important since the mechanism of base alteration within CDR3 that creates the vast diversity in T-cell specificity yields a sequence for which no reference exists. Errors were reported by comparing raw single-pass Illumina reads, double-strand coverage, and high-quality sequences to reference TRBJ sequences (Flicek et al. 2010). The latter set was further scrutinized to establish a sequence validity threshold by computing the proportion of J segments that did not match known TRBJs perfectly, as described in the Results section above.
HLA typing
HLA class I alleles were identified by sequence based typing. gDNA was extracted from patient granulocytes, and exons two and three from HLA class I genes (A, B, and Cw) were amplified by PCR using TAKARA polymerase (NEB), using primer sequences published previously (Cereb et al. 1995). PCR amplicons were inserted into a PCR-4-TOPO vector (Invitrogen) and cloned. Numerous clones for each locus were sequenced using an ABI 3730XL instrument according to standard procedures. Clone sequences were assembled using phred/phrap/Consed (http://www.phrap.org). The resulting sequence data were aligned against all available exon 2 and 3 nucleotide sequences from the 3.1.0 release of the IMGT/HLA database (Robinson et al. 2003) using ClustalW (Larkin et al. 2007). Allele assignments (four-digit codes) (Marsh et al. 2010) were based on high-quality exact or synonymous matches at informative nucleotide positions. Any PCR or cloning-based discrepancies were resolved manually, guided by zygosity, proportional coverage, and the low likelihood that any low coverage sporadic variant represents a novel allele.
Acknowledgments
This work was funded by the Canadian Institutes of Health Research, Genome Canada, and Genome British Columbia. We thank Karen Lambie (BCCA) for assistance with phlebotomy, Winnie Sun (BCCA) for expert help with cell sorting, and Dr. Brad Nelson (BCCA) for helpful discussion of T-cell biology. We thank Carl Schwarz and Anne Chao for helpful discussion of statistical approaches to repertoire size estimation.
Footnotes
[Supplemental material is available for this article. The sequencing data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA020989. A file containing all distinct TCRB sequences observed after all quality filtering is available at ftp://ftp.bcgsc.ca/supplementary/TCRb2010/.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.115428.110.
References
- Akondy RS, Monson ND, Miller JD, Edupuganti S, Teuwen D, Wu H, Quyyumi F, Garg S, Altman JD, Del Rio C, et al. 2009. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol 183: 7919–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arstila TP, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P 1999. A direct estimate of the human T cell receptor diversity. Science 286: 958–961 [DOI] [PubMed] [Google Scholar]
- Bassing CH, Swat W, Alt FW 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109(Suppl): S45–S55 [DOI] [PubMed] [Google Scholar]
- Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, et al. 2009. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med 1: 12ra23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereb N, Maye P, Lee S, Kong Y, Yang SY 1995. Locus-specific amplification of HLA class I genes from genomic DNA: Locus-specific sequences in the first and third introns of HLA-A, -B, and -C alleles. Tissue Antigens 45: 1–11 [DOI] [PubMed] [Google Scholar]
- Davis MM, Bjorkman PJ 1988. T-cell antigen receptor genes and T-cell recognition. Nature 334: 395–402 [DOI] [PubMed] [Google Scholar]
- Deans JP, Boyd AW, Pilarski LM 1989. Transitions from high to low molecular weight isoforms of CD45 (T200) involve rapid activation of alternate mRNA splicing and slow turnover of surface CD45R. J Immunol 143: 1233–1238 [PubMed] [Google Scholar]
- Ewing B, Green P 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194 [PubMed] [Google Scholar]
- Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. 2010. Ensembl's 10th year. Nucleic Acids Res 38: D557–D562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JD, Warren RL, Webb JR, Nelson BH, Holt RA 2009. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res 19: 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty JT, Badovinac VP 2008. Shaping and reshaping CD8 T cell memory. Nat Rev Immunol 8: 107–119 [DOI] [PubMed] [Google Scholar]
- Johannisson A, Festin R 1995. Phenotype transition of CD4+ T cells from CD45RA to CD45R0 is accompanied by cell activation and proliferation. Cytometry 19: 343–352 [DOI] [PubMed] [Google Scholar]
- Khor B, Sleckman BP 2002. Allelic exclusion at the TCRbeta locus. Curr Opin Immunol 14: 230–234 [DOI] [PubMed] [Google Scholar]
- Klarenbeek PL, Tak PP, van Schaik BD, Zwinderman AH, Jakobs ME, Zhang Z, van Kampen AH, van Lier RA, Baas F, de Vries N 2010. Human T-cell memory consists mainly of unexpanded clones. Immunol Lett 133: 42–48 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Fernández-Viña M, Geraghty DE, Holdsworth R, Hurley CK, et al. 2010. Nomenclature for factors of the HLA system. 2010. Tissue Antigens 75: 291–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolich-Zugich J, Slifka MK, Messaoudi I 2004. The many important facets of T-cell repertoire diversity. Nat Rev Immunol 4: 123–132 [DOI] [PubMed] [Google Scholar]
- Ozawa T, Tajiri K, Kishi H, Muraguchi A 2008. Comprehensive analysis of the functional TCR repertoire at the single-cell level. Biochem Biophys Res Commun 367: 820–825 [DOI] [PubMed] [Google Scholar]
- Peters DG, Kassam AB, Yonas H, O'Hare EH, Ferrell RE, Brufsky AM 1999. Comprehensive transcript analysis in small quantities of mRNA by SAGE-lite. Nucleic Acids Res 27: e39 doi: 10.1093/nar/27.24.e39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS 2009. Comprehensive assessment of T-cell receptor β-chain diversity in αβ T cells. Blood 114: 4099–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins HS, Srivastava SK, Campregher PV, Turtle CJ, Andriesen J, Riddell SR, Carlson CS, Warren EH 2010. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med 2: 47ra64 doi: 10.1126/scitranslmed.3001442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SGE 2003. IMGT/HLA and IMGT/MHC: Sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res 31: 311–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GS, Birney E 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31 doi: 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sanders CM, Yang Q, Schroeder HW Jr, Wang E, Babrzadeh F, Gharizadeh, Myers B, Hudson RM Jr JR, Davis RW, et al. 2010. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci 107: 1518–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]