Abstract
Emerging evidence suggests that microRNAs (miRNAs), an abundant class of ∼22-nucleotide small regulatory RNAs, play key roles in controlling the post-transcriptional genetic programs in stem and progenitor cells. Here we systematically examined miRNA expression profiles in various adult tissue-specific stem cells and their differentiated counterparts. These analyses revealed miRNA programs that are common or unique to blood, muscle, and neural stem cell populations and miRNA signatures that mark the transitions from self-renewing and quiescent stem cells to proliferative and differentiating progenitor cells. Moreover, we identified a stem/progenitor transition miRNA (SPT-miRNA) signature that predicts the effects of genetic perturbations, such as loss of PTEN and the Rb family, AML1-ETO9a expression, and MLL-AF10 transformation, on self-renewal and proliferation potentials of mutant stem/progenitor cells. We showed that some of the SPT-miRNAs control the self-renewal of embryonic stem cells and the reconstitution potential of hematopoietic stem cells (HSCs). Finally, we demonstrated that SPT-miRNAs coordinately regulate genes that are known to play roles in controlling HSC self-renewal, such as Hoxb6 and Hoxa4. Together, these analyses reveal the miRNA programs that may control key processes in normal and aberrant stem and progenitor cells, setting the foundations for dissecting post-transcriptional regulatory networks in stem cells.
Stem cells (SCs) have the ability to self-renew and to give rise to committed progenitors of a single lineage or of multiple lineages. Elucidating the genetic circuits that govern SCs to self-renew and to differentiate is essential to understanding the roles of SCs in animal development and to realizing the promise of these cells in regenerative medicine. Extensive efforts have been made to determine these regulatory circuits through profiling of messenger RNA (mRNA) expression in various SCs and their corresponding differentiated progenitors; these studies have yielded critical information on the key regulatory and surface molecules in SCs (Chen et al. 2002, 2003; Ivanova et al. 2002; Ramalho-Santos et al. 2002; Akashi et al. 2003; Chambers et al. 2007). However, since mRNA profiles largely reflect the consequences of transcriptional regulation, these studies do not take into account the extensive post-transcriptional programs that control SC functions, particularly the ones controlled by an abundant class of noncoding RNAs, the microRNAs (miRNAs).
miRNAs are ∼22-nucleotide (nt) regulatory RNAs that repress translation and/or initiate transcript degradation via imperfect base pairing with cognate target mRNAs (Bartel 2009). miRNA-coding genes represent ∼1%–5% of the predicted genes in worms, flies, mice, and humans. Each miRNA can potentially regulate several hundred target genes (Bartel 2009; Friedman et al. 2009). Thus, miRNA-mediated gene regulation may have profound effects on gene expression and constitutes a fundamental layer of post-transcriptional regulation in animals. Abundant evidence demonstrates that these small noncoding RNAs play diverse roles in normal development and in the pathogenesis of human diseases by controlling cellular processes such as proliferation, morphogenesis, apoptosis, and differentiation. Specific miRNAs (e.g., let-7b) and post-transcriptional regulators that control miRNA activities (e.g., TRIM-NHL proteins like mouse TRIM32 and fly Brat and Mei-P26) regulate the self-renewal of tissue-specific SCs (TSCs) (Neumuller et al. 2008; Hammell et al. 2009; Schwamborn et al. 2009). Loss of DICER1 or Drosha (also known as RNASEN), enzymes essential for miRNA biogenesis, affects the proliferation of mouse embryonic stem (ES) cells and fly germline SCs (Wang et al. 2007; Yu et al. 2009). These results strongly indicate that miRNA-mediated post-transcriptional programs are integral components of the genetic circuits that govern SC functions. However, few studies have been carried out to compare miRNA profiles in multiple adult TSCs and their differentiated progenies.
In this study we systematically analyzed miRNA expression in normal and aberrant adult TSCs and their differentiated progeny. We identified miRNAs unique to various adult TSCs and those shared by multiple TSCs. Furthermore, we uncovered miRNA programs that mark the transition from self-renewing and slow-cycling SCs to differentiating and rapidly proliferating transit amplifying cells in muscle and blood, and a miRNA program that predicts the effects of genetic mutations on SC self-renewal and differentiation. Finally, we provide functional evidence on the roles of stem/progenitor transition miRNAs in SCs. Together, these results set the foundations for dissecting miRNA networks in SCs.
Results
miRNA expression profiles in adult tissue stem and progenitor cells
To dissect miRNA programs in stem/progenitor cells, we carried out global analyses of miRNA expression in various TSCs and their corresponding differentiated progenitors from normal and mutant mice (See Table 1 for sample list and abbreviated sample names). We analyzed miRNA expression in SCs of the blood, skeletal muscle, and neural systems: long-term hematopoietic SCs (LT-HSCs), skeletal muscle SCs (MuSCs), and neural stem/progenitor cells (NSPCs), respectively (Chen et al. 2003; Sacco et al. 2008; Renault et al. 2009). Such analyses allowed us to determine both shared and unique miRNA programs in TSCs. Further, we profiled miRNA expression in the differentiating progenitors of the blood and muscle SCs: Kit+/Sca-1+/Lin- (KSLs) bone marrow cells, and myoblasts. By comparing miRNA profiles in SCs and their immediate progeny, we identified the miRNA programs that mark the critical transition from SCs to differentiating progenitors. Finally, we examined how miRNA expression was altered by various genetic perturbations, including loss of PTEN or Rb family genes in HSCs, ectopic expression of AML1-ETO9a in HSCs, and the MLL-AF10 transformation (de Guzman et al. 2002; Yan et al. 2006; Zhang et al. 2006; Viatour et al. 2008; Somervaille et al. 2009). These mutations affect the self-renewal, differentiation, and oncogenic potential of stem and/or progenitor cells. Such analyses may reveal miRNA programs that control the self-renewal and differentiation of stem/progenitor cells.
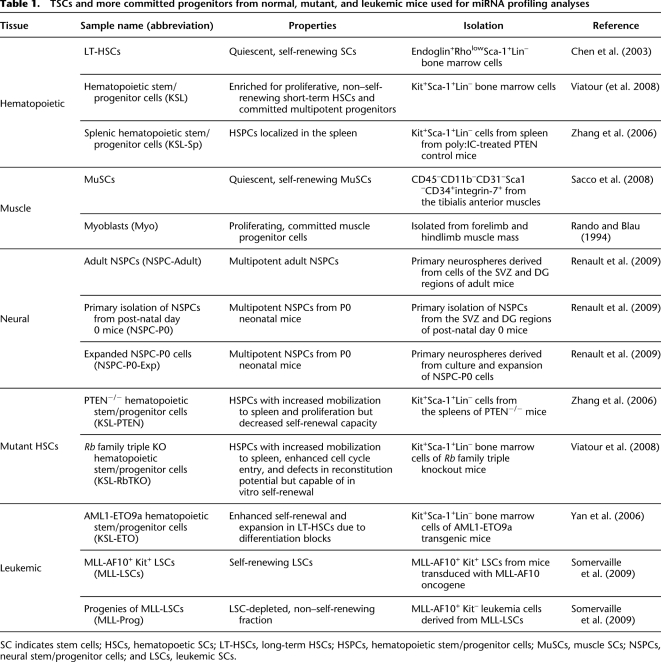
Table 1.
TSCs and more committed progenitors from normal, mutant, and leukemic mice used for miRNA profiling analyses
SC indicates stem cells; HSCs, hematopoetic SCs; LT-HSCs, long-term HSCs; HSPCs, hematopoietic stem/progenitor cells; MuSCs, muscle SCs; NSPCs, neural stem/progenitor cells; and LSCs, leukemic SCs.
We used a multiplex protocol to amplify miRNAs from 20–1000 sorted stem and/or progenitor cells and then analyzed the expression of 425 mature miRNAs using TaqMan miRNA quantitative PCR (qPCR) analyses (Chen et al. 2005, 2007). This method is specific and has been extensively utilized in quantifying miRNA expression in various cell types. Moreover, the combination of pre-amplification and multiplex qPCR increases the sensitivity of miRNA detection to a single cell level without noticeable biases (Mestdagh et al. 2008). Compared to other methods for miRNA expression analyses, such as miRNA microarray and small RNA deep sequencing, which require large amounts of starting material, the miRNA qPCR method can be used to quantify miRNA expression in a single cell or low numbers of cells. Moreover, deep-sequence methods for analyzing small RNA abundance have intrinsic limitations, such as ligation biases and inconsistent levels of contamination with other ribosomal RNAs or tRNA degradation products. The latter issue complicates the use of number of tags per million reads as quantitative readouts. miRNA microarrays seem to have the least sensitivity and specificity because of the difficulties in design of probes with similar melting temperatures and specificities for closely related miRNAs. Most importantly, a recent study established that the results obtained from miRNA qPCR analyses and deep-sequence analyses are largely in agreement (Kuchen et al. 2010). Therefore, multiplex miRNA qPCR assay is a suitable choice for analyzing miRNA expression in rare SC samples.
Using this method, we detected a total of 150 miRNAs [critical threshold (Ct) < 35] in the 13 samples analyzed (Supplemental Table S1). The number of miRNAs detected in various stem/progenitor cell types varied significantly, ranging from about 50 to 100 (Supplemental Fig. S1), and miRNA expression levels varied considerably in stem/progenitor cell types as indicated by median Ct values and inter-quartile ranges (IQRs) of detectable miRNAs (Supplemental Fig. S2A). About 20 LT-HSCs were used in the profiling analyses, and about 1000 MuSCs, KSL-Sps, and KSL-RbTKOs were used. Thus, the low numbers of miRNAs detected in MuSCs, LT-HSCs, KSL-Sps, and KSL-RbTKOs were not because of fewer cells used in profiling analyses. Since we analyzed miRNA expression in a defined number of cells, it is possible that variations in the numbers of miRNAs detected will be influenced by the differences in cell sizes and total RNA content in these cell types, and therefore miRNA numbers are not directly comparable. Thus, it is important not to equate the number of miRNAs detected as the absolute number of miRNAs expressed in those cell types.
We used the median Ct values of expressed miRNAs to normalize the data (Supplemental Fig. S2B; Supplemental Tables S1, S2). Given that miRNA expression profiles have small data sets with highly skewed distributions, a median scaling method is an appropriate method for the normalization of the data collected from SCs and progenitors from different tissues. The most commonly used normalization methods based on all genes on the array would be skewed by a highly disproportional representation of small number of miRNAs. Another alternative, normalization to levels of snoRNA, is complicated by variation in snoRNA expression across multiple tissue types. For example, U6 snoRNA varies as much as 6.5-fold across tissues (Castle et al. 2010), suggesting that normalization methods based on levels of housekeeping genes would be inappropriate.
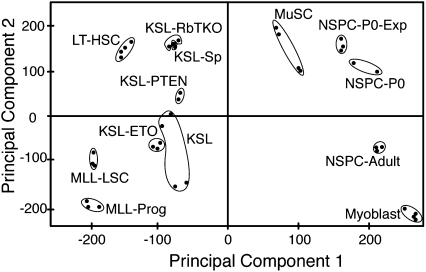
miRNA expression profiles effectively segregated samples by tissue of origin, grouping together hematopoietic, muscle, and neural samples as indicated by principal component analyses (PCAs) (Fig. 1) and hierarchical clustering (HCL) (Supplemental Fig. S3). Sample replicates generally clustered together, although larger variations were observed among the five KSL cell samples isolated by the different laboratories. Within the hematopoietic sample cluster, miRNA expression signatures separated mutant stem and progenitor cell samples from their wild-type counterparts. Interestingly, miRNA signatures of KSL-Sp and KSL-RbTKO clustered closely together, perhaps reflecting the splenic mobilization phenotype observed upon loss of the Rb family in HSCs (Viatour et al. 2008). Clearly, miRNA profiles effectively distinguish SCs and progenitors of the different tissue origins, developmental stages, and genetic modifications. This observation is consistent with previous studies that have shown that miRNA profiles effectively classify cancer cell samples (Lu et al. 2005).
Figure 1.
Principal component analyses indicating the relative distances between the miRNA profiles of various stem and progenitor cell populations.
Common and TSC-related miRNA signatures
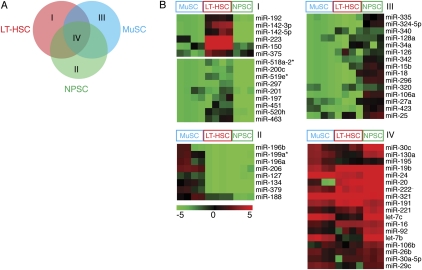
To identify common and TSC-related miRNA signatures, we compared miRNA profiles in adult TSC populations from blood, muscle, and neural tissues (Fig. 2A). We used one-way ANOVA analyses and K-means clustering (KMC) to reveal shared and unique TSC-related miRNA signatures. Three TSC-related miRNA clusters, designated as TSC-miRNAs, were enriched in one TSC type but were undetectable or expressed at very low levels in the other two (Fig. 2B). Of these clusters, 15 miRNAs are LT-HSC miRNAs; these were expressed preferentially in LT-HSCs but were mostly undetectable in NSPCs and MuSCs (Fig. 2B, cluster I). LT-HSC miRNAs were further divided into high and low expression groups, consisting of six and nine miRNAs, respectively. We found eight MuSC-miRNAs (Fig. 2B, cluster II) and 16 NSPC-miRNAs (Fig. 2B, cluster III). Significant nonoverlapping miRNA expression profiles were noted between NSPC-P0, NSPC-P0-exp, and NSPC-Adult cells (Supplemental Fig. S4). miRNA profiles from adult NSPCs were used in comparative analyses. The TSC-miRNAs may play roles in regulating unique functions of TSCs, such as lineage-specific functions, developmental potentials, and commitment events.
Figure 2.
Identification of tissue-specific stem cell–related miRNA signatures. (A) Schematic diagrams illustrate the comparisons made to reveal the tissue-specific SC-related miRNA signatures: I, miRNAs enriched in LT-HSCs (LT-HSC miRNAs); II, miRNAs enriched in NSPCs (NSPC miRNAs); III, miRNAs enriched in MuSCs (MuSC miRNAs); and IV, the common SC miRNA signatures (SC-related miRNAs). They are depicted as the single-colored regions (I, II, III) and a triple-colored region (IV) in the Venn diagram. (B) Heatmaps depicting common and tissue-specific miRNAs derived from KMC analyses. A false color scale was used to indicate normalized arbitrary expression intensity (ΔCt) with “−5” for the lowest expression, “0” for median expression, and “5” for the highest expression.
Finally, we identified 18 common SC-related miRNAs that were highly expressed in all three of the TSC types (Fig. 2B, cluster IV, twofold above median expression for at least two TSC types). It is important to note that the above definition for TSC-miRNAs and common SC-miRNAs did not require that these signature miRNAs be absent in the differentiated cell types. It is likely that TSC-miRNAs and common SC-miRNAs carry out critical functions in SCs despite their presence in more differentiated cell types. We only identified one LT-HSC–specific miRNA (miR-192), one MuSC-specific miRNA (miR-379), and one NSPC-specific miRNA (miR-135b) absent from all other cell types analyzed in this study (Supplemental Fig. S5).
miRNAs differentially expressed in hematopoietic stem and progenitor cells
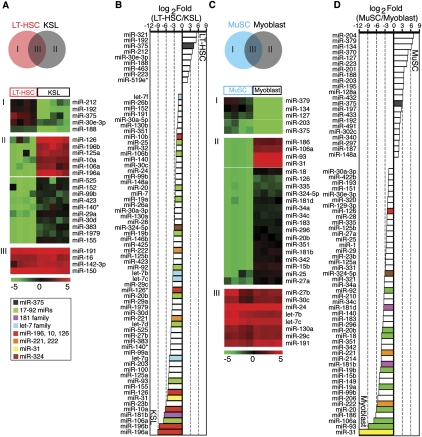
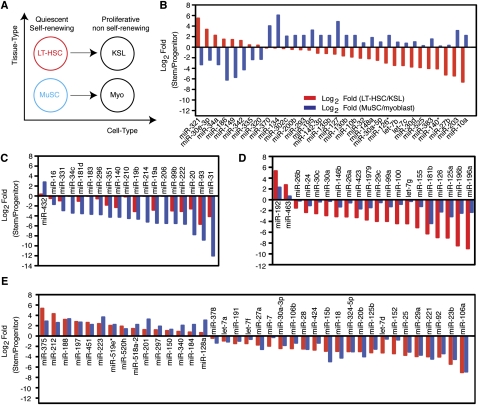
To identify miRNAs that control HSC self-renewal and differentiation, we compared miRNA profiles of LT-HSCs and KSL cells. As isolated, KSL cells consist of 5%–10% LT-HSCs and 90%–95% short-term HSCs (ST-HSCs) and multi-potent progenitors (MPPs). Comparison of miRNA expression profiles in LT-HSCs and KSL cells revealed miRNAs that were turned “on” or “off” or were quantitatively regulated during this transition. We found that five miRNAs (miR-212, miR-192, miR-375, miR-30e-3p, and miR-188) were expressed in LT-HSCs but not in KSL cells (Fig. 3A, cluster I). In contrast, 16 miRNAs were absent in LT-HSCs but turned on in KSL cells (Fig. 3A, cluster II). Moreover, including those miRNAs that exhibited binary expression patterns in LT-HSCs and KSL cells, a total of nine miRNAs were down-regulated and 62 miRNAs were up-regulated (based on statistical analysis of microarrays [SAM], with false discovery rate [FDR] < 0.001) during the LT-HSC to KSL transition (Fig. 3B; Supplemental Table S3). Some of these miRNAs may regulate expression of genes involved in the transition from multipotent, quiescent, and self-renewing LT-HSCs to the differentiating, highly proliferative, non–self-renewing ST-HSCs and committed multipotent progenitors (KSL cells). Intriguingly, miRNAs targeting Hox genes (Yekta et al. 2004; Shen et al. 2008; Woltering and Durston 2008), such as miR-126, miR-196b, miR-196a, and miR-10a (Fig. 3A, cluster II), were nearly undetectable in LT-HSCs but were highly expressed in KSL cells, suggesting that silencing Hox gene expression by these miRNAs may play key roles during this transition.
Figure 3.
miRNAs differentially expressed in LT-HSCs and KSL cells and in MuSCs and myoblasts. (A) Schematic diagram depicting the comparisons made to reveal miRNAs that are expressed in LT-HSCs only (I), expressed in KSL cells only (II), or highly expressed in both (III). SAM analyses were carried out to identify miRNAs that were significantly different or unchanged between LT-HSCs and KSL cells (FDR < 0.001), which were then further classified by KMC analyses as depicted in heatmaps. A false color scale was used to indicate the normalized arbitrary expression intensity (ΔCt). (B) Fold changes in the top 69 miRNAs that differed significantly between LT-HSCs and KSL cells (SAM, FDR < 0.001) are shown (Log2 Fold [LT-HSC/KSL]). (C) Schematic diagram depicting the comparisons made to reveal miRNAs that are expressed in MuSCs only (I), expressed in myoblasts only (II), or highly expressed in both (III). SAM analyses were carried out to identify miRNAs that were significantly different or unchanged between MuSCs and myoblasts (FDR < 0.001), which were then further classified by KMC analyses as depicted in heatmaps. A false color scale was used to indicate the normalized arbitrary expression intensity (ΔCt). (D) Fold changes in the top 69 miRNAs that differed significantly between MuSCs and myoblasts (SAM, FDR < 0.001) are shown (Log2 Fold[MuSC/myoblasts]). Selected miRNAs or groups of miRNA are color-coded. The miR-181 family miRNA consists of miR-181a, miR-181b, miR-181c, and miR-181d. The miR-17-92 family miRNA clusters consist of miR-17-92, miR-106b-25, and miR-106a-363. The let-7 family consists of let-7a-i.
miRNAs differentially expressed in muscle stem and progenitor cells
To identify miRNAs that may control MuSC self-renewal and differentiation, we compared the miRNA profiles of MuSCs and myoblasts. A number of miRNAs were highly differentially regulated during the transition from quiescent and self-renewing MuSCs to differentiating and proliferative myoblasts (Fig. 3C). Five miRNAs (miR-379, miR-134, miR-127, miR-203, and miR-375) were expressed in MuSCs but not myoblasts (Fig. 3C, cluster I), and 20 miRNAs were turned on in myoblasts (Fig. 3C, cluster II). Including those miRNAs that exhibit binary expression patterns in MuSCs and myoblasts, a total of 24 miRNAs were down-regulated and 53 miRNAs were up-regulated during the MuSC to myoblast transition (Fig. 3D; Supplemental Table S3). miR-379 seems to be the only MuSC-specific miRNA based on comparison with miRNA profiles of other stem/progenitor cell types (Supplemental Fig. S5).
Among these differentially regulated miRNAs, miR-181b has been shown to target Hoxa11 during myoblast differentiation (Naguibneva et al. 2006). Many of the miRNAs highly up-regulated (greater than eightfold) during the stem to progenitor transition in muscle and blood lineages are shared (Fig. 3), suggesting that a common set of miRNAs may be induced to facilitate early SC commitment and differentiation. In contrast, many miRNAs that are down-regulated during the stem to progenitor transition in muscle and blood lineages are not shared. For example, miR-204, which is the most highly down-regulated miRNA during the MuSC to myoblast transition, was not detected in LT-HSCs. Such differences may reflect distinctions in lineage origins and potential and unique miRNA functions in muscle and blood SCs.
Coordinated regulation of miRNA programs during stem to progenitor transition
Many key molecular and developmental events, including loss of quiescence and self-renewal potential, increases in proliferation rate, and initiation of commitment programs and differentiation, commence during the transitions from LT-HSCs to KSL cells and from MuSCs to myoblasts. Comparing the changes in miRNA profiles during the LT-HSC to KSL and the MuSC to myoblast transitions may reveal conserved miRNA programs that control these transitions. To this end, we carried out multi-factorial analyses using a bootstrap-based, nonparametric ANOVA (NANOVA) method, and we used a gene classification algorithm to identify miRNA signatures that underlie these critical transitions (Fig. 4A; Supplemental Table S4; Zhou and Wong 2011). We identified a group of miRNAs that are discordantly regulated during the stem to progenitor transition in blood and muscle tissues (Fig. 4B). These miRNAs are likely to contribute to tissue-specific and/or cell type–specific regulatory functions, such as the commitment and/or differentiation to blood and muscle cell types. We also found a group of miRNAs that are concordantly regulated (Fig. 4C–E). These miRNAs are likely to control common stem/progenitor cell programs initiated during the transition from LT-HSCs to KSL cells and MuSCs to myoblasts, such as the regulation of self-renewal and proliferation.
Figure 4.
miRNA programs underlie the stem to progenitor transition. Multi-factorial analyses revealed various miRNA programs that control the transitions from blood and muscle stem cells (LT-HSCs and MuSCs) to the corresponding immediate differentiating progenies (KSL cells and myoblasts, respectively). (A) The comparisons performed in multi-factorial analyses to yield functional miRNA groups identified. (B) miRNAs discordantly regulated during stem to progenitor transition in blood and muscle. (C,D) miRNAs concordantly regulated during stem to progenitor transition but more drastically regulated in either muscle (C) or blood (D). (E) miRNAs concordantly regulated during stem to progenitor transition in blood and muscle.
Among the concordantly regulated miRNAs, some are preferentially altered in muscle development (Fig. 4C), whereas some others are changed in blood development (Fig. 4D). For instance, miR-31 was up-regulated about 17-fold in hematopoietic progenitors versus about 4000-fold in the muscle progenitors (Fig. 4C), and miR-196a and miR-196b were up-regulated over 250-fold during blood SC differentiation versus approximately fivefold during MuSC differentiation (Fig. 4D). These differences during the parallel transitions may reflect tissue-specific regulation. Of interest are those miRNAs that are identically modulated during the stem to progenitor transition in both tissues since these may function in conserved SC regulatory programs (Fig. 4E). Among these (Supplemental Table S4), 15 miRNAs were down-regulated concordantly during stem to progenitor transition, and 23 miRNAs were up-regulated concordantly during stem to progenitor transition. These 15 miRNAs may contribute to the self-renewal and quiescence properties of SCs by suppressing the differentiation and fast proliferation programs; down-regulation of these miRNAs during stem to progenitor transition may permit activation of these programs. In contrast, up-regulation of these 23 miRNAs during stem to progenitor transition may inactivate the self-renewal and quiescence programs in SCs and allow for differentiation and rapid proliferation.
Altered miRNA programs in mutant stem and progenitor cells
To further narrow down the miRNAs that may play critical roles in SCs, we examined the changes in miRNA expression caused by genetic mutations that affect the self-renewal and differentiation potentials of stem and progenitor cells (Table 1). Specifically, we examined miRNA expression profiles in KSL cells with the following genetic modifications: (1) ectopic expression of AML1-ETO9a, which expands HSC compartment and causes acute myeloid leukemia (de Guzman et al. 2002; Yan et al. 2006); (2) loss of Rb family genes, which results in an increase in HSC proliferation and mobilization without comprising self-renewal programs (Viatour et al. 2008); and (3) loss of PTEN, which causes an increase in HSC proliferation and concomitant decrease of self-renewal (Zhang et al. 2006). We also examined miRNA expression in leukemia SCs induced by the MLL-AF10 transformation, which enables self-renewal of differentiated progenitor cells (Somervaille et al. 2009). Comparing miRNA profiles of the mutant and the relevant wild-type stem/progenitor cells revealed miRNAs that potentially function during SC self-renewal and differentiation (Fig. 5; Supplemental Table S3).
Figure 5.
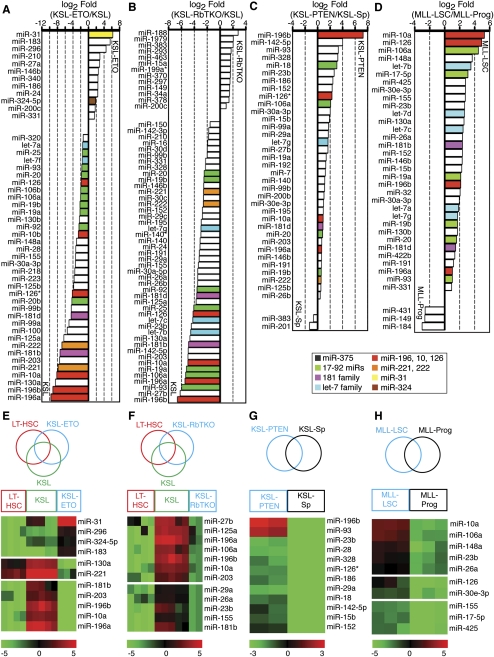
Dysregulation of miRNA programs by genetic mutations altering the functional properties of stem/progenitor cells. Global changes of miRNA expression in mutant stem and progenitor cells: (A) KSL-ETO, (B) KSL-RbTKO, (C) KSL-PTEN, and (D) MLL-LSC. miRNAs that are significantly different between mutant cells and their control cells (SAM, FDR < 0.001) are shown as fold of changes (Log2 Fold Mutant/Control). Heatmaps depict the specific miRNA clusters that were turned on/off in mutant stem and/or progenitor cells: (E) KSL-ETO; (F) KSL-RbTKO; (G) KSL-PTEN; and (H) MLL-LSC. False color scales were used to indicate normalized expression intensity.
These genetic modifications resulted in global changes in miRNA expression (Fig. 5A-D). Relative to the wild-type KSL cells, expression of AML1-ETO9a resulted in down-regulation of 36 miRNAs and up-regulation of 12, whereas loss of the three Rb family genes resulted in down-regulation of 46 miRNAs and up-regulation of 13 (Fig. 5A, B). In contrast, loss of PTEN resulted in an up-regulation of 35 miRNAs and a down-regulation of only two (Fig. 5C). Comparison of MLL-LSCs to non–self-renewing leukemic progeny revealed that 33 miRNAs were up-regulated and three miRNAs were down-regulated (Fig. 5D). It is likely that changes in miRNA expression in part contribute to the effects of these genetic modifications on the self-renewal and differentiation potentials of the targeted cells. Indeed, utilizing the miRNAs identified in this profiling study as a basis for investigation, miR-17-92 was recently shown to promote MLL-LSC potential by modulating p21 expression (Wong et al. 2010).
To further evaluate the miRNA programs controlled by AML1-ETO9a, Rb family genes, PTEN, and MLL-AF10, we determined which miRNAs were turned on or off by these genetic modifications using one-way ANOVA analyses and KMC analyses (Fig. 5E–H). We found that miR-31, miR-296, miR-324-5p, and miR-183 were highly expressed in KSL-ETO cells but were expressed at low or undetectable levels in both LT-HSCs and KSL cells (Fig. 5E). Some miRNAs expressed in KSL cells (miR-196a, miR-196b, miR-10a, miR-203, and miR-181b) or LT-HSCs and KSL cells (miR-130a and miR-221) were turned off in KSL cells expressing AML1-ETO9a. In contrast, the loss of Rb family genes turned off 12 KSL miRNAs but did not turn on any miRNAs that were absent in KSL cells and/or LT-HSCs (Fig. 5F). Loss of PTEN and the MLL-AF10 transformation turned on specific sets of miRNAs but did not turn off any miRNAs present in the corresponding control cell populations (Fig. 5G,H).
Some of these differentially regulated miRNAs may be direct transcriptional targets of the genetic modifications and may contribute to the altered self-renewal potential of mutant HSCs. For example, miR-223, which has previously been shown to be transcriptionally repressed by AML-ETO and to contribute to tumorigenesis, was down-regulated in KSL-ETO cells (Fig. 5A; Fazi et al. 2007). Moreover, in cells with these genetic mutations, the expression of the miRNAs that target Hox genes is altered. Among the five KSL miRNAs that were turned off by AML1-ETO9a, three (miR-196a, miR-196b, and miR-10a) target Hox genes. Intriguingly, loss of Rb family genes also resulted in down-regulation of “Hox-targeting” miRNAs (miR-196a, miR-196b, and miR-10a), whereas loss of PTEN and the MLL-AF10 transformation resulted in up-regulation of some “Hox-targeting” miRNAs (miR-196b and miR-10a). Given that many Hox genes play critical roles in controlling HSC self-renewal (Argiropoulos and Humphries 2007), these results strongly suggest that coordinated miRNA-mediated regulation of Hox genes may control the self-renewal potentials of SCs. Collectively, these analyses revealed diverse and distinct effects of these genetic modifications on miRNA expression and their potential roles in altering the self-renewal and differentiation potentials of stem and progenitor cells.
An miRNA signature predicting the effects of developmental and genetic perturbations
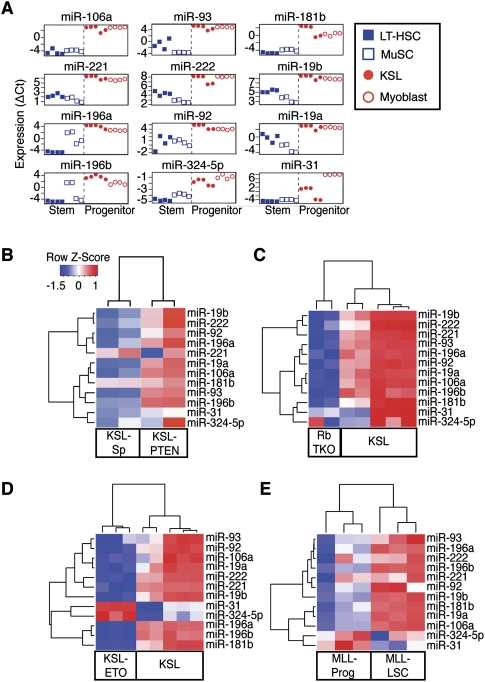
The above analyses indicate that coordinated regulation of a selected set of miRNAs may contribute to the altered self-renewal and proliferation potentials of SCs and progenitor cells during normal stem to progenitor transition (Fig. 4) and to specific genetic perturbations (Fig. 5). To identify those miRNAs that were co-regulated by developmental perturbations (i.e., stem to progenitor transition) and genetic perturbations (i.e., loss of PTEN and Rb family, AML1-ETO9a expression, and MLL-AF10 transformation), we carried out predictive analysis of microarrays (PAM; FDR < 0.001) (Tibshirani et al. 2002). We identified an miRNA signature including 12 miRNAs that characterizes the normal stem to progenitor cell transition in blood and muscle (Fig. 6A); this signature was largely predictive of the effects of the aforementioned genetic modifications on the stem or progenitor cells (Fig. 6B–E). The miRNAs identified are likely to be the key players among all the differentially regulated miRNAs that contribute to the changes in SC functional properties during these genetic and developmental perturbations. All 12 miRNAs in the signature are up-regulated during stem to progenitor transition in blood and muscle (Figs. 4, 6A), indicating that cells expressing this miRNA signature have functional properties akin to progenitors (i.e., KSLs or myoblasts) rather than SCs (i.e., LT-HSCs or MuSCs). Supporting this notion, this miRNA signature classified KSL-PTEN cells as progenitor-like (Fig. 6B), consistent with the high proliferative and low self-renewal potential of these cells. In contrast, it classified the KSL-ETO and KSL-RbTKO cells as stem-like (Fig. 6C,D), consistent with their self-renewal potential. Finally, it classified MLL-LSCs as progenitor-like (Fig. 6E), consistent with the high proliferative potential of these cells and their transcriptional program that deviates from normal adult SCs (Somervaille et al. 2009).
Figure 6.
A stem/progenitor transition miRNA signature predicts the functional properties of mutant stem/progenitor cells. (A) A stem/progenitor transition miRNA signature that is predictive of stem or progenitor identity/property of mutant stem/progenitor cells was identified from the miRNAs that were concordantly regulated during stem to progenitor transition in muscle and blood by using PAM analyses (FDR < 0.001). Hierarchical clustering analyses showed that the 12 stem/progenitor transition miRNAs predict the functional properties of mutant stem/progenitor cells: (B) KSL-PTEN; (C) KSL-ETO; (D) KSL-RbTKO; and (E) MLL-LSC. False color scale depicts relative changes in expression.
These analyses revealed a subset of miRNAs that mark a conserved stem to progenitor transition process (designated stem/progenitor transition miRNAs, SPT-miRNAs). The fact that changes in SPT-miRNA expression correlates well with the functional effects of corresponding genetic modifications further indicates that these miRNAs are likely to play key roles in controlling the self-renewal and proliferation potentials in normal and mutant stem/progenitor cells (Fig. 6B–E). Indeed, some of these SPT-miRNAs have been shown to regulate key SC regulatory molecules (Supplemental Table S5). Nevertheless, not all the SPT-miRNAs were regulated equally by these developmental and genetic perturbations. For example, the expression of miR-31 and miR-324-5p in KSL-ETO and MLL-LSC cells deviated from the pattern of expression of the other miRNAs up-regulated in progenitor cells (Fig. 6D,E). This observation suggests that miR-31 and miR-324-5p may be functionally incompatible with other SPT-miRNAs expressed in KSL-ETO cells and MLL-LSCs.
Effects of the SPT-miRNAs on SC self-renewal and differentiation
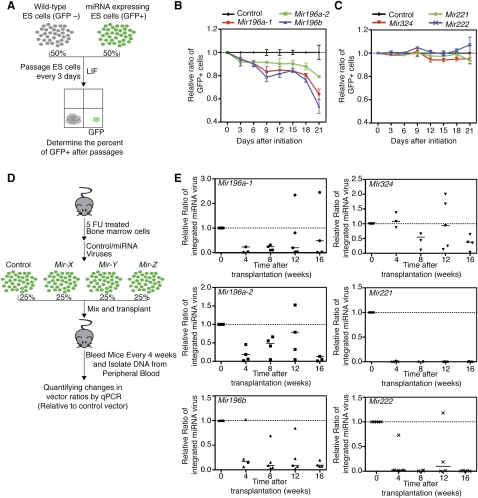
Coordinated regulation of SPT-miRNAs during developmental transitions and genetic perturbations in normal and mutant SCs (Fig. 6) suggests that these miRNAs may mediate the shifts in shared functional properties in normal and aberrant stem/progenitor cells, such as changes in quiescence, self-renewal, and proliferation capacity. We therefore examined the effects of the SPT-miRNAs on ES cell self-renewal and proliferation. ES cells are pluripotent SCs and can be maintained in a self-renewal state in culture; thus quantitative measurements of changes in self-renewal and proliferation potential upon perturbing miRNA expression can be made. To this end we devised a fluorescence-based competition assay (Fig. 7A). ES cells were infected with control virus (no miRNA) or miRNA-expressing virus and then mixed with uninfected ES cells at one-to-one ratio and cultured in the presence of leukemia inhibitory factor (LIF). The infected ES cells are GFP-positive and can be quantified by FACS analyses. The effects of miRNA expression on ES cell self-renewal were determined by measuring the percentage of GFP+ cells at every passage (Fig. 7A). We found that expression of Mir196a-1, Mir196a-2, or Mir196b reduced the relative ratio of GFP+ ES cells by 35%, 20%, and 50%, respectively, after 3 wk (Fig. 7B). In contrast, expression of Mir324, Mir221, or Mir222 had no apparent effects on the relative ratio of GFP+ ES cells (Fig. 7C). Expression of the Mir196 family did not result in premature ES cell differentiation since the mRNA levels of pluripotency genes (Nanog, Pou5f1 [also known as Oct4], Tbx3, and Klf4) are unaffected (Supplemental Fig. S6). These findings demonstrate that the Mir196 family of miRNAs, but not Mir324, Mir221, or Mir222, modulates the self-renewal of ES cells.
Figure 7.
Effects of SPT-miRNAs on ES cell self-renewal and HSC reconstitution. (A) Schematics depicting a competition assay for examining the effects of miRNAs on ES cell self-renewal. (B,C) Relative ratios of miRNA-infected ES cells in a competition assay (n = 3, mean ± SD). (D) Schematics depicting a competition assay for examining the effects of miRNAs on HSC reconstitution. Vector-specific TaqMan qPCR primers and probes were used to determine the relative levels of the miRNA viral integrations relative to that of control viral vector integration. (E) Relative ratios of miRNA-infected cells in a competitive bone marrow transplantation assay (n = 5). Results from individual recipients at various time points (within 5%–95% distribution) after transplantation are shown, and median values of all recipients (horizontal lines) are indicated.
We then examined the effects of SPT-miRNAs in a competitive bone marrow reconstitution assay (Fig. 7D). Lineage-negative hematopoietic stem/progenitor cells were spin-infected with control or miRNA-expressing viruses, pooled, and transplanted into lethally irradiated recipient mice. Viral titers were determined to ensure comparable infection rates by various miRNA-expressing viruses. A fraction of pooled infected cells were set aside, cultured, and used to determine initial levels of integration by control and miRNA-expressing viruses. At various time points after transplantation, we isolated peripheral blood cells from the recipient mice and quantified the levels of viral integrations using qPCR analyses. The relative ratios of integrated viruses at various time points after transplantation were determined by normalizing first to the level of control virus and then to the initial levels of integration by various viral constructs (Supplemental Fig. S7). Two infected pools were generated: (1) control, Mir196a-1, Mir196a-2, and Mir196b and (2) control, Mir324, Mir221, and Mir222 viruses. Each pool of infected cells was independently tested in five recipient mice (Fig. 7E). We found that the number of cells containing Mir324 integration was not significantly different from those with control integration at 4 to 12 wk post-transplantation and only slightly decreased at 16 wk post-transplantation. In contrast, the number of cells containing Mir221 and Mir222 integration decreased drastically, whereas those with Mir196a-1, Mir196a-2, and Mir196b integrations had more modest decreases over the period of 16 wk. These findings are consistent with a previous study that showed that expression of the mature miR-196b had a negative impact on HSC reconstitution potential (O'Connell et al. 2010). These results demonstrate that the Mir196 family, Mir221, and Mir222 miRNAs have negative effects on HSC reconstitution potential. Although further analyses will be necessary to dissect the roles of these miRNAs in homing, survival, self-renewal, proliferation, and differentiation of HSCs, these findings show that a high proportion of SPT-miRNAs (five out of six miRNAs tested) plays roles in HSC self-renewal and differentiation. Finally, the fact that ectopic expression of Mir196a-1, Mir196a-2, and Mir196b had similar effects on ES cells and HSCs indicates that these miRNAs might control conserved programs in distinct SC types.
Combinatorial regulation of Hox genes by the SPT-miRNAs
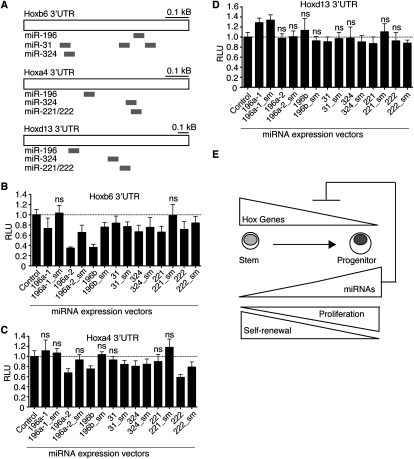
Since the SPT-miRNAs are up-regulated during the stem to progenitor transition, some of them may play roles in down-regulating genes that are important for various functional properties of the multipotent SCs, such as quiescence, proliferation rate, self-renewal, and differentiation. Ectopic expression of the SPT-miRNAs in SCs may prematurely down-regulate the targets that control these properties. In support of our hypothesis that signature miRNAs may function in SC self-renewal, we found that some Hox genes critical for HSC self-renewal are targeted by multiple SPT-miRNAs. For example, Hoxb6, Hoxa4, and Hoxd13 are predicted to contain multiple binding sites for the SPT-miRNAs based on the RNA22 miRNA-target prediction algorithm (Supplemental Figs. S8–S10). RNA22 is an inclusive target prediction program with no requirement for perfect seed (5′ 2–8 nt of mature miRNAs) matches (Miranda et al. 2006).
We used luciferase reporter assays to test whether UTRs from Hox genes can be repressed by the SPT-miRNAs (Fig. 8A). We found that Mir196a-2, Mir196b, and Mir222 specifically repressed expression of reporters with the Hox6 UTR (65%, 60%, and 25%, respectively) (Fig. 8B) and the Hoxa4 UTR (30%, 25%, and 40%, respectively) (Fig. 8C) but did not affect expression of a Hoxd13 UTR reporter (Fig. 8D). Hoxb6 was also specifically repressed by Mir196a-1 (25%) and Mir221 (30%) in a seed-dependent manner and by Mir31 (15%) and Mir324 (30%) in a seed-independent manner. Hoxa4 and Hoxb6 are both down-regulated during the hematopoietic stem to progenitor transition and have been shown to be essential for HSC self-renewal (Georgantas et al. 2004; Fischbach et al. 2005; Lebert-Ghali et al. 2010). Thus, the SPT-miRNAs may coordinately regulate targets that have known functions in self-renewal during the stem to progenitor transition (Fig. 8E). Further characterization of targets of the SPT-miRNAs may help to elucidate molecular networks active during SC self-renewal and differentiation.
Figure 8.
Coordinated regulation of Hox UTRs by the SPT-miRNAs. (A) Schematic diagrams of predicted target sites of SPT-miRNAs on Hoxb6, Hoxa4, and Hoxd13. Repression of luciferase reporters bearing UTRs from Hoxb6 (B), Hoxa4 (C), or Hoxd13 (D) by the SPT-miRNAs and corresponding seed mutant controls (sm) (n = 3, mean ± SD, two-tailed, type 2, Student t-test, compared to the control vector, N.S. for p > 0.05). (E) Model of SPT-miRNA regulation of Hox genes during the stem to progenitor transition and corresponding changes in self-renewal and proliferation potentials.
Discussion
Systematic analyses of miRNA expression profiles in TSCs and their differentiated counterparts revealed miRNAs unique to HSCs, MuSCs, and NSPCs, and those common to all three SC populations. The differential expression of miRNAs in SCs and progenitors suggests an extensive role for miRNAs in regulating self-renewal, proliferation, and quiescence programs in these cells. From these differentially regulated miRNAs, we have identified an SPT-miRNA signature that predicts the effects of developmental and genetic perturbations on the functional properties of stem/progenitor cells. Finally, many SPT-miRNAs have the ability to regulate ES cell and HSC functions and appear to coordinately regulate targets that have established roles in HSC self-renewal, such as Hoxb6 and Hoxa4 (Fig. 8; Supplemental Table S5).
Identification of miRNA programs characterized by differential expression in TSCs or during the transition from TSCs to transit amplifying progenitor cells may shed light on the roles of miRNAs in stem and progenitor cells. For example, miRNAs shared by all TSCs may control general cellular processes that are critical for multiple SCs, such as receptor-mediated signaling pathways, apoptosis, and cell cycle programs. Consistent with this hypothesis, 12 of the 18 SC-miRNAs have previously been shown to regulate the cell cycle: miR-16, miR-19b, miR-20, miR-24, miR-29c, miR-92, miR-106b, miR-195, miR-221, miR-222, let-7b, and let-7c, (Zhang et al. 2007; Lal et al. 2008; Liu et al. 2008b; Medina et al. 2008; Mendell 2008; Park et al. 2009; Xu et al. 2009). In contrast, the TSC-miRNAs—miRNAs that are unique to TSC populations—may regulate lineage-specific functions of these TSCs. Moreover, since the transition from quiescent LT-HSCs into rapidly cycling KSL cells requires a major shift in cell cycle rate, it is likely that some of these miRNAs coordinate programs associated with this transition. Indeed, many miRNAs up-regulated in KSL cells modulate cell cycle progression. These include miR-19a, miR-19b, miR-20, miR-25, miR-92, miR-93, miR-106b, miR-221, miR-222, let-7b, let-7c, and let-7g (Fig. 3B). One of the predicted targets for the LT-HSC–specific miRNA, miR-192, is Noggin, which encodes a BMP4 antagonist (Supplemental Fig. S5). Since BMP4 is a key signal in HSC development and expansion (Sadlon et al. 2004), miR-192 expression may control Noggin expression and fine-tune BMP4 signaling in LT-HSCs. Interestingly, few SC-specific miRNAs were found despite the relatively relaxed standard that was used to define SC-related miRNAs (Supplemental Fig. S5).
It is important to note that it may not be straightforward to assign miRNA function in SCs based on function in other cell types. The same miRNA may have distinct functions in different cells types depending on the target milieu of the cell. For example, some SC-related miRNAs have been shown to promote passage through cell cycle in certain cell types, although other SC-related miRNAs have been shown to inhibit it (Liu et al. 2008b; Medina et al. 2008). Interestingly, among the SC-related miRNAs, the miR-17-92 cluster acts as an oncogene to potentiate the MYC activity in a mouse B-cell lymphoma model, whereas it antagonizes the effects of MYC oncogene in human B-cell lymphoma cells, acting as a classic tumor-suppressor gene (He et al. 2005; O'Donnell et al. 2005). These observations suggest that the same miRNA can have distinct biological activities under different cellular contexts. Such puzzling observations can be explained if miRNA function depends on the unique target milieu of each cell type.
One should be cautious about inferring miRNA function solely based on their relative levels in different cell types. Although it is thought that miRNAs function through base-pairing with their targets, little is known about the correlation between the levels of mature miRNAs and their efficacy in target repression and whether miRNAs function through stoichiometric binding or a catalytic mechanism in vivo. It is also not known whether the relative abundance of the miRNA and corresponding targets affects repression efficacy. In some cases, low abundance miRNAs may play critical roles in controlling expression of low abundance targets. Furthermore, even though an miRNA is maintained at a steady level during a cell fate transition, its ability to regulate its target may be abolished by drastic increases in transcription of the cognate target mRNA.
Clearly, our analyses revealed broad and dynamic differences in miRNA profiles among normal and aberrant TSCs and their differentiated counterparts. Future implementation of deep sequencing analyses on the rare SC populations may further improve this SC miRNA atlas and permit the discovery of novel miRNAs in SCs. Nevertheless, given that each miRNA can regulate hundreds of targets, these results strongly suggest that miRNA-controlled post-transcriptional programs modulate extensive networks that define the functional properties of SC and progenitors. Compared to other forms of gene regulation, such as chromatin remodeling and transcriptional regulation, miRNA-mediated post-transcriptional regulation sits at the step immediately preceding protein synthesis and dictates the levels of proteins synthesized from large numbers of genes—the final outputs of cellular genetic networks. Thus, linking the functions of miRNAs in SC self-renewal, quiescence, and differentiation to the cognate target networks provides an opportunity to unravel the evolutionarily selected molecular networks that control critical biological processes in SCs. The stem and progenitor cell miRNA-expression atlas described here will provide a resource for dissecting the post-transcriptional genetic networks in normal and mutant stem and progenitor cells.
Methods
Stem and progenitor cell samples and miRNA qPCR
Stem and progenitor cell samples and methods of isolation are listed in Table 1 and were performed as described in the original references listed. Defined numbers of cells were FACS sorted or aliquoted into each tube. The samples were then lysed, amplified, and quantified using multiplex RT for TaqMan MicroRNA Assays according to manufacturer's instructions from Applied Biosystems and the method described by Chen et al. (2005, 2007). For all FACS-sorted cell populations, the purity was ensured by double sorting and subsequent FACS analyses (>90%). One thousand cells were used as starting material for all samples except for LT-HSCs; as few as 20 LT-HSCs were utilized. Multiplex miRNA qPCR does not significantly bias miRNA expression profiles after varied cycles of amplification (data not shown). A total of 459 functional miRNA probes were used to profile and quantify miRNA expression in normal and aberrant SC and progenitor cells (Supplemental Table S6).
Comprehensive analyses were carried out to ensure data quality. Nontemplate control analyses (NTCs) were carried out for all samples to remove probes that would lead to nonspecific amplification. Probe sets specific for housekeeping and SC-specific mRNAs, such as HPRT, snoRNAs, and CD34, were included in the miRNA probe sets and co-amplified. Drastic deviation in the amplification of housekeeping gene HPRT was used an indicator of low quality amplification. We have also carried out extensive analyses of Megaplex PreAmp TaqMan MicroRNA Assays with a range of input cell numbers (1000, 100, 10, and single cells). The miRNA expression profiles were strongly correlated with R2 = 0.93 or higher and P < 0.01, and technical variability of single cell miRNA profiling with Megaplex PreAmp TaqMan MicroRNA Assays averaged about 9% (C. Chen, pers. comm.).
Data analyses
Ct values of miRNA probes were used to indicate corresponding miRNA levels within the cell sample. Redundant and overlapping probes were also removed. miRNAs with Ct > 35 were considered undetectable and transformed to Ct = 35. For comparative analysis, miRNA expression within each sample was normalized by subtracting the median Ct value of detectable miRNAs within the sample from the miRNA Ct value to obtain a ΔCt value (Supplemental Fig. S2B; Supplemental Table S2). Mean, SEM, median, and IQR were calculated with Prism software.
Statistical analyses, PCAs, HCL, and KMC analyses were carried out using TM4 microarray software suites (Saeed et al. 2006). One-way ANOVA analyses (95% confidence interval) or SAM analyses (FDR < 0.001) identified miRNAs differentially expressed for multi-sample and pairwise comparisons, respectively. HCL and KMC analyses were then used to generate heatmaps depicting differentially expressed miRNA clusters. Results of PCA analyses were centered across all samples. HCL analyses showed that all replicates clustered together except for three (KSL-Sp sample 3, KSL-PTEN sample 3, and NSPC-P0 sample 3). Sample-specific miRNAs (Supplemental Fig. S5) were determined using the following criteria: an unpaired t-test with P < 0.05 relative to the NTC samples and mean Ct < 32.
A multi-factorial analysis classified miRNAs into five groups (FDR < 0.05) based on their ANOVA structure via step-wise significance tests (for more detailed explanation, see Supplemental material). PAM (FDR < 0.001) was used to identify the predictive miRNA signature from miRNAs differentially expressed in blood and muscle stem and progenitor samples and capable of predicting the functional properties of the following sample sets: LT-HSC vs. KSL; MuSC vs. myoblast; KSL-PTEN vs. KSL-Sp; KSL-ETO vs. KSL; MLL-LSC vs. MLL-Prog; KSL-RbTKO vs. KSL.
ES cell competition assay
Murine CGR8 ES cells were cultured on irradiated mouse embryonic fibroblasts in 15% ES FBS (Omega Scientific, lot no. 104100), 0.1 mM β-mercaptoethanol, NEAA, penicillin, streptomycin, and 103 U/mL LIF. ES cells were infected with control or miRNA vector viruses via spin inoculation as previously described (Chen et al. 2004; Liu et al. 2008a). ES cells were passaged every 3 d, and the percentage of cells expressing GFP was measured on FACSCalibur (BD Biosciences).
HSC reconstitution assay
Bone marrow cells were isolated from C57/BL6J mice (Jackson Laboratory) treated with 5-fluorouracil, fractionated with a Ficoll gradient, and infected with control or miRNA viruses by spinoculation (Chen et al. 2004). Viral titers were determined by infecting ES cells. Titers were normalized to ensure comparable infection efficiencies by control and miRNA viruses. Equal proportions of infected cells were pooled to make two groups containing the following viruses: (1) control, Mir196a-1, Mir196a-2, and Mir196b and (2) control, Mir324, Mir221, and Mir222. About 250,000 pooled cells were injected into each recipient mouse. Five recipient mice were generated for each group. A portion of infected cells from each group were cultured for 48 h. Genomic DNA samples were prepared from the cultured cells or peripheral blood isolated at various time points after transplantation. Relative ratios of miRNA integration (compared to control viral integration) were determined by qPCR using TaqMan assays specific for control and miRNA vectors and GAPDH (as a control for DNA input). The ratios of miRNA integration relative to control viral integration at a specific time point after transplantation were calculated by determining the ΔΔCt[(CtmiR vector − CtGAPDH) − (CtControl Vector − CtGAPDH)] and then compared to the corresponding ΔΔCt at time zero.
Target prediction and luciferase reporter assays
Target predictions were performed using RNA22 (Miranda et al. 2006) with the following criteria: zero unpaired bases in the six seed nucleotides, 14 minimum paired bases, and −25 kcal/mol folding energy in the heteroduplex. Luciferase assays were performed as previously described (Trujillo et al. 2010). A list of primer sequences is available in the Supplemental material.
Acknowledgments
We thank the members of the Chen laboratory and Drs. Michael Longaker and Wing H. Wong for helpful discussions and/or comments on the manuscript. This work was supported by NIH R01, the Distinguished Young Scholar Award from the W.M. Keck foundation, an NIH Director's Pioneer Award, and Baxter and Terman faculty awards to C.-Z.C, Leukemia and Lymphoma Society Scholar awards to J.S. and J.M.P., and a CIRM training grant to C.P.A.
Footnotes
[Supplemental material is available for this article. The microRNA expression data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE28036.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.111385.110.
References
- Akashi K, He X, Chen J, Iwasaki H, Niu C, Steenhard B, Zhang J, Haug J, Li L 2003. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood 101: 383–389 [DOI] [PubMed] [Google Scholar]
- Argiropoulos B, Humphries RK 2007. Hox genes in hematopoiesis and leukemogenesis. Oncogene 26: 6766–6776 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle JC, Armour CD, Lower M, Haynor D, Biery M, Bouzek H, Chen R, Jackson S, Johnson JM, Rohl CA, et al. 2010. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS ONE 5: e11779 doi: 10.1371/journal.pone.0011779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Boles NC, Lin KY, Tierney MP, Bowman TV, Bradfute SB, Chen AJ, Merchant AA, Sirin O, Weksberg DC, et al. 2007. Hematopoietic fingerprints: An expression database of stem cells and their progeny. Cell Stem Cell 1: 578–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li M, de Graaf D, Monti S, Gottgens B, Sanchez MJ, Lander ES, Golub TR, Green AR, Lodish HF 2002. Identification of endoglin as a functional marker that defines long-term repopulating hematopoietic stem cells. Proc Natl Acad Sci 99: 15468–15473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Li M, Lodish HF 2003. The endoglinpositive sca-1positive rhodaminelow phenotype defines a near-homogeneous population of long-term repopulating hematopoietic stem cells. Immunity 19: 525–533 [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86 [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179 doi: 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon D, Lee CT, Blake J, Sun Y, Strauss WM 2007. Defining embryonic stem cell identity using differentiation-related microRNAs and their potential targets. Mamm Genome 18: 316–327 [DOI] [PubMed] [Google Scholar]
- de Guzman CG, Warren AJ, Zhang Z, Gartland L, Erickson P, Drabkin H, Hiebert SW, Klug CA 2002. Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation. Mol Cell Biol 22: 5506–5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco F, et al. 2007. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell 12: 457–466 [DOI] [PubMed] [Google Scholar]
- Fischbach NA, Rozenfeld S, Shen W, Fong S, Chrobak D, Ginzinger D, Kogan SC, Radhakrishnan A, Le Beau MM, Largman C, et al. 2005. HOXB6 overexpression in murine bone marrow immortalizes a myelomonocytic precursor in vitro and causes hematopoietic stem cell expansion and acute myeloid leukemia in vivo. Blood 105: 1456–1466 [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgantas RW 3rd, Tanadve V, Malehorn M, Heimfeld S, Chen C, Carr L, Martinez-Murillo F, Riggins G, Kowalski J, Civin CI 2004. Microarray and serial analysis of gene expression analyses identify known and novel transcripts overexpressed in hematopoietic stem cells. Cancer Res 64: 4434–4441 [DOI] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V 2009. nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell 136: 926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. 2005. A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR 2002. A stem cell molecular signature. Science 298: 601–604 [DOI] [PubMed] [Google Scholar]
- Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, et al. 2010. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 32: 828–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R Jr, Srikantan S, Subrahmanyam R, Martindale JL, Yang X, Ahmed F, et al. 2008. p16(INK4a) translation suppressed by miR-24. PLoS One 3: e1864 doi: 10.1371/journal.pone.0001864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebert-Ghali CE, Fournier M, Dickson GJ, Thompson A, Sauvageau G, and Bijl JJ 2010. HoxA cluster is haploinsufficient for activity of hematopoietic stem and progenitor cells. Exp Hematol 38: 1074–1086 [DOI] [PubMed] [Google Scholar]
- Liu G, Min H, Yue S, Chen CZ 2008a. Pre-miRNA loop nucleotides control the distinct activities of mir-181a-1 and mir-181c in early T cell development. PLoS ONE 3: e3592 doi: 10.1371/journal.pone.0003592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X 2008b. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res 36: 5391–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. 2005. MicroRNA expression profiles classify human cancers. Nature 435: 834–838 [DOI] [PubMed] [Google Scholar]
- Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, Stein GS 2008. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res 68: 2773–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT 2008. miRiad roles for the miR-17-92 cluster in development and disease. Cell 133: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Feys T, Bernard N, Guenther S, Chen C, Speleman F, Vandesompele J 2008. High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res 36: e143 doi: 10.1093/nar/gkn725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I 2006. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217 [DOI] [PubMed] [Google Scholar]
- Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A 2006. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol 8: 278–284 [DOI] [PubMed] [Google Scholar]
- Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA 2008. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454: 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D 2010. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci 107: 14235–14240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- Park SY, Lee JH, Ha M, Nam JW, Kim VN 2009. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol 16: 23–29 [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA 2002. “Stemness”: Transcriptional profiling of embryonic and adult stem cells. Science 298: 597–600 [DOI] [PubMed] [Google Scholar]
- Rando TA, Blau HM 1994. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol 125: 1275–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, et al. 2009. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5: 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM 2008. Self-renewal and expansion of single transplanted muscle stem cells. Nature 456: 502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlon TJ, Lewis ID, D'Andrea RJ 2004. BMP4: Its role in development of the hematopoietic system and potential as a hematopoietic growth factor. Stem Cells 22: 457–474 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J 2006. TM4 microarray software suite. Methods Enzymol 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136: 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C 2008. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol Cell Biol 28: 4609–4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, Chang HY, Shurtleff SA, Downing JR, Cleary ML 2009. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell 4: 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G 2002. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci 99: 6567–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo RD, Yue SB, Tang Y, O'Gorman WE, Chen CZ 2010. The potential functions of primary microRNAs in target recognition and repression. EMBO J 29: 3272–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viatour P, Somervaille TC, Venkatasubrahmanyam S, Kogan S, McLaughlin ME, Weissman IL, Butte AJ, Passegue E, Sage J 2008. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell 3: 416–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R 2007. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39: 380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltering JM, Durston AJ 2008. MiR-10 represses HoxB1a and HoxB3a in zebrafish. PLoS ONE 3: e1396 doi: 10.1371/journal.pone.0001396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P, Iwasaki M, Somervaille TC, Ficara F, Carico C, Arnold C, Chen CZ, Cleary ML 2010. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res 70: 3833–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM 2009. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology 50: 113–121 [DOI] [PubMed] [Google Scholar]
- Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, Wang Y, Chen IM, Chen Z, Rowley JD, Willman CL, et al. 2006. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med 12: 945–949 [DOI] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304: 594–596 [DOI] [PubMed] [Google Scholar]
- Yu JY, Reynolds SH, Hatfield SD, Shcherbata HR, Fischer KA, Ward EJ, Long D, Ding Y, Ruohola-Baker H 2009. Dicer-1-dependent Dacapo suppression acts downstream of Insulin receptor in regulating cell division of Drosophila germline stem cells. Development 136: 1497–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, et al. 2006. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 441: 518–522 [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA 2007. microRNAs as oncogenes and tumor suppressors. Dev Biol 302: 1–12 [DOI] [PubMed] [Google Scholar]
- Zhou B, Wong WH 2011. A bootstrap-based non-parametric ANOVA method with applications to factorial microarray data. Statistica Sinica 21: 495–514 [Google Scholar]