Abstract
Activation of intracellular transient receptor potential vanilloid-1 (TRPV1) in human lung cells causes endoplasmic reticulum (ER) stress, increased expression of proapoptotic GADD153 (growth arrest- and DNA damage-inducible transcript 3), and cytotoxicity. However, in cells with low TRPV1 expression, cell death is not inhibited by TRPV1 antagonists, despite preventing GADD153 induction. In this study, chemical variants of the capsaicin analog nonivamide were synthesized and used to probe the relationship between TRPV1 receptor binding, ER calcium release, GADD153 expression, and cell death in TRPV1-overexpressing BEAS-2B, normal BEAS-2B, and primary normal human bronchial epithelial lung cells. Modification of the 3-methoxy-4-hydroxybenzylamide vanilloid ring pharmacophore of nonivamide reduced the potency of the analogs and rendered several analogs mildly inhibitory. Correlation analysis of analog-induced calcium flux, GADD153 induction, and cytotoxicity revealed a direct relationship for all three endpoints in all three lung cell types for nonivamide and N-(3,4-dihydroxybenzyl)nonanamide. However, the N-(3,4-dihydroxybenzyl)nonanamide analog also produced cytotoxicity through redox cycling/reactive oxygen species formation, shown by inhibition of cell death by N-acetylcysteine. Molecular modeling of binding interactions between the analogs and TRPV1 agreed with data for reduced potency of the analogs, and only nonivamide was predicted to form a “productive” ligand-receptor complex. This study provides vital information on the molecular interactions of capsaicinoids with TRPV1 and substantiates TRPV1-mediated ER stress as a conserved mechanism of lung cell death by prototypical TRPV1 agonists.
Introduction
The capsaicin receptor, TRPV1, is an emerging pharmacological target with many hypothesized roles in mammalian physiology, pathology, and disease (Jia and Lee, 2007; Szallasi et al., 2007). Endogenous and exogenous noxious stimuli including temperature, acid, proinflammatory mediators, and chemical irritants activate TRPV1 (Caterina et al., 1997; Tominaga et al., 1998; Szallasi et al., 2007). The prototypical agonist of TRPV1 is capsaicin (8-methyl-N-vanillyl-trans-6-nonenamide), the pungent compound in chili peppers.
TRPV1 is expressed by sensory neurons and non-neuronal cells. Activation of TRPV1-expressing nerves in the airways elicits cough, dyspnea, and neurogenic inflammation (Geppetti et al., 2006, 2008; Jia and Lee, 2007; Nassini et al., 2010). In lung epithelial cells, coupling of TRPV1 and cellular responses is less understood, but studies suggest that TRPV1 regulates host defenses through the regulation of proinflammatory cytokine and chemokine genes (Veronesi et al., 1999a,b; Oortgiesen et al., 2000; Veronesi and Oortgiesen, 2001; Reilly et al., 2003b, 2005).
Rats exposed to capsaicin aerosols exhibited cell loss in the conducting airways and bronchial and alveolar cell damage (Reilly et al., 2003b). In cultured human lung cells, capsaicin and other TRPV1 agonists activated TRPV1 and promoted both time- and dose-dependent cytokine release, ER stress, and cell death (Reilly et al., 2003b, 2005; Thomas et al., 2007). Increased expression of interleukin-6 and interleukin-8 were associated with activation of cell surface TRPV1, and these molecules contribute to pulmonary inflammation and neutrophilia in the lung (Reilly et al., 2005; Johansen et al., 2006; Thomas et al., 2007). Activation of ER-associated/intracellular TRPV1 causes ER stress, activation of eukaryotic translation initiation factor 2α kinase, induction of GADD153 mRNA and protein, and cell death (Reilly et al., 2003b, 2005; Johansen et al., 2006; Thomas et al., 2007). This latter process is proposed to be the mechanism by which capsaicin damages epithelial cells in the respiratory tract.
Previous studies of TRPV1 agonist toxicity in lung cells demonstrated that blocking TRPV1 with the cell-permeable antagonist N-(4-tert-butylbenzyl)-N′-(1-[3-fluoro-4-(methylsulfonylamino)-phenyl]ethyl)thiourea (LJO-328) ameliorated ER stress and increases in GADD153 expression, which correlated with inhibition of cytotoxicity in TRPV1-overexpressing, but not in BEAS-2B, A549, or normal human bronchial epithelial (NHBE) cells that express only low levels of TRPV1; rather antagonists exacerbated toxicity (Reilly et al., 2003b, 2005; Thomas et al., 2007). These results raised concerns regarding the importance of TRPV1 in mediating cell death in “normal” lung cells and encouraged this structure-activity relationship (SAR) approach to evaluate the requirement for TRPV1 binding, activation, and ER stress in lung death by capsaicinoids.
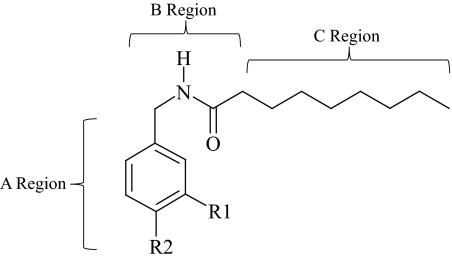
A series of capsaicin/nonivamide analogs with modified vanilloid ring pharmacophores (Fig. 1 and Table 1) were synthesized to differentiate between TRPV1-mediated and nonspecific cell death in lung cells with vastly different TRPV1 expression levels and sensitivity to capsaicinoids. Previous SAR studies have revealed that a specific aromatic A-ring configuration, the presence of a hydrogen bond-donating group in the B region, and a hydrophobic hydrocarbon C-region tail of 8 to 12 carbons are required for maximum potency at TRPV1 (Fig. 1) (Walpole et al., 1993a,b,c). For the A region the 3-methoxy-4-hydroxybenzyl analogs were the most potent, and removal, alkylation, or switching of the 4-OH or 3-MeO moieties and modification of positions 2, 5, and 6 of the ring reduced activity, indicating stringent binding interactions between TRPV1 and the A-ring region. This notion is supported by the finding that 6-iodo-nordihydrocapsaicin is a potent TRPV1 antagonist (Appendino et al., 2003) and indicates a requirement for a hydrogen-bonding moiety at the 4-position. However, the 3,4-dihydroxy catechol analogs of capsaicin exhibited approximately equal potency to the 3-MeO-4-OH cognates in vitro using calcium flux assays, but were less potent in animal-based analgesia models. Finally, site-directed mutagenesis studies of TRPV1 to identify species-specific determinants of differential sensitivity to capsaicin and resiniferatoxin and molecular modeling of the capsaicin binding region have identified amino acid residues and structural features important for binding and activation including Tyr511, Ser512, Met547 (in rat), Trp549, Thr550, Met 552, and Leu553 and a hydrophobic pocket composed of Trp549, Met552, and Leu553 (Jordt and Julius, 2002; Gavva et al., 2004; Ryu et al., 2008). These studies generally concur with the chemical features identified by Walpole et al. (1993a,b,c) and have facilitated efforts to design more potent and selective TRPV1 agonists and antagonists. These studies also provided guidance for analogs that should be prepared and evaluated in the current study.
Fig. 1.
Annotated structure of nonivamide.
TABLE 1.
Structural composition of the capsaicinoid analogs used in this study
| Analog Name | Abbreviation | R1 | R2 |
|---|---|---|---|
| N-(3-Methoxy-4-hydroxybenzyl)nonanamide | Nonivamide | OCH3 | OH |
| N-Benzylnonanamide | Benzyl | H | H |
| N-(3-Methoxybenzyl)nonanamide | 3-MeO | OCH3 | H |
| N-(3,4-Dimethoxybenzyl)nonanamide | 3,4-DiMeO | OCH3 | OCH3 |
| N-(3-Hydroxy-4-methoxybenzyl)nonanamide | 3-OH-4-MeO | OH | OCH3 |
| N-(3,4-Dihydroxybenzyl)nonanamide | 3,4-DiOH | OH | OH |
| 3-Methoxy-4-(nonamidomethyl)phenyl sulfate | Sulfate | OCH3 | SO4 |
| N-(4-Trifluoromethylbenzyl)nonanamide | CF3 | H | CF3 |
| N-(4-Hydroxybenzyl)nonanamide | 4-OH | H | OH |
Materials and Methods
Chemicals.
Nonivamide (N-vanillylnonanamide or N-(4-hydroxy-3-methoxybenzyl)nonanamide), ionomycin, N-acetylcysteine (NAC), dimethyl sulfoxide, 3-hydroxy-4-methoxybenzylamine-HCl, 3,4-dihydroxybenzylamine-HBr, 3,4-dimethoxybenzylamine 4-triflouromethybenzylamine, 3-methoxybenzylamine, nonanoyl chloride, glutathione, and sulfinpyrazone were purchased from Sigma-Aldrich (St. Louis, MO). 4-Hydroxybenzylamine was from Matrix Scientific (Columbia, SC). Nonivamide and capsaicin have indistinguishable toxicity profiles in the human lung cells used in this study, thus nonivamide was used as the prototype TRPV1 agonist because of its facile synthesis and availability of reagents to prepare variants. LJO-328 (Reilly et al., 2005) was synthesized by J.L. PCR primers for TRPV1, GADD153, and β2 macroglobulin (β2M) were synthesized at the University of Utah Core Facilities, and cell culture media were purchased from Invitrogen (Carlsbad, CA) unless otherwise stated. All other reagents were purchased form established suppliers.
Synthesis of Capsaicinoid Analogs.
All analogs, except the sulfate analog, were synthesized as described previously for N-benzylnonanamide (Thomas et al., 2007). Analogs containing a hydroxyl group were deacetylated by reacting with sodium methoxide. Product purity and structure were determined by high-performance liquid chromatography with UV detection at 230 nm, liquid chromatography/mass spectrometry, and 1H NMR. Detailed synthetic methods and NMR and mass spectrometry data of the analogs are provided as Supplemental Data Section 1.
Cell Culture.
Immortalized human bronchial epithelial cells (BEAS-2B) were purchased from the American Type Culture Collection (Manassas, VA), and TRPV1 overexpressing BEAS-2B (TRPV1-OE) cells were generated as described previously (Reilly et al., 2003b). BEAS-2B and TRPV1-OE cells were cultured in LHC-9 media. NHBE cells were purchased from Lonza Walkersville, Inc. (Walkersville, MD) and cultured in bronchial epithelial cell growth medium. Culture flasks were coated with LHC basal media fortified with 30 μg/ml collagen, 10 μg/ml fibronectin, and 10 μg/ml bovine serum albumin. Cells were maintained between 30 and 90% maximum density and were subcultured every 2 to 4 days depending on cell growth rates.
Cytotoxicity/Dose-Response Assays.
Cells were subcultured into 96-well plates, grown to ∼90% confluence, and treated for 24 h at 37°C with increasing concentrations of chemical. Treatment solutions were prepared using LHC-9 media for BEAS-2B and TRPV1-OE cells and Opti-MEM I for NHBE cells. Treatment solutions contained ≤0.5% dimethyl sulfoxide. Cell viability was assessed using the Dojindo cell counting kit-8 (Dojindo Laboratories, Gaithersburg MD).
Fluorometric Calcium Assays.
TRPV1-OE cells were used to evaluate calcium flux as a consequence of TRPV1 activation. Cells were subcultured into 96-well plates and grown to approximately 90% maximum density. Cells were loaded with Fluo-4-acetoxymethyl ester (2.5 μM) (Invitrogen) for 60 min at room temperature (approximately 22°C), in the presence of 200 μM sulfinpyrazone in LHC-9 media. Cells were washed twice and incubated at room temperature for an additional 20 min in sulfinpyrazone containing media to permit methyl ester hydrolysis and activation of Fluo-4 in cells. Changes in cellular fluorescence were measured over 60 s using a NOVOStar plate reader (BMG Labtech GmbH, Offenburg, Germany) with excitation at 485 nm and emission at 520 nm. Baseline readings were normalized, and the maximum fluorescence intensity was determined (n = 3 for each treatment). Ionomycin (10 μM) was also used to normalize responses.
Quantitative Real-Time PCR Analysis of Gene Expression.
Cells were subcultured into six-well cell culture plates and grown to ∼90% density. Cells were treated with TRPV1 agonists and antagonists for 4 h at 37°C. Total RNA was extracted from cells using the Invitrogen PureLink Micro-to-Midi Total RNA Purification kit, and 1 μg of the total RNA was transcribed into cDNA using Superscript III (Invitrogen). cDNA was diluted 1:100 in sterile H2O, and gene-specific cDNA was amplified by PCR using RT2 SYBR Green qPCR Master Mix from SA Biosciences (Frederick, MD) in 25-μl reaction volumes on a Chromo 4 Real-Time Detection System (Bio-Rad Laboratories, Hercules, CA) using MJ Opticon Monitor 3 software (Bio-Rad Laboratories). The PCR program used consisted of an initial 10-min incubation at 95°C, followed by 40 cycles of 95°C for 15 s, 55°C for 30 s, then 72°C for 30 s. Experiments were performed in triplicate with a copy number standard curve for both the normalization gene (β2M) and the genes of interest. Primer sequences were (5′ → 3′): β2M, sense GATGAGTATGCCTGCCGTGTG and antisense CAATCCAAATGCGGCATCT; TRPV1, sense CTGCGGACCCACTCCAAAA and antisense CCTCGTGAGGGCAATCCAC; GADD153, sense AGAACCAGGAAACGGAAACAGA and antisense TCTCCTTCATGCGCTGCTTT.
Molecular Modeling.
The structures of nonivamide and the analogs were drawn using Gaussview 3.0 (Gaussian, Inc., Wallingford, CT) and optimized in the gas-phase using Gaussian 03 software at the HF/6-31G* level of theory. All ligands were treated as closed shell neutral species, except the sulfate analog, which was assigned −1 net charge. The stability of minima were confirmed using frequency analysis. The quantum mechanics optimized analog structures and a model of TRPV1 transmembrane helices 3 and 4 (residues Ser510–Ile569), provided as a Protein Data Bank-formatted file by Dr. Narender Gavva (Amgen Inc., Thousand Oaks, CA) (Gavva et al., 2004), were prepared for docking with Autodock Tools (The Scripps Research Institute, La Jolla, CA). Hydrogen atoms were added, Kollman charges were assigned to the protein template, and torsions and charges were assigned with Autodock tools for each ligand. The active search space for docking was defined and prepared using Autogrid 3.0 (The Scripps Research Institute), and a search grid box was constructed centered at coordinates −5.572, 4.504, and 3.77 with dimensions 80, 58, and 58 and the default grid spacing of 0.375 Å. This active space included the cleft previously identified to be important for ligand/receptor interactions. Each ligand was docked into the TRPV1 model using the same parameters with AutoDock 3.05 software (The Scripps Research Institute). These searches were performed with 200 runs using the Lamarckian genetic algorithm for each ligand structure, and the remaining Lamarckian genetic algorithm search parameters were left at default values. The docking results were then clustered based on docking energy. The clustering results based on 2-Å root mean square deviation tolerance of the predicted substrate geometry were then visualized using Autodock tools for qualitative comparison of binding interactions for the analogs with the TRPV1 helices. Representative structures of each of the most highly populated and lowest energy conformation clusters for the docked structures were saved as Protein Data Bank files and used to generate figures for publication with the Chimera package 1.4.1 from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco. Hydrogen bonding was predicted using Chimera with default cutoff criteria of 5-Å bond length and a 20° bond angle.
Statistical Analysis.
Statistical analysis was performed using 95% confidence intervals as the limit for significance. All results are represented as mean ± S.E.M. unless otherwise indicated. For comparisons of two groups, unpaired Student's t test was used. To compare more than two groups, one-way ANOVA analysis with Newman-Keuls multiple comparison test for post hoc testing was used, as indicated in figure legends. Statistical analyses were performed using Prism version 4.02 for Windows (GraphPad Software Inc., San Diego, CA).
Results
Structural attributes of each analog are presented in Fig. 1 and Table 1. Synthetic methods and analytical data are provided as supplemental data.
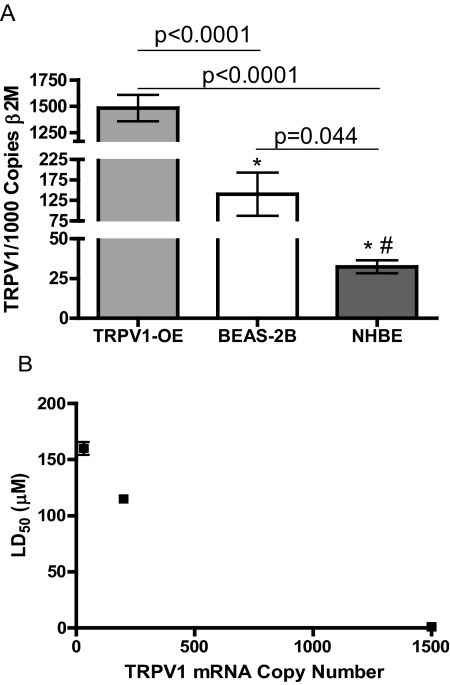
TRPV1 mRNA was quantified using RT-qPCR in TRPV1-OE, BEAS-2B, and NHBE cells. TRPV1-OE cells expressed ∼1.5 copies of TRPV1 mRNA/copy β2M, which was ∼10 times more than BEAS-2B cells (0.14 copies/copy β2M), and ∼45 times more than NHBE cells (0.03 copies/copy β2M). (Fig. 2A). Correlation of the mean copy number for TRPV1 mRNA with cytotoxicity for the three cell types (Fig. 2B) produced a Pearson R value of −0.99; however, this was not significant with the limited number of samples (p = 0.1) (Supplemental Data Section 2).
Fig. 2.
Quantification of TRPV1 mRNA in TRPV1-OE, BEAS-2B, and NHBE cells by RT-qPCR (A) and correlation of copy number with the LD50 of nonivamide for each cell type (B). Copy numbers values were compared using Student's t test. Correlation values are in the order of NHBE, BEAS-2B, and TRPV1-OE cells and were analyzed using the correlation plot analysis feature provided by GraphPad Prism 5.0 software.
The cytotoxicity of each analog was determined, and the LC50 values for each analog in TRPV1-OE, BEAS-2B, and NHBE cells are presented in Table 2. TRPV1-OE cells were most sensitive to nonivamide and N-(3,4-dihydroxybenzyl)nonanamide with LC50 values of 1 and 3.4, respectively. All other analogs produced LC50 values >63 μM, with N-benzylnonanamide and N-(3,4-dimethoxybenzyl)nonanamide exhibiting the lowest toxicity with LC50 values >250 μM. BEAS-2B cells were also most sensitive to nonivamide, N-(3,4-dihydroxybenzyl)nonanamide, and 3-methoxy-4-(nonamidomethyl)phenyl sulfate with LC50 values of 115, 16, and 87 μM, respectively, but they were also sensitive to N-(3-methoxybenzyl)nonanamide at ∼100 μM, similar to the TRPV1-OE cells (LC50 116 μM) and NHBE cells (LC50 106 μM), suggesting a reduction in TRPV1-mediated signaling in cell death. All other analogs produced LC50 values >164 μM, and N-benzylnonanamide and N-(3,4-dimethoxybenzyl)nonanamide were again the least toxic with LC50 values >250 μM. NHBE cells exhibited comparable sensitivity to the analogs as BEAS-2B cells, with the exception that they were markedly less sensitive to nonivamide and N-(3,4-dihydroxybenzyl)nonanamide. These results indicate that the abundance of TRPV1 expression is generally predictive of the relative sensitivity of lung cells to N-(3-hydroxy-4-methoxybenzyl)nonanamide, N-(3,4-dihydroxybenzyl)nonanamide, and N-(4-trifluoromethybenzyl)nonanamide, as presented above for nonivamide, but not for the other analogs, indicating that TRPV1 binding and cytotoxicity were not directly coupled for the lowest potency analogs.
TABLE 2.
LC50 values for capsaicinoid analogs in TRPV1-OE, BEAS-2B, and NHBE cells
Data represent the calculated LC50 for each analog in each cell type with the 95% confidence interval (CI).
| Analog | LC50 (95% CI) |
||
|---|---|---|---|
| TRPV1-OE | BEAS-2B | NHBE | |
| μM | |||
| Nonivamide | 1.0 (0.7–1.4) | 115 (104–127) | 160 (156–164) |
| N-Benzylnonanamide | >250 | >250 | >200 |
| N-(3-Methoxybenzyl)nonanamide | 116 (102–133) | 101 (95–106) | 106 (46–171) |
| N-(3,4-Dimethoxybenzyl)nonanamide | >250 | >250 | >200 |
| N-(3-Hydroxy-4-methoxybenzyl)nonanamide | 75.0 (62–91) | 217 (165–286) | 181 (121–270) |
| N-(3,4-Dihydroxybenzyl)nonanamide | 3.4 (2.6–4.3) | 16 (13–19) | 157 (121–203) |
| 3-Methoxy-4-(nonamidomethyl)phenyl sulfate | 63 (47–86) | 87 (84–91) | 58 (46–73) |
| N-(4-Trifluoromethylbenzyl)nonanamide | 113 (85–150.0) | 184 (145–234) | 174 (109–277) |
| N-(4-Hydroxybenzyl)nonanamide | 148 (122–180) | 164 (135–198) | 165 (113–242) |
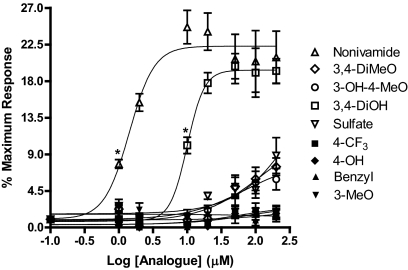
TRPV1 activation by the analogs was quantified using calcium flux (Fig. 3). TRPV1-OE cells treated with nonivamide and N-(3,4-dihydroxybenzyl)nonanamide exhibited dose-dependent, up to 25-fold, increases in cytosolic calcium content, with EC50 values of 1.4 and 10 μM, respectively. All other analogs were markedly less potent with maximum changes in fluorescence between 0 and 9 at 200 μM. The EC50 values for N-(3-hydroxy-4-methoxybenzyl)nonanamide, N-(3,4-dimethoxybenzyl)nonanamide, and 3-methoxy-4-(nonamidomethyl)phenyl sulfate were 70, 120, and 470 μM, respectively, whereas EC50 values for the other analogs could not be estimated because of little, if any, response. Comparisons of calcium flux at 2, 20, 100, and 200 μM for all of the analogs are presented in Table 3. Analogs exhibiting the greatest toxicity to cells (Table 2), having a rank order of TRPV1-OE > BEAS-2B ≥ NHBE, were also the most potent in the calcium flux assay, indicating a functional relationship for these two endpoints.
Fig. 3.
Dose response curves for calcium flux in TRPV1-OE cells treated with capsaicinoid analogs: nonivamide (▵), N-(3,4-dihydroxybenzyl)nonanamide (□), N-(3-hydroxy-4-methoxybenzyl)nonanamide (○), 3-methoxy-4-(nonamidomethyl)phenyl sulfate (▿), N-benzylnonanamide (▴), N-(3-methoxybenzyl)nonanamide (▾), N-(3,4-dimethoxybenzyl)nonanamide (◊), N-(4-trifluoromethylbenzyl)nonanamide (●), and N-(4-hydroxybenzyl)nonanamide (♦). Statistically significant calcium flux, p < 0.05 by one-way ANOVA, was observed for nonivamide and the 3,4-DiOH analog at concentrations equal to and greater than as indicated by *. Data are represented as percentage of response relative to ionomycin ± S.E.M. (n ≥3).
TABLE 3.
EC50 values and calcium flux by capsaicinoid analogs at 2, 20, and 200 μM
Data represent mean and S.D.
| Analog | EC50 | Calcium Flux (% Ionomycin) |
|||
|---|---|---|---|---|---|
| 2 μM | 20 μM | 100 μM | 200 μM | ||
| μM | |||||
| Nonivamide | 1.4 | 15 ± 3* | 24 ± 7* | 20 ± 9 | 21 ± 9* |
| N-Benzylnonanamide | —a | N.D. | N.D. | N.D. | N.D. |
| N-(3-Methoxybenzyl)nonanamide | — | N.D. | N.D. | N.D. | N.D. |
| N-(3,4-Dimethoxybenzyl)nonanamide | 120 | N.D. | N.D. | 6 ± 4 | 7 ± 3 |
| N-(3-Hydroxy-4-methoxybenzyl)nonanamide | 70 | N.D. | N.D. | 6 ± 4 | 6 ± 4 |
| N-(3,4-Dihydroxybenzyl)nonanamide | 10.0 | N.D. | 18 ± 4* | 19 ± 5 | 19 ± 3* |
| 3-Methoxy-4-(nonamidomethyl)phenyl sulfate | 470 | N.D. | 4 ± 1 | 5 ± 3 | 9 ± 7 |
| N-(4-Trifluoromethylbenzyl)nonanamide | — | N.D. | N.D. | N.D. | N.D. |
| N-(4-Hydroxybenzyl)nonanamide | — | N.D. | N.D. | 2 ± 1 | 2 ± 1 |
N.D., no calcium flux (i.e., the deviation exceeded the mean value).
—, EC50 value could not be calculated.
Statistical significance vs. untreated control using one-way ANOVA with Neuman-Kuels post-test (p < 0.05).
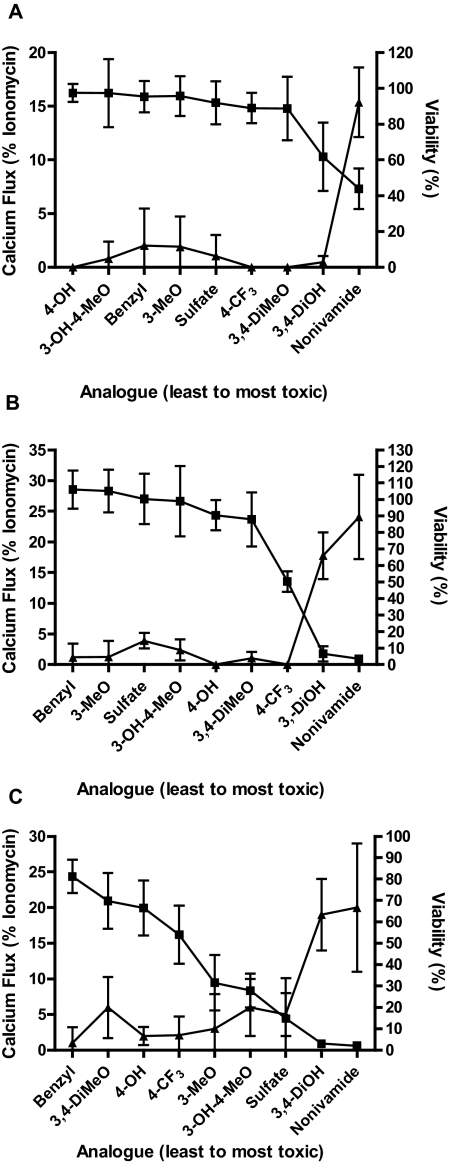
Correlation analysis between calcium flux elicited by the analogs and cytotoxicity in TRPV1-OE cells (Fig. 4 and Supplemental Data Section 3) demonstrated significant negative correlations with Pearson R values = −0.78, p = 0.01; −0.87, p = 0.003, and −0.75, p = 0.02 at low, intermediate, and high concentrations of analog, respectively. Correlation analyses for BEAS-2B and NHBE cells were not evaluated because calcium flux was not detectable.
Fig. 4.
Comparison of TRPV1-OE cell viability at 2.5 μM (A), 25 μM (B), and 100 μM (C) and analog-induced calcium flux at 2 μM (A), 20 μM (B), and 100 μM (C) analog concentration. Cell viability was assayed after 24-h treatment, and calcium flux was measured in real time over 1 min using a BMG Labtech NOVOstar plate reader as described under Materials and Methods. Data (mean ± S.E.M.) (left axis) are percentage of maximum attainable calcium response (▴) and percentage of cell viability relative to untreated controls (■). Analogs are arranged on the x-axis in order of lowest to highest cytotoxicity (left to right). Correlations between these two indices of TRPV1 activation are reported as the Pearson R value and two-tailed p value under Results and in the supplemental data.
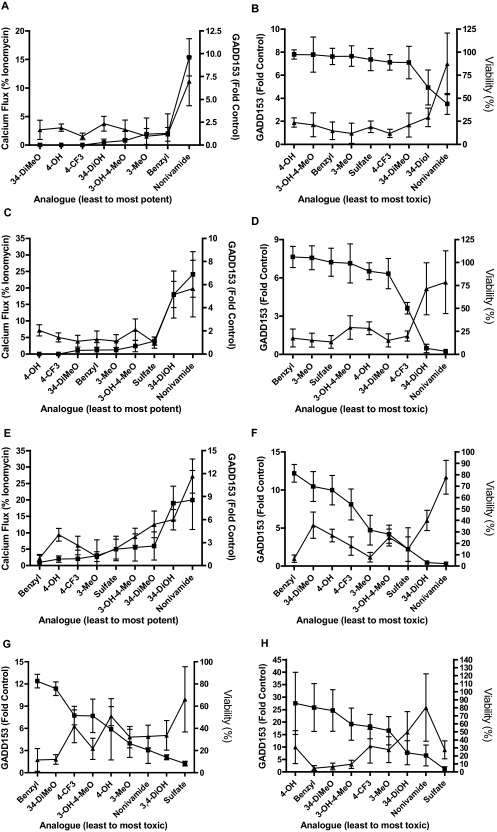
Changes in GADD153 gene expression were quantified as a maker of ER stress and precursor to cytotoxicity (Thomas et al., 2007) (Table 4). Treatment of TRPV1-OE cells with 2 or 20 μM nonivamide for 4 h produced up to 7-fold increases in GADD153 mRNA, whereas 2 and 5-fold increases were observed for N-(3,4-dihydroxybenzyl)nonanamide. All other analogs failed to induce GADD153 at these concentrations, consistent with their low cytotoxicity and low potency (Figs. 3 and Table 3). However, at 100 μM, increases in GADD153 mRNA were observed with most analogs, again consistent with data in Tables 2 and 3, but the relationship between toxicity, potency, and GAdd153 became insignificant (Fig. 5, E and F) because of an apparent cell-specific effect of the less potent analogs on GADD153 expression. BEAS-2B cells exhibited increased GADD153 mRNA after 4-h treatment with all analogs (200 μM), except N-benzylnonanamide and N-(3,4-dimethoxybenzyl)nonanamide. Significant increases in GADD153 mRNA were observed in NHBE cells with nonivamide (26-fold) and N-(3,4-dihydroxbenzyl)nonanamide (16-fold), similar to TRPV1-OE cells. Increases with other analogs were not statistically significant.
TABLE 4.
GADD153 mRNA expression in TRPV1, BEAS-2B, and NHBE cells after treatment with capsaicinoid analogs
Cells were treated with capsaicinoid analogs for 4 h, previously determined to be the peak induction for nonivamide. Data are the mean ± S.D.
| Analog | GADD153 mRNA |
||||
|---|---|---|---|---|---|
| TRPV1-OE (2 μM) | TRPV1-OE (20 μM) | TRPV1-OE (100 μM) | BEAS-2B (200 μM) | NHBE (200 μM) | |
| fold | |||||
| Nonivamide | 7 ± 3* | 6 ± 3* | 12 ± 2* | 5 ± 2* | 26 ± 20* |
| N-Benzylnonanamide | 1.2 ± 0.8 | 1.3 ± 0.7 | 1.0 ± 0.5 | 2 ± 2 | 1.6 ± 1.0 |
| N-(3-Methoxybenzyl)nonanamide | 1.0 ± 0.9 | 1.1 ± 0.5 | 1.2 ± 0.6 | 5 ± 1* | 9 ± 5 |
| N-(3,4-Dimethoxybenzyl)nonanamide | 2 ± 1 | 1.1 ± 0.5 | 5 ± 2* | 1.8 ± 0.6 | 2 ± 1 |
| N-(3-Hydroxy-4-methoxybenzyl)nonanamide | 2 ± 1.0 | 2.1 ± 0.9 | 4 ± 1* | 3 ± 1 | 3 ± 2 |
| N-(3,4-Dihydroxybenzyl)nonanamide | 2.3 ± 0.8 | 5 ± 2* | 6 ± 1* | 5 ± 2* | 16 ± 8* |
| 3-Methoxy-4-(nonamidomethyl)phenyl sulfate | 1.5 ± 0.5 | 1.0 ± 0.5 | 2 ± 2 | 10 ± 4* | 9 ± 4 |
| N-(4-Trifluoromethylbenzyl)nonanamide | 0.9 ± 0.4 | 1.4 ± 0.4 | 3 ± 1 | 6 ± 2* | 10 ± 7 |
| N-(4-Hydroxybenzyl)nonanamide | 1.9 ± 0.4 | 2.0 ± 0.5 | 4.0 ± 0.8* | 8 ± 1* | 10 ± 7 |
Statistical significance using one-way ANOVA with Neuman-Keuls post-test (p < 0.05).
Fig. 5.
A to E, comparison of calcium flux (▴) and GADD153 induction (■) (A, C, and E) in TRPV1-OE cells at 2 μM (A), 20 μM (B), and 100 μM (C) analog concentration and GADD153 (▴) and cell viability (■) (B, D, and E) using 2 and 2.5 μM (B), 20 and 25 μM (D), and 100 μM (F) analog. G and H, comparison of GADD153 expression (▴) and cytotoxicity (■) for BEAS-2B (G) and NHBE (H) cells treated with 200 μM analog. Percentage of maximum calcium flux was measured in real time for 1 min using a NOVOStar plate reader, GADD153 mRNA expression was measured using RT-qPCR (4-h treatment), and cell viability (24-h treatment) was measured using the Dojindo assay, as described under Materials and Methods. Analogs are arranged on the x-axis in all graphs from least to most potent (left to right) in calcium flux and cytotoxicity assays. Correlations between the indices of TRPV1 activation are reported as the Pearson R value and two-tailed p value under Results.
Correlation analysis between calcium flux and GADD153 expression and GADD153 and cytotoxicity for TRPV1-OE cells (Fig. 5, A–F and Supplemental Data Section C) demonstrated a relationship between all three phenomena. For calcium flux and GADD153 (Fig. 4, A, C, and E and supplemental data) Pearson R values of 0.94, p = 0.0002 (low dose); 0.95, p < 0.0001 (intermediate dose); and 0.91, p = 0.001 (high dose) were obtained. For GADD153 and cytotoxicity (Fig. 4, B, D, and F and supplemental data), Pearson R values of −0.87, p = 0.002 (low dose); −0.9, p = 0.001 (intermediate dose); and −0.5, p = 0.17 (high dose) were obtained. For BEAS-2B and NHBE cells comparison of cell viability and GADD153 expression also demonstrated significant correlations with Pearson R values of −0.73, p = 0.02 and −0.81, p = 0.01, respectively, when the sulfate analog was omitted from the NHBE data set (Fig. 5, G and H and supplemental data). Collectively these results demonstrate that TRPV1 activation/calcium flux, GADD153 expression, and cytotoxicity are coupled and stringent structural requirements exist for agonists to elicit these specific responses.
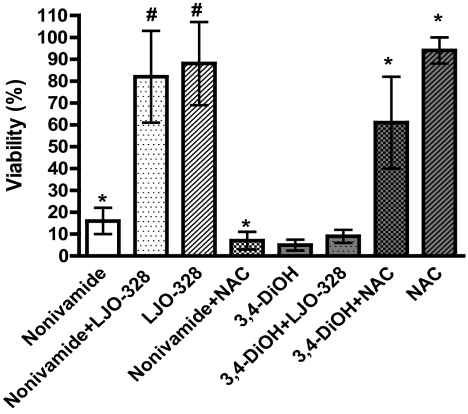
Table 2 shows that N-(3,4-dihydroxybenzyl)nonanamide exhibited similar toxicity in TRPV1-OE and BEAS-2B cells, but reduced toxicity in NHBE cells, suggesting a decrease in potency at TRPV1 and the potential involvement of parallel processes that lead to cytotoxicity. Cotreatment of TRPV1-OE cells with N-(3,4-dihydroxybenzyl)nonanamide and LJO-328 had little effect on toxicity, whereas cotreatment with NAC significantly reduced cytotoxicity, implicating redox cycling/electrophile formation as a major factor for cytotoxicity for this analog (Fig. 6). However, similar protective results by NAC were not observed for either BEAS-2B or TRPV1-OE cells using a 25 μM dose of N-(3,4-dihydroxybenzyl)nonanamide (data not shown), indicating a limitation in NAC to attenuate toxicity, thus preventing us from completely ruling out a role for TRPV1 in cytotoxicity, as suggested by the shorter-term calcium flux and GADD153 expression studies. Likewise, 3-methoxy-4-(nonamidomethyl)phenyl sulfate, N-(3-methoxybenzyl)nonanamide, and N-(4-hydroxybenzyl)nonanamide exhibited equal cytotoxicity in the three cell types, suggesting that cell death was also partially independent of TRPV1. Bioactivation of N-(3-methoxybenzyl)nonanamide and N-(4-hydroxybenxyl)nonanamide by cytochrome P450 enzymes to either nonivamide or N-(3,4-dihydroxybenzyl)nonanamide via hydroxylation was investigated by preincubating and cotreating BEAS-2B and NHBE cells with the cytochrome P450 inhibitor 1-aminobenzotriazole. Inhibition of P450s by 1-aminobenzotriazole using the protocol of Weems and Yost (2010) had no effect on cytotoxicity, indicating that P450 metabolism was not a substantial factor (data not shown). The toxicity of 3-methoxy-4-(nonamidomethyl)phenyl sulfate was not investigated further, but production of nonivamide via sulfate ester hydrolysis was discounted because the cytotoxicity was not reduced by LJO-328 cotreatment in any cell type (data not shown).
Fig. 6.
Assessment of TRPV1 dependence and non-TRPV1-mediated/redox stress-mediated toxicity for nonivamide and N-(3,4-dihydroxybenzyl)nonanamide in TRPV1-OE cells. *, statistically significant difference (p < 0.05) from untreated controls. #, statistical difference between treatment with analog alone or with LJO-328 or NAC (p < 0.05) using one-way ANOVA. Data are represented as the mean ± S.D. (n = 3).
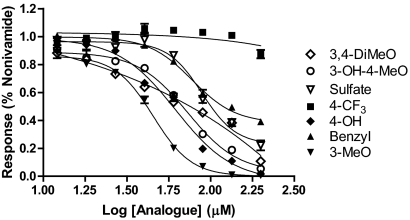
The least toxic analogs were evaluated as protective agents for cytotoxicity via inhibition of TRPV1 by nonivamide. Pretreatment of TRPV1-OE cells with all analogs except N-(3,4-dihydroxybenzyl)nonanamide inhibited nonivamide-induced calcium flux, indicating competitive inhibition of TRPV1 (Fig. 7), but N-(3,4-dihydroxybenzyl)nonanamide cotreatment increased basal fluorescence consistent with the results in Fig. 3. The IC50 values were 45, 62, 73, 78, 89, 120, and >200 μM for N-(3-methoxybenzyl)nonanamide, N-(4-hydroxybenzyl)nonanamide, N-(3-hydroxy-4-methoxybenzyl)nonanamide, N-benzylnonanamide, 3-methoxy-4-(nonamidomethyl)phenyl sulfate, N-(3,4-dimethoxybenzyl)nonanamide, and N-(4-trifluoromethybenzyl)nonanamide, respectively. Cotreatment of TRPV1-OE cells with nonivamide and increasing concentrations of N-benzylnonanamide also demonstrated an attenuation of cell death from 53 ± 8% viability to 85 ± 10 and 78 ± 8% at concentrations of 50 and 100 μM, respectively. Protection (∼15–20% decrease) was also observed with N-(3-methoxybenzyl)nonanamide at 50 μM, but all other analogs failed to protect and produced additive toxicity at higher concentrations. These results were consistent with the results in Figs. 4 and 5 and Tables 2 and 3 in that these analogs failed to elicit significant calcium flux at 100 μM and were the least potent inducers of GADD153 and the least toxic.
Fig. 7.
Dose response curves for inhibition of nonivamide- (2.5 μM) induced calcium flux in TRPV1-OE cells treated by capsaicinoid analogs: N-(3-hydroxy-4-methoxybenzyl)nonanamide (○), 3-methoxy-4-(nonamidomethyl)phenyl sulfate (▿), N-benzylnonanamide (▴), N-(3-methoxybenzyl)nonanamide (▾), N-(3,4-dimethoxybenzyl)nonanamide (◊), N-(4-trifluoromethylbenzyl)nonanamide (■), and N-(4-hydroxybenzyl)nonanamide (♦). Data are normalized to nonivamide only and were fit using the sigmoidal dose response (variable slope) equation to estimate the IC50 value. *, concentration at which statistically significant inhibition, p < 0.05 by one-way ANOVA, was observed. Data are the mean and S.E.M. (n = 3).
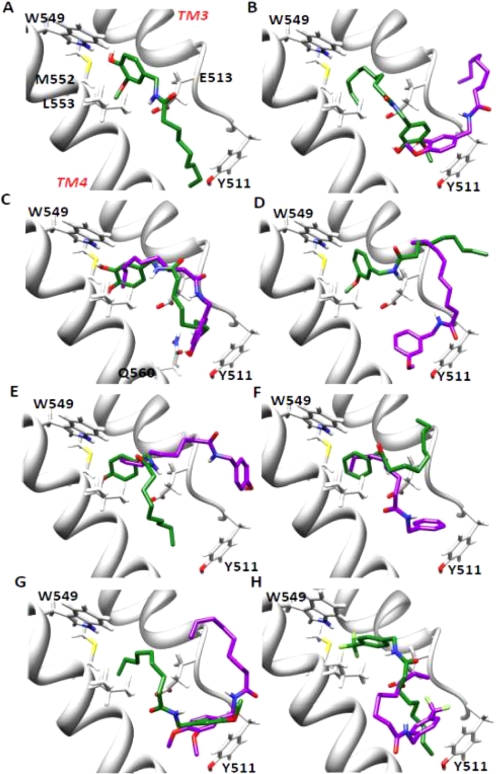
Finally, molecular modeling was used to explore bonding interactions between capsaicinoid analogs and TRPV1 as a basis for differential potency and toxicity. Representative poses for nonivamide (the prototype), N-(3-hydroxy-4-methoxybenzyl)nonanamide, N-(3,4-dihydroxybenzyl)nonanamide, N-(3-methoxybenzyl)nonanamide, N-(4-hydroxybenzyl)nonanamide, N-benzylnonanamide, N-(3,4-dimethoxybenzyl)nonanamide, and N-(4-trifluoromethylbenzyl)nonanamide are shown in Fig. 8. For nonivamide (Fig. 8A), the lowest energy pose (green) was most consistent with the model proposed by Gavva et al. (2004) and the presumed binding location suggested from site-directed mutagenesis studies. In this pose, the oxygen atom of the 4-OH moiety was predicted to hydrogen-bond with the hydrogen on the imidazole nitrogen of Trp549 (distance 1.9 Å), and the hydrogen on the amide nitrogen of nonivamide was predicted to hydrogen-bond with the carboxylate group of Glu513 (2.3 Å). The alkyl terminus of nonivamide was localized ∼2.4 Å from the phenol ring of Tyr511 where hydrophobic interactions probably occur and the 3-MeO group was localized in the hydrophobic “hole-A” pocket composed of Trp549, Met552, Leu553, and likely Phe516 and Leu521, as identified by Ryu et al. (2008). This “network” of bonding is consistent with a mechanism of activation for TRPV1 by capsaicinoids whereby the ligand acts as a “molecular tether” between TM3 and TM4 that induces a change in protein structure and/or protein dynamics that ultimately translate into pore opening. Analysis of N-(3-hydroxy-4-methoxybenzyl)nonanamide, N-(3-methoxybenzyl)nonanamide, N-(4-hydroxybenzyl)nonanamide, and N-benzylnonanamide failed to show similar intermolecular interactions, regardless of binding energy or occupancy. These results correspond with the low potency of these analogs in the functional assays. However, the lowest energy confirmation cluster for N-(3,4-dihydroxybenzyl)nonanamide (Fig. 8C) did exhibit similarities to nonivamide binding, in that the oxygen atom of the 3-OH moiety was predicted to be within hydrogen-bonding distance (1.9 Å) with the hydrogen of Trp549. However, hydrogen bonding was not predicted because of bond angle constraints presumably resulting from decreased ligand stabilization in the hydrophobic hole-A pocket caused by conversion of the 3-MeO moiety to the more hydrophilic 3-OH moiety. In addition, hydrogen bonding between the carboxylate of Glu513 and the amide hydrogen (distances of 3.9–5.5 Å) and the alkyl terminus with Tyr511 (∼3.1–4 Å) were not predicted to be as favorable, because of an apparent “kinking” of the substrate molecule at the A/B region compared with nonivamide (Fig. 8C), supporting a decrease in potency for the diol in the functional assays. Similarly hydrogen-bonding interactions between the 4-OH and amide hydrogen of N-(4-hydroxybenzyl)nonanamide (Fig. 8E) with Trp549 and Glu513 were not observed, emphasizing the requirement for localization of the 3-MeO group in hole A for proper ligand binding and maximum potency. Finally, the most highly populated pose for N-(3,4-dihydroxybenzyl)nonanamide (Fig. 8C, purple) revealed the association of the ligand in an opposite orientation compared with nonivamide, similar to orientations predicted for all of the other inactive analogs, further supporting the observed decrease in potency in the functional assays.
Fig. 8.
Images depicting predicted binding interactions between TRPV1 transmembrane helices 3 and 4 (residues Ser510–Ile569) (Gavva et al., 2004) and capsaicinoid analogs. Lowest energy (green) and most highly populated (purple) poses for nonivamide (A), N-(3-hydroxy-4-methoxybenzyl)nonanamide (B), N-(3,4-dihydroxybenzyl)nonanamide (C), N-(3-methoxybenzyl)nonanamide (D), N-(4-hydroxybenzyl)nonanamide (E), N-benzylnonanamide (F), N-(3,4-dimethoxybenzyl)nonanamide (G), and N-(4-trifluoromethybenzyl)nonanamide (H) are shown. For nonivamide, the structure represents both the lowest energy and highest populated structure.
Discussion
Characterization of cellular responses to TRPV1 agonists is essential to understanding how TRPV1 regulates normal and aberrant cellular processes. TRPV1 is expressed in many cell types, and it is often difficult to translate results from one model to another because of differences in its expression level, subcellular distribution and coupling, physiological regulation, and variability in antagonist efficacy, as observed in previous studies of BEAS-2B and NHBE cells (Reilly et al., 2003b, 2005; Thomas et al., 2007). Here, a set of capsaicinoid analogs (Fig. 1 and Table 1) with minimal structural modifications were developed to study the relationship between TRPV1 binding, activation, and ER stress/GADD153 induction in lung cell death. The collective data demonstrate that TRPV1 binding, activity, ER stress, and cytotoxicity highly depend on the presence of a vanilloid ring pharmacophore and that when this optimum configuration is present all of these processes are coupled in lung cells.
The SAR for capsaicinoid analogs was evaluated in four ways (TRPV1 expression, TRPV1-mediated calcium flux, changes in proapoptotic GADD153 expression, and cytotoxicity) in three different lung cell lines with vastly different levels of TRPV1 expression. Quantification of TRPV1 mRNA (Fig. 2) and the cytotoxicity of various nonivamide analogs (Table 2) in TRPV1-OE, BEAS-2B, and NHBE cells demonstrated that the relative sensitivity of lung cells to nonivamide, and to some extent N-(3,4-dihydroxybenzyl)nonanamide, were predicted by the level of TRPV1 expression. However, for structural analogs with substantially reduced potency at TRPV1, this relationship was less significant. These results support a role for TRPV1 in lung cell death by TRPV1 agonists, but also show that there is a concentration (∼200–250 μM) at which all analogs become cytotoxic independent of TRPV1 expression or function.
This conclusion was further explored by comparing calcium flux, GADD153 induction, and cytotoxicity for the various analogs because these endpoints each represent a step in a proposed mechanism of TRPV1-mediated cell death for lung epithelial cells (Thomas et al., 2007). Analysis of TRPV1 activation using calcium flux demonstrated that nonivamide and N-(3,4-dihydroxybenzyl)nonanamide were most potent (Fig. 3 and Table 3). Calcium flux was also detected with N-(3,4-dimethoxybenzyl)nonanamide, N-(3-hydroxy-4-methoxybenzyl)nonanamide, and 3-methoxy-4-(nonamidomethy)phenyl sulfate, but the magnitude was reduced and occurred only at concentrations >50 times the EC50 of nonivamide. These data agree with previous structure-activity relationship studies where the 3-MeO-4-OH vanilloid ring motif was shown to be essential for maximum activation of TRPV1 by capsaicin-like molecules (Walpole et al., 1993c). The link between calcium flux, GADD153 expression, and cell death was subsequently evaluated using correlation analysis. Figures 4 and 5, A to E (and Supplemental Data Section 3) indicate that analog-induced calcium flux was associated with increases in GADD153 expression and cell death in TRPV1-OE cells at 2 and 20 μM. However, at 100 μM, the relationship for GADD153 and cytotoxicity became less significant, but was still quite evident (Fig. 4, E and F). Using analog potency data for GADD153 induction and cell death, significant correlations also were found for BEAS-2B and NHBE cells, but only when the sulfate analog was omitted from the NHBE data set (Fig. 5, G and H and Supplemental Data Section 3). These data confirm that calcium flux, GADD153 induction, and cell death are coupled in lung cells for TRPV1 ligands that optimally bind TRPV1 and activate TRPV1, regardless of TRPV1 expression.
Parallel and/or alternative pathways of toxicity for N-(3,4-dihydroxybenzyl)nonanamide, N-(4-hydroxybenzyl)nonanamide, and N-(3-methoxybenzyl)nonanamide were also investigated. Similar to nonivamide, N-(3,4-dihydroxybenzyl)nonanamide elicited significant calcium flux in TRPV1-OE cells and induced GADD153 in all three cell types. However, because the EC50 value for N-(3,4-dihydroxybenzyl)nonanamide was ∼5-fold that of nonivamide in TRPV1-OE cells and the LD50 for BEAS-2B cells was lower than was predicted (Table 2), we hypothesized that the diol analog toxicity was not fully TRPV1-mediated. This notion was supported by results showing that the TRPV1 antagonist LJO-328 did not attenuate cell killing in TRPV1-OE cells, but that cell death was almost completely inhibited by N-acetylcysteine (Fig. 6). It is known that oxidative stress and binding of electrophiles to cellular nucleophiles can trigger ER stress, eukaryotic translation initiation factor 2α kinase 3 activation, and GADD153 induction through activation of the Nrf2/Keap-1 pathway (Cullinan et al., 2003; Li and Kong, 2009; Copple et al., 2010). This provides a plausible explanation for the toxicity of this analog, but lack of attenuation of cell death at 25 μM in both TRPV1-OE and BEAS-2B cells by NAC suggests that TRPV1 must still be involved in some way, as suggested by the correlation studies. The substantially lower toxicity of the diol in NHBE cells also suggests a fundamental difference in reactive oxygen species/electrophile detoxification capacity between immortalized and primary lung cells, which is important because N-(3,4-dihydroxybenzyl)nonanamide is efficiently produced by CYP1A2 and CYP2C19 (Reilly et al., 2003a; Reilly and Yost, 2006) and, thus, it may be difficult to modulate capsaicinoid toxicity in the intact lung using TRPV1 antagonists alone. Currently, an explanation for the toxicity of N-(4-hydroxybenzyl)nonanamide, 3-methoxy-4-(nonamidomethy)phenyl sulfate, and N-(3-methoxybenzyl)nonanamide is not apparent, but metabolism by cytochromes P450 by hydroxylation to produce nonivamide or N-(3,4-dihydroxybenzyl)nonanamide does not seem to be operative.
Molecular modeling was used to evaluate TRPV1 binding by the different analogs and provide a theoretical explanation for the observed differences in agonist potency and toxicity. Using a homology model of TRPV1 transmembrane segments 3 and 4 (residues Ser510–Ile569) (Gavva et al., 2004), it was predicted that nonivamide formed a “productive” binding interaction with TRPV1 (Fig. 8A) wherein the 4-OH moiety of the vanilloid ring was hydrogen-bonded with Trp549, the carboxylate group of Glu513 hydrogen-bonded with the amide hydrogen, the 3-MeO group resided in a hydrophobic pocket composed of Trp549, Met552, Leu553, and probably Phe516 and Leu521, and the alkyl terminus interacted with Tyr511 via hydrophobic interactions. Unique from the model proposed by Jordt and Julius (2002), the alkyl terminus of nonivamide was predicted to interact with Tyr511, as shown by Gavva et al. (2004) using capsaicin. However, direct roles for Leu547 (in human TRPV1; Met in rat) and Thr550, were not indicated. Because most of the residues implicated in our model are conserved residues between both highly sensitive (humans and rats) and minimally sensitive (rabbit) species, our results suggest that changes of residues adjacent to these key residues alters the geometry of the binding site of TRPV1 such that binding is either favored or disfavored, rather than specific interactions of these residues with agonists, as discussed previously by Gavva et al. This conclusion is supported by data showing that the propensity to form the “productive” pose of nonivamide was reduced for the other analogs. Nonivamide binding seemed to be highly driven by the combined interactions of the 4-OH and 3-MeO moieties and TM3/4 side chain residues because neither N-(3-methoxy-4-hydroxybenzyl)nonanamide (Fig. 8B), N-(3-methoxybenzyl)nonanamide (Fig. 8D), nor N-(4-hydroxybenzyl)nonanamide (Fig. 8E) formed comparable poses. Although the dihydroxy analog (Fig. 8C) was predicted to adopt a similar pose in TRPV1, hydrogen bonding between either the 3- or 4-OH groups with Trp549 was not predicted and the orientation of the A/B region was skewed relative to nonivamide, presumably caused by decreased anchoring by the 3-MeO group in the hole-A pocket, thus decreasing interaction with Glu513 and effectively reducing the length of the analog, such that interactions with Tyr511 were less favorable. However, if the alkyl terminus of the dihydroxy analog was lengthened, perhaps TRPV1 would be more potently activated, as suggested by studies with arvanil, linvanil, anandamide, and the arachidonic acid esters of acetaminophen (AM404) (Högestätt et al., 2005) and dopamine (Huang et al., 2002). The removal of the 3-MeO group in the TRPV1 antagonist capsazepine and constraint of the A/B region (Walpole et al., 1994), as well as increased potency of N-(2-benzyl-3-pivaloyloxypropyl) 2-[4-(methylsulfonylamino)phenyl]propionamide antagonist analogs by the addition of a 4-tert-butyl group to the 2-phenylproionate moiety (Ryu et al., 2008), also support the importance of the 3-MeO and amide groups in TRPV1 activation by agonists. Finally, N-benzylnonanamide (Fig. 8F), as well as the other inactive analogs, were predicted to form essentially opposite binding orientations within TRPV1 compared with nonivamide, as predicted previously (Jordt and Julius, 2002), suggesting that such an orientation may be inhibitory, idea conjecture that was confirmed (Fig. 7) by the ability of N-benzylnonanamide and N-(3-methoxybenzyl)nonanamide to reduce cytotoxicity.
In conclusion, the data provide consistent evidence that TRPV1-mediated ER calcium release, ER stress, and GADD153 expression are major components of the chain of events that cause lung cell death by capsaicin and nonivamide, but that additional cellular processes such as biotransformation and detoxification modulate the disposition of less potent analogs that must be considered when using these modified substances to probe TRPV1 function in different models. Finally, molecular modeling supports the functional data and previous SAR and site-directed mutagenesis studies and further emphasizes the critical relationship between agonist structure, TRPV1 function, ER stress, and cytotoxicity.
Supplementary Material
This work was supported in part by the National Institutes of Health National Heart, Lung, and Blood Institute [Grant HL069813] (to C.A.R.), the National Institutes of Health National Institute of Environmental Health Sciences [Grant ES017431] (to C.A.R.), and a seed grant from the University of Utah Department of Anesthesiology (to C.A.R.). Support for the molecular modeling studies was provided by the National Institutes of Health National Institute for General Medical Sciences [Grant GM079383] (to T.E.C.). Computer time was provided by the Center for High Performance Computing at the University of Utah and supported by the National Science Foundation TeraGrid [Grant MCA01S027]. J.L. was supported by the National Research Foundation of Korea [Grant R11-2007-107-02001-0].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.110.178491.
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
ABBREVIATIONS:
- TRPV1
- transient receptor potential vanilloid 1
- GADD153
- growth arrest- and DNA damage-inducible transcript 3
- NHBE
- normal human bronchial epithelial
- SAR
- structure-activity relationship
- RT
- reverse transcription
- PCR
- polymerase chain reaction
- qPCR
- quantitative PCR
- LC50
- concentration at which 50% loss in viability (lethality) is observed
- MeO
- methoxy
- OH
- hydroxyl
- ER
- endoplasmic reticulum
- LJO-328
- N-(4-tert-butylbenzyl)-N′-(1-[3-fluoro-4-(methylsulfonylamino)-phenyl]ethyl)thiourea
- ANOVA
- analysis of variance
- TM
- transmembrane
- NAC
- N-acetylcysteine
- β2M
- β2 macroglobulin.
Authorship Contributions
Participated in research design: Thomas, Shahrokh, Sun, Yost, and Reilly.
Conducted experiments: Thomas, Ethirajan, Shahrokh, Sun, and Reilly.
Contributed new reagents or analytic tools: Ethirajan and Lee.
Performed data analysis: Thomas, Ethirajan, Shahrokh, Sun, and Reilly.
Wrote or contributed to the writing of the manuscript: Thomas, Ethirajan, Shahrokh, Sun, Cheatham, Yost, and Reilly.
Other: Lee, Cheatham, Yost, and Reilly obtained funding for the research.
References
- Appendino G, Harrison S, De Petrocellis L, Daddario N, Bianchi F, Schiano Moriello A, Trevisani M, Benvenuti F, Geppetti P, Di Marzo V. (2003) Halogenation of a capsaicin analogue leads to novel vanilloid TRPV1 receptor antagonists. Br J Pharmacol 139:1417–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D. (1997) The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389:816–824 [DOI] [PubMed] [Google Scholar]
- Copple IM, Goldring CE, Kitteringham NR, Park BK. (2010) The keap1-nrf2 cellular defense pathway: mechanisms of regulation and role in protection against drug-induced toxicity. Handb Exp Pharmacol 233–266 [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. (2003) Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol 23:7198–7209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavva NR, Klionsky L, Qu Y, Shi L, Tamir R, Edenson S, Zhang TJ, Viswanadhan VN, Toth A, Pearce LV, et al. (2004) Molecular determinants of vanilloid sensitivity in TRPV1. J Biol Chem 279:20283–20295 [DOI] [PubMed] [Google Scholar]
- Geppetti P, Materazzi S, Nicoletti P. (2006) The transient receptor potential vanilloid 1: role in airway inflammation and disease. Eur J Pharmacol 533:207–214 [DOI] [PubMed] [Google Scholar]
- Geppetti P, Nassini R, Materazzi S, Benemei S. (2008) The concept of neurogenic inflammation. BJU Int 101(Suppl 3):2–6 [DOI] [PubMed] [Google Scholar]
- Högestätt ED, Jönsson BA, Ermund A, Andersson DA, Björk H, Alexander JP, Cravatt BF, Basbaum AI, Zygmunt PM. (2005) Conversion of acetaminophen to the bioactive N-acylphenolamine AM404 via fatty acid amide hydrolase-dependent arachidonic acid conjugation in the nervous system. J Biol Chem 280:31405–31412 [DOI] [PubMed] [Google Scholar]
- Huang SM, Bisogno T, Trevisani M, Al-Hayani A, De Petrocellis L, Fezza F, Tognetto M, Petros TJ, Krey JF, Chu CJ, et al. (2002) An endogenous capsaicin-like substance with high potency at recombinant and native vanilloid VR1 receptors. Proc Natl Acad Sci USA 99:8400–8405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Lee LY. (2007) Role of TRPV receptors in respiratory diseases. Biochim Biophys Acta 1772:915–927 [DOI] [PubMed] [Google Scholar]
- Johansen ME, Reilly CA, Yost GS. (2006) TRPV1 antagonists elevate cell surface populations of receptor protein and exacerbate TRPV1-mediated toxicities in human lung epithelial cells. Toxicol Sci 89:278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordt SE, Julius D. (2002) Molecular basis for species-specific sensitivity to “hot” chili peppers. Cell 108:421–430 [DOI] [PubMed] [Google Scholar]
- Li W, Kong AN. (2009) Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog 48:91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassini R, Materazzi S, De Siena G, De Cesaris F, Geppetti P. (2010) Transient receptor potential channels as novel drug targets in respiratory diseases. Curr Opin Investig Drugs 11:535–542 [PubMed] [Google Scholar]
- Oortgiesen M, Veronesi B, Eichenbaum G, Kiser PF, Simon SA. (2000) Residual oil fly ash and charged polymers activate epithelial cells and nociceptive sensory neurons. Am J Physiol Lung Cell Mol Physiol 278:L683–L695 [DOI] [PubMed] [Google Scholar]
- Reilly CA, Ehlhardt WJ, Jackson DA, Kulanthaivel P, Mutlib AE, Espina RJ, Moody DE, Crouch DJ, Yost GS. (2003a) Metabolism of capsaicin by cytochrome P450 produces novel dehydrogenated metabolites and decreases cytotoxicity to lung and liver cells. Chem Res Toxicol 16:336–349 [DOI] [PubMed] [Google Scholar]
- Reilly CA, Johansen ME, Lanza DL, Lee J, Lim JO, Yost GS. (2005) Calcium-dependent and independent mechanisms of capsaicin receptor (TRPV1)-mediated cytokine production and cell death in human bronchial epithelial cells. J Biochem Mol Toxicol 19:266–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly CA, Taylor JL, Lanza DL, Carr BA, Crouch DJ, Yost GS. (2003b) Capsaicinoids cause inflammation and epithelial cell death through activation of vanilloid receptors. Toxicol Sci 73:170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly CA, Yost GS. (2006) Metabolism of capsaicinoids by P450 enzymes: a review of recent findings on reaction mechanisms, bio-activation, and detoxification processes. Drug Metab Rev 38:685–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Jin MK, Kim SY, Choi HK, Kang SU, Kang DW, Lee J, Pearce LV, Pavlyukovets VA, Morgan MA, et al. (2008) Stereospecific high-affinity TRPV1 antagonists: chiral N-(2-benzyl-3-pivaloyloxypropyl) 2-[4-(methylsulfonylamino)phenyl]propionamide analogues. J Med Chem 51:57–67 [DOI] [PubMed] [Google Scholar]
- Szallasi A, Cortright DN, Blum CA, Eid SR. (2007) The vanilloid receptor TRPV1: 10 years from channel cloning to antagonist proof-of-concept. Nat Rev Drug Discov 6:357–372 [DOI] [PubMed] [Google Scholar]
- Thomas KC, Sabnis AS, Johansen ME, Lanza DL, Moos PJ, Yost GS, Reilly CA. (2007) Transient receptor potential vanilloid 1 agonists cause endoplasmic reticulum stress and cell death in human lung cells. J Pharmacol Exp Ther 321:830–838 [DOI] [PubMed] [Google Scholar]
- Tominaga M, Caterina MJ, Malmberg AB, Rosen TA, Gilbert H, Skinner K, Raumann BE, Basbaum AI, Julius D. (1998) The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 21:531–543 [DOI] [PubMed] [Google Scholar]
- Veronesi B, Carter JD, Devlin RB, Simon SA, Oortgiesen M. (1999a) Neuropeptides and capsaicin stimulate the release of inflammatory cytokines in a human bronchial epithelial cell line. Neuropeptides 33:447–456 [DOI] [PubMed] [Google Scholar]
- Veronesi B, Oortgiesen M. (2001) Neurogenic inflammation and particulate matter (PM) air pollutants. Neurotoxicology 22:795–810 [DOI] [PubMed] [Google Scholar]
- Veronesi B, Oortgiesen M, Carter JD, Devlin RB. (1999b) Particulate matter initiates inflammatory cytokine release by activation of capsaicin and acid receptors in a human bronchial epithelial cell line. Toxicol Appl Pharmacol 154:106–115 [DOI] [PubMed] [Google Scholar]
- Walpole CS, Bevan S, Bovermann G, Boelsterli JJ, Breckenridge R, Davies JW, Hughes GA, James I, Oberer L, Winter J. (1994) The discovery of capsazepine, the first competitive antagonist of the sensory neuron excitants capsaicin and resiniferatoxin. J Med Chem 37:1942–1954 [DOI] [PubMed] [Google Scholar]
- Walpole CS, Wrigglesworth R, Bevan S, Campbell EA, Dray A, James IF, Masdin KJ, Perkins MN, Winter J. (1993a) Analogues of capsaicin with agonist activity as novel analgesic agents; structure-activity studies. 2. The amide bond “B-region.” J Med Chem 36:2373–2380 [DOI] [PubMed] [Google Scholar]
- Walpole CS, Wrigglesworth R, Bevan S, Campbell EA, Dray A, James IF, Masdin KJ, Perkins MN, Winter J. (1993b) Analogues of capsaicin with agonist activity as novel analgesic agents; structure-activity studies. 3. The hydrophobic side-chain “C-region.” J Med Chem 36:2381–2389 [DOI] [PubMed] [Google Scholar]
- Walpole CS, Wrigglesworth R, Bevan S, Campbell EA, Dray A, James IF, Perkins MN, Reid DJ, Winter J. (1993c) Analogues of capsaicin with agonist activity as novel analgesic agents; structure-activity studies. 1. The aromatic “A-region.” J Med Chem 36:2362–2372 [DOI] [PubMed] [Google Scholar]
- Weems JM, Yost GS. (2010) 3-Methylindole metabolites induce lung CYP1A1 and CYP2F1 enzymes by AhR and non-AhR mechanisms, respectively. Chem Res Toxicol 23:696–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.