Abstract
NarE is a 16 kDa protein identified from Neisseria meningitidis, one of the bacterial pathogens responsible for meningitis. NarE belongs to the family of ADP-ribosyltransferases (ADPRT) and catalyzes the transfer of ADP-ribose moieties to arginine residues in target protein acceptors. Many pathogenic bacteria utilize ADP-ribosylating toxins to modify and alter essential functions of eukaryotic cells. NarE is further the first ADPRT which could be shown to bind iron through a Fe-S center, which is crucial for the catalytic activity. Here we present the NMR solution structure of NarE, which shows structural homology to other ADPRTs. Using NMR titration experiments we could identify from Chemical Shift Perturbation data both the NAD binding site, which is in perfect agreement with a consensus sequence analysis between different ADPRTs, as well as the iron coordination site, which consists of 2 cysteines and 2 histidines. This atypical iron coordination is also capable to bind zinc. These results could be fortified by site-directed mutagenesis of the catalytic region, which identified two functionally crucial residues. We could further identify a main interaction region of NarE with antibodies using two complementary methods based on antibody immobilization, proteolytic digestion, and mass spectrometry. This study combines structural and functional features of NarE providing for the first time a characterization of an iron-dependent ADPRT.
Keywords: ADP-ribosylation, Antibodies, Bacterial Toxins, Mass Spectrometry (MS), NMR, Protein Structure
Introduction
Neisseria meningitidis is a Gram-negative bacterium best known for its role in meningitis and septicemia. This human-specific pathogen is considered as a major cause of morbidity and mortality during childhood in industrialized countries and is responsible for epidemics in Africa and in Asia (1). Like most bacterial pathogens, N. meningitidis synthesizes a number of toxic products believed to target and kill their host cells. Among these toxins, proteins that exert ADP-ribosylation activity represent a large family of potentially toxic enzymes able to modify or disrupt essential functions of eukaryotic cells (2). ADP-ribosyltransferases (ADPRTs)3 catalyze the transfer of a single ADP-ribosyl group from β-nicotinamide adenine dinucleotide (NAD+) onto specific amino acid residues of host cell proteins with the simultaneous release of nicotinamide (3). Many bacterial pathogens use ADP-ribosylating enzymes to block protein synthesis or to alter signal transduction by inactivating key target proteins such as GTP-binding proteins (4).
The putative ADP-ribosylating NarE protein from N. meningitidis was recently identified on the basis of a profile-based computational approach (5). NarE shares structural features with toxins from Vibrio cholerae and Escherichia coli, and, like cholera toxin and heat-labile enterotoxins, retains the capacity to ADP-ribosylate arginine and to hydrolyze NAD in ADP-ribose and nicotinamide. Another feature of NarE is its ability to bind iron through an iron-sulfur center (Fe-S) (6). Interestingly, site-directed mutagenesis and enzymatic assays showed that correct assembly of the iron-binding site is essential for transferase but not for hydrolase activity (6), suggesting for the first time an implication of a Fe-S cluster in catalysis within the ADP-ribosyltransferase family. By using solution state NMR spectroscopy we solved the atomic structure of the apo-form of NarE, which is structurally homologous to other ADPRTs. We could further characterize the NAD binding site using 15N-HSQC titration experiments. The resulting Chemical Shift Perturbation (CSP) data were used to model the first NAD-bound complex of an ADPRT using the docking software HADDOCK (7). This data is in agreement with results from site directed mutagenesis identifying two functionally crucial residues within the substrate binding site. Moreover we could identify the iron coordination site of NarE consisting of two cysteines and two histidines using CSP experiments as well as paramagnetic effects of the bound iron. We could further identify a main epitope of NarE using four different monoclonal antibodies and two complementary methods based on antibody immobilization, proteolytic digestion, and mass spectrometry. Because of the eminent implication of ADPRTs in pathogenicity the presented results will stimulate drug design for a wide variety of pathogens.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification for NMR
The NarE gene was PCR amplified from the chromosomal DNA of N. meningitidis MC58 strain and cloned into a pET21b+ plasmid (Novagen) under the control of the T7 promoter. The resulting expression vector encodes a 17 kDa fusion protein that contains the wild-type NarE sequence (145 residues) followed by an 8-residue C-terminal histidine tag (LEHHHHHH) used for purification purposes. The pET21b+ plasmid was transformed into an E. coli BL21 (DE3) expression strain. Recombinant protein was prepared from cells grown to an A600 of 0.5–0.6 at 37 °C in a bacterial culture supplemented by 100 μg/ml ampicillin. Protein expression was then induced by addition of 1 mm isopropyl-1-thio-β-d-galactopyranoside (IPTG), and the culture was further grown for 3 h at 37 °C. The cells were harvested by centrifugation (8000 × g for 30 min), resuspended in 50 mm phosphate buffer (pH 8.0) containing 300 mm NaCl, and lysed with a French press cell (SLM, Aminco). The soluble fraction enriched with the NarE protein was separated from the inclusion bodies by centrifugation at 39,200 × g for 45 min, and loaded on a nickel affinity chromatography column (GE Healthcare). The protein was purified using IMAC standard protocols and an imidazole concentration of 125 mm was required to elute recombinant NarE. A second step of purification was performed using anion exchange QHP chromatography (GE Healthcare). Fractions containing the target protein were collected at a NaCl concentration of 300 mm and finally concentrated by ultrafiltration using a 3-kDa cutoff membrane (Millipore). Expression of uniformly labeled 15N and 15N/13C proteins was carried out by growing BL21 strains in ISOGRO-15N and ISOGRO-15N,13C rich media (Sigma), respectively.
NMR Measurements/Structure Calculation
NMR spectra were acquired at 300 K on Bruker Avance-II 600 and 900 spectrometer equipped with cryoprobes and on an Avance-II 750 spectrometer. All experiments were performed with solutions of 0.6 mm NarE protein in 25 mm phosphate buffer (pH 7.5), 75 mm NaCl, complete protease inhibitor EDTA free (Roche), 0.01% NaN3 in either 90% H2O/10% D2O or 100% D2O. Trimethylsilyl-[2,2,3,3-2H4]propionate (TSP) was added as an internal 1H chemical shift reference. 13C and 15N chemical shifts were referenced indirectly to TSP, using the absolute frequency ratios (8).
All spectra were processed using Topspin 2.1 (Bruker Biospin GmbH). Backbone resonance assignment was carried out based on the triple-resonance experiment sets CBCA(CO)NH/CBCANH and HN(CA)CO/HNCO (9, 10). Side chain resonances were identified using HBHA(CO)NH (11) and HCCH-TOCSY (12) experiments recorded in 90% H2O/10% D2O or 100% D2O, respectively, supplemented by the analysis of NOE spectra. The extent of the resonance assignment was described recently (13). Interproton distance information for structure calculations was derived from 15N-NOESY-HSQC spectra as well as 13C-NOESY-HMQC (14) spectra recorded in 90% H2O/10% D2O or 100% D2O, respectively. All NOESY spectra were acquired using a mixing time of 90 ms. Sparky (15) was used for resonance assignment of the protein. Dihedral angle restraints were predicted using the chemical shift-based dihedral angle prediction software TALOS+ (16). Automated assignment of NOESY spectra was performed with CYANA 2.1 (17). The resulting assigned distance restraints were imported back to Sparky and manually refined by iterative assignment corrections and structure calculations using CNS1.2 (18). For these calculations the assigned distance restraints were categorized into three classes: (I, < 4 Å; II, < 5 Å; III, < 6 Å) according to the signal volume. The 20 lowest energy structures were subjected to a refinement in water using CNS1.2 (18) and the parallhdg5.3 force field (19). The quality of the final ensemble was analyzed with PSVS 1.3 (20).
Relaxation rate measurements were performed as a series of 1H-15N-HSQC-type spectra (21). Relaxation rates of the amide nitrogens were extracted from eleven spectra with delays of 10, 20, 40, 80, 100, 200, 400, 600, 800, 1000, and 2000 ms for R1 and 5, 10, 20, 30, 40, 50, 60, 80, 100, 150, and 200 ms for R2 by fitting the signal intensities to an exponential decay as implemented in the Sparky software. The 15N heteronuclear NOE was measured interleaved with a reference spectrum without NOE transfer. Information to internal dynamics were extracted from the ratio of the signal intensities between both spectra. Structural statistics are presented in Table 1.
TABLE 1.
NMR and refinement statistics for the ensemble of 20 NarE conformers obtained from PSVS [20]
| NMR distance and dihedral restraints | |
| Number of distance restraints | |
| Total NOE | 661 |
| Intra-residue | 0 |
| Inter-residue | 661 |
| Sequential (|i–j| = 1) | 265 |
| Medium-range (|i–j| ≤ 4) | 139 |
| Long-range (|i–j| > 4) | 257 |
| Hydrogen bonds | 82 |
| Number of dihedral angle restraints (Talos+ [16]): Total | 198 |
| ϕ | 99 |
| ψ | 99 |
| Structure statistics | |
| Violations | |
| No. distance restraints (>0.5 Å) | 0 |
| No. dihedral angle restraints (>5°) | 0 |
| Max. dihedral angle violation (°) | 4.8 |
| Max. distance restraint violation (Å) | 0.36 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 0.7 |
| Average pairwise RMSD among 20 refined structures (Å) (L5-S10,Y68-E143) | |
| Heavy | 1.4 |
| Backbone | 0.8 |
| Ramachandran Plot Analysis from Procheck (L5-S10,Y68-E143) | |
| Most favored region | 90.1% |
| Additionally allowed | 9.9% |
| MolProbity Clashscore (Raw score/Z-score) | |
| Raw score | 26.90 |
| Z-score | −3.09 |
NMR Measurements/CSP Experiments
CSP were measured between a reference NMR sample containing 0.1 mm NarE protein in the previously described buffer and test samples additionally containing (I) 1 mm DTT (II) 1 mm DTT, 0.1 mm FeCl3 (Sigma-Aldrich) (III) 1 mm DTT, 0.2 mm β-nicotinamide-adenine-dinucleotide (NAD; Sigma) (IV) 1 mm DTT, 0.1 mm FeCl3, 0.2 mm NAD (V) 1 mm DTT, 0.1 mm ZnCl2 (Merck). Residues showing chemical shift perturbations were assumed to be less affected by the ligand binding than residues showing a signal attenuation. In this regard perturbations of chemical shifts were assumed to be minor and assignments for well dispersed signals were transferred from the reference spectrum. The extracted changes of 15N and 1H chemical shifts were used to calculate the combined chemical shift perturbation in Equation 1 (22).
HADDOCK Calculations
Models of the NAD-NarE complex were calculated using HADDOCK2.1 (23) making use of the eNMR (24) Grid-enabled web server version of HADDOCK (7). Residues affected by the binding of NAD as revealed by CSP experiments and further conserved within the ADPRT group were considered as directly participating in ligand binding (active residues). All remaining affected residues were categorized as indirectly involved in the ligand binding (passive residues). Active and passive residues were only discriminated during the rigid body docking (it0). During semiflexible docking (it1) and water refinement all affected residues on NarE were categorized as passive while NAD was kept active. To account for the flexibility of the loop region of NarE residues 10–66 were defined as fully flexible. One additional ambiguous restraint was defined from the Nitrogen atom of the nicotinamide cleavage site to any protein side-chain glutamate oxygen.
Site-directed Mutagenesis
To generate the NarE-R7K mutant as histidine fusion protein in E. coli, the gene coding sequence, devoid of the STOP codon, was amplified by PCR from N. meningitidis strain MC58, using mutated oligonucleotides containing an NdeI or XhoI site in the forward or reverse primers, respectively, which were used to clone the mutant gene into the pET21b+ vector (Table 2). To generate NarE E111D and E120D mutants, internal primers containing codon changes were designed according to the QuikChange II XL site-directed mutagenesis instruction manual. 100 ng of the pET-NarE-His plasmid were used as template in a methylation reaction, then 12.5 ng of methylated plasmid were employed as substrate in a mutagenesis reaction, using the primer pairs listed in the Table 2. All the mutants were expressed and purified as soluble forms at 37 °C.
TABLE 2.
Primer sequences for the site-directed mutagenesis
Underlined nucleotides correspond to NdeI or XhoI restriction sites. The changed base-triplet is in bold with the mutated nucleotide in lower case.
| Primer | Sequence | Codon change |
|---|---|---|
| R7K forward | CGCGGATCCCATATGGGAAATTTCTTATATAaAGGCATTAG | AGA - AaA |
| R7K reverse | CCCGCTCGAGGTTAATTTCTATCAACTCTTTAGCA | |
| E120D forward | CCAGAAAACCCAAATGAGAAGGAcGTAACAATCAGAGC | GAA - GAc |
| E120D reverse | AGCTCTGATTGTTACgTCCTTCTCATTTGGGTTTTCTGG | |
| E111D forward | GAATATGAGGTTGAcCATCCAGAAAACCCAAATGAGAAGG | GAA - GAc |
| E111D reverse | CTTCTCATTTGGGTTTTCTGGATGgTCAACCTCATATTC |
ADP-ribosyltransferase Assay
The ADP-ribosyltransferase activity of NarE was assayed in vitro by monitoring the enzymatic transfer of ADP-ribose to agmatine as described previously (25). NarE was added to a 0.3 ml assay mixture containing 50 mm potassium phosphate, pH 7.5, 20 mm agmatine as ADP-ribose acceptor and 0.1 mm [adenine-U-14C]NAD (0.05 μCi). After incubation at 30 °C, duplicate samples (100 μl) were applied to 1-ml columns of Dowex AG 1-X2 followed by eluting the product [adenine- 14C]ADP-ribosylagmatine with 5 ml of H2O.
NAD-glycohydrolase Assay
An analog assay based on nicotinamide release was employed to measure the NAD-glycohydrolase (NADase) activity of NarE (26). This assay was carried out in 50 mm potassium phosphate, pH 7.5, 0.1 mm [carbonyl-14C]NAD (0.05 μCi) replacing [adenine-U-14C]NAD in a total volume of 0.3 ml in the presence of the appropriate concentrations of chloride salts. Samples (100 μl) after incubation at 30 °C were applied to a 1-ml column of Dowex AG 1-X2 and 14C-nicotinamide eluted with 5 ml of H2O for liquid scintillation counting. NADase activity was expressed as nanomol/h of [14C]nicotinamide released.
Epitope Mapping with Monoclonal Antibodies
The epitope mapping protocols are based on the approach described by Peter and Tomer (27), which we adapted in two different protocols used here (28).
1) Immunocapturing of peptides from antigen partial digestion. Peptide mixtures were obtained by digestion of NarE with trypsin and LysC (independently) in 50 mm ammonium bicarbonate buffer in a ratio 10:1 at 37 °C for 3 h. To capture the epitope-containing peptide, a 25-μl suspension of Dyanbeads Pan Mouse IgG (uniform, superparamagnetic polystyrene beads of 4.5-μm diameter coated with monoclonal human anti-mouse IgG antibodies) was used. The beads were washed twice with PBS using a magnet and resuspended in the initial volume. 1 μg of the probe (murine) mAb was added and incubated for 30 min at room temperature, after which the beads were washed twice with PBS to remove mAb excess. 0.5 μl of Protease Inhibitor Mix (GE Healthcare) was added prior to the addition of the peptide mixture to avoid potential degradation of the antibodies. The sample was incubated for 30 min at room temperature with gentle tilting and rotation. After incubation, the beads were washed three times with 1 ml of PBS, and the bound peptide was then eluted with 50 μl of 0.2% TFA. The elute fraction was concentrated and washed with C18 ZipTips (Millipore) and eluted in 3 μl of 50% ACN and 0.1% TFA. For MALDI-MS analysis, 1 μl of sample was mixed with the same volume of a solution of α-cyano-4-hydroxy-transcinnamic acid matrix (0.3 mg/ml in H2O:ACN:TFA at 6:3:1), spotted onto the MALDI target plate and allowed to air-dry at room temperature. MALDI-mass spectra were recorded in the positive ion mode on an UltrafleXtreme MALDI TOF/TOF instrument (Bruker Daltonics). Ion acceleration was set to 25 kV. All mass spectra were externally calibrated using a standard peptide mixture. For MS/MS analysis, the MASCOT search engine (Matrix Science. London, UK) was used with the following parameters: one missed cleavage permission, 20 ppm measurement for MS, and 0.3 Da for MS/MS tolerance. Positive identification was accepted with p values lower than 0.05. In the searches, methionine residues modified to methionine sulfoxide were allowed.
2) Partial digestion of immunocaptured antigens. To allow the capture of conformational epitopes, the order of the steps in the previous protocol was inverted. The intact protein (20 μg) was added to the beads, allowing binding to the immobilized mAbs. The protease was then added to the sample in a ratio 50:1, with incubation at 37 °C for 3 h. After proteolysis, the beads were washed ten times with 1 ml of PBS, and the bound peptide was then eluted as previously described. To avoid the analysis of proteolyzed antibody fragments within the elute fraction, NarE was substituted by PBS in negative controls.
RESULTS
NarE Is Structurally Homologous to Other ADPRTs
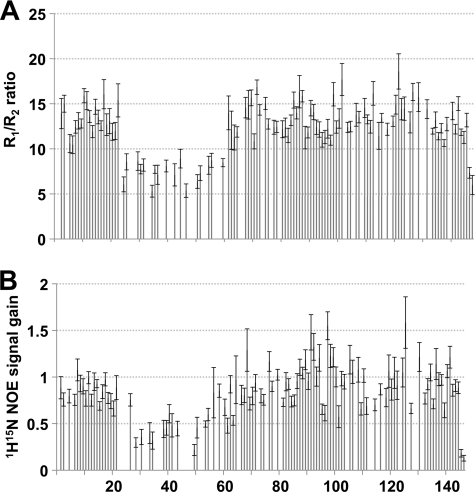
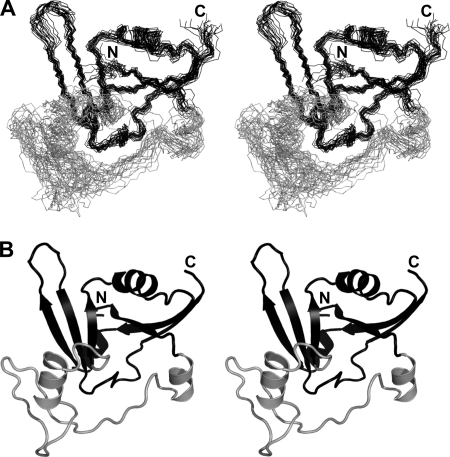
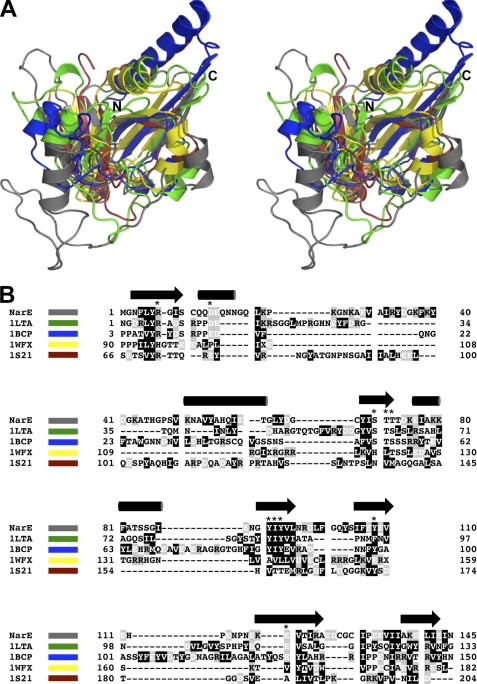
As previously described, the family of ADPRTs shares a similarly folded active site region consisting of three antiparallel β-strands with an α-helix adjacent to the central strand (5). Using solution state nuclear magnetic resonance (NMR) spectroscopy we determined the structure of apo-NarE and characterized its dynamics. Table 1 summarizes the structural statistics of the ensemble of NarE conformers. From R1 and R2 relaxation rates as well as heteronuclear Nuclear Overhauser Effects (NOE) we identified a loop region corresponding to residues 20–60, which is characterized by fast motions in the range of μs-ms within the otherwise rigid protein (Fig. 1, A and B). A lack of NMR signals detected for this loop region also suggests exchange effects as a result of motions in a range of ms-s. The rigid part forms a β-α-α-β-α-β-β-β-β fold with two groups of each three antiparallel β-strands interrupted by three α-helices (Fig. 2, A and B). This domain is commonly found within the ADPRT group as indicated by a DALI (29) search for structurally similar proteins within the Protein Data Bank. Fig. 3A shows some of the highest-ranking hits superposed onto NarE. Surprisingly, besides classical ADPRTs like the heat-labile enterotoxin from E. coli (PDB-ID: 1LTA (30)) or the pertussis toxin (PDB-ID: 1BCP (31)), other proteins without a ribosyltransferase activity like the RNA 2′-phosphotransferase from Aeropyrum pernix K1 (PDB-ID: 1WFX; (32)) or the type III effector protein AvrPphF, which is part of the operon from Pseudomonas syringae (PDB-ID:1S21; (33)), share structural homology to NarE. The RNA 2′-phosphotransferase is, however, still able to bind the substrate NAD (32) while the operon protein is stated to lack this activity (33).
FIGURE 1.
NarE contains a highly flexible loop region. Ratio of R1 and R2 relaxation rates (A) and the ratio of signal intensities between a heteronuclear 1H-15N-NOE spectrum and a reference spectrum without NOE transfer (B) mapped to the residue number. Low R1/R2 ratio and signal gain due to the heteronuclear NOE indicate high flexibility.
FIGURE 2.
The solution NMR structure of NarE. A, backbone stereo view of the 20 lowest energy conformers of NarE. B, lowest energy conformer of NarE in cartoon representation. The N and C termini are indicated. The flexible loop region is shown in gray. Stereoview is shown.
FIGURE 3.
NarE is structurally homologous to other ADP-ribosyl-transferases. A, superposition of NarE (gray) with the heat labile enterotoxin from E. coli (1LTA; green), pertussis toxin (1BSP; blue), the RNA 2′-phosphotransferase from Aeropyrum pernix K1 (1WFX; yellow) and the type III effector protein AvrPphF from Pseudomonas syringae (1S21; red) in stereo view. B, structure-based alignment of the various sequences shown in A. NarE, 1LTA, and 1BCP belong to the group of ADP-ribosyl-transferases while 1WFX and 1S21 do not have this enzyme activity. Secondary structure elements are indicated on top of the sequence. Amino acids are color-coded according to their chemical properties. Residues affected by the binding of NAD, as revealed by NMR Chemical Shift Perturbation (CSP) data, and further only conserved within the ADPRT family are indicated by a star.
NAD Is Well Coordinated within the Active Site Region
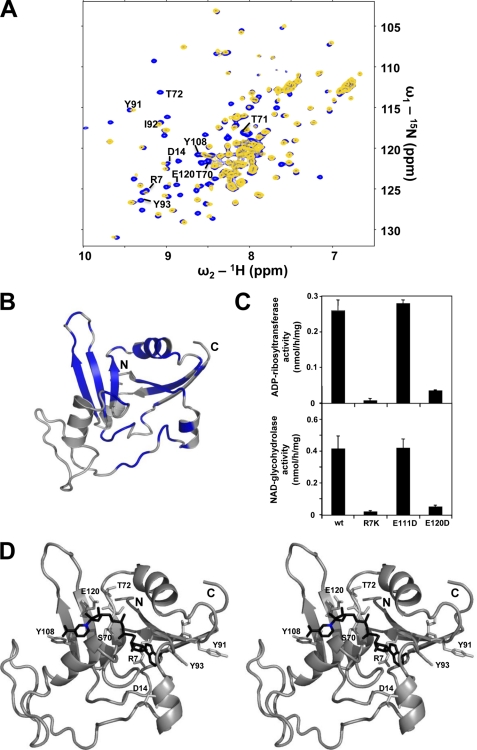
The active site region of NarE could be characterized by measuring chemical shift perturbations between a reference sample of NarE and a sample containing an excess of β-nicotinamide adenine dinucleotide (NAD+). As shown in Fig. 4A, the addition of NAD results in the disappearance of several NMR signals, which could be explained by induced fit motions or cleavage of NAD during the measurement. These slow exchange processes lead to a constantly changing system resulting in increasing linewidth of the NMR signals. In Fig. 4B the affected residues are mapped onto the NarE structure, highlighting the expected NAD binding site as found in other ADPRTs (5). To determine which residues are directly involved in the NAD binding, an analysis of conserved regions between different ADPRTs was carried out. Fig. 3B presents a structure-based alignment of the sequences of the 5 proteins shown in Fig. 3A. Residues showing a perturbation of NMR signals after addition of NAD and that are conserved within the group of ADPRTs are marked with a star (Fig. 4B). These residues, namely Arg-7, Asp-14, Ser-70, Thr-71, Thr-72, Tyr-91, Ile-92, Tyr-93, Tyr-108, and Glu-120, are considered to actively interact with NAD, whereas all other affected residues are thought to be only indirectly involved in NAD binding. To fortify these results we performed a site-directed mutagenesis on two potentially crucial residues, namely Arg-7 and Glu-120. As a control we further analyzed a mutation of Glu-111, which is predicted to have a minor influence in catalysis. To maintain the fold of the protein, residues were substituted by chemically similar amino acids, which leads to the mutations R7K, E111D, and E120D. The activity of the three purified NarE mutants was tested by an in vitro ADP-ribosylation assay as described previously (25).
FIGURE 4.
Characterization of the NAD binding site. A, superposition of the 1H-15N-HSQC spectra of NarE (blue) and NarE containing an excess of β-Nicotinamide-adenine-dinucleotide (NAD) (yellow). B, residues with NMR signals attenuated by more than 50% or showing a combined chemical shift perturbation Δδcomb > 0.03 ppm corresponding to a remarkable signal shift after addition of NAD are mapped in blue onto the NarE. C, site-directed mutagenesis of Arg-7 and Glu-120 residues results in a drastic reduction of both ADP-ribosylation activity (upper panel) and NADase activity (lower panel). The enzymatic activity of NarE was assayed by monitoring both the transfer of ADP-ribose to agmatine and the release of nicotinamide, as described under “Experimental Procedures.” Bars indicate the standard deviations of three individual chambers. Repeated experiments gave similar results. D, model of NAD (black) bound to NarE obtained with the HADDOCK webserver in stereo representation (7). Residues affected by NAD binding and conserved within the ADPRT family are highlighted. The nicotinamide nitrogen nucleophilically attacked by Glu-120 is marked blue.
Whereas the activity of the R7K and E120D mutants is almost completely abolished, the substitution of Glu-111 did not affect it (Fig. 4C), which is in perfect agreement with the described NMR data. In addition, to test whether NarE mutations not only influence the binding to NAD but also its ability to hydrolyze the substrate, we measured the NADase activity of NarE. As shown in Fig. 4C, the enzymatic activity of each mutant well correlated with their capacity to hydrolyze NAD.
This information was used to model the complex between NarE and NAD using the HADDOCK web-server (7). The resulting best scoring model is shown in Fig. 4D. Crucial contacts between NAD and NarE include hydrophobic interactions between Tyr-91 (and possibly Tyr-93) and the adenine ring with possible hydrogen bonding to Asp-14. The branched positively charged Arg-7 side-chain coordinates both NAD phosphates while the ribose ring is fixed via hydrogen bonding to the polar residues Ser-70/Thr-71/Thr-72 that are centrally located within the active site. The nicotinamide ring forms hydrophobic contacts to Tyr-108 while Glu-120 is in the proper position to attack the positively charged nicotinamide nitrogen that will lead to the splitting of the nicotinamide from the ADP-ribose. Similar contacts can be found, for example, within the NAD-complexes of the ADP-ribosyl-cyclase from Aplysia californica (PDB-ID: 3I9K (34)) or the human CD38 protein (PDB-ID: 3I9N (34)).
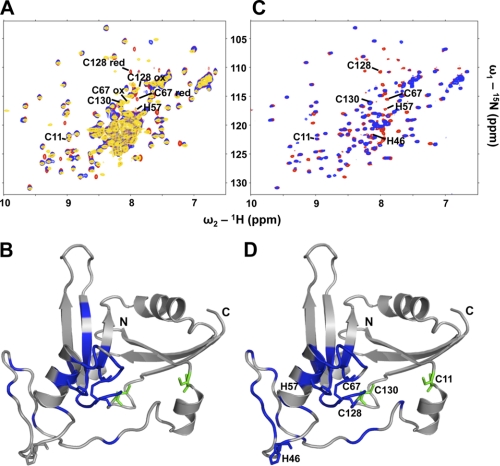
NarE Coordinates a Single Iron via 2 Cysteines and 2 Histidines
As described previously NarE coordinates a single iron via at least 2 of the 4 cysteines present, namely Cys-67 and Cys-128, which were shown, by mutational analysis, to be crucial for the activity of the enzyme (6). In contrast to the yellowish protein samples described by Del Vecchio et al. (6) our highly concentrated NMR samples were colorless. The recorded NMR spectra further showed no evidence for the presence of paramagnetic iron-III, which would strongly increase the linewidth of NMR signals. Hence, it is likely that the NMR structure of NarE described here refers to the apo-form. For that reason Cys-67 and Cys-128 that are crucial for the coordination of iron are forming a disulfide bridge as evidenced from their characteristic 13Cβ side-chain chemical shifts of 46 ppm for Cys-67 and 44 ppm for Cys-128. In contrast, the 13Cβ chemical shifts of C11 (26 ppm) and C130 (28 ppm) indicate these cysteines are in their reduced form (13). To characterize the iron binding site, an equivalent amount of FeCl3, together with 1 mm DTT to open the disulfide bridge, was added to the NMR sample of NarE. Fig. 5A shows the superposed 15N-HSQC spectra of NarE (yellow), NarE with 1 mm DTT (red), and NarE with 1 mm DTT and 1 mm FeCl3 (blue). The paramagnetic effect of iron-III is likely the cause for the disappearance of NMR signals from residues located in the vicinity of the bound iron: the amide signals of Cys-67 and Cys-128 vanish while those of Cys-11 and Cys-130 perfectly match in all 3 spectra. Another residue affected by iron binding is His-57. In Fig. 5B all affected residues are mapped onto the NarE structure. A metal binding site including cysteines and histidines could also coordinate zinc, as found in zinc fingers. As expected, the addition of ZnCl2 to the NarE sample affects similar residues as FeCl3 (Fig. 5, C and D). Because of the diamagnetic nature of zinc-II, in contrast to paramagnetic iron-III, the spectra still show a good resolution, which allows the identification of the fourth metal-coordinating residue within NarE namely His-46. The high flexibility of the loop region in which His-46 is located, allows it to position in proximity to Cys-67, Cys-128, and His-57. These data are taken as evidence that NarE is coordinating a single iron or zinc via Cys-67, Cys-128, His-46, and His-57.
FIGURE 5.
Iron or zinc is coordinated by C67-C128-H46-H57. A, superposition of the 1H-15N-HSQC spectra of NarE (yellow), NarE containing 1 mm DTT (red), and NarE with 1 mm DTT and an equivalent amount of FeCl3 (blue). B, residues whose NMR signals are attenuated by more than 50% or show a combined chemical shift dispersion Δδcomb > 0.03 after addition of FeCl3 are mapped in blue onto the NarE structure. NarE four cysteines as well as His-46/His-57 are highlighted. Cys-67 and Cys-128 are affected by the binding of iron while Cys-11 and Cys-130 (green) remain unaffected. C, superposition of the 1H-15N-HSQC spectra of NarE containing 1 mm DTT (red) and NarE with 1 mm DTT and an equivalent amount of ZnCl2 (blue). D, residues whose NMR signals where attenuated more than 50% or show a combined chemical shift dispersion Δδcomb > 0.03 after addition of ZnCl2 are mapped in blue onto the NarE structure. The NarE four cysteines as well as His-46/His-57 are highlighted. Cys-67 and Cys-128 as well as His-46/His-57 are affected by the binding of zinc while Cys-11 and Cys-130 (green) are unaffected.
Mapping the Antibody Interaction Interface of NarE
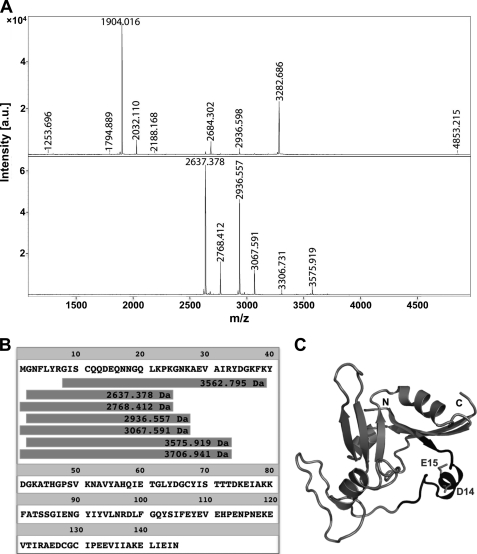
Epitope mapping was performed with four different monoclonal antibodies, labeled 27G1/C11, 21E6/D1, 8E10/A1a, and 8E10/A1b. Two different approaches were used, one of them allowing the identification of conformational epitopes (see “Experimental Procedures”). With either approach, the experiments were performed in triplicate, using the proteases trypsin, LysC, and GluC. The results indicate that the four monoclonal antibodies recognize the same region of NarE, within the sequence 8-GISCQQDEQNNGQLKPK-24. Fig. 6A shows the mass spectrum of the total tryptic digestion of NarE (upper spectrum). The mass spectrum of the peptides immunocaptured with mAb 27G1/C11 (Fig. 6A, lower spectrum) refers to N-terminal fragments of NarE sharing the described epitope (Fig. 6B) To confirm the sequence of these peptides, MS/MS spectra were also obtained for some of the peaks (not shown). Digestion with LysC leads to the same results as with trypsin, albeit with different missed cleavage sites. When using GluC, however, no immunocaptured peptide fraction could be detected. Because the peptides retained by the antibodies after trypsin or LysC digestion contained the residues Asp-14 and Glu-15 which are potential cleavage sites for GluC, these results suggest Glu-14 and Glu-15 to be at the core of the epitope region, such that cleavage at either site would prevent binding. In particular, the side chain of Glu-15 is well accessible at the surface of the protein (Fig. 6C), making it a clear candidate for recognition and binding. The fact that the two approaches lead to the same result indicates that epitope recognition is primarily based on sequence.
FIGURE 6.
Epitope mapping of NarE. A, upper spectrum: total digestion of NarE with LysC; lower spectrum: peptides captured with the monoclonal antibody 27G1/C11 (using protocol 1 as described under “Experimental Procedures”). B, sequences corresponding to the captured peptides appearing in the lower spectrum of panel A, aligned with the entire NarE sequence. C, determined epitope is mapped to the NarE structure. The centrally located Asp-14 and Glu-15 are highlighted.
DISCUSSION
The generation of structural and functional information on bacterial determinants is a fundamental step in unraveling their role in bacterial pathogenesis. In this context, the data reported in this study may help to uncover important aspects of meningococcal physiology, as this protein belongs to the family of ADPRTs, which constitutes a class of functionally conserved enzymes displaying toxic activity in a variety of bacterial pathogens (35). Interestingly, although lacking the canonical N-terminal leader peptide, NarE was found in the periplasmic space of meningococcus (5), suggesting that in order to be active, it may require a further processing.
The NMR structure determination of NarE reveals a domain with β-α-α-β-α-β-β-β-β fold with a Gln-20 to Ala-53 loop region between the first and second α-helix which shows both fast flexibilities as well as intermediate conformational motions (Fig. 2A). The six antiparallel β-strands are arranged in two groups of each 3 strands. This domain is typically found within the ADPRT group (Fig. 3). As described previously the active site region of ADPRTs contains a central β-strand characterized by several polar residues. This central strand is followed by an α-helix and flanked by two antiparallel β-strands containing a crucial arginine and glutamate residue (5). A further histidine located within α-helix 2, which covers the active site, is considered as crucial for the recognition of, and interaction with, the target protein (36, 37, 38). By measuring chemical shift perturbations after addition of NAD to the NarE sample we could determine those residues that are affected by the interaction with NAD (Fig. 4B). These data perfectly match with the former proposed active site (5). In agreement with these data, site-directed mutagenesis of Arg-7 and Glu-120 resulted in a drastic reduction of ADP-ribosylation activity, indicating that these two residues play a crucial role in catalysis. We could further characterize a previously described iron binding site within NarE, a novelty within the ADPRT family (6): the addition of an equivalent amount of FeCl3 to the NarE sample affects only Cys-67 and Cys-128 but not Cys-11 nor Cys-130. This is consistent with a mutational analysis of these 4 cysteines describing the crucial role of Cys-67 and Cys-128 for the activity of the protein (6). The iron binding site is composed further of two histidines, namely His-46 and His-57 that could be identified to coordinate the single iron (Fig. 5A). This atypical Cys2His2 iron binding site, which is more characteristic for the coordination of zinc, could be confirmed by CSP data after addition of ZnCl2 to the NarE sample: the same residues are affected by zinc binding as by iron (Fig. 5C). As described previously, iron binding is only crucial for the transferase but not for the glycohydrolase activity of NarE (6). This is consistent with our CSP data showing that the addition of NAD to both apo NarE and the iron-bound protein leads to the same perturbations of NMR signals of active site residues (data not shown). Hence, the apo form of NarE is still able to bind and hydrolyze NAD but unable to transfer the ADP-ribose to the target protein. For that reason it is very likely that the iron coordination is crucial for the recognition and binding of the target protein. This is further supported by the fact that His-57, which was recently shown to be crucial for the transferase activity (36, 37, 38), is part of the Cys2His2 iron site. Intriguingly His-57 is part of an α-helix which is found in proteins with ADPRT activity but not within structurally similar NAD-binding proteins (Fig. 3A). Because NarE natively coordinates iron (6) the relevance of zinc coordination remains unclear. Even though zinc binding in vitro is evident from the CSP data, we do not have any evidence for this interaction in vivo. Most intriguingly, helix α2 together with β-strands 2 and 5 are mimicking structural features of a zinc finger domain, which could play a crucial role in target protein recognition.
A number of surface associated and secreted determinants produced by N. meningitidis are responsible for bacterial uptake by host cells and have been reported to cause major consequences to intracellular signaling cascade (39). Modifying small GTP-binding proteins activates specific pathways, required for the efficient internalization of meningococcus into host cells. The fact that several ADP-ribosylating toxins exert their cytotoxic effects on the cytoskeleton through an inactivation of the Rho GTPases of the Ras superfamily would suggest that also NarE may be involved in the mechanism of meningococcal internalization. ADP-ribosyltransferases have been extensively characterized and successfully used as vaccine components and mucosal adjuvants (35). In this context, the attempt to test the capacity of antisera against the wild-type and mutant toxins to induce bactericidal titers failed due to the low activity measured (data not shown). These data are in agreement with the fact that NarE is localized at the bacteria periplasmic space (5), making antibody recognition and consequent complement deposition very unlikely. However, in terms of the NarE immunogenic properties, generation of monoclonal antibodies against this antigen revealed the propensity for all of them to recognize a specific region (Gly-8 to Lys-24), largely corresponding to α-helix 1. The epitope mapping results suggest that Glu-15 could be one of the essential residues for recognition and interaction with antibodies. These data suggest that the catalytic site is not particularly immunogenic, which makes NarE to an unlikely candidate for an antibody-based preventive strategy. The data reported in this study provide a useful link between structural and biochemical features of NarE and define the basis for future studies on its role in meningococcal pathogenesis.
Accession Numbers
The structure coordinates of NarE have been deposited into the Protein Data Bank under accession code 2KXI. Assigned resonances as well as all restraints used for structure calculation have been deposited into the BMRB under accession code 16737. The extent of resonance assignment was described previously (13).
Acknowledgments
We thank the Dutch BiG Grid project (Netherlands Organization for Scientific Research) for the use of computing and storage facilities.
This project was supported by funding under the Sixth Research Framework Programme of the European Union (Ref. LSHB-CT-2006-037325, BacAbs). The eNMR Project (European FP7 e-Infrastructure Grant, Contract no. 213010), supported by the National GRID Initiatives of Italy, Germany, and The Netherlands.
The atomic coordinates and structure factors (code 2KXI) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Assigned resonances as well as all restraints used for structure calculation have been deposited into the BMRB under Accession Code 16737.
- ADPRT
- ADP-ribosyltransferase
- NOE
- Nuclear Overhauser Effects
- CSP
- Chemical Shift Perturbation
- NADase
- NAD-glycohydrolase.
REFERENCES
- 1. Caroline Genco, Lee Wetzler. (ed) (2010) Neisseria: Molecular Mechanisms of Pathogenesis, Caister Academic Press, Norwich, UK [Google Scholar]
- 2. Moss J., Vaughan M. (1988) Adv. Enzymol. Relat. Areas Mol. Biol. 61, 303–379 [DOI] [PubMed] [Google Scholar]
- 3. Althaus F. R., Richter C. (1987) Mol. Biol. Biochem. Biophys. 37, 1–237 [PubMed] [Google Scholar]
- 4. Locht C., Keith J. M. (1986) Science 232, 1258–1264 [DOI] [PubMed] [Google Scholar]
- 5. Masignani V., Balducci E., Di Marcello F., Savino S., Serruto D., Veggi D., Bambini S., Scarselli M., Aricò B., Comanducci M., Adu-Bobie J., Giuliani M. M., Rappuoli R., Pizza M. (2003) Mol. Microbiol. 50, 1055–1067 [DOI] [PubMed] [Google Scholar]
- 6. Del Vecchio M., Pogni R., Baratto M. C., Nobbs A., Rappuoli R., Pizza M., Balducci E. (2009) J. Biol. Chem. 284, 33040–33047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. deVries S. J., van Dijk M., Bonvin A. M. (2010) Nat. Protoc. 5, 883–897 [DOI] [PubMed] [Google Scholar]
- 8. Wishart D. S., Bigam C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. (1995) J. Biomol. NMR 6, 135–140 [DOI] [PubMed] [Google Scholar]
- 9. Grzesiek S., Bax A. (1992) J. Am. Chem. Soc. 114, 6291–6293 [Google Scholar]
- 10. Grzesiek S., Bax A. (1993) J. Biomol. NMR 3, 185–204 [DOI] [PubMed] [Google Scholar]
- 11. Montelione G., Lyons B., Emerson S., Tashiro M. (1992) J. Am. Chem. Soc. 114, 10974–10975 [Google Scholar]
- 12. Kay L., Ikura M., Bax A. (2002) J. Am. Chem. Soc. 112, 888–889 [Google Scholar]
- 13. Carlier L., Koehler C., Veggi D., Pizza M., Soriani M., Boelens R., Bonvin A. M. (2011) Biomol. NMR Assign. 5, 35–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clore G., Gronenborn A. (1992) Prog. Nucl. Magn. Reson. Spectrosc. 23, 43–92 [Google Scholar]
- 15. Goddard T., Kneller D. Sparky 3, University of California, San Francisco [Google Scholar]
- 16. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guentert P. (2004) in Automated NMR Protein Structure Calculation with Cyana, Methods in Molecular Biology (Downing A. K. ed), pp. 353–378, Humana Press; [DOI] [PubMed] [Google Scholar]
- 18. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 19. Linge J., Williams M., Spronk C., Bonvin A., Nilges M. (2003) Proteins-Structure Function Genetics 50, 496–506 [DOI] [PubMed] [Google Scholar]
- 20. Bhattacharya A., Tejero R., Montelione G. T. (2007) Proteins 66, 778–795 [DOI] [PubMed] [Google Scholar]
- 21. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 22. Mulder F. A., Schipper D., Bott R., Boelens R. (1999) J. Mol. Biol. 292, 111–123 [DOI] [PubMed] [Google Scholar]
- 23. Dominguez C., Boelens R., Bonvin A. M. (2003) J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 24. Bonvin A. M., Rosato A., Wassenaar T. A. (2010) J. Struct. Funct. Genomics 11, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moss J., Stanley S. J. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 4809–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moss J., Manganiello V. C., Vaughan M. December 1976. Proc. Natl. Acad. Sci. U.S.A. 73, 4424–4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peter J. F., Tomer K. B. (2001) Anal. Chem. 73, 4012–4019 [DOI] [PubMed] [Google Scholar]
- 28. Soriani M., Petit P., Grifantini R., Petracca R., Gancitano G., Frigimelica E., Nardelli F., Garcia C., Spinelli S., Scarabelli G., Fiorucci S., Affentranger R., Ferrer-Navarro M., Zacharias M., Colombo G., Vuillard L., Daura X., Grandi G. (2010) J. Biol. Chem. 285, 30126–30138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Merritt E. A., Sixma T. K., Kalk K. H., van Zanten B. A., Hol W. G. (1994) Mol. Microbiol. 13, 745–753 [DOI] [PubMed] [Google Scholar]
- 31. Hazes B., Boodhoo A., Cockle S. A., Read R. J. (1996) J. Mol. Biol. 258, 661–671 [DOI] [PubMed] [Google Scholar]
- 32. Kato-Murayama M., Bessho Y., Shirouzu M., Yokoyama S. (2005) J. Mol. Biol. 348, 295–305 [DOI] [PubMed] [Google Scholar]
- 33. Singer A. U., Desveaux D., Betts L., Chang J. H., Nimchuk Z., Grant S. R., Dangl J. L., Sondek J. (2004) Structure 12, 1669–1681 [DOI] [PubMed] [Google Scholar]
- 34. Liu Q., Graeff R., Kriksunov I. A., Jiang H., Zhang B., Oppenheimer N., Lin H., Potter B. V., Lee H. C., Hao Q. (2009) J. Biol. Chem. 284, 27637–27645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Masignani V., Balducci E., Serruto D., Veggi D., Aricò B., Comanducci M., Pizza M., Rappuoli R. (2004) Int. J. Med. Microbiol. 293, 471–478 [DOI] [PubMed] [Google Scholar]
- 36. Pizza M., Covacci A., Bartoloni A., Perugini M., Nencioni L., De Magistris M. T., Villa L., Nucci D., Manetti R., Bugnoli M. (1989) Science 246, 497–500 [DOI] [PubMed] [Google Scholar]
- 37. Xu Y., Barbançon-Finck V., Barbieri J. T. (1994) J. Biol. Chem. 269, 9993–9999 [PubMed] [Google Scholar]
- 38. Jobling M. G., Holmes R. K. (2001) J. Bacteriol. 183, 4024–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hill D. J., Griffiths N. J., Borodina E., Virji M. (2010) Clin. Sci. 118, 547–564 [DOI] [PMC free article] [PubMed] [Google Scholar]