Abstract
GTP-bound forms of Ras family small GTPases exhibit dynamic equilibrium between two interconverting conformations, “inactive” state 1 and “active” state 2. A great variation exists in their state distribution; H-Ras mainly adopts state 2, whereas M-Ras predominantly adopts state 1. Our previous studies based on comparison of crystal structures representing state 1 and state 2 revealed the importance of the hydrogen-bonding interactions of two flexible effector-interacting regions, switch I and switch II, with the γ-phosphate of GTP in establishing state 2 conformation. However, failure to obtain both state structures from a single protein hampered further analysis of state transition mechanisms. Here, we succeed in solving two crystal structures corresponding to state 1 and state 2 from a single Ras polypeptide, M-RasD41E, carrying an H-Ras-type substitution in residue 41, immediately preceding switch I, in complex with guanosine 5′-(β,γ-imido)triphosphate. Comparison among the two structures and other state 1 and state 2 structures of H-Ras/M-Ras reveal two new structural features playing critical roles in state dynamics; interaction of residues 31/41 (H-Ras/M-Ras) with residues 29/39 and 30/40, which induces a conformational change of switch I favoring its interaction with the γ-phosphate, and the hydrogen-bonding interaction of switch II with its neighboring α-helix, α3-helix, which induces a conformational change of switch II favoring its interaction with the γ-phosphate. The importance of the latter interaction is proved by mutational analyses of the residues involved in hydrogen bonding. These results define the two novel functional regions playing critical roles during state transition.
Keywords: Crystal Structure, NMR, Oncogene, Ras, Signal Transduction, Conformational Dynamics, Switch Region
Introduction
H-Ras, K-Ras, and N-Ras, collectively called Ras, are the products of the ras proto-oncogenes and belong to the Ras family of small GTPases, which also includes Rap1, Rap2, R-Ras, R-Ras2/TC1, M-Ras/R-Ras3, Ral etc. (1, 2). They function as guanine nucleotide-dependent molecular switches by cycling between the GTP-bound active and GDP-bound inactive forms in intracellular signaling pathways controlling cell growth and differentiation (3). The interconversion between the GDP-bound and GTP-bound forms is catalyzed by guanine nucleotide exchange factors and GTPase-activating proteins (4, 5). X-ray crystallographic and NMR analyses of H-Ras and Rap1A, alone or in complex with their effectors, revealed that the exchange of GTP for GDP results in allosteric conformational changes in two adjacent regions, termed switch I (residues 32–38) and switch II (residues 60–75), consisting of a single loop and a loop and an α-helix (α2-helix), respectively, and enables Ras to execute downstream signaling through direct interaction with its effectors, such as Raf kinases, phosphoinositide 3-kinases, and phospholipase Cϵ (3, 4) (see supplemental Fig. S1). Switch I almost overlaps with the effector region (residues 32–40), which forms a principal binding interface for effector recognition (6–9).
31P NMR studies revealed that H-Ras and K-Ras in complex with Mg2+ and a non-hydrolyzable GTP analog, GppNHp,3 exhibit equilibrium between the two conformational states, termed state 1 and state 2 (10). The two states are characterized by different chemical shift values for the resonances of the phosphorous atoms of the α- and γ-phosphate groups of GppNHp. The chemical shift values are mainly influenced by the distance of the phosphate groups from the aromatic ring of Tyr-32 in switch I, which exerts a “ring current shift” effect (10). The interconversion between the two states occurs in a millisecond time scale and appears to be a general property shared by members of the Ras family small GTPases irrespective of the nature of the bound guanine nucleotide triphosphate: GTP, GppNHp, or GTPγS (10–13). However, the state distribution exhibited a great variation even among closely related GTPase species; the state 1 population occupies 36 ± 2, 15 ± 1, and 93 ± 2% for H-Ras, Rap1A, and M-Ras, respectively (12), which possess the identical switch I residues and share some of the effectors such as c-Raf-1 (14, 15). Because association of H-Ras-GppNHp with its effectors such as c-Raf-1 induced a shift of the equilibrium toward state 2, state 1 and state 2 were presumed to represent inactive and active conformations, respectively (10).
Although the crystal structures corresponding to state 2 had been solved with H-Ras-GppNHp alone or in complex with the effectors (8, 9, 16, 17), those corresponding to state 1 remained unsolved until our determination of the crystal structure of M-Ras-GppNHp (18). So far, state 1 structures were solved with the crystals of the GppNHp-bound forms of M-Ras (18), H-RasG60A (19), H-RasG60A/K147A (20), M-RasP40D (21), and H-RasT35S (21). H-RasT35S yielded two distinct structures, termed form 1 and form 2. Comparison of these structures with state 2 structures indicated that the most fundamental feature distinguishing state 1 from state 2 is the loss of the direct and Mg2+-coordinated indirect hydrogen-bonding interactions of Thr-35/45 (H-Ras/M-Ras) in switch I with the γ-phosphate of GppNHp. This results is in marked deviation of the switch I loop away from the guanine nucleotide and increases the distance between Tyr-32/42 and the γ-phosphate, accounting for at least a major part of the observed 31P NMR chemical shift changes. Moreover, the structures of M-Ras-GppNHp and H-RasT35S-GppNHp form 1 revealed another key structural feature of state 1, the loss of the hydrogen-bonding interaction between Gly-60/70 in switch II and the γ-phosphate, whose importance was also supported by the adoption of the state 1 conformation by H-RasG60A-GppNHp and H-RasG60A/K147A-GppNHp (19, 20). Thus, the state 2 structure is characterized by stabilization of the switch I and switch II loops to the nucleotide through interactions of Thr-35/45 and Gly-60/70 with the γ-phosphate. The results also indicated the importance of the nucleotide-mediated interdependence between the two switch regions because a mutation in one switch region induced a gross conformational change of the other switch region (19–21). The structures of M-RasP40D-GppNHp and H-RasT35S-GppNHp form 2, retaining the Gly-60/70-γ-phosphate interaction whereas losing the Thr-35/45-γ-phosphate interaction, were presumed to represent the intermediate between the two states (21). However, these analyses were carried out by comparison of the crystal structures representing either state derived from different mutant polypeptides, which tended to be compromised by the secondary effects of the mutations. Thus, further analysis of the state transition mechanisms needed determination of both state structures from a single Ras polypeptide.
In the present study, we utilize the M-RasD41E polypeptide to solve both state 1 and state 2 crystal structures in the GppNHp-bound forms. The D41E mutation, where Asp-41 of M-Ras was substituted by the corresponding Glu-31 of H-Ras, was known to increase state 1 occupancy of M-Ras most efficiently as a single mutation from our previous study (21). Comparison of the resulting two crystal structures corresponding to state 1 and state 2 reveal two new structural features pertaining to the state transition; the interaction among the residues immediately preceding switch I including Glu-31/Asp-41 and the hydrogen-bonding interaction of switch II with the α3-helix. The two interactions facilitate establishment of the Thr-35/45-γ-phosphate and Gly-60/70-γ-phosphate interactions, respectively, and thereby induce the shift of state equilibrium toward state 2. Applicability of this mechanism to other members of the Ras family small GTPases, H-Ras, and more distantly related RalA, will be discussed.
EXPERIMENTAL PROCEDURES
Protein Purification
Mouse M-RasD41E (residues 1–178) was expressed as fusions with glutathione S-transferase in Escherichia coli BL21(DE3) using pGEX-6P-1 vector (GE Healthcare, Buckinghamshire, UK), immobilized on glutathione-agarose, and eluted by cleavage with PreScission protease (GE Healthcare). After further purification by ion exchange chromatography to the final purity of >95%, it was loaded with GppNHp and used for crystallization or NMR spectroscopy as described before (18). Human c-Raf-1 RBD (residues 51–130) was purified as described (12).
NMR Spectroscopy
31P NMR spectra were recorded in the presence or absence of c-Raf-1 RBD on a Brucker AVANCE-500 NMR spectrometer (18). The 31P spectra were referenced as described (22).
Crystallization of Ras Proteins in Complex with Mg2+ and GppNHp
M-RasD41E-GppNHp was dissolved in buffer (50 mm Tris-HCl, pH 7.4, 50 mm NaCl, 5 mm MgCl2, and 1 mm DTT). Crystals of M-RasD41E-GppNHp were grown by the sitting drop vapor diffusion method at 20 °C in drops containing 1 μl of protein solution (19 mg/ml) and 1 μl of reservoir 1 (100 mm Tris-HCl, pH 8.5, 200 mm MgCl2, and 20% (w/v) PEG 8000) for 1 week, or in drops containing 0.8 μl of protein solution 2 (21.5 mg/ml) and 1 μl of reservoir 2 (100 mm Tris-HCl, pH 6.5, 200 mm MgCl2, and 25% (w/v) PEG 8000) for 3 weeks, respectively. Crystals with space group P622 grew in reservoir 1, whereas those with space group P212121 grew in reservoir 2.
Data Collection and Structure Determination
The data collections at 100 K were carried out at BL38B1 using Jupiter 210 (Rigaku corporation) or Quantum 210 (ADSC) CCD detectors in SPring-8. Distinct structures, type 1 and type 2, were solved from the crystals with space groups P622 and P212121, respectively. The data for M-RasD41E-GppNHp type 1 were processed using the program HKL2000 (23), whereas those for type 2 were processed with the program MOSFLM (24) and scaled with the program SCALA in the CCP4 program suite (25). The crystal structures were determined by the molecular replacement method with MOLREP (26) using M-Ras-GppNHp (Protein Data Bank code 1X1S) as a search model. After the two models were refined with programs CNS (27) and REFMAC (28), their stereochemical quality was checked with PROCHECK in the CCP4 program suit (29). The data collection and refinement statistics are summarized in supplemental Table S1. The number of detected water molecules in type 1 (resolution = 2.75 Å) was substantially smaller than that in type 2 (resolution = 1.55 Å). Thus, the assignment of water-mediated hydrogen bonds in type 1 was done with great care only in reference to those found in type 2, when the corresponding water molecules were invisible.
Graphics
The figures were prepared with PyMOL (DeLano Scientific).
PDB Codes of the Coordinates Used in this Paper
M-RasD41E-GppNHp type 1, PDB code 3PIR; M-RasD41E-GppNHp type 2, PDB code 3PIT; M-Ras-GppNHp, PDB code 1X1S; M-RasP40D-GppNHp, PDB code 3KKP; M-RasP40D/D41E/L51R-GppNHp, PDB code 3KKO; H-Ras-GppNHp, PDB code 1CTQ; H-RasT35S-GppNHp form 2, PDB code 3KKM; Rap2A-GTP, PDB code 3RAP chain R; RalA-GppNHp, PDB code 1U8Y chain B were used.
RESULTS
Determination of Two Distinct Crystal Structures of M-RasD41E-GppNHp, Corresponding to State 1 and State 2
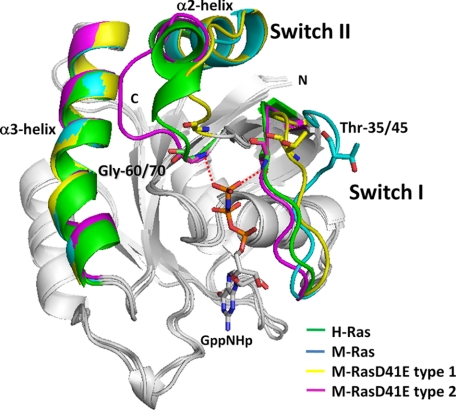
We previously showed that the residues immediately preceding switch I (hereafter termed pre-switch I residues) of M-Ras (supplemental Fig. S1), particularly Pro-40 and Asp-41, play critical roles in its state 1 predominance because H-Ras-type substitutions for these residues, P40D and D41E, caused a significant increase in the state 2 population (21). In particular, M-RasD41E-GppNHp exhibited the most prominent increase of the state 2 population to 21 ± 2 over 7 ± 2% of M-Ras-GppNHp (supplemental Table S2). This led us to examine crystallization conditions of M-RasD41E-GppNHp for crystals representing either state. With the use of two distinct conditions different in pH and PEG8000 concentrations (see “Experimental Procedures”), we obtained two crystals with a different space group, which yielded two distinct tertiary structures, type 1 and type 2 (supplemental Table S1). Their overall structures superimposed very well with those of M-Ras-GppNHp and H-Ras-GppNHp, representing state 1 and state 2 structures, respectively, except the two switch regions (Fig. 1). Electron density of residues 69–73 in switch II, which were invisible in the M-Ras-GppNHp structure (18), was completely clear in both structures. The effect of crystal packing on the determined structures seemed negligible. Although we observed dot-like contacts only at residues 74 and 112 with a neighboring molecule in type 1, such extremely faint contacts were unlikely to affect the conformation of the corresponding regions. Indeed, the temperature factor values for the two residues were kept high, suggesting that these residues were not fixed by crystal packing.
FIGURE 1.
Comparison of the crystal structures of M-RasD41E-GppNHp types 1 and 2 with those of M-Ras-GppNHp and H-Ras-GppNHp with a special emphasis on switch I, switch II, and the α3-helix. Superimposition of the backbone structures of M-Ras-GppNHp, M-RasD41E-GppNHp type 1, M-RasD41E-GppNHp type 2, and H-Ras-GppNHp. Only switch I, switch II, and the α3-helix are colored as indicated. The structure of GppNHp is excerpted from the model of M-RasD41E-GppNHp type 2. GppNHp and the side chains of Thr-35/45 and Gly-60/70 (H-Ras/M-Ras) are shown in the stick model (red, oxygen; blue, nitrogen; deep pink, phosphorus). Direct hydrogen bonds of Thr-45 and Gly-70 with the γ-phosphate in type 2 are shown by red dotted lines, whereas Mg2+- or water-mediated hydrogen bonds (see supplemental Fig. 2) are not shown. The models were generated with PyMOL, based on the least square fittings of the Cα atoms of the residues excluding those of the two switch regions.
M-RasD41E-GppNHp type 1 exhibited the loss of the interactions of both Thr-45 in switch I and Gly-70 in switch II with the γ-phosphate of GppNHp (see also supplemental Fig. S2) and deviation of the switch I loop away from the nucleotide, indicating that it corresponded to state 1. Indeed, switch II residues of type 1 superimposed very well with the visible part of switch II residues of M-Ras-GppNHp (Fig. 1) and the type 1 crystal possessed similar unit cell dimensions and the same space group with the M-Ras-GppNHp crystal, implying similar crystal packing. Although no interaction was recognized between the switch I loop and γ-phosphate, Thr-45 of type 1 showed a significant positional shift approaching the γ-phosphate compared with that of M-Ras-GppNHp and was located in the middle positions of Thr-45 of M-Ras-GppNHp (state 1) and Thr-35 of H-Ras-GppNHp (state 2) (Fig. 1). This positional change of Thr-45 of type 1 may facilitate establishment of a hydrogen-bonding interaction with the γ-phosphate and account for the increase of the state 2 population in M-RasD41E-GppNHp.
On the other hand, M-RasD41E-GppNHp type 2 corresponded to state 2 because the interactions of both Thr-45 and Gly-70 with the γ-phosphate were retained as commonly observed in state 2 conformers such as H-Ras-GppNHp and M-RasP40D/D41E/L51R (21) (Fig. 1 and supplemental Figs. S2 and S3). Indeed, the backbone structures of the switch I loop and α2-helix in switch II of type 2 superimposed very well with those of state 2 conformers, except switch II loop residues, which exhibited a minor conformational change (Fig. 1 and supplemental Fig. S3). Thus, M-RasD41E-GppNHp yielded both state 1 and state 2 crystal structures for the first time as Ras family small GTPases.
Structural Differences Around Switch I and Switch II between Type 1 and Type 2
We made a detailed comparison of the structures of the switch I and switch II regions, which exhibited marked differences between M-RasD41E-GppNHp type 1 and type 2 (Fig. 1). Because of the relatively low resolution of type 1, we predicted the existence of water-mediated hydrogen bonds in type 1 only for those actually detected in type 2 when the corresponding water molecules were invisible.
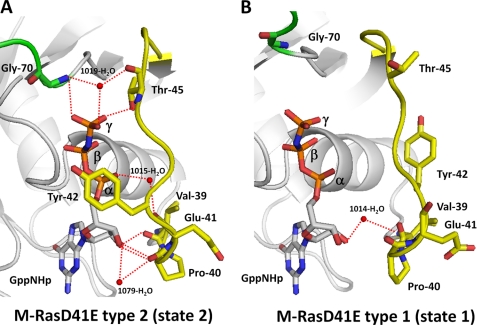
Comparison of the structures of the switch I and pre-switch I regions indicated that Tyr-42 exhibited the most drastic positional changes (Fig. 2). In type 2, where the Thr-45-γ-phosphate hydrogen-bonding interaction was present, the side chain of Tyr-42 was located very close to the nucleotide (Fig. 2A). In contrast, it was pulled far away from the nucleotide in type 1, which lacked the Thr-45-γ-phosphate interaction (Fig. 2B). Consistent with this, the main chain amide of the adjacent Glu-41 formed a water-mediated hydrogen bond with the α-phosphate of GppNHp in type 2, whereas the corresponding interaction was absent in type 1 (Fig. 2 and supplemental Fig. S2). Moreover, the modes of interaction with the ribose ring of GppNHp showed a marked difference; Val-39 formed a direct hydrogen bond and Pro-40 formed both direct and water-mediated hydrogen bonds in type 2, whereas type 1 formed only a water-mediated hydrogen bond via Val-39. Thus, the hydrogen-bonding network in switch I and the pre-switch I regions of type 2 showed closer resemblance to that of H-Ras-GppNHp than that of M-Ras-GppNHp (supplemental Fig. S2). These results hinted at an intimate relationship of the composition of the hydrogen-bonding networks in these regions with the formation of the Thr-45/35-γ-phosphate (M-Ras/H-Ras) hydrogen bond, the determinant of the conformational states. Mechanisms underlying this relationship will be discussed (see “Discussion”).
FIGURE 2.
Comparison of the hydrogen-bonding networks around the pre-switch I and switch I regions between M-RasD41E-GppNHp types 1 and 2. The pre-switch I and switch I residues (yellow) and the switch II residue Gly-70 (green) are shown for M-RasD41E-GppNHp type 2 (A) and type 1 (B). Oxygen (red) and nitrogen (blue) atoms are shown only for the main chains and side chains of Val-39, Pro-40, Glu-41, Tyr-42, Thr-45, and Gly-70, and GppNHp. Water molecules mediating hydrogen bonds are shown by red balls, whereas hydrogen bonds are shown by red dotted lines. The models were generated with PyMOL.
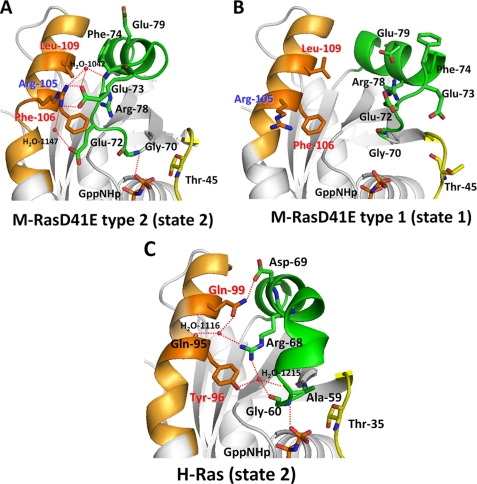
Comparison of the structures of the switch II region indicated that α2-helix was located close to the α3-helix in type 2 compared with that in type 1 (Fig. 1). We observed a clear difference in the mode of interaction between switch II and the α3-helix. In type 2, where the Gly-70-γ-phosphate interaction was present, Glu-72, Glu-73, and Phe-74 in switch II formed direct and/or water-mediated hydrogen bonds with Arg-105 in the α3-helix (Fig. 3A). In sharp contrast, residues 70–79 including these 3 residues in switch II failed to form any hydrogen bonds with residues 105–109 in the α3-helix (Fig. 3B). Intriguingly, H-Ras-GppNHp exhibited hydrogen-bonding interactions between corresponding residues 60–69 in switch II and residues 95–99 in the α3-helix, which were similar to those observed in type 2 (Fig. 3C). However, the interactions were more intensive in the case of H-Ras-GppNHp; they were composed of a direct hydrogen bond of Asp-69 with Gln-99, and water-mediated hydrogen bonds of Arg-68 with Gln-95, Tyr-96, and Gln-99, and Gly-60 with Tyr-96. The α2-helices of type 2 and H-Ras-GppNHp exhibited a similar rotational change compared with that of type 1, and their Gly-70/60 (M-Ras/H-Ras) formed hydrogen bonds with the γ-phosphate of GppNHp (21) (Fig. 3, A and C).
FIGURE 3.
Comparison of the hydrogen-bonding interactions between switch II and the α3-helix among M-RasD41E-GppNHp types 1 and 2 and H-Ras-GppNHp. The tertiary structures around switch II and the α3-helices are shown for M-RasD41E-GppNHp type 2 (A), M-RasD41E-GppNHp type 1 (B), and H-Ras-GppNHp type 2 (C). Switch I, switch II, and the α3-helix are shown in yellow, green, and orange, respectively. Hydrogen bonds are shown by red dotted lines. The models were generated as described in the legend to Fig. 2.
Crucial Role of the Switch II-α3-Helix Interaction in Facilitating Adoption of State 2
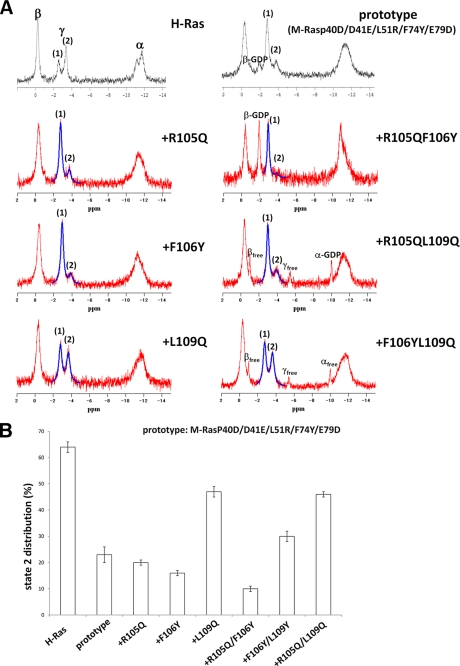
The results described above suggested that the mode of the switch II-α3-helix interaction, which depends on the nature of the relevant residues, may play a critical role in state transition. Comparison of amino acid sequences showed that some of the residues involved in the switch II-α3-helix interactions were not conserved between H-Ras and M-Ras, such as Tyr-64 versus Phe-74 and Asp-69 versus Glu-79, respectively, in switch II, and Gln-95 versus Arg-105, Tyr-96 versus Phe-106, Glu-98 versus Gln-108, and Gln-99 versus Leu-109, respectively, in the α3-helix (supplemental Fig. S1). We knew from our previous study (21) that H-Ras-type amino acid substitutions for Phe-74 and Glu-79, F74Y and E79D, in M-Ras, alone or in combination with the P40D/D41E/L51R mutation (see supplemental Table S2), had no significant effect on state distribution. Accordingly, we introduced H-Ras-type amino acid substitutions for Arg-105, Phe-106, and Leu-109, i.e. R105Q, F106Y, and L109Q, into M-RasP40D/D41E/L51R/F74Y/E79D, and the resulting mutant polypeptides in complex with GppNHp were examined for the state distribution by 31P NMR spectroscopy (Fig. 4). We used M-RasP40D/D41E/L51R/F74Y/E79D as the prototype for mutation because it possessed the hydrogen-bonding partner residues such as Tyr-74 and Asp-79 in switch II. The result clearly showed that only the L109Q substitution caused a substantial increase in the state 2 population; the state 2 peak of the γ-phosphate resonance line at −3.7 ppm increased to 47 ± 2% in M-RasP40D/D41E/L51R/F74Y/E79D/L109Q-GppNHp compared with 23 ± 3% at −3.68 ppm in M-RasP40D/D41E/L51R/F74Y/E79D-GppNHp (Fig. 4B and supplemental Table S2). The state assignment of the two peaks of the γ-phosphate resonance line for M-RasP40D/D41E/L51R/F74Y/E79D/L109Q-GppNHp was confirmed by addition of c-Raf-1 RBD (supplemental Fig. S3 and Table S2). The results suggested that restoration of the hydrogen-bonding interaction between Gln-109 and Asp-79, corresponding to the Gln-99—Asp-69 interaction in H-Ras, facilitated the adoption of state 2 in this M-Ras mutant. Collectively, our results implied that the higher density or proper arrangement of the hydrogen-bonding network between switch II and the α3-helix facilitates the adoption of state 2, resulting in the higher state 2 population.
FIGURE 4.
31P NMR spectra of the GppNHp-bound M-Ras mutants bearing H-Ras-type amino acid substitutions in the α3-helix. A, the spectra were recorded using 1 mm solution of the indicated M-Ras mutant and H-Ras proteins in complex with GppNHp. α, β, and γ represent the α-, β- and γ-phosphate resonances, respectively. (1) and (2) represent the γ-phosphate resonance peaks corresponding to states 1 and 2, respectively. αfree, βfree, and γfree represent the α-, β-, and γ-phosphate resonances of the free GppNHp, respectively. α-GDP and β-GDP represent the α- and β-phosphate resonances, respectively, of GDP bound to the M-Ras mutants. B, bar graph representation of state 2 populations of the M-Ras mutants in complex with GppNHp. The state 1 and state 2 peaks in the γ-phosphate resonance were fitted by Lorentz curves (blue lines in A), and the state 2 population was calculated as [state 2]/([state 1]+[state 2]), where [state 1] and [state 2] represent the relative concentrations obtained as the integrals of the corresponding peaks. Error bars, S.D. derived from the Lorentz curve fitting. The data of H-Ras-GppNHp and M-RasP40D/D41E/L51R/F74Y/E79D-GppNHp were adopted from our previous reports (18, 21).
Another H-Ras-type substitution, F106Y, in the α3-helix of M-RasP40D/D41E/L51R/F74Y/E79D, whether introduced alone or in combination with L109Y, caused a significant decrease in state 2 population even though the hydrogen-bonding partners of Tyr-106, Gly-70, and Arg-78 existed (Fig. 4, A and B). This unexpected effect of the F106Y substitution may be accounted for by the notion that the addition of a hydroxyl group to Phe-106 of M-RasD41E-GppNHp type 2 is likely to bring collision with the side chain of Asp-21 in the P-loop according to the calculation using the program PyMOL (supplemental Fig. S5). In the case of H-Ras-GppNHp, the less massive side chain of Ala-11, corresponding to Asp-21 of M-Ras, leads to an escape from the collision with Tyr-96. This finding suggests that the α3-helix-P-loop interaction may have a role in state transition.
DISCUSSION
Comparison of the Pre-switch I and Switch I Structures and Its Implication in the State Transition Mechanism of M-Ras
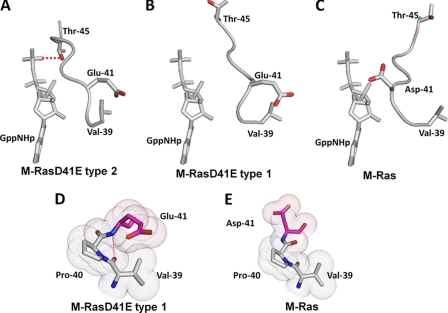
The positional change of Thr-45 of M-RasD41E-GppNHp type 1 approaching the nucleotide (Fig. 1) has prompted us to clarify its underlying mechanism focusing on the role of residue 41 on the switch I conformational changes during the state transition of M-Ras. To this end, we make a detailed comparison of the structures of the pre-switch I (residues 39–41) (Fig. 5) and switch I (residues 42–48) regions (Figs. 5 and 6) among M-Ras-GppNHp (state 1) and M-RasD41E-GppNHp types 1 (state 1) and 2 (state 2).
FIGURE 5.
Main chain and side chain configurations of the pre-switch I and switch I residues of M-Ras-GppNHp and M-RasD41E-GppNHp types 1 and 2. The backbone structures of the pre-switch and switch I regions and the structure of GppNHp are shown for M-RasD41E-GppNHp type 2 (A), M-RasD41E-GppNHp type 1 (B), and M-Ras-GppNHp (C), on which only the side chains of Val-39, Glu/Asp-41, and Thr-45 are depicted (red, oxygen). The hydrogen bond between Thr-45 and the γ-phosphate in M-RasD41E-GppNHp type 2 is shown by a red broken line. The van der Waals distances for Val-39, Pro-40, and Glu/Asp-41 of M-RasD41E-GppNHp type 1 (D) and M-Ras-GppNHp (E) are highlighted, where a red dotted line represents a hydrogen bond. The models were generated by PyMOL.
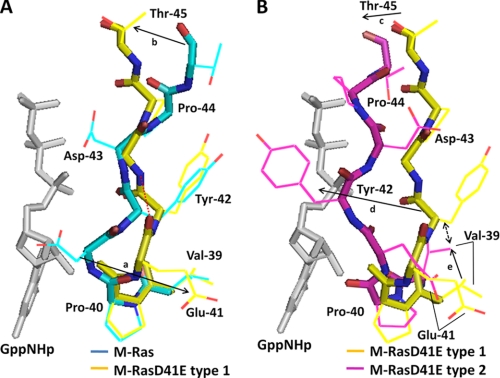
FIGURE 6.
Comparison of the structures of the pre-switch I and switch I regions among M-Ras-GppNHp and M-RasD41E-GppNHp types 1 and 2. Shown are superimpositions of the structures of the residues 39–45 of M-Ras-GppNHp (light blue) and M-RasD41E-GppNHp type 1 (yellow) (A) and M-RasD41E-GppNHp type 1 (yellow) and type 2 (pink) (B) are shown. The structure of GppNHp (white) is excerpted from the model of M-RasD41E-GppNHp type 1. Main chains and side chains are shown by thick and thin bars, respectively, on which oxygen and nitrogen atoms are shown by red and blue colors, respectively. Arrows a and b in A represent the conformational changes of Glu-41 and Thr-45, respectively, whereas arrows c, d, and e in B represent those of Thr-45, Tyr-42, and Val-39, respectively. A red dotted line in A shows the hydrogen bond between the main chains of Glu-41 and Asp-43. A bidirectional broken arrow represents the collision of the side chain of Tyr-42 with that of Val-39. The models were generated as described in the legend to Fig. 2.
The configuration of residue 41 exhibits a clear difference among the three structures (Fig. 5, A-C). In M-RasD41E-GppNHp type 2, the side chain of Glu-41 is arranged face to face with that of Val-39 and pulled away from the nucleotide (Fig. 5A). A hydrophobic interaction of Cδ of Glu-41 with the Cγ of Val-39 appears to play a role in establishing this constellation. In contrast, Asp-41 of M-Ras-GppNHp fails to establish any interactions with Val-39 and Pro-40, leading to marked displacement of Asp-41 from Val-39 (Fig. 5C), which could be accounted for by the fact that the shorter side chain of Asp-41 prohibits the formation of a van der Waals contact with the side chain of Val-39 or Pro-40 (Fig. 5E). Intriguingly, M-RasD41E-GppNHp type 1 exhibits a constellation of Glu-41 and Val-39 very similar to that of type 2 as characterized by a flip of Glu-41 away from the nucleotide (Fig. 5B). The underlying mechanism could be that the longer side chain of the Glu-41 substitute restored the van der Waals interactions with the side chain of Val-39 (Fig. 5D).
Superimposition of the main chain and side chain structures of residues 39–45 of M-Ras-GppNHp and M-RasD41E-GppNHp type 1 (Fig. 6A) reveals that the inside-out flip of Glu-41 (arrow a) causes substantial rotational and positional changes of the backbone structure of the switch I loop and pre-switch I residues starting at Pro-40. For example, the (ϕ, ψ) dihedral angles of Pro-40 and residue 41 exhibit significant changes from (−51°, 147°) and (41°, 44°), respectively, in M-Ras-GppNHp to (−54°, −31°) and (−84°, −5°), respectively, in M-RasD41E-GppNHp type 1. These conformational changes, resulting from the D41E substitution, induce a gross regional rearrangement of the hydrogen-bonding network involving the switch I loop and pre-switch I residues: both the direct interaction of the Asp-41 side chain with the ribose and the water-mediated interaction of the main chain carbonyl group of Tyr-42 with the α-phosphate in M-Ras-GppNHp are lost and replaced by the newly formed hydrogen bond between the main chains of Glu-41 and Asp-43 in M-RasD41E-GppNHp type 1 (Fig. 6A and supplemental Fig. S2). Thus, the rotational and positional changes initiated at the pre-switch I residues are conveyed to the switch I loop, resulting in the positional shift of Thr-45 approaching the γ-phosphate (arrow b). As already discussed, this positional shift of Thr-45 is likely to facilitate the formation of a hydrogen bond with the γ-phosphate and may account for the increase of the state 2 population in M-RasD41E-GppNHp. The reason why M-RasD41E-GppNHp type 1 failed to establish Thr-45-γ-phosphate hydrogen-bonding interaction despite the adoption of the type 2-like constellation of Val-39 and Glu-41 could be accounted for by the presence of Pro-40, which exerts a rotational constraint on the backbone structure.
Determination of both the state 1 and state 2 structures from M-RasD41E-GppNHp enables us to analyze the state transition mechanisms through comparison of the two structures. Superimposition of residues 39–45 of type 1 and type 2 (Fig. 6B) reveals drastic changes in the main chain and side chain structures of the switch I loop and pre-switch I residues, resulting in establishment of the Thr-45-γ-phosphate hydrogen-bonding interaction in type 2 (arrow c). Type 2, but not type 1, forms both direct and water-mediated hydrogen bonds between Pro-40 and the ribose, and a water-mediated hydrogen bond between Glu-41 and the α-phosphate (Fig. 2 and supplemental Fig. S2). Above all, Tyr-42 exhibits the most drastic positional and rotational change toward the nucleotide (arrow d). This is presumably caused by collision of Tyr-42 with the side chain of Val-39 (bidirectional broken arrow), which undergoes a positional change (arrow e) during the state 1 to state 2 transition.
Collectively, the results demonstrate that having Glu at residue 41 of M-Ras plays a pivotal role in the adoption of the state 2 conformation for the following two reasons. 1) The interaction of Glu-41 with the neighboring Val-39 causes a rotational change of the pre-switch I residues, leading to the switch I loop conformational change in combination with the rearrangement of the hydrogen-bonding network involving the switch I loop, pre-switch I residues, and the nucleotide. As a result, Thr-45 undergoes a positional shift approaching the γ-phosphate, facilitating the formation of a hydrogen bond with the γ-phosphate. 2) The remarkable displacement of Tyr-42 through the positional change of Val-39 brings about a drastic conformational change of the switch I loop toward the nucleotide, leading to the Thr-45-γ-phosphate hydrogen bond formation (see Fig. 2A). On the contrary, having Asp at residue 41 as in the case of M-Ras-GppNHp totally abolishes the corresponding interactions and facilitates the adoption of state 1.
State-specific Constellations of the Pre-switch I Residues Conserved Among M-Ras, H-Ras, and Their Mutants
The critical role of residue 41 in the state transition of M-Ras-GppNHp and M-RasD41E-GppNHp lead us to examine intramolecular interactions involving pre-switch I residues in H-Ras-GppNHp, H-RasT35S-GppNHp, and M-RasP40D/D41E/L51R-GppNHp. The configuration of the residues 31/41 (H-Ras/M-Ras) showed a clear difference between state 1 and state 2 conformers (Fig. 7). In state 2 conformers, H-Ras-GppNHp and M-RasP40D/D41E/L51R-GppNHp, their Glu-31/41 residues establish interactions with neighboring residues 29/39 and/or 30/40, leading to adoption of a constellation of residues 29/39 and 31/41, which is equivalent to that of M-RasD41E-GppNHp type 2 (Figs. 5A and 7A). However, the mode of interactions is different among the three proteins. In H-Ras-GppNHp, the side chain carboxyl group of Glu-31 forms a water-mediated hydrogen bond with the main chain amide of Asp-30, which is presumably assisted by a van der Waals contact of the Cδ of Glu-31 with the Cγ of Val-29 (Fig. 7C). In the case of M-RasP40D/D41E/L51R-GppNHp (PDB code 3KKO, chain A), both the main chain and side chain of Asp-40 formed water-mediated hydrogen bonds with the side chain of Glu-41 (data not shown). As already discussed, in the case of M-RasD41E-GppNHp type 2, a hydrophobic interaction of Cδ of Glu-41 with the Cγ of Val-39 appears to be involved. In contrast, Glu-31 of the state 1 conformer, H-RasT35S-GppNHp form 2, fails to establish any interactions with Val-29 and Asp-30, leading to marked displacement of Glu-31 from Val-29 as observed in M-RasGppNHp (Figs. 5C and 7B), whose shorter side chain of Asp-41 prohibits interaction with Val-39 or Pro-40 as already discussed (Fig. 5E). In this case, the loss of the Glu-31—Val-29 interaction presumably occurs by the marked deviation of the switch I loop following the loss of the Ser-35-γ-phosphate interaction. As already pointed out, M-RasD41E-GppNHp type 1 (state 1) represents an exceptional case; it exhibits a constellation of Glu-41 and Val-39 very similar to that of the state 2 conformers (Fig. 5B), which can be explained that the longer side chain of the Glu-41 substitute restores the van der Waals interactions with the side chain of Val-39 (Fig. 5D). These results collectively indicate that the critical role of the Glu-31/41—Val-29/39 interaction in adoption of the state 2 conformation is conserved between M-Ras and H-Ras. It is also supported by the observation that H-RasV29G-GppNHp exhibits a vast increase in the state 1 population as assessed from its 31P NMR spectrum (30).
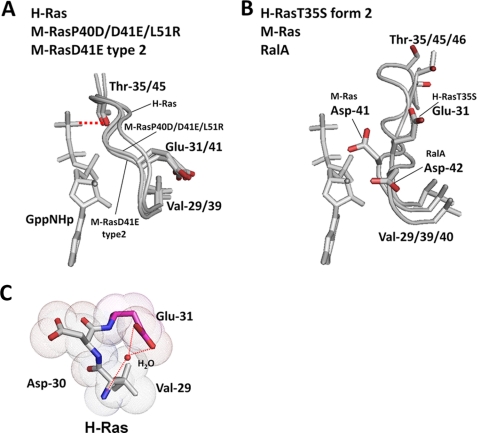
FIGURE 7.
Comparison of the structures of the pre-switch I and switch I regions among M-Ras, H-Ras, RalA, and their mutants. Shown are superimpositions of the structures of the residues 29–35/39–45/40–46 (H-Ras/M-Ras/RalA) of the state 2 conformers, M-RasD41E-GppNHp type 2, M-RasP40D/D41E/L51R-GppNHp, and H-Ras-GppNHp (A), and of the state 1 conformers, M-Ras-GppNHp, H-RasT35S-GppNHp form 2, and RalA-GppNHp (B). The structures of GppNHp are excerpted from the models of H-Ras-GppNHp (A) and M-Ras-GppNHp (B). The side chains of residues 29/39/40, 31/41/42, and 35/45/46 (H-Ras/M-Ras/RalA) are highlighted with the stick model (red, oxygen). The hydrogen bond between Thr-35 and the γ-phosphate in H-Ras is shown by a red broken line. The models were generated as described in the legend to Fig. 1. C, van der Waals distances for Val-29, Asp-30, and Glu-31 of H-Ras-GppNHp are highlighted. Red dotted lines represent hydrogen bonds. The models were generated by the program PyMOL.
The study on M-RasD41E-GppNHp types 1 and 2 also has indicated the importance of the displacement of Tyr-42 through the positional change of Val-39 in the conformational change of Thr-45 leading to the establishment of its interaction with the γ-phosphate. Comparison of the structures of the pre-switch I and switch I regions between H-RasT35S-GppNHp form 2 and H-Ras-GppNHp shows the existence of a similar displacement of Tyr-32 toward the nucleotide in H-Ras-GppNHp, which is presumably caused by the positional change of Val-29 (supplemental Fig. S6). Thus, this mechanism may also be shared between M-Ras and H-Ras.
Conservation of the Roles of the Novel Intramolecular Interactions in a More Distantly Related Ras Family Member, RalA
Conservation of the critical roles of the two interactions, around the pre-switch I residues and between switch II and the α3-helix, in the state transition of M-Ras and H-Ras leads us to test whether a similar mechanism is working in more distantly related members of Ras family small GTPases. To this end, we focus on RalA, whose switch I residues are poorly conserved (supplemental Fig. S1), because it is the only Ras family member whose state 1 crystal structure has been determined other than M-Ras, H-Ras, and their mutants (31). RalA-GppNHp mainly adopts state 1 in solution with a state 1 population of 53 ± 7% (12). The crystal structure of RalA-GppNHp corresponds to state 1 judging from the loss of the interaction of Thr-46 (corresponding to Thr-35/45 of H-Ras/M-Ras) with the γ-phosphate (31) and its backbone structure of switch I superimposes very well with that of M-Ras-GppNHp (data not shown) despite the existence of non-conserved residues, Glu-44, Lys-47, and Ala-48 (corresponding to Asp-33/43, Ile-36/46, and Glu-47 of H-Ras/M-Ras) (see supplemental Fig. S1). In RalA-GppNHp, Asp-42, equivalent to Asp-41 in M-Ras, fails to interact with Val-40, equivalent to Val-39 in M-Ras, adopting a constellation of the pre-switch I residues analogous to those of state 1 structures of M-Ras and H-Ras (Fig. 7B).
On the other hand, RalA-GppNHp retains the hydrogen-bonding interaction of Gly-71 (corresponding to Gly-60/70 of H-Ras/M-Ras) with the γ-phosphate (31) (supplemental Fig. S7). In RalA-GppNHp, Gln-110 in the α3-helix, equivalent to Gln-99 of H-Ras, formed hydrogen bonds with Ile-78 and Asp-80 in switch II, like the case of H-Ras-GppNHp, whereas Phe-107, equivalent to Phe-106 of M-Ras, fails to interact with switch II residues, such as M-RasD41E-GppNHp type 1. This is consistent with our notion that the higher density or the proper arrangement of the hydrogen-bonding network between switch II and the α3-helix facilitates the adoption of state 2, resulting in the higher state 2 population. These results suggest that RalA shares a similar mechanism of state transition with M-Ras and H-Ras.
In conclusion, our study has demonstrated the critical roles of the two novel intramolecular interactions in the state 1 to state 2 transition; one around the pre-switch I residues and the other between switch II and the α3-helix, which appear to facilitate the formation of hydrogen bonds of Thr-35/45 (H-Ras/M-Ras) in switch I and Gly-60/70 in switch II with the γ-phosphate of GTP, respectively. Although it is evident that the four interactions are intimately related with one another, the sequence of these various events cannot be determined from the present study, which deals with static structures. This mechanism of state transition seems to be conserved among M-Ras, H-Ras, and more distantly related RalA. Thus, the amino acid sequence diversity of the residues involved in the relevant interactions seems to be responsible for determination of the intrinsic state distribution of Ras family small GTPases. Our work also suggests that the P-loop may have a role in the state transition via interaction with the α3-helix. This is supported at least in part by the observation that P-loop mutants of H-Ras, such as H-RasG12V and H-RasG12D, showed an increase in the state 1 population in their GppNHp-bound forms (10, 30). The α3-helix and P-loop were thought to be rather immobile from crystallographic studies. However, the backbone amide 15N spin relaxation rcCPMG measurements by O'Connor et al. (32) suggested that they exhibit a significant mobility upon state transition, which is further supported by our latest study4 of 15N relaxation times and heteronuclear nuclear Overhauser effects of H-RasT35S-GppNHp. As already proposed (21), the various events relevant to the state transition could be targeted for development of Ras inhibitors, i.e. inhibition of the state 1 to state 2 transition leads to inactivation of the Ras function.
Supplementary Material
Acknowledgments
The synchrotron radiation experiments were performed at BL38B1 and BL41XU in SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (Proposal number 2009B1117). We thank Kazuya Hasegawa, Nobutaka Shimizu, Seiki Baba, Nobuhiro Mizuno, and Masatomo Makino of JASRI/SPring-8 for data collection in the SPring-8 and for technical advice. We thank Tomoko Inoue and Shinya Ichikawa for excellent technical assistance.
This work was supported by Grants-in-aid for Scientific Research in Priority Areas 17014061 and 18057014, and Global COE Program A08 from the Ministry of Education, Science, Sports and Culture of Japan, and by a grant from the Program for Promotion of Fundamental Studies of Health Sciences 06-3 from the National Institute of Biomedical Innovation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S7.
M. Araki, F. Shima, Y. Yoshikawa, S. Muraoka, Y. Ijiri, Y. Nagahara, T. Shirono, T. Kataoka, and A. Tamura, unpublished results.
- GppNHp
- guanosine 5′-(β,γ-imido)triphosphate
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- RBD
- Ras-binding domain.
REFERENCES
- 1. Takai Y., Sasaki T., Matozaki T. (2001) Physiol. Rev. 81, 153–208 [DOI] [PubMed] [Google Scholar]
- 2. Downward J. (2003) Nat. Rev. Cancer 3, 11–22 [DOI] [PubMed] [Google Scholar]
- 3. Karnoub A. E., Weinberg R. A. (2008) Nat. Rev. Mol. Cell Biol. 9, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Corbett K. D., Alber T. (2001) Trends Biochem. Sci. 26, 710–716 [DOI] [PubMed] [Google Scholar]
- 5. Vetter I. R., Wittinghofer A. (2001) Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 6. Nassar N., Horn G., Herrmann C., Scherer A., McCormick F., Wittinghofer A. (1995) Nature 375, 554–560 [DOI] [PubMed] [Google Scholar]
- 7. Geyer M., Herrmann C., Wohlgemuth S., Wittinghofer A., Kalbitzer H. R. (1997) Nat. Struct. Biol. 4, 694–699 [DOI] [PubMed] [Google Scholar]
- 8. Huang L., Hofer F., Martin G. S., Kim S. H. (1998) Nat. Struct. Biol. 5, 422–426 [DOI] [PubMed] [Google Scholar]
- 9. Pacold M. E., Suire S., Perisic O., Lara-Gonzalez S., Davis C. T., Walker E. H., Hawkins P. T., Stephens L., Eccleston J. F., Williams R. L. (2000) Cell 103, 931–943 [DOI] [PubMed] [Google Scholar]
- 10. Geyer M., Schweins T., Herrmann C., Prisner T., Wittinghofer A., Kalbitzer H. R. (1996) Biochemistry 35, 10308–10320 [DOI] [PubMed] [Google Scholar]
- 11. Spoerner M., Nuehs A., Herrmann C., Steiner G., Kalbitzer H. R. (2007) FEBS J. 274, 1419–1433 [DOI] [PubMed] [Google Scholar]
- 12. Liao J., Shima F., Araki M., Ye M., Muraoka S., Sugimoto T., Kawamura M., Yamamoto N., Tamura A., Kataoka T. (2008) Biochem. Biophys. Res. Commun. 369, 327–332 [DOI] [PubMed] [Google Scholar]
- 13. Fenwick R. B., Prasannan S., Campbell L. J., Nietlispach D., Evetts K. A., Camonis J., Mott H. R., Owen D. (2009) Biochemistry 48, 2192–2206 [DOI] [PubMed] [Google Scholar]
- 14. Quilliam L. A., Castro A. F., Rogers-Graham K. S., Martin C. B., Der C. J., Bi C. (1999) J. Biol. Chem. 274, 23850–23857 [DOI] [PubMed] [Google Scholar]
- 15. Rodriguez-Viciana P., Sabatier C., McCormick F. (2004) Mol. Cell. Biol. 24, 4943–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pai E. F., Krengel U., Petsko G. A., Goody R. S., Kabsch W., Wittinghofer A. (1990) EMBO J. 9, 2351–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scheidig A. J., Burmester C., Goody R. S. (1999) Structure 7, 1311–1324 [DOI] [PubMed] [Google Scholar]
- 18. Ye M., Shima F., Muraoka S., Liao J., Okamoto H., Yamamoto M., Tamura A., Yagi N., Ueki T., Kataoka T. (2005) J. Biol. Chem. 280, 31267–31275 [DOI] [PubMed] [Google Scholar]
- 19. Ford B., Skowronek K., Boykevisch S., Bar-Sagi D., Nassar N. (2005) J. Biol. Chem. 280, 25697–25705 [DOI] [PubMed] [Google Scholar]
- 20. Ford B., Boykevisch S., Zhao C., Kunzelmann S., Bar-Sagi D., Herrmann C., Nassar N. (2009) Biochemistry 48, 11449–11457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shima F., Ijiri Y., Muraoka S., Liao J., Ye M., Araki M., Matsumoto K., Yamamoto N., Sugimoto T., Yoshikawa Y., Kumasaka T., Yamamoto M., Tamura A., Kataoka T. (2010) J. Biol. Chem. 285, 22696–22705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spoerner M., Herrmann C., Vetter I. R., Kalbitzer H. R., Wittinghofer A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4944–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24. Leslie A. G. W. (1992) Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography, No. 26, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 25. Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D 50, 760–76315299374 [Google Scholar]
- 26. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 27. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunsteleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 28. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29. Laskowski R. A., McArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 30. Spoerner M., Wittinghofer A., Kalbitzer H. R. (2004) FEBS Lett. 578, 305–310 [DOI] [PubMed] [Google Scholar]
- 31. Nicely N. I., Kosak J., de Serrano V., Mattos C. (2004) Structure 12, 2025–2036 [DOI] [PubMed] [Google Scholar]
- 32. O'Connor C., Kovrigin E. L. (2008) Biochemistry 47, 10244–10246 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.