Abstract
The FS0 [4Fe-4S] cluster of the catalytic subunit (DmsA) of Escherichia coli dimethyl sulfoxide reductase (DmsABC) plays a key role in the electron transfer relay. We have now established an additional role for the cluster in directing molybdenum cofactor assembly during enzyme maturation. EPR spectroscopy indicates that FS0 has a high spin ground state (S = ) in its reduced form, resulting in an EPR spectrum with a peak at g ∼ 5.0. The cluster is predicted to be in close proximity to the molybdo-bis(pyranopterin guanine dinucleotide) (Mo-bisPGD) cofactor, which provides the site of dimethyl sulfoxide reduction. Comparison with nitrate reductase A (NarGHI) indicates that a sequence of residues (18CTVNC22) plays a role in both FS0 and Mo-bisPGD coordination. A DmsAΔN21 mutant prevented Mo-bisPGD binding and resulted in a degenerate [3Fe-4S] cluster form of FS0 being assembled. DmsA belongs to the Type II subclass of Mo-bisPGD-containing catalytic subunits that is distinguished from the Type I subclass by having three rather than two residues between the first two Cys residues coordinating FS0 and a conserved Arg residue rather than a Lys residue following the fourth cluster coordinating Cys. We introduced a Type I Cys group into DmsA in two stages. We changed its sequence from 18CATVNCBGSRCCP27 to 18CATYCBGVGCCG26 (similar to that of formate dehydrogenase (FdnG)) and demonstrated that this eliminated both Mo-bisPGD binding and EPR-detectable FS0. We then combined this change with a DmsAR61K mutation and demonstrated that this additional change partially rescued Mo-bisPGD insertion.
Keywords: Electron Paramagnetic Resonance (EPR), Electron Transfer, Iron-Sulfur Protein, Molybdenum, Protein Assembly, Dimethyl Sulfoxide Reductase, Maturation, Molybdo-bis(pyranopterin Guanine Dinucleotide) Cofactor, Molybdopterin, Nitrate Reductase
Introduction
Escherichia coli is a facultative anaerobe able to respire with oxygen or alternative respiratory oxidants such as nitrate, nitrite, fumarate, and dimethyl sulfoxide (1, 2). Dimethyl sulfoxide reductase (DmsABC) is a terminal reductase that reduces dimethyl sulfoxide to dimethyl sulfide (3). It is a member of the bacterial CISM (complex iron-sulfur molybdoenzyme) family that includes E. coli formate dehydrogenases (FdnGHI and FdoGHI) (2, 4, 5), E. coli nitrate reductases (NarGHI and NarZYV) (6, 7), Salmonella typhimurium thiosulfate reductase (PhsABC) (8), and the Wolinella succinogenes and Thermus thermophilus (PsrABC) polysulfide reductases (9, 10). This family of enzymes contributes to the broad metabolic diversity that permits bacterial growth utilizing a wide range of respiratory substrates. Each structurally characterized enzyme of this family consists of a catalytic subunit with a molybdo-bis(pyranopterin guanine dinucleotide) (Mo-bisPGD)3 cofactor and an FS0 [4Fe-4S] cluster, an electron transfer subunit containing four [Fe-S] clusters, and a membrane anchor subunit containing a quinol/quinone-binding site (Q-site). Each enzyme catalyzes an overall reaction that involves transferring two electrons, through an electron transfer relay connecting the Q-site and the Mo-bisPGD cofactor, and reduction (or oxidation) of substrate at the catalytic molybdenum active site of the enzyme (see Fig. 1A). Although DmsABC does not contribute to the transmembrane proton electrochemical potential, it can support anaerobic growth with dimethyl sulfoxide as respiratory oxidant when coupled with an appropriate proton-translocating dehydrogenase (11). DmsABC couples the oxidation of menaquinol within the hydrophobic membrane milieu to the reduction of water-soluble dimethyl sulfoxide in the membrane-extrinsic periplasmic domain of the enzyme (1, 12).
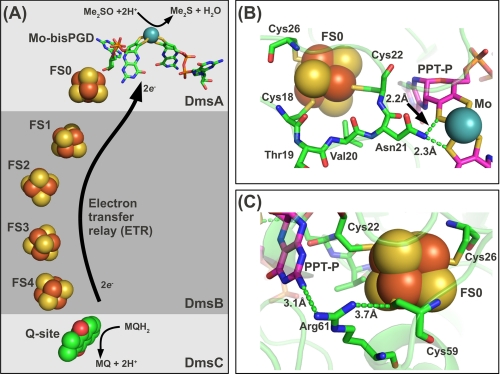
FIGURE 1.
Electron transfer relay architecture and predicted DmsA structure around FS0. A, predicted electron transfer relay in DmsABC connecting a menaquinol-binding site in the DmsC subunit with the Mo-bisPGD cofactor in the DmsA subunit. In this model, note that DmsC is predicted to be membrane-intrinsic and anchors to DmsA and DmsB to the periplasmic side of the E. coli cytoplasmic membrane (2). B, a structural model of DmsA was generated using the ESyPred3D server (29) with the structure of the NarG subunit of NarGHI as a template (Protein Data Bank code 1Q16) (40). The predicted position of the 18CTVNC22 loop is shown. The distances shown are arbitrary but indicate a possible H-bonding interaction between Asn-21 and one dithiolene sulfur from each pterin. C, the predicted position of Arg-61 between FS0 and the proximal pterin (PPT-P).
In E. coli DmsABC, it is the DmsA catalytic subunit that contains the Mo-bisPGD cofactor and an FS0 [4Fe-4S] cluster. The FS0 cluster is the last stepping stone of the electron transfer relay and transfers electrons from FS1 in DmsB to the cofactor in the active site of DmsA (see Fig. 1A). The N-terminal sequence of DmsA contains four highly conserved Cys residues, which form a ferredoxin-like Cys motif that, as we show below, coordinates FS0. In the CISM enzymes, the Cys group has the following consensus sequence: (CA/HA)X2–3CBX3CCX27–34CDX(K/R) (1, 2). Sequences with two residues between CA and CB are classified as Type I sequences and typically have a Lys residue located after CD (13, 14). DmsA is classified as a Type II sequence because it has three residues between CA and CB and an Arg residue after CD. The third residue after CA in DmsA is an Asn, which is highly conserved in DmsABC-type dimethyl sulfoxide reductases and NarGHI-type nitrate reductases across bacterial species (2). The interplay between the Type I and II sequences and molybdoenzyme assembly and function remains poorly understood.
Previous studies on the FS0-binding motif of DmsA indicated that mutating the second Cys residue to Ser or Ala results in assembly of a [3Fe-4S] cluster into DmsA (13) at the FS0 position. Careful analysis of the Mo(V) EPR properties of the Mo-bisPGD cofactor indicated that there is a spin-spin interaction between the molybdenum center and a paramagnetic center with a midpoint potential of around −140 mV (15), but in none of these studies was the EPR spectrum of wild-type FS0 observed. In the case of another Type II enzyme, E. coli NarGHI, the FS0 spectrum was observed at low temperature at g ∼ 5.0 (16, 17). Further studies revealed that FS0 insertion was closely linked to that of the Mo-bisPGD cofactor (18). An important distinction between the Type II Cys group sequence of DmsA and that of NarG is that there is a His in the CA position of the latter and a Cys at this position in the former. It was not known whether the observation of a high spin FS0 EPR spectrum is related to the presence of His at the CA position.
Maturation of the catalytically competent DmsABC requires efficient cofactor insertion into DmsA (19), association of DmsA and DmsB to form the DmsAB “catalytic dimer” (20), translocation of DmsAB across the cytoplasmic membrane via the tat translocon (3, 21, 22), and anchoring of DmsAB to the membrane through association with DmsC (20). However, assembly and targeting of the apoenzyme are not cofactor insertion-dependent (19, 23). In the case of NarGHI, NarJ functions as a system-specific chaperone participating in cofactor insertion (24, 25). In the case of DmsABC, the system-specific chaperone DmsD plays a role in targeting DmsAB to the tat translocon and may have additional roles in maturation (26). We have recently demonstrated by protein crystallography that, in NarGHI, FS0 assembly is a prerequisite for Mo-bisPGD insertion (18). It is therefore of great interest to see if a similar sequence of cluster assembly and cofactor insertion occurs in the maturation of DmsABC.
In this study, we establish that DmsA contains a novel [4Fe-4S] FS0 cluster that has a high spin ground state (S = ) in its reduced form. By generating mutants of the DmsA N-terminal Cys group, we show that there is critical interplay between correct assembly of FS0 and molybdenum cofactor insertion. These results point toward a new paradigm for Mo-bisPGD cofactor-containing subunit maturation in which FS0 must be assembled prior to cofactor insertion.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
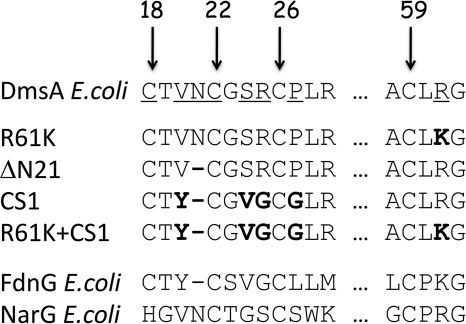
E. coli strains DSS301 (20) and TOPP2 were used in this study (Table 1). DSS301 was used for the growth experiments. TOPP2 was used for enzyme expression and spectroscopic studies, as we found that it was optimal for DmsABC overexpression and assembly (27). The plasmids (Table 1) used for cloning and expression included pBR322, pDMS160 (28), and pATZ (EcoRI-EcoRV fragment of pDMS160 (28) cloned into pTZ18R (AmpRlacZ′ (Pharmacia)) (laboratory collection)). The sequence alignment for the residues mutated in this study is shown in Fig. 2.
TABLE 1.
Bacterial strains and plasmids
| Description | Source | |
|---|---|---|
| Strain | ||
| DSS301 | TG1, kanR ΔdmsABC | Lab collection (20) |
| TOPP2 | rifR (F′, proAB, lacIqZΔM15, Tn10 (tetR)) | Stratagene |
| Plasmid | ||
| pBR322 | TetR AmpR | Pharmacia |
| pDMS160 | dmsABC cloned into the EcoRI-SalI fragment of pBR322, AmpR | Lab collection (28) |
| R61K | pDMS160-dmsAR61KBC | This study |
| ΔN21 | pDMS160-dmsAΔN21BC | This study |
| CS1a | pDMS160-dmsACS1BC | This study |
| CS1 + R61K | pDMS160-dmsACS1+R61KBC | This study |
a For simplicity, this plasmid is named CS1. The actual mutations are V20Y, ΔN21, S24V, R25G, and P43G (Fig. 1).
FIGURE 2.
Sequences of DmsA mutants in comparison with Cys group sequences of FdnG and NarG. Numbers indicate the positions of four conserved Cys residues in the mature protein sequence. The amino acid residues that were mutated for this study are underlined. In the sequences of the mutants, the resultant residues of mutation are shown in boldface. The sequences of E. coli FdnG, a typical Type I enzyme, and E. coli NarG, a typical Type II enzyme, are also shown for comparison.
Design of Mutants
A model for the structure of DmsA was generated using the ESyPred3D server (29) and the E. coli NarGHI structure as template (Protein Data Bank code 1Q16) (18). The PyMOL molecular graphics program (Version 1.2.r1, DeLano Scientific LLC) was used to generate the images presented in Fig. 1.
Cloning and Site-directed Mutagenesis
Construction of the site-directed mutants was carried out following the QuikChange method (Stratagene) (27) using pATZ as the template for mutagenesis. Mutants were verified by DNA sequencing carried out by the Department of Biochemistry DNA Core Facility at the University of Alberta. The EcoRI-EcoRV fragments of DmsA containing the mutations were cloned back into the pDMS160 expression vector (30). The mutant plasmids were then transformed into E. coli DSS301 and TOPP2.
Growth of Bacteria in Glycerol/Dimethyl Sulfoxide Minimum Medium
The ability of the mutant DmsABC enzymes described herein to support respiratory growth on dimethyl sulfoxide was evaluated as follows. Appropriate plasmids were transformed into E. coli DSS301, and anaerobic growth was evaluated in sealed Klett flasks at 37 °C using glycerol/dimethyl sulfoxide minimum medium (27, 31). Culture turbidity was monitored using a Klett-Summerson spectrophotometer equipped with a No. 66 filter (31).
Growth of Bacteria and Membrane Preparation
Wild-type and mutant plasmids were transformed into TOPP2 cells. The cells were then allowed to grow anaerobically for 24 h in 10 or 19 liters of glycerol/fumarate medium (28). Cells were harvested by centrifugation, washed, and resuspended in buffer containing 100 mm MOPS and 5 mm EDTA (pH 7). Membranes were prepared by cell lysis using an Avestin microfluidizer and differential centrifugation. PMSF (2 mm final concentration) was added to the cell suspension prior to cell lysis. Membranes were washed, pelleted by ultracentrifugation, resuspended in 100 mm MOPS and 5 mm EDTA (pH 7), and flash-frozen with liquid N2 before being stored at −70 °C prior to use (27).
Lowry Protein Assay
Protein concentrations of the membrane preparations were determined with the Lowry assay (32) modified by including 1% (w/v) SDS (33).
SDS-PAGE
45 μg of each protein was analyzed on a 12% SDS-polyacrylamide slab gel (34). 30 μg of purified DmsABC was loaded as a standard. Gel electrophoresis was run at 200 V for 45 min, and the gel was stained with Coomassie Brilliant Blue.
Measurement of DmsABC Enzyme Concentration by 2-n-Heptyl-4-hydroxyquinoline N-Oxide (HOQNO) Fluorescence Quench Titration
HOQNO is a menaquinol analog that binds to the DmsC quinol-binding site of DmsABC with 1:1 stoichiometry (35). The assay is based on the method of van Ark and Berden (36) and can be used to estimate the concentration of overexpressed DmsABC in membrane samples (27). Fluorescence was monitored at an excitation wavelength of 341 nm and an emission wavelength of 479 nm as aliquots (2 μl) of 0.1 mm HOQNO were added to a membrane sample with final protein concentrations of 0.25, 0.5, 0.75, and 1.0 mg/ml in a total volume of 3.5 ml. Fluorescence readings were taken using a PerkinElmer LS 50B luminescence spectrophotometer. The concentration of enzyme was estimated as the concentration at which HOQNO fluorescence became detectable in the sample cuvette (27).
Purification of DmsABC
Proteins were extracted from the membrane with 1% n-dodecyl β-d-maltoside and 0.5 mm DTT in 100 mm MOPS, 5 mm EDTA, 10% glycerol, and 160 μm PMSF (pH 7.0) for 1.5 h with gentle shaking on ice. The extraction mixture was then centrifuged at 40,000 rpm for 1.5 h to remove insoluble material, and DmsABC was separated from other proteins in the supernatant by FPLC using a Mono Q HR 10/10 anion exchange column and a GE Healthcare ÄKTA FPLC system. DmsABC was eluted from the column with a linear salt gradient (0–400 mm KCl) in 50 mm MOPS, 0.5 mm EDTA, 10% glycerol, 0.05% n-dodecyl β-d-maltoside, and 0.5 mm DTT. Protein samples were concentrated by ultrafiltration.
Benzyl Viologen (BV)/Trimethylamine N-Oxide Oxidoreductase Activity Assay
The specific activities of the wild-type and mutant enzymes were measured as the rate of trimethylamine N-oxide-dependent oxidation of dithionite reduced BV by following the decrease in BV absorption at 570 nm (37). Enzyme activity is expressed as micromoles of BV oxidized per min/mg of total protein. The final concentration of trimethylamine N-oxide was 72 mm in 50 mm MOPS and 5 mm EDTA. The extinction coefficient for BV is 7.4 mm−1 cm−1.
Lapachol/Trimethylamine N-Oxide Oxidoreductase Activity Assay
Quinol-dependent enzyme activity was determined as the rate of trimethylamine N-oxide-dependent anaerobic oxidation of reduced lapachol (LPCH2) (38). Lapachol was reduced by zinc in hydrochloric acid to form LPCH2, which was added to degassed 100 mm MOPS buffer containing 5 mm EDTA and 70 mm trimethylamine N-oxide in a closed cuvette. The reaction was initiated by the addition of enzyme. Lapachol absorbance at 481 nm was monitored and used to calculate the activity as micromoles of LPCH2 oxidized per min/mg of total protein. The extinction coefficient of lapachol is 2.66 mm−1 cm−1 (38).
Form A Fluorescence
The Form A fluorescence assay was used to estimate the relative amount of Mo-bisPGD in each DmsA enzyme preparation (23, 39). Form A molybdopterin derivatives were prepared by acidification of membrane preparations (10 mg/sample) with HCl and oxidation by boiling with 2% potassium iodide and 4% iodine (w/v). 200 μl of the acid-denatured iodine-oxidized extracts was added to 3.5 ml of 1 m NH4OH to measure the fluorescence (23). Excitation spectra were recorded at 240–420 nm (emission at 448 nm), and the emission spectra were recorded at 410–520 nm (excitation at 397 nm) using a PerkinElmer LS 50 luminescence spectrometer. Fluorescence spectra were corrected by subtraction of the spectrum of 1 m ammonium hydroxide as well as the spectrum of a membrane preparation of E. coli harboring only the pBR322 vector.
EPR Spectroscopy
Reduced samples were prepared by anaerobic incubation of purified DmsABC or membrane samples enriched in DmsABC with 6 mm sodium dithionite for 5 min at room temperature. All samples were prepared in 3-mm internal diameter quartz EPR tubes, rapidly frozen in liquid nitrogen-chilled ethanol, and stored in liquid nitrogen until used. For analysis of [Fe-S] clusters, EPR spectra were recorded using a Bruker ELEXSYS E500 spectrometer equipped with a Bruker SHQE cavity and an Oxford Instruments ESR900 flowing helium cryostat.
RESULTS AND DISCUSSION
Structural Relationship between FS0 and the Mo-bisPGD Cofactor
The catalytic subunit (NarG) of E. coli NarGHI is currently the best characterized Type II molybdoenzyme subunit (2, 16, 18, 40, 41). We used its structure (Protein Data Bank code 1Q16) to generate a model of DmsA in the FS0 region (Fig. 1, B and C). The segment comprising three residues between CA and CB, 19TVN21, is predicted to place the side chain of Asn-21 in close proximity to one dithiolene sulfur from each pyranopterin (Fig. 1B). The Arg residue that follows CD in the FS0-coordinating motif (Arg-61) is predicted to be located between the proximal pterin and FS0 (Fig. 1C). Site-directed mutagenesis has demonstrated that this residue plays a critical role in electron transfer and intercenter interactions in E. coli DmsA (14, 15), and similar results have been obtained for the structurally characterized E. coli NarGHI (18) and Ralstonia eutropha NapA (42). We exploited our structural model to investigate the interplay between the FS0 [4Fe-4S] cluster and the Mo-bisPGD cofactor during the final stages of DmsABC maturation.
We generated mutants of residues predicted to surround the FS0 cluster of DmsA (i) to identify the EPR signature of FS0, (ii) to investigate the effect of converting the DmsA Cys group from a Type II to a Type I sequence, and (iii) to investigate the interplay between FS0 assembly and Mo-bisPGD insertion. Fig. 2 shows the sequences of the mutant Cys groups generated herein. Arg-61 was mutated to Lys to emulate this feature of the Type I sequences. Asn-21 was deleted to investigate the effect of decreasing the CA-CB gap from three to two residues. Mutant enzyme CS1 contains multiple changes (V20Y, ΔAsn-21, S25V, and R25G) to render the DmsA Cys group similar to that of E. coli formate dehydrogenase N (FdnGHI) (43, 44). R61K and CS1 complete the conversion of the DmsA Cys group from a Type II to a Type I sequence.
Direct Spectroscopic Evidence for the Presence of a High Spin [4Fe-4S] Cluster in DmsABC
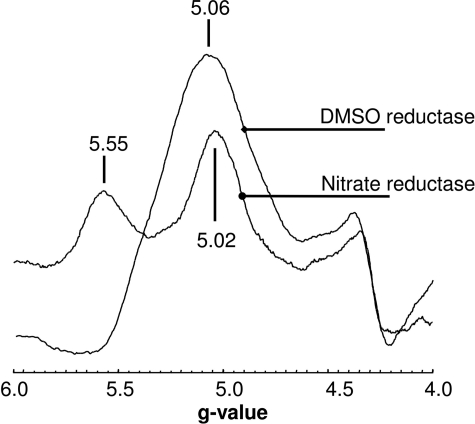
EPR spectra recorded at g ∼ 2.0 have provided no direct evidence for the existence of FS0 in DmsA (13, 15, 28, 37, 45). The emergence of a range of protein structures of molybdoenzyme subunits with N-terminal Cys groups allows us to predict that such a cluster must also exist in DmsA. In the case of the Type I enzymes, EPR spectra for FS0 have been reported and demonstrate that, in its reduced form, it has an S = ½ ground state with well resolved EPR features at g ∼ 2.0 (46, 47). EPR evidence for an FS0 cluster in DmsA is limited to the discovery that the Mo(V) form of the Mo-bisPGD cofactor is able to participate in a spin-spin interaction with an adjacent paramagnetic center during enzyme reduction (15). NarGHI, which is also a Type II enzyme, has been demonstrated to contain an FS0 cluster, which in its reduced form has a high spin ground state (S = ) (16–18). With the availability of highly purified DmsABC coupled with improved EPR instrumentation, we reinvestigated the EPR spectrum of reduced DmsABC with the aim of identifying additional spectral features in the g = 5.0 region. Fig. 3 shows the spectrum at g ∼ 5.0 of dithionite-reduced purified DmsABC. The spectrum of the reduced enzyme has a distinct peak at g = 5.06, consistent with it arising from a [4Fe-4S]+ cluster with an S = ground state. Spectra of other [4Fe-4S]+ systems with S = ground states typically exhibit two visible peaks, one at g ∼ 5.0 corresponding to a ΔS = ±½ transition and another at a slightly higher g value corresponding to a ΔS = ± transition (16, 17, 48, 49). The existence of a single peak in EPR spectra of systems with an S = ground state is not unusual, as the optimum temperature for the observation of the ΔS = ± transition can be below those reachable with commonly used cryostats. Examples of [4Fe-4S]+ clusters with an S = ground state that exhibit a single peak in the g = 5.0 region include those of E. coli fumarase A (50), component 1 from the iron-only nitrogenase of Rhodobacter capsulatus (51), and the 8Fe form of ferredoxin III from Desulfovibrio africanus (52). The intensity of the g = 5.06 feature of the DmsABC spectrum has a peak intensity at <4.5 K and is almost undetectable at temperatures above ∼20 K (data not shown). Overall, the observation of a low temperature peak at g ∼ 5 in EPR spectra of reduced DmsABC is consistent with the presence of a high spin form (S = ground state) of FS0. To confirm this, we studied the EPR properties of the range of mutants described in Fig. 2.
FIGURE 3.
Low field EPR spectrum of purified DmsABC and NarGHI. Samples were reduced by anaerobic incubation with 6 mm sodium dithionite for 5 min. EPR conditions were as follows: temperature, 9 K; microwave power, 100 milliwatts at 9.387 GHz; and modulation amplitude, 20 Gpp (EPR modulation amplitude intensity, peak to peak, in units of Gauss) at 100 KHz.
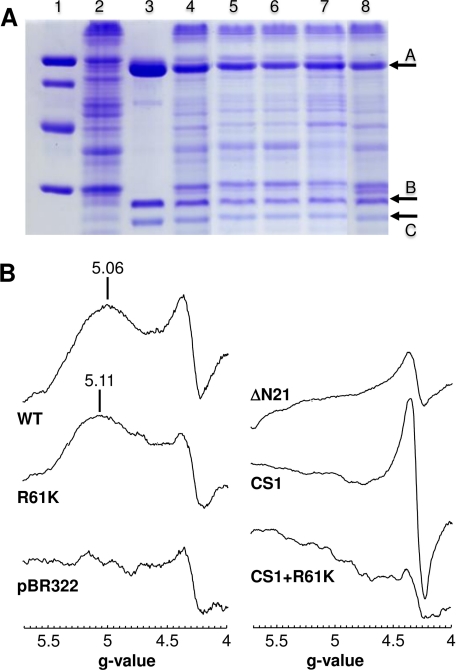
Assignment of the g = 5.06 Signal to FS0
Fig. 4A shows the effects of the mutations generated herein on DmsABC expression and assembly into the E. coli inner membrane. Each of the mutant enzymes was expressed to a level comparable with that of the wild-type enzyme (cf. lane 4 with lanes 5–8). Fig. 4B also shows EPR spectra of the mutant enzymes in the g = 5 region, providing evidence for assignment of the g = 5.06 peak to the FS0 cluster of DmsA. The R61K mutant retained a high spin signal centered at g = 5.11, indicating that, as is the case in NarGHI (18), mutation of the conserved basic residue C-terminal to the FS0-coordinating Cys group perturbs the FS0 EPR signal. Deletion of Asn-21 resulted in elimination of the signal, as did both the CS1 multimutant and the CS1 plus R61K mutant (Fig. 4B). Clearly, mutations of residues within the N-terminal Cys group of DmsA have significant effects on the EPR spectrum of reduced enzyme at g ∼ 5.0. The spectrum of DmsABC in the g = 2 region is complicated by spectral overlap and spin-spin interactions between the four [4Fe-4S] clusters of DmsB (28, 37, 45), and for this reason, our analyses of the mutant spectra did not reveal evidence for additional features corresponding to a modified form of FS0 with an S = ½ ground state in its reduced form (data not shown). One of the mutants, ΔAsn-21, assembled a [3Fe-4S] cluster at the FS0 position (data not shown), which exhibited a spectrum in its oxidized state similar to those of the [3Fe-4S] clusters in mutants of DmsA previously reported by Trieber et al. (13, 15). Overall, the EPR properties of the mutant enzymes allowed us to assign the g = 5.06 signal of reduced wild-type DmsABC to the FS0 cluster, which has an S = ground state in its reduced form.
FIGURE 4.
A, SDS-polyacrylamide gel of DmsABC and its mutants. Lane 1, low molecular weight markers; lane 2, membranes from the negative control TOPP2/pBR322; lane 3, purified wild-type DmsABC; lane 4, TOPP2 membranes containing overexpressed wild-type DmsABC; lane 5, DmsAR61KBC; lane 6, DmsACS1BC; lane 7, DmsAΔN21BC; lane 8, DmsACS1+R61KBC. Arrow A marks DmsA (85.8 kDa), arrow B marks DmsB (22.7 kDa), and arrow C marks DmsC (30.8 kDa). 45 μg of total membrane protein was added per lane, except for lane 3, in which 30 μg of purified enzyme was used. B, effect of mutations of residues close to FS0 or the proximal pterin on the low field DmsABC EPR spectrum. EPR conditions were as described in the legend to Fig. 3, except that spectra are of membrane samples normalized to a protein concentration of 30 mg/ml.
Interplay between FS0 and Mo-bisPGD Insertion during DmsABC Maturation
A principal aim of this work was to examine the interplay between assembly of FS0 and insertion of Mo-bisPGD during DmsABC maturation. In the case of NarGHI, it is clear that Mo-bisPGD is not inserted in enzymes that lack FS0 (18, 25), and it is therefore important to determine whether this is an emerging paradigm for bacterial molybdoenzyme assembly. To address this question, we compared the relative amounts of Mo-bisPGD assayed using the Form A method (23, 39, 53) with estimates of DmsABC concentration measured by fluorescence quench titration (35, 45, 54, 55). The fluorophore used in this second assay is HOQNO, the fluorescence of which in free solution is completely quenched when bound to the Q-site of DmsABC (35) and of other enzymes such as E. coli fumarate reductase and NarGHI (54–56).
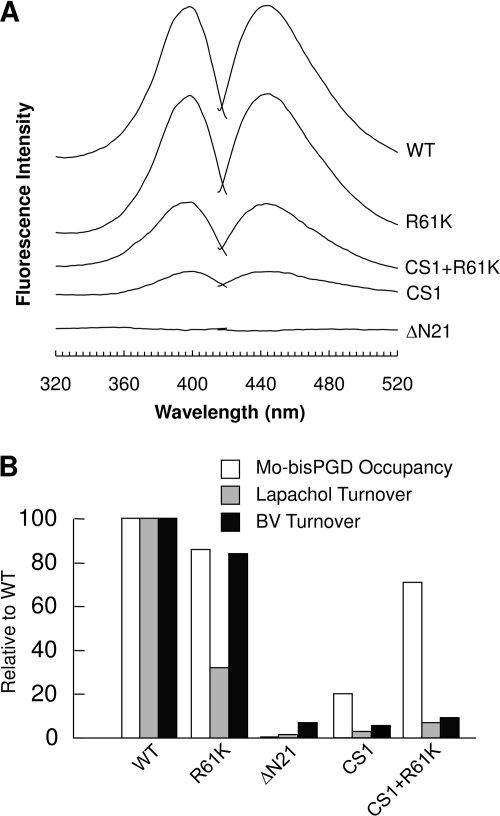
Fig. 5A shows fluorescence spectra of Form A preparations derived from membrane samples containing overexpressed wild-type and mutant DmsABC. The conservative mutation R61K had little effect on Mo-bisPGD assembly as judged by Form A preparation fluorescence levels. Membranes containing the multimutation CS1 enzyme yielded Form A preparations with only ∼13% of the fluorescence levels of those from membranes containing overexpressed wild-type enzyme. Interestingly, when the CS1 multimutant was combined with the R61K mutation, the Form A preparation fluorescence level returned to ∼44% of that of the wild-type enzyme. Finally, removal of Asn-21 eliminated detectable Form A fluorescence (Fig. 5A) but did not prevent enzyme assembly into the E. coli inner membrane (Fig. 4A). This latter observation is notable for two reasons: (i) the CS1/R61K multimutant also has only two residues between the CA and CB positions yet is able to assemble cofactor, and (ii) the ΔAsn-21 mutant assembles a [3Fe-4S] cluster at the FS0 position. Taken together, these observations indicate that the side chain of Asn-21 plays a critical role in facilitating Mo-bisPGD insertion into DmsA. These results complement those recently reported by us for cofactor insertion into NarG (18).
FIGURE 5.
Mo-bisPGD cofactor occupancies and enzyme turnover rates. A, Form A fluorescence spectra of extracts of membrane enriched in wild-type or mutant DmsABC variants. The fluorescence of the TOPP2/pBR322 membrane has been subtracted from each spectrum to eliminate the contribution from chromosomal DmsABC. B, relative cofactor occupancies and enzyme turnover rates. All values were normalized to the wild type, being 100. White bars are relative Mo-bisPGD cofactor occupancies in mutant DmsA enzymes calculated using DmsABC concentrations and relative Form A fluorescence. Gray bars are relative enzyme turnover rates calculated using enzyme-specific activities with LPCH2 as the substrate and DmsABC concentrations. Black bars are relative enzyme turnover rates calculated using enzyme-specific activities with reduced BV as the substrate and DmsABC concentrations.
Table 2 summarizes the effects of the mutants studied herein on enzyme assembly and Mo-bisPGD insertion. Of the mutants studied, the ΔAsn-21 mutant had the most dramatic effect on cofactor content, effectively eliminating detectable Form A derivative while retaining ∼50% of wild-type levels of assembly into the cytoplasmic membrane (cf. Figs. 4A and 5A). Asn-21 is part of the loop connecting CA and CB of the N-terminal Cys group, and its side chain carboxamide nitrogen is predicted to be within H-bonding distance of dithiolene sulfurs from the two pterins of the Mo-bisPGD cofactor (Fig. 1B). It is therefore likely that Asn-21 side chain positioning is critical for Mo-bisPGD insertion.
TABLE 2.
Enzyme and Mo-bisPGD cofactor quantifications and enzyme activities determined by in vitro reduction assays and in vivo growth experiments
| Enzyme | [DmsABC]a | Mo-bisPGD contentb | Specific activityc |
Growthd | |
|---|---|---|---|---|---|
| BV | LPCH2 | ||||
| nmol/mg | |||||
| pBR322 | 0 | 0 | 11.8 | 0.44 | None |
| DmsABC | 0.94 | 100 | 141 | 7.72 | High |
| R61K | 0.99 | 90 | 125 | 2.63 | High |
| ΔN21 | 0.57 | 0 | 5.91 | 0.07 | None |
| CS1 | 0.62 | 13 | 5.22 | 0.14 | None |
| CS1 + R61K | 0.58 | 44 | 7.97 | 0.33 | None |
a The concentrations of wild-type and mutant DmsABC in each membrane sample were determine by HOQNO fluorescence quench titration. pBR322 is an empty vector, so no DmsABC could be detected.
b The Mo-bisPGD cofactor content determined by the Form A fluorescence is qualitative and reported in terms of relative fluorescence intensities. The peak emission fluorescence for wild-type DmsABC was normalized to 100, and the relative fluorescence for the mutant enzymes was calculated using their peak emission fluorescence. The relative cofactor occupancies were then calculated using relative cofactor content and DmsABC concentrations and are shown in Fig. 5B.
c Specific enzyme activities of wild-type and mutant DmsABC were assayed by BV- or LPCH2-dependent reductions of trimethylamine N-oxide. Assays were carried out in triplicate, and average enzyme specific activities are reported. The specific enzyme activities are given in micromoles of BV or LPCH2/min/mg of total protein. The turnover rates (s−1) were then calculated using the specific activities and DmsABC concentrations, and the relative rates with the wild type normalized to 100 are reported in Fig. 5B.
d DSS301 cells transformed with plasmid encoding wild-type DmsABC and its variants were grown anaerobically on glycerol/dimethyl sulfoxide minimum medium.
Table 2 also summarizes the effects of the DmsA mutants on enzyme activity. Two assays were used, the BV/trimethylamine N-oxide oxidoreductase assay and the LPCH2/trimethylamine N-oxide oxidoreductase assay. The BV assay addresses the ability of the enzyme to catalyze substrate reduction with a nonspecific electron donor (BV), whereas the LPCH2 assay addresses the ability of the enzyme to catalyze electron transfer through its electron transfer relay to the Mo-bisPGD cofactor. With the exception of the R61K mutant, enzyme activity was essentially eliminated in all of the mutants studied herein. In the case of CS1 plus R61K, a small amount of activity was rescued, but this did not correlate well with the amount of cofactor insertion detected in Form A fluorescence assays. A likely explanation for this is that the H-bonding contacts between the Asn-21 carboxamide nitrogen and the Mo-bisPGD dithiolene sulfurs are also critical in defining a catalytically competent molybdenum coordination environment.
Role of FS0 in DmsABC Maturation
As is the case for E. coli NarGHI (18), it is clear that the N-terminal Cys group of DmsA plays a critical role in controlling enzyme maturation by coordinating FS0 and Mo-bisPGD assembly. The results presented herein point to an emerging paradigm for Type II molybdoenzyme maturation: FS0 insertion is a prerequisite for enzyme maturation but is not a prerequisite for final assembly of the enzyme into the cytoplasmic membrane.
The mechanism of FS0 and Mo-bisPGD assembly likely involves [4Fe-4S] cluster insertion, generating a cofactor binding-competent conformation of DmsA. In the context of the work presented herein, correctly assembled FS0 dictates a correct conformation for the 18CTVNC22 sequence, which allows subsequent Mo-bisPGD binding.
An important distinction between the two most characterized Type II enzymes is that whereas NarGH is anchored to the inner surface of the cytoplasmic membrane by NarI, DmsAB is translocated to the periplasm by the tat translocon and is anchored to the outer surface of the cytoplasmic membrane by DmsC (2, 22). In both cases, system-specific chaperones are involved, NarJ for NarGHI (24, 25) and DmsD for DmsABC (26, 57). It has been proposed in the case of NarGHI that NarJ is essential for Mo-bisPGD and FS0 insertion and binds to two sites on NarG, the first being the 50-amino acid pseudo tat leader at its N terminus (58, 59) and the second being elsewhere in the protein. In the case of DmsABC, DmsD has been shown to bind to the tat leader, directing the fully folded cofactor-containing DmsAB dimer to the periplasm via the tat translocon without an obligatory role in cofactor insertion (21). The apparently different roles of NarJ and DmsD have to be reconciled with their similar structures. When NarJ from Archaeoglobus fulgidus (60) is compared with DmsD from E. coli (61) using the secondary structure matching server (62), 132 amino acid residues align, corresponding to an overlap of 89% of NarJ residues and 65% of DmsD residues. It is therefore notable that NarJ appears to coordinate insertion of both FS0 and Mo-bisPGD (25), whereas DmsD has a more specific role in directing DmsAB to the periplasm (21). Chan et al. (63) recently demonstrated a possible role of the tat translocon in enhancing the targeting of NarGH to the cytoplasmic membrane. The maturation of DmsABC is an example of one in which the apoenzyme can be translocated by the tat translocon across the cytoplasmic membrane and assembled into its correct location in the absence of its Mo-bisPGD cofactor and/or its FS0 [4Fe-4S] cluster (19, 21, 23, 64). Further studies will be required to clarify the role of DmsD in DmsABC maturation.
We investigated the N-terminal Cys group of NarG using a combination of site-directed mutagenesis and protein crystallography (18, 41). The CA residue of the NarG Cys group is in fact a His residue (His-49), and when mutated to a Cys, FS0 is still assembled into the enzyme along with the Mo-bisPGD cofactor without a detectable EPR signal in the g = 5 region. However, the observed high spin form of FS0 in DmsABC makes it clear that the presence of His at the CA position in NarG is not the source of the EPR signal with an S = ground state. When His-49 is mutated to Ser, neither FS0 nor Mo-bisPGD is assembled. In the crystal structure of this mutant, sequence 49SGVNCTG55 (equivalent to 18CTVNCGS24 in DmsA) is unresolved, suggesting that the correct positioning of this sequence is critical for Mo-bisPGD cofactor insertion (18). In this study, disruption of the DmsA Cys group by generating the CS1 multimutant eliminated most of the Mo-bisPGD cofactor and prevented detection of FS0 by EPR. Combining this multimutant with an R61K mutant to generate a Type I Cys group sequence rescued Mo-bisPGD insertion (44% of the wild-type level) and a small amount of enzyme activity (Fig. 5B and Table 2). The most dramatic effect on cofactor insertion was elicited by the ΔAsn-21 mutant. In this case, Mo-bisPGD insertion was completely eliminated, suggesting that the Asn-21 side chain plays a critical role in facilitating Mo-bisPGD insertion.
Conclusions
Overall, we have used a combination of site-directed mutagenesis and a range of assays to identify the FS0 EPR signature of DmsABC. We have clearly demonstrated that sequence 18CATVNCBG23 plays a critical role in facilitating Mo-bisPGD binding, with deletion of Asn-21 having the most catastrophic effects. When the wild-type sequence 18CATVNCBGSRCCP27 is converted to a sequence similar to that of the FdnG subunit of E. coli formate dehydrogenase N (18CATYCBGVGCCG26), cofactor binding is almost completely eliminated, but when the 18CATYCBGVGCCG26 multimutant is combined with an R61K mutant, generating a true Type I sequence, cofactor insertion is rescued. These results shed additional light on the maturation of members of the CISM family of bacterial molybdoenzymes.
Acknowledgment
We thank Nasim Boroumand for help and expertise with the purification of DmsABC.
This work was supported in part by Canadian Institutes of Health Research Grant MOP15292.
- Mo-bisPGD
- molybdo-bis(pyranopterin guanine dinucleotide)
- HOQNO
- 2-n-heptyl-4-hydroxyquinoline N-oxide
- BV
- benzyl viologen
- LPCH2
- reduced lapachol.
REFERENCES
- 1. Weiner J. H., Rothery R. A., Sambasivarao D., Trieber C. A. (1992) Biochim. Biophys. Acta 1102, 1–18 [DOI] [PubMed] [Google Scholar]
- 2. Rothery R. A., Workun G. J., Weiner J. H. (2008) Biochim. Biophys. Acta 1778, 1897–1929 [DOI] [PubMed] [Google Scholar]
- 3. Weiner J. H., Bilous P. T., Shaw G. M., Lubitz S. P., Frost L., Thomas G. H., Cole J. A., Turner R. J. (1998) Cell 93, 93–101 [DOI] [PubMed] [Google Scholar]
- 4. Plunkett G., 3rd, Burland V., Daniels D. L., Blattner F. R. (1993) Nucleic Acids Res. 21, 3391–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berg B. L., Baron C., Stewart V. (1991) J. Biol. Chem. 266, 22386–22391 [PubMed] [Google Scholar]
- 6. Blasco F., Iobbi C., Giordano G., Chippaux M., Bonnefoy V. (1989) Mol. Gen. Genet. 218, 249–256 [DOI] [PubMed] [Google Scholar]
- 7. Blasco F., Iobbi C., Ratouchniak J., Bonnefoy V., Chippaux M. (1990) Mol. Gen. Genet. 222, 104–111 [DOI] [PubMed] [Google Scholar]
- 8. Heinzinger N. K., Fujimoto S. Y., Clark M. A., Moreno M. S., Barrett E. L. (1995) J. Bacteriol. 177, 2813–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krafft T., Bokranz M., Klimmek O., Schröder I., Fahrenholz F., Kojro E., Kröger A. (1992) Eur. J. Biochem. 206, 503–510 [DOI] [PubMed] [Google Scholar]
- 10. Jormakka M., Yokoyama K., Yano T., Tamakoshi M., Akimoto S., Shimamura T., Curmi P., Iwata S. (2008) Nat. Struct. Mol. Biol. 15, 730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bogachev A. V., Murtazina R. A., Skulachev V. P. (1996) J. Bacteriol. 178, 6233–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sambasivarao D., Scraba D. G., Trieber C., Weiner J. H. (1990) J. Bacteriol. 172, 5938–5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Trieber C. A., Rothery R. A., Weiner J. H. (1996) J. Biol. Chem. 271, 4620–4626 [DOI] [PubMed] [Google Scholar]
- 14. Trieber C. A., Rothery R. A., Weiner J. H. (1994) J. Biol. Chem. 269, 7103–7109 [PubMed] [Google Scholar]
- 15. Rothery R. A., Trieber C. A., Weiner J. H. (1999) J. Biol. Chem. 274, 13002–13009 [DOI] [PubMed] [Google Scholar]
- 16. Rothery R. A., Bertero M. G., Cammack R., Palak M., Blasco F., Strynadka N. C., Weiner J. H. (2004) Biochemistry 43, 5324–5333 [DOI] [PubMed] [Google Scholar]
- 17. Lanciano P., Savoyant A., Grimaldi S., Magalon A., Guigliarelli B., Bertrand P. (2007) J. Phys. Chem. B 111, 13632–13637 [DOI] [PubMed] [Google Scholar]
- 18. Rothery R. A., Bertero M. G., Spreter T., Bouromand N., Strynadka N. C., Weiner J. H. (2010) J. Biol. Chem. 285, 8801–8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sambasivarao D., Turner R. J., Bilous P. T., Rothery R. A., Shaw G., Weiner J. H. (2002) Biochem. Cell Biol. 80, 435–443 [DOI] [PubMed] [Google Scholar]
- 20. Sambasivarao D., Weiner J. H. (1991) J. Bacteriol. 173, 5935–5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sambasivarao D., Turner R. J., Simala-Grant J. L., Shaw G., Hu J., Weiner J. H. (2000) J. Biol. Chem. 275, 22526–22531 [DOI] [PubMed] [Google Scholar]
- 22. Stanley N. R., Sargent F., Buchanan G., Shi J., Stewart V., Palmer T., Berks B. C. (2002) Mol. Microbiol. 43, 1005–1021 [DOI] [PubMed] [Google Scholar]
- 23. Rothery R. A., Grant J. L., Johnson J. L., Rajagopalan K. V., Weiner J. H. (1995) J. Bacteriol. 177, 2057–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blasco F., Dos Santos J. P., Magalon A., Frixon C., Guigliarelli B., Santini C. L., Giordano G. (1998) Mol. Microbiol. 28, 435–447 [DOI] [PubMed] [Google Scholar]
- 25. Lanciano P., Vergnes A., Grimaldi S., Guigliarelli B., Magalon A. (2007) J. Biol. Chem. 282, 17468–17474 [DOI] [PubMed] [Google Scholar]
- 26. Oresnik I. J., Ladner C. L., Turner R. J. (2001) Mol. Microbiol. 40, 323–331 [DOI] [PubMed] [Google Scholar]
- 27. Cheng V. W., Rothery R. A., Bertero M. G., Strynadka N. C., Weiner J. H. (2005) Biochemistry 44, 8068–8077 [DOI] [PubMed] [Google Scholar]
- 28. Rothery R. A., Weiner J. H. (1991) Biochemistry 30, 8296–8305 [DOI] [PubMed] [Google Scholar]
- 29. Lambert C., Léonard N., De Bolle X., Depiereux E. (2002) Bioinformatics 18, 1250–1256 [DOI] [PubMed] [Google Scholar]
- 30. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31. Bilous P. T., Weiner J. H. (1985) J. Bacteriol. 162, 1151–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 33. Markwell M. A., Haas S. M., Bieber L. L., Tolbert N. E. (1978) Anal. Biochem. 87, 206–210 [DOI] [PubMed] [Google Scholar]
- 34. Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. (1977) J. Biol. Chem. 252, 1102–1106 [PubMed] [Google Scholar]
- 35. Zhao Z., Weiner J. H. (1998) J. Biol. Chem. 273, 20758–20763 [DOI] [PubMed] [Google Scholar]
- 36. Van Ark G., Berden J. A. (1977) Biochim. Biophys. Acta 459, 119–127 [DOI] [PubMed] [Google Scholar]
- 37. Cammack R., Weiner J. H. (1990) Biochemistry 29, 8410–8416 [DOI] [PubMed] [Google Scholar]
- 38. Rothery R. A., Chatterjee I., Kiema G., McDermott M. T., Weiner J. H. (1998) Biochem. J. 332, 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson J. L., Hainline B. E., Rajagopalan K. V., Arison B. H. (1984) J. Biol. Chem. 259, 5414–5422 [PubMed] [Google Scholar]
- 40. Bertero M. G., Rothery R. A., Palak M., Hou C., Lim D., Blasco F., Weiner J. H., Strynadka N. C. J. (2003) Nat. Struct. Mol. Biol. 10, 681–687 [DOI] [PubMed] [Google Scholar]
- 41. Magalon A., Asso M., Guigliarelli B., Rothery R. A., Bertrand P., Giordano G., Blasco F. (1998) Biochemistry 37, 7363–7370 [DOI] [PubMed] [Google Scholar]
- 42. Hettmann T., Siddiqui R. A., von Langen J., Frey C., Romão M. J., Diekmann S. (2003) Biochem. Biophys. Res. Commun. 310, 40–47 [DOI] [PubMed] [Google Scholar]
- 43. Berg B. L., Stewart V. (1990) Genetics 125, 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jormakka M., Törnroth S., Byrne B., Iwata S. (2002) Science 295, 1863–1868 [DOI] [PubMed] [Google Scholar]
- 45. Rothery R. A., Weiner J. H. (1996) Biochemistry 35, 3247–3257 [DOI] [PubMed] [Google Scholar]
- 46. Gladyshev V. N., Boyington J. C., Khangulov S. V., Grahame D. A., Stadtman T. C., Sun P. D. (1996) J. Biol. Chem. 271, 8095–8100 [DOI] [PubMed] [Google Scholar]
- 47. Breton J., Berks B. C., Reilly A., Thomson A. J., Ferguson S. J., Richardson D. J. (1994) FEBS Lett. 345, 76–80 [DOI] [PubMed] [Google Scholar]
- 48. Duderstadt R. E., Brereton P. S., Adams M. W., Johnson M. K. (1999) FEBS Lett. 454, 21–26 [DOI] [PubMed] [Google Scholar]
- 49. Kowal A. T., Werth M. T., Manodori A., Cecchini G., Schröder I., Gunsalus R. P., Johnson M. K. (1995) Biochemistry 34, 12284–12293 [DOI] [PubMed] [Google Scholar]
- 50. Flint D. H., Emptage M. H., Guest J. R. (1992) Biochemistry 31, 10331–10337 [DOI] [PubMed] [Google Scholar]
- 51. Müller A., Schneider K., Knüttel K., Hagen W. R. (1992) FEBS Lett. 303, 36–40 [DOI] [PubMed] [Google Scholar]
- 52. George S. J., Armstrong F. A., Hatchikian E. C., Thomson A. J. (1989) Biochem. J. 264, 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Johnson J. L., Indermaur L. W., Rajagopalan K. V. (1991) J. Biol. Chem. 266, 12140–12145 [PubMed] [Google Scholar]
- 54. Rothery R. A., Seime A. M., Spiers A. M., Maklashina E., Schröder I., Gunsalus R. P., Cecchini G., Weiner J. H. (2005) FEBS J. 272, 313–326 [DOI] [PubMed] [Google Scholar]
- 55. Rothery R. A., Weiner J. H. (1998) Eur. J. Biochem. 254, 588–595 [DOI] [PubMed] [Google Scholar]
- 56. Rothery R. A., Blasco F., Magalon A., Asso M., Weiner J. H. (1999) Biochemistry 38, 12747–12757 [DOI] [PubMed] [Google Scholar]
- 57. Ray N., Oates J., Turner R. J., Robinson C. (2003) FEBS Lett. 534, 156–160 [DOI] [PubMed] [Google Scholar]
- 58. Vergnes A., Pommier J., Toci R., Blasco F., Giordano G., Magalon A. (2006) J. Biol. Chem. 281, 2170–2176 [DOI] [PubMed] [Google Scholar]
- 59. Li H., Turner R. J. (2009) Can. J. Microbiol. 55, 179–188 [DOI] [PubMed] [Google Scholar]
- 60. Kirillova O., Chruszcz M., Shumilin I. A., Skarina T., Gorodichtchenskaia E., Cymborowski M., Savchenko A., Edwards A., Minor W. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 348–354 [DOI] [PubMed] [Google Scholar]
- 61. Stevens C. M., Winstone T. M., Turner R. J., Paetzel M. (2009) J. Mol. Biol. 389, 124–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Krissinel E., Henrick K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 63. Chan C. S., Chang L., Winstone T. M., Turner R. J. (2010) FEBS Lett. 584, 4553–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sambasivarao D., Dawson H. A., Zhang G., Shaw G., Hu J., Weiner J. H. (2001) J. Biol. Chem. 276, 20167–20174 [DOI] [PubMed] [Google Scholar]