Abstract
The Clostridium botulinum neurotoxins (BoNTs) cleave SNARE proteins, which inhibit binding and thus fusion of neurotransmitter vesicles to the plasma membrane of peripheral neurons. BoNTs comprise an N-terminal light chain (LC) and C-terminal heavy chain, which are linked by a disulfide bond. There are seven serotypes (A–G) of BoNTs based upon immunological neutralization. Although the binding and entry of BoNT/A into neurons has been subjected to considerable investigation, the intracellular events that allow BoNT/A to efficiently cleave SNAP-25 within neurons is less well understood. Earlier studies showed that intracellular LC/A bound to the plasma membrane of neurons. In this study, intracellular LC/A is shown to directly bind SNAP-25 on the plasma membrane. Solid phase binding showed that the N-terminal residues of LC/A bound residues 80–110 of SNAP-25, which was also observed in cultured neurons. Association of the N-terminal 8 amino acids of LC/A and residues 80–110 of SNAP-25 also enhanced substrate cleavage. These findings explain how LC/A associates with SNAP-25 on the plasma membrane and provide a basis for LC/A cleavage of SNAP-25 within the SNARE complex.
Keywords: Bacterial Toxins, Enzymes, Intracellular Trafficking, Plasma Membrane, Protease, Botulinum Neurotoxin, SNAP-25
Introduction
The botulinum neurotoxins (BoNTs)3 are the most toxic proteins for humans (1, 2). The potency and longevity of BoNT intoxication have facilitated use of BoNTs as therapeutic agents (3–5) and as potential biological weapons (6, 7). There are seven BoNT serotypes (A–G), which are organized as dichain proteins, where the N-terminal catalytic light chain (LC) and C-terminal heavy chain are linked by a disulfide bond (8). Neurotrophic activity is based upon two properties of the BoNTs, binding to a neuron-specific receptor and cleavage of neuron-specific soluble NSF attachment protein receptor (SNARE) protein(s).
BoNTs are zinc proteases that cleave SNARE proteins (9). BoNT/A cleaves the plasma membrane-associated SNARE protein synaptosome-associated protein of 25 kDa (SNAP-25) (10) between residues 197 and 198. This cleavage inhibits SNAP-25-mediated neurotransmitter vesicle fusion to the plasma membrane (11, 12). LC/A recognizes multiple sites within SNAP-25: an extended surface on SNAP-25 distanced from the site of cleavage (13, 14) and residues adjacent to the scissile bond that are discontinuous and appear as pockets surrounding the cleavage site (15). This implicates multistep recognition of SNAP-25 for cleavage by LC/A.
In addition to substrate recognition to facilitate neurotrophic intoxication, BoNTs bind to dual host receptors that are specific to neurons (1). BoNT/A first binds a ganglioside on resting neurons, and upon fusion of synaptic vesicles to the plasma membrane, it binds to the luminal domain of the synaptic vesicle (SV) protein 2 (SV2) (16, 17). The BoNT/A-receptor complex is endocytosed upon recycling of SV proteins from the plasma membrane. As the SV matures and acidifies, the translocation domain of the heavy chain (HCT) inserts into the SV membrane. Membrane-inserted HCT facilitates LC translocation into the cytosol, where LC cleaves plasma membrane-associated SNAP-25 (18–20). Although earlier studies reported the intracellular localization of ectopic expressed BoNT/A LC to the host plasma membrane (21, 22), there is limited understanding of the intracellular events that lead to plasma membrane localization. In this study, a new interaction between LC/A and SNAP-25 is identified that facilitates high affinity binding of LC/A to SNAP-25 on the plasma membrane of neurons.
EXPERIMENTAL PROCEDURES
Materials
Neuro-2A cells were purchased from the ATCC (CCL131). pEGFP, pERFP, and pEYFP were purchased from Invitrogen. α-SNAP-25 IgG and α-myelin IgG were purchased from Santa Cruz Biotechnology. Lipofectamine LTX was purchased from Invitrogen. Rat cerebral cortex was purchased from Pel-Freez Biologicals and stored at −80 °C prior to processing. SuperSignal, Ultra TMB, and α-3xFLAG-HRP were purchased from Pierce Biochemicals.
Plasmid Construction for Protein Expression
LC/A was expressed in Neuro-2A cells, as a GFP fusion protein within pEGFP. LC/A derivatives were constructed by PCR amplification of the indicated regions of LC/A and subcloning into pEGFP. YFP-SNAP-25 was engineered by subcloning the full length of SNAP-25(1–206) into pEYFP. RFP-LC/E was constructed by subcloning DNA encoding LC/E(1–418) into pERFP. Expression plasmids encoding LC/A, SNAP-25, and the respective truncated derivatives were constructed by PCR-amplifying DNA encoding LC/A, SNAP-25, and their different truncation mutations into pET-15b. Plasmids encoding these constructs were transformed into Escherichia coli BL21(DE3) RIL or BL21(DE3) (Stratagene). Protein expression and purification were achieved as described previously (13).
Cell Culture, Transfection, Fractionation, and Confocal Microscopy
Neuro-2A cells were cultured in minimum essential medium supplemented with 10% newborn calf serum, 1.4% sodium bicarbonate, and 0.5% penicillin-streptomycin at 37 °C in 5% CO2. Subconfluent cells were transfected with the indicated plasmids by using Lipofectamine LTX as suggested by the manufacturer. Cells were fractionated as described previously (22). For confocal microscopy (Leica), cells were cultured on 12-mm coverslips and transfected as indicated. Cells were fixed and subjected for immunostaining or imaged directly. Fluorescence was captured in grayscale; imaged colors may not match fusion protein fluorescence.
Brain Lysate Binding
Rat brain membranes were prepared as described previously (23). Membranes were centrifuged at 48,000 × g for 30 min to pellet rat brain cell membranes, which were suspended in PBS or extracted with detergent.
Far Western Assay
Rat brain membrane was extracted in PBS containing 2% Triton X-100 overnight at 4 °C. Proteins in the insoluble pellet were separated by SDS-PAGE and stained with Coomassie Blue or transferred to PVDF membranes. Membranes were stained with Ponceau S to confirm transfer and blocked in PBS with 2% BSA for 1 h at room temperature. Membranes were washed three times in PBS with 0.1% Tween 20 and incubated with 0.1 μm 3xFLAG-LC/A or 3xFLAG-LC/A(8–438) for 2 h at room temperature. After washes, membranes were incubated with mouse α-3xFLAG-HRP antibody (1:10,000 final dilution) and developed with SuperSignal.
Solid Phase Binding Assay
Recombinant SNAP-25 derivatives (100 μl at 5 μg/ml) in 50 mm Na2CO3 (pH 9.6) were added to 96-well plates and incubated overnight at 4 °C. Wells were washed three times in PBS and then blocked with 50 mm Na2CO3 (pH 9.6) containing 1% (w/v) BSA for 1 h at room temperature. After washes, 100 μl of 3xFLAG-LC/A or 3xFLAG-LC/A(8–438) (0.4–800 nm in PBS + 1% BSA) were added and incubated for 1 h at room temperature. Wells were washed three times and incubated with mouse α-3xFLAG-HRP antibody (1:10000 final dilution) for 1 h at room temperature, washed three times, and developed with 100 μl of Ultra TMB for 20 min at room temperature. Absorbance (450 nm) was read after quenching with 100 μl of 1 m H2SO4. A450 was plotted versus LC/A, and the k50 (amount of LC/A that bound 50% of SNAP-25) was determined.
Linear Velocity Assay
Linear velocity of SNAP-25 cleavage by LC/A and LC/A(8–438) was performed in a 20-μl reaction containing: 2 μm SNAP-25 derivatives and the indicated amounts of LC in 10 mm Tris-HCl (pH 7.6) and 20 mm NaCl. Reactions were incubated at 37 °C for 15 min and then subjected to SDS-PAGE. Coomassie Blue-stained gels were assayed for the cleavage of substrate by densitometry. The percentage of substrate cleavage and the concentration of LCs were plotted, and the LCs required to cleave 50% of substrate were derived.
RESULTS
Multiple Domains of LC/A Confer High Affinity Binding to the Neuronal Plasma Membrane
Deletion mapping showed that N- and C-terminal regions of LC/A facilitated localization to the plasma membrane of Neuro-2A cells (Fig. 1). Upon ectopic expression, LC/A(1–438) localized primarily to the plasma membrane, whereas LC/A(1–425) localized to the plasma membrane and showed a diffuse cytoplasmic presence. Although this indicated that the C terminus (residues 426–438) contributed to membrane localization, deletions to the N terminus of LC/A had a greater effect on the plasma membrane association where LC/A(8–438) and LC/A(17–438) were present primarily within the cytoplasm. This indicated that the N-terminal 17 amino acids of LC/A contributed membrane localization. Subcellular fractionation showed that ∼90% of LC/A(1–438), 75% of LC/A(1–425), 35% of LC/A(8–438), and 5% of LC/A(17–438) were membrane-bound, supporting a primary role for the N terminus in plasma membrane localization (Fig. 1). Protein modeling showed that residues Lys6 and Lys11 of LC/A were surface-exposed and thus candidate residues for interactions with the plasma membrane (Fig. 1). Upon ectopic expression, LC/A(K6A,K11A) was present as a diffuse cytoplasmic protein in Neuro-2A cells with ∼50% of LC/A(K6A,K11A) located in the cytoplasm. Because LC/A(17–438) was not catalytically active, LC/A(8–438) was used as the representative N-terminal deletion protein.
FIGURE 1.
Mapping regions of LC/A that contribute to intracellular localization in Neuro-2A cells. Upper panel, pGFP-LC/A derivatives (inset, LC/A(K6A,K11A) is termed LC/A(DKM) were transfected into Nero-2A cell. After an overnight incubation, LC/A localization phenotypes were visualized using confocal microscope, and representative images are shown. Middle panel, cells were also lysed, and membrane and cytosolic fractions were generated by centrifugation and subjected to SDS-PAGE. LC/A was detected by Western blotting using anti-GFP antibody and quantified. Data are the average of three independent experiments. Error bars indicate S.D. Lower panel, the N terminus of LC/A was modeled in PyMOL, and surface lysine residues (Lys6 and Lys11) were highlighted in blue. An alignment of the N termini of LC/A and LC/E is shown with surface-exposed residues that include Lys6 and Lys11 highlighted.
The N Terminus of LC/A Confers High Affinity Binding to SNAP-25
Far Western analysis of detergent-extracted rat brain membranes showed that LC/A bound the membrane as a tight band at ∼25 kDa, whereas LC-A(8–438) did not show detectable binding to the membrane (Fig. 2). The background signal from LC/A was greater than LC/A(8–438). LC/MS-MS analysis of the band of membrane where LC/A bound identified two rat-specific proteins: SNAP-25 and myelin (Table 1). The membrane was next probed with α-SNAP-25 and α-myelin antibodies. α-SNAP-25 antibody bound at the site where LC/A bound the membrane, whereas myelin antibody bound to a faster migrating band and the signal was more diffuse than bound LC/A (Fig. 2). These results indicate that the N terminus of LC/A facilitates high affinity binding to SNAP-25, a previously unrecognized interaction.
FIGURE 2.
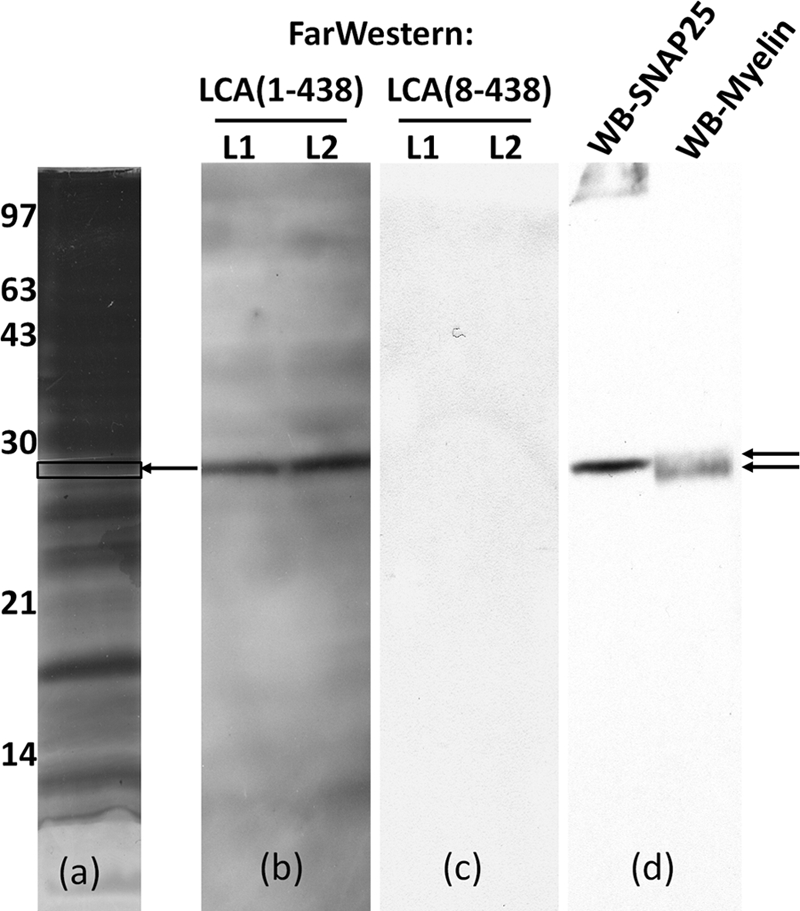
Far Western detection of LC/A bound to an ∼25-kDa protein in rat brain plasma membrane lysates. a, Triton X-100-extracted rat brain plasma membrane lysates were subjected to SDS-PAGE. Gel was stained with Coomassie Blue. The boxed area of the gel (arrow) was cut out and subjected to MS analysis as described in Table 1. b and c, the gel was transferred to PVDF membranes, blocked, and subjected to Far Western analysis, incubating with 0.1 μm 3xFLAG-LC/A(1–438) (b) or 3xFLAG-LC/A(8–438) (c) for 2 h. Bound LC/A was detected with α-FLAG-HRP antibody followed by SuperSignal analysis; an image of the x-ray film is shown (lanes L1 and L2 are identical lysate samples). The left blot was stripped, and the L1 and L2 lanes were divided and probed for SNAP-25 (α-SNAP-25 antibody) or myelin (α-myelin antibody), respectively. Bound antibody was detected with an HRP secondary antibody followed by SuperSignal analysis. d, an image of the x-ray film is shown. WB, Western blot.
TABLE 1.
Identification of LC/A binding proteins in the rat brain plasma membrane lysates
Proteins were in-gel-digested with trypsin, and extracted peptides were subjected to LC/MS-MS analysis. Peptides were aligned with Mascot screening Rattus norvegicus proteins. Two rat-derived proteins identified are shown.
| Protein identified | Mass | Peptides matched |
|---|---|---|
| Da | ||
| gi number 149023409, synaptosomal-associated protein 25 | 27291 | 3 |
| gi number 8381631, myelin/oligodendrocyte glycoprotein | 24948 | 1 |
A solid phase binding assay showed that LC/A bound SNAP-25 with a higher affinity (k50 ∼13 nm) than LC/A(8–438), which showed a k50 of ∼100 nm for SNAP-25 (Fig. 3). These data support a role for the N terminus of LC/A for high affinity binding to SNAP-25.
FIGURE 3.

Binding of LC/A to the recombinant SNAP-25. SNAP-25 was coated in an ELISA plate and incubated with the indicated 3xFLAG-LC/A derivative for 1 h at room temperature. Bound LC/A was detected with mouse α-FLAG-HRP antibody (Pierce, 1:10,000 final dilution) for 1 h at room temperature and developed with Ultra TMB (Pierce) for 20 min at room temperature. The reaction was quenched with H2SO4, and A450 was determined. The amount of LC/A needed to bind 50% SNAP-25 was determined.
Two Regions of SNAP-25 Contribute to LC/A High Affinity Binding
Deletion mapping also localized the region(s) in SNAP-25 that contributed to high affinity binding to LC/A. Although truncation of the N-terminal 80 amino acids of SNAP-25 did not influence LC/A affinity, extending the truncation to the N-terminal 93 amino acids decreased LC/A affinity by ∼6-fold, and truncation of the N-terminal 110 amino acids had a greater effect on LC/A affinity (Fig. 4). This indicated that residues 80–110 of SNAP-25 contributed to high affinity binding to LC/A. The contribution of the C terminus of SNAP-25 for high affinity LC/A binding was also determined. Although LC/A bound SNAP-25(1–197) and SNAP-25(1–206) with similar affinity, LC/A had a lower affinity for SNAP-25(1–180) (Fig. 4). This indicated that contact with residues on adjacent sites to the scissile bond contributed to the high affinity of LC/A for SNAP-25. Thus, two regions (80–110 and 180–197) of SNAP-25 contribute to the high affinity binding to LC/A.
FIGURE 4.
High affinity binding of LC/A to SNAP-25. a, schematic of SNAP-25 (SN25) bound to the plasma membrane. SNAP-25 is a peripheral, plasma membrane-binding protein that comprises two helixes that bind the membrane through four palmitoylated cysteine interactions. b and c, solid phase binding assays; SNAP-25 (SN) derivatives (0.5 μg) were coated in 96-well plates and incubated with the indicated amounts of 3xFLAG-LC/A (in nm). Bound LC/A was detected with mouse α-FLAG-HRP antibody and Ultra TMB. A450 was plotted versus [LC/A] to determine k50, the amount of LC/A needed to bind 50% SNAP-25. d, schematic of LC/A interactions with membrane-bound SNAP-25. (Panel 1) At the cell plasma membrane, syntaxin 1a (Syn) and SNAP-25 form a SNARE complex (31). (Panel 2) The N terminus of LC/A binds residues 80–110 of SNAP-25 (panel 3) which facilitates substrate binding in competition with syntaxin 1a. (Panel 4) After SNAP-25 cleavage, LC/A has a high affinity for membrane-bound SNAP-25(1–197).
Association of LC/A with SNAP-25 within Neurons
Ectopic expression characterized LC localization within Neuro-2A cells. Initial experiments showed that GFP-LC/A colocalized with endogenous SNAP-25 or YFP-SNAP-25 on the plasma membrane (Fig. 5). A 4-amino acid mutation of SNAP-25(C85A,C88A,C90A,C92A) was engineered and observed to be expressed in the cytosol, consistent with palmitoylation at these cysteines being responsible for plasma membrane localization of SNAP-25 (24). Coexpression of SNAP-25(C85A,C88A,C90A,C92A) with GFP-LC/A changed the localization of LC/A to the cytoplasm. Quantification showed that ∼90% of GFP-LC/A was plasma membrane-bound when coexpressed with SNAP-25, whereas ∼80% of GFP-LC/A was cytosolic when coexpressed with SNAP-25(C85A,C88A,C90A,C92A) (Fig. 5). These data support the direct intracellular binding of LC/A to SNAP-25 on the plasma membrane of neurons.
FIGURE 5.
Coexpression of SNAP-25(C85A,C88A,C90A,C92A) disrupts LC/A plasma membrane localization. Upper panel, Neuro-2A cells were transfected with pLC/A (blue) alone (SN-Endo) or with pSNAP-25 (SN-YFP) or pSNAP-25(C85A,C88A,C90A,C92A)-YFP (SN(4CM)). Cells were washed, fixed, permeabilized, and probed with α-SNAP-25 antibody (SN-Endo) and subjected to confocal microscopy alone or merged. Lower panel, greater than 100 cells were visually scored for cytosolic LC/A upon each transfection treatment. Numbers are the average of three independent experiments. Error bars indicate S.D. SN, SNAP-25.
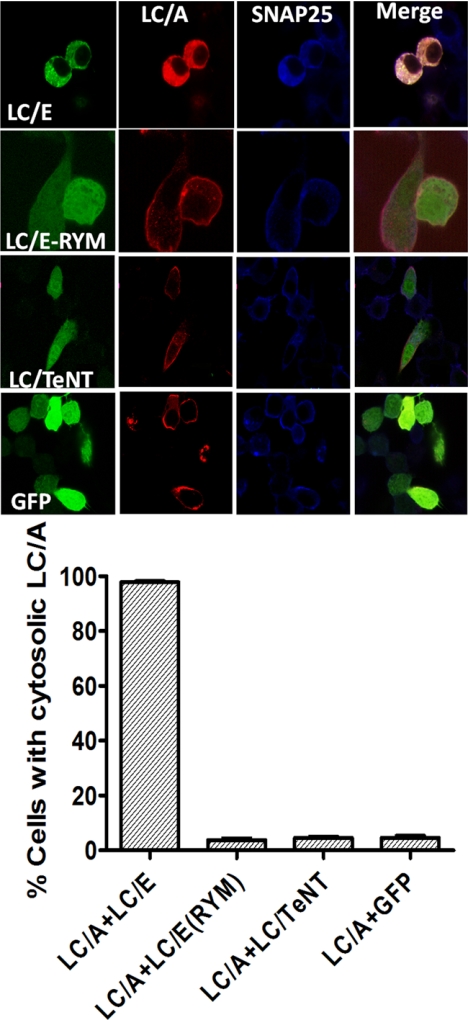
Another experiment utilized the LC of BoNT/E (LC/E) to determine how SNAP-25 influenced the localization of LC/A in Neuro-2A cells. LC/E cleaves SNAP-25 between residues 180 and 181, which releases SNAP-25(1–180) into the cytosol (25). Upon coexpression with LC/E, LC/A localization changed from the plasma membrane to the cytoplasm (Fig. 6). Controls showed that the movement of LC/A from the cytoplasm was due to the enzymatic activity of LC/E because upon coexpression with LC/E(R347A,R349F), a catalytically inactive form of LC/E (26, 27), LC/A remained plasma membrane-localized. Other controls showed that upon coexpression with the LC of Tetanus toxin or GFP, LC/A remained plasma membrane-localized. These experiments also supported the direct interaction of LC/A with SNAP-25 within cells.
FIGURE 6.
Coexpression of LC/E disrupts LC/A plasma membrane localization. Upper panel, Neuro-2A cells were cotransfected with pLC/E, pLC/E(R347A,Y349F) (LC/E(RYM) pLC/Tetanus toxin (LC/TeNT), or pGFP derivative (green) and pLC/A (red). Cells washed, fixed, and probed for SNAP-25, using α-SNAP-25 IgG (blue). Cells were visualized by confocal microscopy, and images were merged. Lower panel, greater than 100 cells were scored for LC/A cytosolic phenotype under each transfection treatment. Results are the average of three independent experiments. Error bars indicate S.D.
High Affinity Binding of LC/A to SNAP-25 Enhances Substrate Cleavage
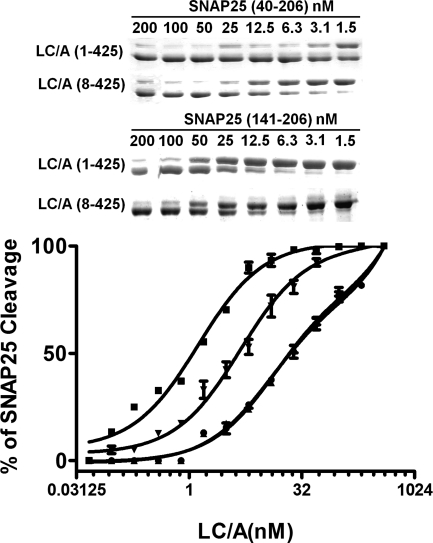
Experiments determined the influence of N terminus of LC/A on SNAP-25 cleavage. SNAP-25(40–206) was used as a substitute for full-length SNAP-25 because SNAP-25(40–206) is readily purified as a recombinant protein in E. coli (13). LC/A cleaved SNAP-25(40–206) ∼20-fold more efficiently than LC/A(8–438), whereas LC/A showed an ∼10-fold higher rate of SNAP-25(141–206) cleavage than LC/A(8–438) (Fig. 7). Thus, the N terminus of LC/A facilitates SNAP-25 cleavage through interactions with the N-terminal coiled residues of SNAP-25, with some N-terminal LC/A interactions with SNAP-25(146–206) also contributing to catalysis.
FIGURE 7.
Cleavage of SNAP-25 by LC/A and LC/A(8–438). Two μm SNAP-25(1–206) was incubated with the indicated amounts of LC/A(8–425) (●) or LC/A (1–425) (■), or 2 μm SNAP-25(146–206) was incubated with the indicated amounts of LC/A(8–425) (▴) or LC/A (1–425) (▾) in 10 mm Tris-HCl and 20 mm NaCl for 15 min at 37 °C and stopped with sample buffer. The gel was stained for protein, and SNAP-25 cleavage was determined by densitometry. Stained gels shown are from a representative experiment analyzing 1.5 and 200 nm LCs. The percentage of substrate cleaved was plotted versus [LC]. Results are the average of at least three independent experiments. Error bars indicate S.D.
DISCUSSION
Although earlier studies reported the intracellular localization of the LC of BoNT/A to the plasma membrane (21, 22), the basis for plasma membrane association was not determined. The current study shows that LC/A binds directly to SNAP-25 on the plasma membrane of neurons. The LC/A-SNAP-25 interaction appears stable, with little detectable LC/A present in the cytoplasm. The N terminus of LC/A contributes to high affinity binding of LC/A to SNAP-25 on the plasma membrane and ultimately to the efficient cleavage of SNAP-25. Two interactions were detected, and high affinity binding of LC/A to SNAP-25 required the presence of residues 80–110 and 180–197 of SNAP-25.
Identification of a SNAP-25 interaction with the N terminus of LC/A was not expected because BoNT/A or LC/A was previously to bind and utilize SNAP-25(146–206) as an efficient substrate for cleavage (13, 15, 28). The role of the N terminus of LC/A in SNAP-25 binding/cleavage may not have been detected in the earlier studies due to the experimental conditions used. The interaction between the N terminus of LC/A and SNAP-25 residues 80–110 increased the efficiency of SNAP-25 cleavage, implicating a functional consequence for this interaction. The significance of the interaction between the N terminus of LC/A and SNAP-25 residues 80–110 may be related to the SNARE complex of which SNAP-25 is a component (Fig. 4d). Within neurons, SNAP-25 is associated with syntaxin 1a forming a pre-SNARE complex for VAMP-2 recruitment (29, 30), and the C terminus of SNAP-25 may not be accessible to LC/A because SNAP-25(1–87) and SNAP-25(131–206) form a double helix complex (31). Thus, interactions of LC/A with the non-SNARE complex region of SNAP-25, residues 80–110, may allow LC/A initial access to SNAP-25 within the complex and compete with syntaxin 1a for SNAP-25 binding. The finding that LC/A localizes to the plasma membrane through direct interactions with SNAP-25 also provides a basis for the potency of BoNT/A within neurons. The recent finding that an N-terminal deletion to BoNT/A has reduced toxicity supports a role of the N terminus in substrate recognition (32). This study also reported that the deletion of the N-terminal 7 amino acids of BoNT/A did not affect the longevity of SNAP-25 cleavage or neuromuscular paralysis, indicating that intracellular targeting to the plasma membrane does not influence LC half-life within a neuron. These studies are consistent with the recent determination that recognition by the proteasome contributes to LC stability within neurons (33).
The intracellular trafficking and localization of bacterial toxins within host cells have been reported for several exotoxins and type III effectors but remain a relatively under investigated field of study. The limited focus on intracellular trafficking of toxins may be due to the high sensitivity needed to detect this activity and earlier observations that both bacterial toxin and viruses, which often utilize similar entry strategies, can utilize multiple pathways to enter and traffic in host cells (34–38). For example, exotoxin A of Pseudomonas aeruginosa possesses a KDEL-like endoplasmic reticulum retention sequence that is utilized to retrograde traffic from the early endosome to the endoplasmic reticulum where the catalytic domain is translocated into the cytosol to ADP-ribosylate elongation factor II (39, 40). Possibly more analogous to LC/A, we have reported that ExoS, a type III-delivered cytotoxin, traffics on the cytoplasmic face of endosomes to retrograde traffic to the endoplasmic reticulum. This trafficking facilitates the targeting of Ras and Rho GTPases (41, 42). The unique property of the interaction between LC/A and SNAP-25 was that the interaction also enhanced the rate of substrate cleavage.
This study provides insight toward understanding the mechanism of action of LC/A inside the neuron. These findings help explain the potency of BoNT/A by utilizing the substrate, SNAP-25, as an intracellular receptor to increase the efficiency of substrate cleavage. This is a layer of sophistication of BoNT action that has not been previously appreciated. Understanding the intracellular localization and substrate recognition of LC/A may provide insight into designing new therapeutics to neutralize BoNT intoxications.
Acknowledgment
We acknowledge John Leszyk, Core Laboratory for Proteomic Mass Spectrometry, University of Massachusetts Medical School (Worcester, MA), for LC-MS/MS analysis.
This work was supported, in whole or in part, by National Institutes of Health Grant U54 AI057153 through the Great Lakes Regional Center of Excellence (to J. T. B.).
- BoNT
- botulinum neurotoxin
- LC
- light chain
- SV
- synaptic vesicle.
REFERENCES
- 1. Montecucco C., Schiavo G. (1994) Mol. Microbiol. 13, 1–8 [DOI] [PubMed] [Google Scholar]
- 2. Gill D. M. (1982) Microbiol. Rev. 46, 86–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhidayasiri R., Tarsy D. (2006) Expert Rev. Neurother. 6, 863–886 [DOI] [PubMed] [Google Scholar]
- 4. Carruthers A., Carruthers J. (2008) Skin Therapy Lett. 13, 1–4 [PubMed] [Google Scholar]
- 5. Mahajan S. T., Brubaker L. (2007) Am. J. Obstet. Gynecol. 196, 7–15 [DOI] [PubMed] [Google Scholar]
- 6. Josko D. (2004) Clin. Lab. Sci. 17, 30–34 [PubMed] [Google Scholar]
- 7. Bossi P., Bricaire F. (2003) Presse Med. 32, 463–465 [PubMed] [Google Scholar]
- 8. Turton K., Chaddock J. A., Acharya K. R. (2002) Trends Biochem. Sci. 27, 552–558 [DOI] [PubMed] [Google Scholar]
- 9. Dolly J. O., Aoki K. R. (2006) Eur. J. Neurol. 13, Suppl. 4, 1–9 [DOI] [PubMed] [Google Scholar]
- 10. Schiavo G., Santucci A., Dasgupta B. R., Mehta P. P., Jontes J., Benfenati F., Wilson M. C., Montecucco C. (1993) FEBS Lett. 335, 99–103 [DOI] [PubMed] [Google Scholar]
- 11. Rossetto O., Seveso M., Caccin P., Montecucco C. (2002) Curr. Probl. Dermatol. 30, 117–125 [DOI] [PubMed] [Google Scholar]
- 12. Jahn R., Scheller R. H. (2006) Nat. Rev. Mol. Cell Biol. 7, 631–643 [DOI] [PubMed] [Google Scholar]
- 13. Chen S., Barbieri J. T. (2006) J. Biol. Chem. 281, 10906–10911 [DOI] [PubMed] [Google Scholar]
- 14. Breidenbach M. A., Brunger A. T. (2004) Nature 432, 925–929 [DOI] [PubMed] [Google Scholar]
- 15. Chen S., Kim J. J., Barbieri J. T. (2007) J. Biol. Chem. 282, 9621–9627 [DOI] [PubMed] [Google Scholar]
- 16. Dong M., Yeh F., Tepp W. H., Dean C., Johnson E. A., Janz R., Chapman E. R. (2006) Science 312, 592–596 [DOI] [PubMed] [Google Scholar]
- 17. Mahrhold S., Rummel A., Bigalke H., Davletov B., Binz T. (2006) FEBS Lett. 580, 2011–2014 [DOI] [PubMed] [Google Scholar]
- 18. Fischer A., Montal M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10447–10452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fischer A., Montal M. (2007) J. Biol. Chem. 282, 29604–29611 [DOI] [PubMed] [Google Scholar]
- 20. Koriazova L. K., Montal M. (2003) Nat. Struct. Biol. 10, 13–18 [DOI] [PubMed] [Google Scholar]
- 21. Fernández-Salas E., Ho H., Garay P., Steward L. E., Aoki K. R. (2004) Mov. Disord. 19, Suppl. 8, S23–S34 [DOI] [PubMed] [Google Scholar]
- 22. Fernández-Salas E., Steward L. E., Ho H., Garay P. E., Sun S. W., Gilmore M. A., Ordas J. V., Wang J., Francis J., Aoki K. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3208–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baldwin M. R., Barbieri J. T. (2007) Biochemistry 46, 3200–3210 [DOI] [PubMed] [Google Scholar]
- 24. Veit M., Söllner T. H., Rothman J. E. (1996) FEBS Lett. 385, 119–123 [DOI] [PubMed] [Google Scholar]
- 25. Bajohrs M., Rickman C., Binz T., Davletov B. (2004) EMBO Rep. 5, 1090–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Agarwal R., Binz T., Swaminathan S. (2005) Biochemistry 44, 8291–8302 [DOI] [PubMed] [Google Scholar]
- 27. Binz T., Bade S., Rummel A., Kollewe A., Alves J. (2002) Biochemistry 41, 1717–1723 [DOI] [PubMed] [Google Scholar]
- 28. Vaidyanathan V. V., Yoshino K., Jahnz M., Dörries C., Bade S., Nauenburg S., Niemann H., Binz T. (1999) J. Neurochem. 72, 327–337 [DOI] [PubMed] [Google Scholar]
- 29. Pérez-Brangulí F., Ruiz-Montasell B., Blasi J. (1999) FEBS Lett. 458, 60–64 [DOI] [PubMed] [Google Scholar]
- 30. Washbourne P., Cansino V., Mathews J. R., Graham M., Burgoyne R. D., Wilson M. C. (2001) Biochem. J. 357, 625–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. (1998) Nature 395, 347–353 [DOI] [PubMed] [Google Scholar]
- 32. Wang J., Zurawski T. H., Meng J., Lawrence G., Olango W. M., Finn D. P., Wheeler L., Dolly J. O. (2011) J. Biol. Chem. 286, 6375–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsai Y. C., Maditz R., Kuo C. L., Fishman P. S., Shoemaker C. B., Oyler G. A., Weissman A. M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 16554–16559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watson P., Spooner R. A. (2006) Adv. Drug Deliv. Rev. 58, 1581–1596 [DOI] [PubMed] [Google Scholar]
- 35. Iglesias-Bartolomé R., Trenchi A., Comín R., Moyano A. L., Nores G. A., Daniotti J. L. (2009) Biochim. Biophys. Acta. 1788, 2526–2540 [DOI] [PubMed] [Google Scholar]
- 36. Wälchli S., Skånland S. S., Gregers T. F., Lauvrak S. U., Torgersen M. L., Ying M., Kuroda S., Maturana A., Sandvig K. (2008) Mol. Biol. Cell. 19, 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salinas S., Schiavo G., Kremer E. J. (2010) Nat. Rev. Microbiol. 8, 645–655 [DOI] [PubMed] [Google Scholar]
- 38. Spooner R. A., Smith D. C., Easton A. J., Roberts L. M., Lord J. M. (2006) Virol. J. 3, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith D. C., Spooner R. A., Watson P. D., Murray J. L., Hodge T. W., Amessou M., Johannes L., Lord J. M., Roberts L. M. (2006) Traffic 7, 379–393 [DOI] [PubMed] [Google Scholar]
- 40. Jackson M. E., Simpson J. C., Girod A., Pepperkok R., Roberts L. M., Lord J. M. (1999) J. Cell Sci. 112, 467–475 [DOI] [PubMed] [Google Scholar]
- 41. Zhang Y., Deng Q., Barbieri J. T. (2007) J. Biol. Chem. 282, 13022–13032 [DOI] [PubMed] [Google Scholar]
- 42. Zhang Y., Deng Q., Porath J. A., Williams C. L., Pederson-Gulrud K. J., Barbieri J. T. (2007) Cell Microbiol. 9, 2192–2201 [DOI] [PubMed] [Google Scholar]