Abstract
Several studies have clearly established the importance of the interaction between macrophages and CX3CL1 in the progression of disease. A previous study demonstrated that Syk was required for CX3CL1-mediated actin polymerization and chemotaxis. Here, we delineated the signaling cascade of Syk-mediated cell migration in response to CX3CL1. Inhibition of Syk in bone marrow-derived macrophages or reduction of Syk expression using siRNA in RAW/LR5 cells indicated that Syk was required for the activation of PI3K, Cdc42, and Rac1. Also, reduction in WASP or WAVE2 levels, common downstream effectors of Cdc42 or Rac1, resulted in impaired cell migration to CX3CL1. Syk indirectly regulated WASP tyrosine phosphorylation through Cdc42 activation. Altogether, our data identify that Syk mediated chemotaxis toward CX3CL1 by regulating both Rac1/WAVE2 and Cdc42/WASP pathways, whereas Src family kinases were required for proper WASP tyrosine phosphorylation.
Keywords: Chemotaxis, G protein-coupled receptors (GPCR), Macrophage, Signal Transduction, Tyrosine Protein Kinase (Tyrosine Kinase)
Introduction
Chemokines were first described as chemoattractant cytokines synthesized at sites of inflammation that stimulate the directional migration of leukocytes and mediate inflammation (1, 2). Among chemokines, CX3CL1 (also called fractalkine) is the only chemokine that functions not only as a chemoattractant but also as an adhesion molecule, and it is likely that CX3CL1 is involved in the extravasation of leukocytes into inflamed tissues (2–4). Inappropriate expression or function of CX3CL1 has also been implicated in inflammatory conditions leading to vascular and tissue damage (2), and CX3CL1 has now emerged as important mediator in the pathogenesis of various clinical diseases, including rheumatoid arthritis (5–7), atherosclerosis, and cardiovascular disease (8–11).
The recruitment of monocyte/macrophages in response to CX3CL1 has been shown to significantly correlate with the extent of disease (12). The direct effects of CX3CL1 at the wound site has been tested using disruption of the receptor for CX3CL1, CX3CR1, and included marked reduction of macrophages and macrophage products such as TGF-β1 and vascular endothelial growth factor (13). Indeed, ApoE−/−CX3CR1−/− mice have significantly reduced atherosclerotic lesion size and macrophage recruitment (9). In addition, deletion of CX3CL1 in CCR2 (−/−) mice dramatically reduced macrophage accumulation in the artery wall and the subsequent development of atherosclerosis (14). These multiple studies have clearly established the importance of the interaction between macrophages and CX3CL1. However, the mechanistic details underlying macrophage to chemotaxis toward CX3CL1 are not well understood.
CX3CL1 exerts its activities upon interaction with a G protein-coupled receptor, CX3CR1, that is highly expressed on different lymphocyte populations, including monocytes, T cells, and natural killer cells (15, 16). Cambien et al. (1) have identified activation of both Src and Syk tyrosine kinases under soluble CX3CL1 stimulation in MonoMac6 cells. Also, Syk was rapidly activated and required for proper formation of membrane protrusions and cell chemotaxis in response to CX3CL1 in RAW/LR5 monocyte/macrophage cells (17). Furthermore, Stabile et al. (18) show that Wiskott-Aldrich syndrome protein (WASP)3 was associated with Fyn and Pyk-2 tyrosine kinases and that Cdc42 and WASP could play a crucial role in the regulation of natural killer cell migration by CX3CL1. These observations indicate that chemotaxis toward CX3CL1 could be regulated by the coordinate action of both Cdc42/WASP and tyrosine kinases. However, the functional role of these proteins in the signal transduction cascade downstream of CX3CL1 has not been investigated.
The RAW/LR5 monocyte/macrophage cell line has been reported to be a valid model to study CX3CL1-induced chemotaxis (17). Therefore, we investigated the signaling mechanisms required for cell migration to soluble CX3CL1 using this cell line. Here, we show that Syk mediated chemotaxis toward CX3CL1 by the activation of both Cdc42/WASP and Rac1/WAVE2 (WASP family verprolin-homologous protein 2) pathways, which are required for proper macrophage cell migration in response to CX3CL1.
EXPERIMENTAL PROCEDURES
Cells, Antibodies, Reagents
RAW/LR5 cells, derived from the murine monocyte/macrophage RAW 264.7 cell line described previously (19), were cultured in RPMI 1640 medium (Mediatech, Inc.) supplemented with 10% heat-inactivated fetal bovine serum (Sigma) and antibiotics (100 units/ml penicillin/100 μg/ml streptomycin). The Myc-tagged H246D or Y291F WASP clonal cell lines used in this study were generated previously to stably express the indicated constructs in WASPsh RAW/LR5 cells at levels comparable to noninfected cells and Ctrlsh cells (20). Murine bone marrow-derived macrophages (BMMs) were isolated as described previously (21) and were grown in α-minimum essential medium containing 15% FBS, 360 ng/ml recombinant human CSF-1 (Chiron, Emeryville, CA), and antibiotics. Recombinant mouse CX3CL1 (amino acids 25–105) was purchased from R&D Systems. Monoclonal anti-Myc, anti-HA, anti-phospho-Syk (Tyr519/520 in mouse, Tyr525/526 in human), and rabbit anti-phospho-Akt (Ser473) and anti-Akt was from Cell Signaling Technology (Beverly, MA). Rabbit anti-WASP (H250), anti-Cdc42 (SC87), anti-Syk (N19), and protein A/G plus-agarose beads were from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal anti-Rac1 (23A8) was from UpState/Millipore (Temecula, CA) and HRP mouse anti-phosphotyrosine (PY20) was from BD Transduction Laboratories. Anti-β-actin (AC-15) antibody was from Sigma. Rabbit polyclonal anti-WAVE2 antibody (22) was from Tadaomi Takenawa (University of Tokyo). Alexa Fluor 568-phalloidin and all secondary antibodies conjugated to Alexa Fluor 488 or 568 were from Molecular Probes (Eugene, OR), and secondary antibodies conjugated to HRP were from Jackson ImmunoResearch Laboratories (West Grove, PA). LY294002 was from Biomol (Plymouth Meeting, PA). Wortmannin, piceatannol, and PP2 were from Calbiochem/EMD Bioscience (Germany).
Constructs, RNA-mediated Interference, and Cell Transfection
Reduction of Cdc42, WASP, Syk, and WAVE2 expression in RAW/LR5 cells was performed using the pSUPER RNAi system from Oligoengine (Seattle, WA), as described previously (17, 20, 23, 24). In brief, the shRNA plasmids were transfected into a HEK293T-based packaging cell line, and the cell culture supernatant was used to retrovirally infect RAW/LR5 cells. Stable heterogeneous populations with reduced protein expression (for a period of at least 3 weeks) were obtained after puromycin selection (contained in the pSUPER.retro.puro plasmid). Reduction in protein expression was routinely monitored by Western blotting. Myc-tagged human Rac1N17, Rac2N17, and Myc-tagged L270P WASP were constructed as described (19, 20). HA-tagged IRSp53ΔSH3 (25) was from Jim Casanova (University of Virginia). Transient transfections were performed using SuperFect, according to the manufacturer's instructions (Qiagen, Valencia, CA).
Immunofluorescence Microscopy
Adherent RAW/LR5 cells were serum-starved for at least 3 h before stimulation. For stimulation, cells were treated with buffer with divalent cation (BWD) (20 mm HEPES, 125 mm NaCl, 5 mm KCl, 5 mm dextrose, 10 mm NaHCO3, 1 mm KH2PO4, 1 mm CaCl2, and 1 mm MgCl2, pH 7.4) in the presence or absence of CX3CL1 at 37 °C for 1 min. Cells were then fixed with 3.7% formaldehyde in BWD, permeabilized with 0.2% Triton X-100 in BWD, stained with phalloidin and the indicated antibodies, and mounted on slides using a medium containing 100 mm n-propyl gallate. Images were taken using the 60× oil/1.40 phase 3 objective of an Olympus IX71 microscope coupled to a Sensicam cooled CCD camera. For quantitation, a protrusion was defined by the presence of F-actin-rich submembranous folds. The extent of CX3CL1-induced F-actin-rich membrane protrusions (CX3CL1 ruffles) was scored using a scale of 0–3 (modified from Ref. 19), where 0 = no protrusion, 1 = protrusions in one area of the cell, 2 = protrusions in two distinct areas of the cell, and 3 = protrusions in more than two distinct areas of the cell. The protrusion index was calculated as the average protrusion score of at least 80 cells and was expressed as a percentage of control. For cells expressing Myc- and HA-tagged proteins, at least 50 cells positive for the epitope were analyzed, and the extent of protrusions was quantified and expressed as a percentage of nonexpressing cells on the same coverslip.
Chemotaxis Assay
Chemotaxis was measured using 8-μm pore size inserts (Falcon; BD Biosciences) according to the manufacturer's instructions. Briefly, the inserts were placed into 24-well plates containing RPMI 1640 in the presence or the absence of CX3CL1. Serum-starved cells (n = 500,000) were then loaded onto the inserts and incubated at 37 °C for 4 h. Cell migration was quantified by counting the number of cells that migrated through the insert (at least 10 different randomly selected fields/well) and was compared with the number of migrated cells in the absence of any stimulant (fold increase). The maximum number of migrated cells in the absence of any stimulant was <1%, and there was no difference in the basal level of migration in any of the cells tested.
Immunoprecipitation and Western Blotting
Cells were lysed in ice-cold buffer A containing 25 mm Tris, 137 mm NaCl, 1% Nonidet P-40, 2 mm EDTA, 1 mm orthovanadate, 1 mm benzamidine, 10 μg/ml aprotinin, 10 μg/ml leupeptin, pH 7.4. Whole cell lysates were either used for immunoprecipitation or mixed with 5× Laemmli buffer and boiled for 5 min. Immunoprecipitations were performed by incubation of cell lysates at 4 °C with control IgG prebound to protein A/G-agarose beads (preclear step) followed by incubation with specific antibody prebound to beads. Total cell lysates and/or immunoprecipitates were resolved by SDS-PAGE, transferred onto PVDF membranes (Immobilon-P, Millipore) and after blocking, incubated with primary antibodies overnight at 4 °C followed by secondary antibodies conjugated to HRP. Signals were visualized using the SuperSignal West Pico Chemiluminescent Substrate from Pierce, and images were acquired using a Kodak Image Station 440 in triplicate.
Cdc42 and Rac Activity Assay
GST-PBD binding assay was done essentially as described previously (26). Briefly, GST-PAK-PBD (p21-binding domain of PAK1) expressed in Escherichia coli was purified using glutathione-Sepharose beads and added to cell lysates. Bound proteins were collected by centrifugation and suspended in SDS sample buffer. Proteins were fractionated by SDS-PAGE and transferred to PVDF membranes, and the amount of precipitated Cdc42 or Rac1 was estimated by Western blotting with an anti-Cdc42 or anti-Rac1 antibody.
Data Analysis
All results were calculated as the mean ± S.E. Data were analyzed using Student's t test, and differences with a p value < 0.05 were regarded as significant. Error bars represent S.E.
RESULTS
Requirement of Syk in CX3CL1-induced Small GTPase Activity
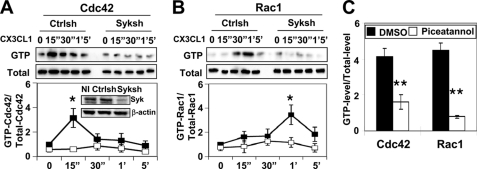
In a previously published study, reduced expression of Syk in macrophages inhibited cell migration and F-actin-enriched cell protrusions in response to CX3CL1 (17). As Syk was known to regulate Rac and Cdc42 during phagocytosis in macrophages (27, 28), we examined whether Syk regulated the activity of Cdc42 and/or Rac in response to CX3CL1 using either siRNA to reduce endogeneous levels of Syk in RAW/LR5 macrophages by 75–80% or treatment of BMMs with the Syk inhibitor piceatannol. The effect on Cdc42 or Rac activity of cells was then evaluated by specific binding of the active GTPase to PBD fused to glutathione S-transferase as described in “Experimental Procedures.” Cdc42 was rapidly and maximally activated at 15 s, whereas Rac1 was activated at 1 min upon CX3CL1 treatment in Ctrlsh cells (Fig. 1, A and B). However, CX3CL1-induced Cdc42 or Rac1 activation was abolished in Syk shRNA-treated cells (Fig. 1, A and B). Similar results were also obtained by treatment of BMMs with the Syk inhibitor piceatannol (Fig. 1C). Because macrophages express both Rac1 and Rac2, although Rac1 is the major isoform (29), the level of Rac2 activation also was examined. In contrast to Rac1 activation in response to CX3CL1, no significant increase in Rac2 activity was observed in either control infected (Ctrlsh) RAW/LR5 cells or BMMs (data not shown). Therefore, these data suggest that Syk is required for CX3CL1-induced activation of Cdc42 and Rac1 in both RAW/LR5 cells and BMMs.
FIGURE 1.
CX3CL1-induced Rho GTPases activation requires Syk. Cells were incubated with 50 ng/ml CX3CL1 for the indicated times, and Cdc42 and Rac1 activity was determined as described under “Experimental Procedures.” A, representative blot and quantification of GTP-bound Cdc42 normalized to total Cdc42 (Total) relative to control shRNA-treated (Ctrlsh) cells at time 0 are shown. The inset shows a representative blot of noninfected (NI) and puromycin-resistant stable populations of Ctrlsh or Syk shRNA-treated cells (Syksh) immunoblotted for Syk and β-actin. B, a representative blot and quantification of Rac1 activity relative to Ctrlsh cells at time 0 are shown (n = 3). Data represent mean ± S.E. *, p < 0.05 compared with Ctrlsh cells at time 0. C, BMMs pretreated with DMSO (vehicle) or 50 μm piceatannol were stimulated with CX3CL1 and Cdc42 activity at 15 s (15″) or Rac1 activity at 1 min (1′) was determined (n = 3). Data represent mean ± S.E. **, p < 0.01 compared with DMSO-treated BMMs.
Syk Is Upstream of PI3Ks in Response to CX3CL1
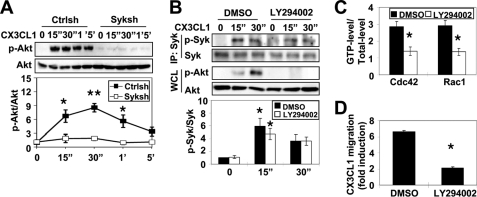
PI3Ks are important regulators of cell migration through the activation of guanine nucleotide exchange factor activity to Rac1 and Cdc42, which are required for CSF-1-elicited protrusions (19, 30, 31) and chemotaxis (32). Interestingly, Syk is known to be required for the translocalization of PI3K to the leading edge in neutrophil-like differentiated HL-60 cells (33). Therefore, we determined whether Syk was required for PI3K activation in response to CX3CL1. Ctrlsh or Syk shRNA-treated cells were tested for the ability to activate PI3K using Akt phosphorylation as a reporter of PI3K activation. As shown in Fig. 2A, CX3CL1 treatment of Ctrlsh cells demonstrated a peak of Akt phosphorylation at 30 s, which gradually decreased. However, there was no significant increase in Akt phosphorylation in response to CX3CL1 in Syk shRNA-treated cells (Fig. 2A). To confirm that Syk was activated upstream of PI3K, cells were preincubated in the presence or absence of PI3K inhibitor, LY294002, before CX3CL1 stimulation for various times and after lysis and immunoprecipitation of Syk. Syk activity was monitored using an antibody that recognizes Syk phosphorylation in the activation site. CX3CL1 treatment of DMSO-incubated cells resulted in a rapid increase in the tyrosine phosphorylation level of Syk (Fig. 2B), consistent with previously published results (17). No difference in the tyrosine phosphorylation level of Syk was observed between DMSO- and LY294002-pretreated cells (Fig. 2B), whereas inhibition of PI3K completely blocked Akt phosphorylation (Fig. 2B, middle panel). These results indicated that PI3K acted downstream of Syk; thus, we tested whether PI3K regulated CX3CL1-induced chemotaxis through the activation of Cdc42 and Rac1. Consistent with reduced expression or inhibition of Syk, CX3CL1-induced Cdc42 or Rac1 activation was abolished in LY294002-pretreated BMMs (Fig. 2C). To confirm the role of PI3K CX3CL1-induced chemotaxis RAW/LR5 cells or BMMs were pretreated with either DMSO or LY294002 and CX3CL1 mediated chemotaxis was examined. PI3K inhibition resulted in significantly reduced chemotaxis to CX3CL1 as compared with DMSO-pretreated cells (Fig. 2D and data not shown). Overall, these data suggested that PI3K is a key downstream mediator of Syk function required for macrophage migration in response to CX3CL1.
FIGURE 2.
PI3K activation by Syk is required for CX3CL1-induced Rho GTPase activation and chemotaxis. A, Ctrlsh and Syk shRNA-treated cells (Syksh) were stimulated with CX3CL1 for the indicated times, and the activation of Akt was assessed by Akt phosphorylation (p-Akt). Representative blots of p-Akt and total Akt are shown. p-Akt levels were quantified by densitometry and normalized to total Akt. Akt activity relative to Ctrlsh cells at time 0 was plotted (n = 3). Data represent mean ± S.E. *, p < 0.05 and **, p < 0.01 compared with Ctrlsh cells at time 0. B, RAW/LR5 cells were preincubated for 30 min with DMSO (vehicle) or 100 μm LY294002 prior to CX3CL1 stimulation for the indicated times. Representative blot of immunoprecipitated Syk (IP) probed for either phospho-specific Syk or Syk (upper panels). LY294002 efficacy was monitored by p-Akt levels in whole cell lysates (WCL; middle panels). Quantification of Syk activity relative to DMSO-treated cells at time 0 is shown (lower panels). C, BMMs pre-treated with DMSO (vehicle) or 100 μm LY294002 were incubated with CX3CL1 for the indicated times, and Cdc42 or Rac1 activity was determined at 15 sec (15″) or 1 min (1′), respectively, as described in the legend to Fig. 1 (n = 3). Mean ± S.E. *, p < 0.05 compared with DMSO-treated BMMs. D, RAW/LR5 cells were preincubated for 30 min with DMSO (vehicle) or 100 μm LY294002 before being subjected to a Transwell migration assay. CX3CL1-induced chemotaxis was expressed as fold induction compared with DMSO-treated cells in the absence of CX3CL1 (n = 3). Mean ± S.E. *, p < 0.05 compared with DMSO-treated cells.
WAVE2 and WASP Are Required for Macrophage Chemotaxis to CX3CL1
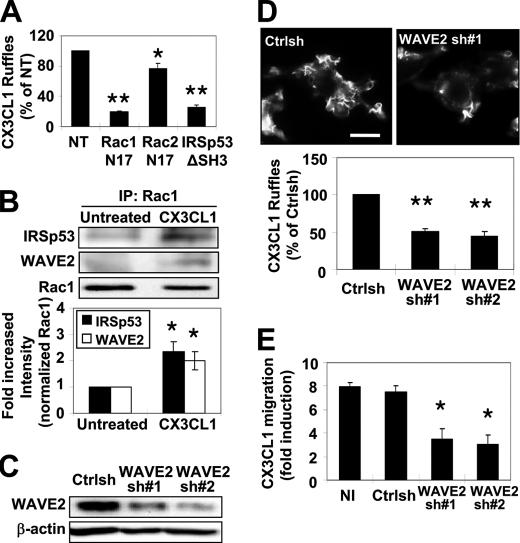
Rho GTPase-dependent assembly of F-actin has been shown to be mediated by the WASP/WAVE family of proteins where WASP proteins act downstream of the Rho GTPase Cdc42, and WAVE proteins are activated by Rac (34–36). Also, WASP and WAVE2, the predominant family members expressed in macrophages, are required for CSF-1-induced chemotaxis in macrophages (20, 24). Therefore, the possible involvement of WASP and WAVE2 in the formation of protrusions and cell migration toward CX3CL1 was investigated. We demonstrated previously that Rac indirectly interacted with WAVE2 through IRSp53 (insulin receptor substrate protein 53), and this interaction was crucial for CSF-1-mediated actin assembly and migration in macrophages (37). Thus, the role of Rac and IRSp53 on F-actin-rich membrane protrusions in response to CX3CL1 was determined. Transient transfection of dominant-negative versions of Rac1 (Rac1N17) or IRSp53 lacking the WAVE2-binding site (IRSp53ΔSH3) in RAW/LR5 cells profoundly inhibited CX3CL1-induced ruffle formation compared with nonexpressing cells on the same coverslip (Fig. 3A). However, consistent with the lack of Rac2 activation in response to CX3CL1, expression of dominant-negative Rac2 (Rac2N17) had only minor effect on ruffle formation (Fig. 3A). Also, CX3CL1-induced Rac1 activation led to the association of WAVE2 to Rac1 through IRSp53 (Fig. 3B). Thus these results suggested a possible role for WAVE2 in the CX3CL1-induced membrane protrusions and cell migration. To confirm this possibility, the effect of reduced expression of endogeneous WAVE2 on CX3CL1 chemotaxis was tested. Similar to previous results (24), retroviral delivery of WAVE2-specific shRNA (WAVE2sh#1 and WAVE2sh#2) in RAW/LR5 cells resulted in a ∼50% reduction of WAVE2 protein expression compared with Ctrlsh-treated cells (Fig. 3C). The ability of cells with reduced WAVE2 expression to exhibit F-actin-rich membrane protrusions in response to CX3CL1 was evaluated and compared with Ctrlsh-treated cells. As expected, reduction of WAVE2 expression resulted in a significant inhibition of CX3CL1-induced membrane protrusions as compared with Ctrlsh cells (Fig. 3D). The 50% reduction in ruffling was consistent with the level of WAVE2 reduction, as seen previously in the case of CSF-1 induced protrusions (24). In addition, the effect of reduced expression of WAVE2 on cell migration toward CX3CL1 was determined. An ∼8-fold increase in cell migration in response to CX3CL1 was observed in both noninfected and Ctrlsh cells (Fig. 3E). However, CX3CL1 chemotaxis was impaired significantly in WAVE2 shRNA-treated cells as compared with both controls (Fig. 3E). Therefore, WAVE2 was required for F-actin membrane protrusions and macrophage migration toward CX3CL1.
FIGURE 3.
WAVE2 activation by Rac1 through IRSp53 is required for ruffle formation and cell migration in response to CX3CL1. A, RAW/LR5 cells expressing dominant-negative Myc-tagged Rac1 (Rac1N17), Myc-tagged Rac2 (Rac2N17), or HA-tagged IRSp53 deficient in WAVE2 binding (IRSp53ΔSH3) were stimulated with CX3CL1 for 1 min. Cells expressing the constructs were identified by epitope staining (Myc for Rac1/2 and HA for IRSp53) and F-actin-rich protrusions (ruffles) were visualized by Alexa Fluor 568 phalloidin staining. The number of CX3CL1-elicited protrusions (CX3CL1 ruffles) was quantified as described under “Experimental Procedures” and expressed as a percentage of the CX3CL1 stimulation observed in nontransfected cells (NT) on the same coverslip (n = 3). Mean ± S.E. *, p < 0.05 and **, p < 0.01 compared with nontransfected cells. B, RAW/LR5 cells were either left untreated or subjected to CX3CL1 stimulation for 1 min prior to lysis and immunoprecipitation (IP) of Rac1 followed by Western blotting with the indicated antibodies. A representative blot is shown with quantification of IRSp53 (black bar) or WAVE2 (white bar) co-immunoprecipitated by Rac (n = 3). Mean ± S.E. *, p < 0.05 compared with untreated cells. C, a representative Western blot of WAVE2 and β-actin expression in RAW/LR5 cells infected with either control sequence (Ctrlsh) or two different shRNA sequences targeting WAVE2 (sh#1 and sh#2). D, representative images of Ctrlsh or WAVE2 shRNA-treated cells stimulated with CX3CL1 for 1 min and stained for F-actin with Alexa Fluor 568 phalloidin staining (upper panels). The number of CX3CL1-elicited protrusions was quantified as described in A and expressed as a percentage of the CX3CL1 stimulation observed in Ctrlsh-treated cells (lower panel) (n = 3). Mean ± S.E. **, p < 0.01 compared with Ctrlsh-treated cells. Scale bar = 10 μm. E, CX3CL1 chemotaxis of the indicated cell lines was determined using a Transwell assay and expressed as fold induction compared with the corresponding condition in the absence of CX3CL1 (n = 3). Mean ± S.E. *, p < 0.05 compared with noninfected (NI) cells.
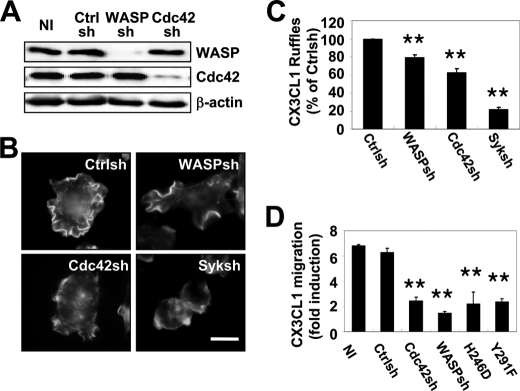
We have shown previously that Cdc42 was required for WASP-dependent chemotaxis to CSF-1 (20), and WASP is required for chemotaxis to several different chemoattractants (20, 38, 39). Therefore, the role of Cdc42 in regulating WASP during CX3CL1 chemotaxis was examined. More than 90% WASP reduction using shRNA had only a minor effect on F-actin-rich membrane protrusions in response to CX3CL1, whereas a 75% reduction in Cdc42 levels had a more dramatic effect (Fig. 4, A–C). However, both Cdc42 and WASP were required for CX3CL1 chemotaxis (Fig. 4D). To confirm that Cdc42 was required for WASP function, the ability of WASPsh cells stably expressing WASP containing a point mutation that abolishes Cdc42 binding (H246D) (20, 23) to chemotax toward CX3CL1 was tested. As with WASP or Cdc42 shRNA-treated cells, reduced migration toward CX3CL1 also was observed in shWASP cells expressing WASP H246D (Fig. 4D). These data suggested that the interaction between Cdc42 and WASP was required for macrophage migration toward CX3CL1. Because tyrosine phosphorylation of residue 291 of WASP affected actin polymerization rates, phagocytosis and CSF-1 mediated chemotaxis in macrophages (20, 23, 40, 41), WASP bearing a point mutation in Tyr291 to abolish phosphorylation (Y291F) was introduced to the WASP-silenced cells (20, 23). Interestingly, Y291F expressing cells also showed significantly reduced migration toward CX3CL1, comparable to H246D mutants (Fig. 4D). These data suggest that both Cdc42 activation and WASP phosphorylation were required for migration toward CX3CL1.
FIGURE 4.
Cdc42 binding and tyrosine phosphorylation of WASP are required for CX3CL1-induced cell migration. A, RAW/LR5 cells were infected with a retrovirus bearing a control sequence (Ctrlsh) or a sequence against WASP (WASPsh) or Cdc42 (Cdc42sh). A representative blot of lysates from noninfected (NI) and puromycin-resistant stable populations of infected cells immunoblotted against WASP, Cdc42, and β-actin are shown (n = 5). B, Ctrlsh, WASPsh, Cdc42sh, and Syk shRNA-treated (Syksh) cells were stimulated with CX3CL1 for 1 min, and ruffles were visualized by Alexa Fluor 568 phalloidin staining. Representative images of three independent experiments are shown. Scale bar, 10 μm. C, the number of CX3CL1-elicited protrusions (CX3CL1 ruffles) was expressed as a percentage of the CX3CL1 stimulation observed in Ctrlsh cells (n = 3). Mean ± S.E. **, p < 0.01 compared with Ctrlsh cells. D, chemotactic ability of the indicated cells was determined using a Transwell assay. In addition, clones of WASPsh cells expressing human Myc-tagged WASP deficient in Cdc42 binding (H246D) or phosphorylation-deficient WASP (Y291F; Ref. 20) were tested. Data were expressed as fold induction compared with the corresponding condition in the absence of CX3CL1 (n = 3). Mean ± S.E. **, p < 0.01 compared with CX3CL1-induced migration in noninfected (NI) cells.
Syk Regulates Tyrosine Phosphorylation of WASP through Cdc42 Activation
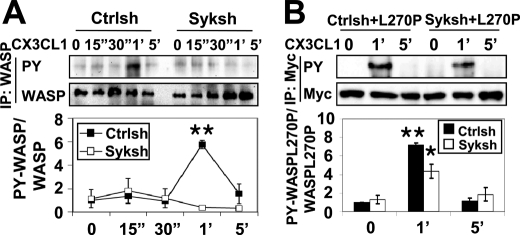
Although Syk is required for Cdc42 activation in response to CX3CL1 (Fig. 1), Syk may also be required for WASP phosphorylation because Syk has been shown to exist in a complex with WASP through the CrkL adaptor protein (42). To test whether Syk was required for WASP tyrosine phosphorylation, WASP was immunoprecipitated after CX3CL1 treatment in Ctrlsh or Syk shRNA-treated cells, and then the level of WASP phosphorylation was determined. Ctrlsh cells showed increased WASP phosphorylation that peaked at 1 min, whereas no WASP phosphorylation was detected Syk shRNA-treated cells (Fig. 5A).
FIGURE 5.
Syk regulates tyrosine phosphorylation of WASP through Cdc42 activation. A, control (Ctrlsh) and Syk shRNA-treated cells (Syksh) were stimulated with CX3CL1 for the indicated times. WASP was immunoprecipitated (IP) using WASP antibody followed by Western blotting for phosphotyrosine (PY) and WASP. Blots were quantified by densitometry and normalized to total WASP. A representative blot and quantification of WASP tyrosine phosphorylation relative to Ctrlsh cells at time 0 are shown (n = 3). Mean ± S.E. **, p < 0.01 compared with Ctrlsh cells at time 0. B, Ctrlsh or Syk shRNA-treated cells were transiently transfected with Myc-tagged constitutively active WASP (L270P) and then stimulated with CX3CL1 for the indicated times. Myc-tagged L270P WASP was immunoprecipitated using Myc antibody following by Western blotting for phosphotyrosine and Myc. Blots were quantified by densitometry and normalized to Myc. A representative blot and quantification of L270P WASP tyrosine phosphorylation relative to Ctrlsh cells at time 0 are shown (n = 3). Mean ± S.E. *, p < 0.05 and **, p < 0.01 compared with Ctrlsh cells at time 0. 15″, 15 sec; 1′, 1 min.
It has been demonstrated that Cdc42 binding to WASP was required for tyrosine phosphorylation of WASP in vitro (43) and in vivo (23). Therefore, defect of tyrosine phosphorylation of WASP in Syk shRNA-treated cells might be due to the inactivation of Cdc42 by reduced Syk expression (Fig. 1). We have demonstrated previously that an open, active form of WASP (L270P) can be tyrosine phosphorylated in a Cdc42-independent manner (23), and when this form of WASP was expressed in Syk shRNA-treated cells, a significant restoration of WASP phosphorylation by CX3CL1 treatment was observed (Fig. 5B). Overall, these data indicated that Syk was required for Cdc42 activation in response to CX3CL1 and does not directly phosphorylate WASP.
WASP Is Tyrosine-phosphorylated by SFKs
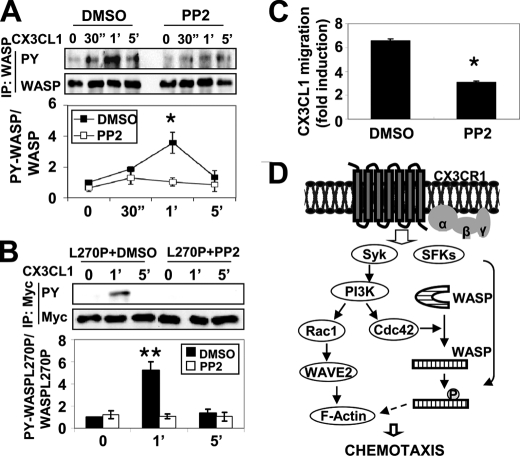
Both Syk and Src family kinases (SFKs) are activated downstream of CX3CL1 stimulation in human monocytic cells (1). Because Syk was not required to directly phosphorylate WASP (Fig. 5B), we determined whether SFKs played a role in tyrosine phosphorylation of WASP. No difference was observed in the ability of DMSO-treated cells to tyrosine-phosphorylate WASP in response to CX3CL1, whereas WASP tyrosine phosphorylation was abolished when either RAW/LR5 cells or BMMs were pretreated with the SFKs inhibitor PP2 or a structurally unrelated pan-SFK inhibitor SU6656 (Fig. 6A and supplemental Fig. S1). Also, PP2 inhibited WASP phosphorylation in cells expressing L270P WASP (Fig. 6B), suggesting that SFKs were acting independently of Cdc42 activation. SFKs have been shown to act upstream of Syk in FcγR-mediated phagocytosis (44, 45), suggesting that SFKs also might regulate Syk-mediated signaling pathways. Contrary to this, inhibition of SFKs using PP2 or SU6656 did not result in any reduction in the activation of Cdc42, Rac1, and Akt in BMMs (supplemental Fig. S2). Finally, we analyzed the chemotactic ability of cells following the inhibition of SFKs. PP2 treatment of RAW/LR5 cells and BMMs inhibited CX3CL1 chemotaxis compared with DMSO-treated cells (Fig. 6C and data not shown), consistent with the results obtained when nonphosphorylated WASP (Y291F) was expressed (Fig. 4D). Therefore, these data indicated that Syk-mediated signaling pathways in response to CX3CL1 are independent of SFKs; however, SFKs are required for tyrosine phosphorylation of WASP- and CX3CL1-induced cell migration in macrophages.
FIGURE 6.
SFKs regulate WASP tyrosine phosphorylation. A, RAW/LR5 cells were pretreated for 1 h with DMSO (vehicle) or 10 μm PP2 prior to CX3CL1 stimulation for the indicated times. WASP was immunoprecipitated (IP) followed by Western blotting for phosphotyrosine (PY) and WASP. Blots were quantified by densitometry and normalized to amount of WASP. A representative blot and quantification of WASP tyrosine phosphorylation relative to DMSO-treated cells at time 0 are shown (n = 3). Mean ± S.E. *, p < 0.05 compared with DMSO-treated cells at time 0. B, RAW/LR5 cells expressing Myc-L270P WASP were preincubated with DMSO (vehicle) or 10 μm PP2 prior to CX3CL1 stimulation for the indicated times. Myc-tagged L270P WASP was immunoprecipitated using Myc antibody followed by Western blotting for phosphotyrosine and Myc. Blots were quantified by densitometry and normalized to Myc. A representative blot and quantification of L270P WASP tyrosine phosphorylation relative to DMSO-treated cells at time 0 are shown (n = 3). Mean ± S.E. **, p < 0.01 compared with DMSO-treated cells at time 0. C, RAW/LR5 cells were preincubated for 1 h with DMSO (vehicle) or 10 μm PP2 before being subjected to a Transwell migration assay. CX3CL1-induced chemotaxis was expressed as fold induction compared with DMSO-treated cells in the absence of CX3CL1 (n = 3). Mean ± S.E. *, p < 0.05 compared with DMSO-treated cells. D, model of Syk-mediated signal pathways in response to CX3CL1. Syk and SFKs are activated independently by CX3CL1, leading to efficient chemotaxis. Syk regulates both Cdc42/WASP and Rac1/WAVE2 pathways through activation of PI3K. Syk is required to activate Cdc42, which induces a conformational change in WASP permitting phosphorylation by SFKs (see “Discussion” for details). 30″, 30 sec; 1′, 1 min.
Taken together, our data identify a CX3CL1-induced signal pathway where Syk and PI3K are required to activate Cdc42, which induces a conformational change in WASP that permits phosphorylation by SFKs (Fig. 6D). These results indicate that both Syk and SFKs participate in different, but essential steps required for proper macrophage chemotaxis to CX3CL1.
DISCUSSION
G protein-coupled receptors activate tyrosine kinases as well as classical heterotrimeric G protein second messenger pathways (46). In this study, we demonstrate a role for the tyrosine kinases Syk and SFKs in CX3CL1-induced macrophage chemotaxis. Previous reports using monocyte/macrophage cell lines demonstrated that Syk and SFKs were activated independently of each other in response to CX3CL1 (1), and Syk activity was required for actin cytoskeletal reorganization and migration to CX3CL1 (17). Here, we provide evidence for the molecular mechanism by which Syk regulates CX3CL1-induced chemotaxis in macrophages. Our results clearly indicated that Syk regulates both Cdc42/WASP and Rac1/WAVE2 pathways in response to CX3CL1 and that these pathways are required for efficient chemotaxis (Fig. 6D).
Syk is highly expressed in cells of hematopoietic origin and plays a crucial role in signaling in most of these cells (47, 48). Syk is known to play a role in actin-dependent processes, such as integrin-mediated signaling (49, 50) and FcγR-mediated phagocytosis (51, 52). Syk is activated downstream of G protein-coupled receptors in B cells, T cells, L1.1 cells, mast cells, and monocyte/macrophages (1, 53–55). Syk has been shown to be required for G protein-coupled receptor-mediated chemotaxis using the Syk inhibitor piceatannol or RNAi approaches with HL60 differentiated neutrophils, THP-1 monocytes or RAW/LR5 monocyte/macrophages (17, 33). However, the role of Syk in neutrophil chemotaxis is controversial (56). Although an earlier study of Syk-deficient neutrophils from bone marrow chimeras did not show any defect in chemotaxis to LTB4, C5a, MIP-1α, MIP-2, and fMLP (57), a later study demonstrated reduced chemotaxis in vitro and extravasation in vivo in response to fMLP in the absence of Syk (58). However, the results from these neutrophil studies may be complicated because the former study used uncoated surfaces and the latter used a fibrinogen-coated surface, and Syk is important for adhesion via β2 integrins (59). Interestingly, one of the molecules identified in this study as being downstream of Syk, WASP, is required for monocyte but not for neutrophil chemotaxis to fMLP (38, 60), suggesting that the signaling requirements may be different between monocyte/macrophages and neutrophils.
Similar to the results shown here for CX3CL1, WAVE2 and WASP are known downstream effectors of Rac and Cdc42 that are required for actin remodeling and chemotaxis in macrophages in response to CSF-1 (20, 24, 30, 32). WAVE2 does not bind directly to Rac and IRSp53 was reported to be an important candidate linking WAVE2 to Rac1 (61, 62). Consistent with IRSp53 in mediating the interaction between Rac and WAVE2, WAVE2 was present in a complex with Rac1 and IRSp53 in response to CX3CL1 and disruption of this interaction using a dominant-negative form of IRSp53-inhibited membrane protrusions in response CX3CL1. These results in combination with a previous study demonstrating the importance of IRSp53 in CSF-1 chemotaxis suggest that IRSp53 plays a vital role in macrophage chemotaxis (37).
WASP also is critical for leukocyte migration in vitro and in vivo (63), supporting the role of the Cdc42/WASP pathway as a key downstream mediator of several receptors in leukocytes migration. Cdc42 is required to relieve the autoinhibition of WASP necessary for subsequent WASP tyrosine phosphorylation both in vitro and in cells (23, 64, 65). In the case of CX3CL1, Syk was required for WASP phosphorylation and migration toward CX3CL1. However, Syk did not directly phosphorylate WASP. Instead, Syk was needed to induce an open conformation of WASP through Cdc42, and SFK activity was required to directly phosphorylate WASP. Interestingly, the Src kinase Hck can phosphorylate WASP at tyrosine 291 and this phosphorylation results in enhanced actin polymerization in vitro (40), suggesting that Hck may be responsible for the phosphorylation of WASP. However, WASP can associate with both the Src kinase Fyn and with focal adhesion kinase Pyk-2 following the triggering of chemokine receptors on NK cells (18). Further studies are required to identify the specific tyrosine kinase required for WASP phosphorylation during CX3CL1chemotaxis.
SFKs have been shown to act as upstream molecules of Syk in FcγR-mediated phagocytosis signaling (44, 45) However, CX3CL1 induces activation of SFKs and Syk independently of each other (1). Similar to this, we demonstrated that SFKs are not required for Syk-mediated PI3K and Rho GTPases activation and suggested that SFKs and Syk might have specific and complex functions during chemotaxis. Interestingly, WASP was required for chemotaxis but was not necessary for the formation of F-actin ruffles in response to CSF-1, whereas reduction of WAVE2 expression resulted in a significant inhibition of CSF-1-induced membrane protrusions (20, 24). Similar to this, reduction of WAVE2 expression was associated with an impaired ability of CX3CL1-induced F-actin-rich membrane protrusions compared with WASP and Cdc42. These observations suggested that F-actin-rich membrane protrusions are regulated mainly by the Rac1/WAVE2 pathway. Despite the minor effect on F-actin ruffling by WASP and Cdc42, silencing of these proteins in RAW/LR5 cells dramatically reduced migration in response to CX3CL1. WASP also is required for the formation of F-actin-rich adhesion structure called podosomes (66), and podosomes are commonly formed at the leading edge of a migrating cell, thereby defining the direction of motility (30). Other studies have demonstrated that WASP phosphorylation regulates podosome turnover as well as chemotaxis to CSF-1 (20). Consistent with this, WASP phosphorylation was also required for CX3CL1 chemotaxis. However, further studies are needed to determine the precise role by which Cdc42/WASP regulates CX3CL1-induced chemotaxis.
Supplementary Material
Acknowledgments
We thank members of the Condeelis and Segall laboratories for helpful discussions and Michael Cammer (New York University) for assistance with figure preparation and editing.
This work was supported, in whole or in part, by National Institutes of Health Grant GM071828 (to D. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- WASP
- Wiskott-Aldrich syndrome protein
- Ctrlsh
- control shRNA-treated
- SFK
- Src-family kinase
- BMM
- bone marrow-derived macrophage
- DMSO
- dimethyl sulfoxide
- fMLP
- formylmethionylleucylphenylalanine
- WASPsh
- WASP shRNA-treated
- WAVE2
- WASP family verprolin-homologous protein 2
- PBD
- p21-binding domain of PAK1
- IRSp53
- insulin receptor substrate protein 53.
REFERENCES
- 1. Cambien B., Pomeranz M., Schmid-Antomarchi H., Millet M. A., Breittmayer V., Rossi B., Schmid-Alliana A. (2001) Blood 97, 2031–2037 [DOI] [PubMed] [Google Scholar]
- 2. Umehara H., Bloom E. T., Okazaki T., Nagano Y., Yoshie O., Imai T. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 34–40 [DOI] [PubMed] [Google Scholar]
- 3. Imai T., Hieshima K., Haskell C., Baba M., Nagira M., Nishimura M., Kakizaki M., Takagi S., Nomiyama H., Schall T. J., Yoshie O. (1997) Cell 91, 521–530 [DOI] [PubMed] [Google Scholar]
- 4. Goda S., Imai T., Yoshie O., Yoneda O., Inoue H., Nagano Y., Okazaki T., Imai H., Bloom E. T., Domae N., Umehara H. (2000) J. Immunol. 164, 4313–4320 [DOI] [PubMed] [Google Scholar]
- 5. Ruth J. H., Volin M. V., Haines G. K., 3rd, Woodruff D. C., Katschke K. J., Jr., Woods J. M., Park C. C., Morel J. C., Koch A. E. (2001) Arthritis Rheum. 44, 1568–1581 [DOI] [PubMed] [Google Scholar]
- 6. Nanki T., Imai T., Nagasaka K., Urasaki Y., Nonomura Y., Taniguchi K., Hayashida K., Hasegawa J., Yoshie O., Miyasaka N. (2002) Arthritis Rheum. 46, 2878–2883 [DOI] [PubMed] [Google Scholar]
- 7. Sawai H., Park Y. W., Roberson J., Imai T., Goronzy J. J., Weyand C. M. (2005) Arthritis Rheum. 52, 1392–1401 [DOI] [PubMed] [Google Scholar]
- 8. Lesnik P., Haskell C. A., Charo I. F. (2003) J. Clin. Invest. 111, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Combadière C., Potteaux S., Gao J. L., Esposito B., Casanova S., Lee E. J., Debré P., Tedgui A., Murphy P. M., Mallat Z. (2003) Circulation 107, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 10. Moatti D., Faure S., Fumeron F., Amara Mel-W., Seknadji P., McDermott D. H., Debré P., Aumont M. C., Murphy P. M., de Prost D., Combadière C. (2001) Blood 97, 1925–1928 [DOI] [PubMed] [Google Scholar]
- 11. McDermott D. H., Halcox J. P., Schenke W. H., Waclawiw M. A., Merrell M. N., Epstein N., Quyyumi A. A., Murphy P. M. (2001) Circ. Res. 89, 401–407 [DOI] [PubMed] [Google Scholar]
- 12. Truman L. A., Ford C. A., Pasikowska M., Pound J. D., Wilkinson S. J., Dumitriu I. E., Melville L., Melrose L. A., Ogden C. A., Nibbs R., Graham G., Combadiere C., Gregory C. D. (2008) Blood 112, 5026–5036 [DOI] [PubMed] [Google Scholar]
- 13. Ishida Y., Gao J. L., Murphy P. M. (2008) J. Immunol. 180, 569–579 [DOI] [PubMed] [Google Scholar]
- 14. Saederup N., Chan L., Lira S. A., Charo I. F. (2008) Circulation 117, 1642–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raffaghello L., Cocco C., Corrias M. V., Airoldi I., Pistoia V. (2009) Semin. Cancer Biol. 19, 97–102 [DOI] [PubMed] [Google Scholar]
- 16. Fraticelli P., Sironi M., Bianchi G., D'Ambrosio D., Albanesi C., Stoppacciaro A., Chieppa M., Allavena P., Ruco L., Girolomoni G., Sinigaglia F., Vecchi A., Mantovani A. (2001) J. Clin. Invest. 107, 1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gevrey J. C., Isaac B. M., Cox D. (2005) J. Immunol. 175, 3737–3745 [DOI] [PubMed] [Google Scholar]
- 18. Stabile H., Carlino C., Mazza C., Giliani S., Morrone S., Notarangelo L. D., Notarangelo L. D., Santoni A., Gismondi A. (2010) Blood 115, 2818–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cox D., Chang P., Zhang Q., Reddy P. G., Bokoch G. M., Greenberg S. (1997) J. Exp. Med. 186, 1487–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dovas A., Gevrey J. C., Grossi A., Park H., Abou-Kheir W., Cox D. (2009) J. Cell Sci. 122, 3873–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stanley E. R. (1997) Methods Mol. Biol. 75, 301–304 [DOI] [PubMed] [Google Scholar]
- 22. Yamazaki D., Suetsugu S., Miki H., Kataoka Y., Nishikawa S., Fujiwara T., Yoshida N., Takenawa T. (2003) Nature 424, 452–456 [DOI] [PubMed] [Google Scholar]
- 23. Park H., Cox D. (2009) Mol. Biol. Cell 20, 4500–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kheir W. A., Gevrey J. C., Yamaguchi H., Isaac B., Cox D. (2005) J. Cell Sci. 118, 5369–5379 [DOI] [PubMed] [Google Scholar]
- 25. Shi J., Scita G., Casanova J. E. (2005) J. Biol. Chem. 280, 29849–29855 [DOI] [PubMed] [Google Scholar]
- 26. Schmitz U., Thömmes K., Beier I., Wagner W., Sachinidis A., Düsing R., Vetter H. (2001) J. Biol. Chem. 276, 22003–22010 [DOI] [PubMed] [Google Scholar]
- 27. Greenberg S. (1999) J. Leukoc. Biol. 66, 712–717 [DOI] [PubMed] [Google Scholar]
- 28. Cox D., Greenberg S. (2001) Semin. Immunol. 13, 339–345 [DOI] [PubMed] [Google Scholar]
- 29. Wells C. M., Walmsley M., Ooi S., Tybulewicz V., Ridley A. J. (2004) J. Cell Sci. 117, 1259–1268 [DOI] [PubMed] [Google Scholar]
- 30. Jones G. E., Zicha D., Dunn G. A., Blundell M., Thrasher A. (2002) Int. J. Biochem. Cell Biol. 34, 806–815 [DOI] [PubMed] [Google Scholar]
- 31. Allen W. E., Jones G. E., Pollard J. W., Ridley A. J. (1997) J. Cell Sci. 110, 707–720 [DOI] [PubMed] [Google Scholar]
- 32. Allen W. E., Zicha D., Ridley A. J., Jones G. E. (1998) J. Cell Biol. 141, 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schymeinsky J., Then C., Sindrilaru A., Gerstl R., Jakus Z., Tybulewicz V. L., Scharffetter-Kochanek K., Walzog B. (2007) PLoS One 2, e1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Millard T. H., Sharp S. J., Machesky L. M. (2004) Biochem. J. 380, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takenawa T., Miki H. (2001) J. Cell Sci. 114, 1801–1809 [DOI] [PubMed] [Google Scholar]
- 36. Miki H., Suetsugu S., Takenawa T. (1998) EMBO J. 17, 6932–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abou-Kheir W., Isaac B., Yamaguchi H., Cox D. (2008) J. Cell Sci. 121, 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zicha D., Allen W. E., Brickell P. M., Kinnon C., Dunn G. A., Jones G. E., Thrasher A. J. (1998) Br. J. Haematol. 101, 659–665 [DOI] [PubMed] [Google Scholar]
- 39. Haddad E., Zugaza J. L., Louache F., Debili N., Crouin C., Schwarz K., Fischer A., Vainchenker W., Bertoglio J. (2001) Blood 97, 33–38 [DOI] [PubMed] [Google Scholar]
- 40. Cory G. O., Garg R., Cramer R., Ridley A. J. (2002) J. Biol. Chem. 277, 45115–45121 [DOI] [PubMed] [Google Scholar]
- 41. Tsuboi S., Meerloo J. (2007) J. Biol. Chem. 282, 34194–34203 [DOI] [PubMed] [Google Scholar]
- 42. Oda A., Ochs H. D., Lasky L. A., Spencer S., Ozaki K., Fujihara M., Handa M., Ikebuchi K., Ikeda H. (2001) Blood 97, 2633–2639 [DOI] [PubMed] [Google Scholar]
- 43. Torres E., Rosen M. K. (2006) J. Biol. Chem. 281, 3513–3520 [DOI] [PubMed] [Google Scholar]
- 44. Crowley M. T., Costello P. S., Fitzer-Attas C. J., Turner M., Meng F., Lowell C., Tybulewicz V. L., DeFranco A. L. (1997) J. Exp. Med. 186, 1027–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Allen L. A., Aderem A. (1996) Curr. Opin. Immunol. 8, 36–40 [DOI] [PubMed] [Google Scholar]
- 46. Hall A. (1994) Annu. Rev. Cell Biol. 10, 31–54 [DOI] [PubMed] [Google Scholar]
- 47. Turner M., Schweighoffer E., Colucci F., Di Santo J. P., Tybulewicz V. L. (2000) Immunol Today 21, 148–154 [DOI] [PubMed] [Google Scholar]
- 48. Navara C. S. (2004) Curr. Pharm. Des. 10, 1739–1744 [DOI] [PubMed] [Google Scholar]
- 49. Clark E. A., Shattil S. J., Ginsberg M. H., Bolen J., Brugge J. S. (1994) J. Biol. Chem. 269, 28859–28864 [PubMed] [Google Scholar]
- 50. Suen P. W., Ilic D., Caveggion E., Berton G., Damsky C. H., Lowell C. A. (1999) J. Cell Sci. 112, 4067–4078 [DOI] [PubMed] [Google Scholar]
- 51. Greenberg S., Chang P., Wang D. C., Xavier R., Seed B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1103–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Strzelecka A., Pyrzyńska B., Kwiatkowska K., Sobota A. (1997) Cell Motil. Cytoskeleton 38, 287–296 [DOI] [PubMed] [Google Scholar]
- 53. Wan Y., Kurosaki T., Huang X. Y. (1996) Nature 380, 541–544 [DOI] [PubMed] [Google Scholar]
- 54. Ganju R. K., Brubaker S. A., Chernock R. D., Avraham S., Groopman J. E. (2000) J. Biol. Chem. 275, 17263–17268 [DOI] [PubMed] [Google Scholar]
- 55. Shefler I., Sagi-Eisenberg R. (2001) J. Immunol. 167, 475–481 [DOI] [PubMed] [Google Scholar]
- 56. Mocsai A., Ruland J., Tybulewicz V. L. Nat. Rev. Immunol. 10, 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mócsai A., Zhang H., Jakus Z., Kitaura J., Kawakami T., Lowell C. A. (2003) Blood 101, 4155–4163 [DOI] [PubMed] [Google Scholar]
- 58. Schymeinsky J., Sindrilaru A., Frommhold D., Sperandio M., Gerstl R., Then C., Mócsai A., Scharffetter-Kochanek K., Walzog B. (2006) Blood 108, 3919–3927 [DOI] [PubMed] [Google Scholar]
- 59. Mócsai A., Zhou M., Meng F., Tybulewicz V. L., Lowell C. A. (2002) Immunity 16, 547–558 [DOI] [PubMed] [Google Scholar]
- 60. Badolato R., Sozzani S., Malacarne F., Bresciani S., Fiorini M., Borsatti A., Albertini A., Mantovani A., Ugazio A. G., Notarangelo L. D. (1998) J. Immunol. 161, 1026–1033 [PubMed] [Google Scholar]
- 61. Miki H., Takenawa T. (2002) Biochem. Biophys. Res. Commun. 293, 93–99 [DOI] [PubMed] [Google Scholar]
- 62. Oda A., Miki H., Wada I., Yamaguchi H., Yamazaki D., Suetsugu S., Nakajima M., Nakayama A., Okawa K., Miyazaki H., Matsuno K., Ochs H. D., Machesky L. M., Fujita H., Takenawa T. (2005) Blood 105, 3141–3148 [DOI] [PubMed] [Google Scholar]
- 63. Snapper S. B., Meelu P., Nguyen D., Stockton B. M., Bozza P., Alt F. W., Rosen F. S., von Andrian U. H., Klein C. (2005) J. Leukoc. Biol. 77, 993–998 [DOI] [PubMed] [Google Scholar]
- 64. Torres E., Rosen M. K. (2003) Mol. Cell 11, 1215–1227 [DOI] [PubMed] [Google Scholar]
- 65. Cammer M., Gevrey J. C., Lorenz M., Dovas A., Condeelis J., Cox D. (2009) J. Biol. Chem. 284, 23302–23311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Linder S., Nelson D., Weiss M., Aepfelbacher M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9648–9653 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.