Abstract
Redoxins are involved in maintenance of thiol redox homeostasis, but their exact sites of action are only partly known. We have applied a combined redox proteomics and transcriptomics experimental strategy to discover specific functions of two interacting redoxins: dually localized glutaredoxin 2 (Grx2p) and mitochondrial peroxiredoxin 1 (Prx1p). We have identified 139 proteins showing differential postranslational thiol redox modifications when the cells do not express Grx2p, Prx1p, or both and have mapped the precise cysteines involved in each case. Some of these modifications constitute functional switches that affect metabolic and signaling pathways as the primary effect, leading to gene transcription remodeling as the secondary adaptive effect as demonstrated by a parallel high throughput gene expression analysis. The results suggest that in the absence of Grx2p, the metabolic flow toward nucleotide and aromatic amino acid biosynthesis is slowed down by redox modification of the key enzymes Rpe1p (d-ribulose-5-phosphate 3-epimerase), Tkl1p (transketolase) and Aro4p (3-deoxy-d-arabino-heptulosonate-7-phosphate synthase). The glycolytic mainstream is then diverted toward carbohydrate storage by induction of trehalose and glycogen biosynthesis genes. Porphyrin biosynthesis may also be compromised by inactivation of the redox-sensitive cytosolic enzymes Hem12p (uroporphyrinogen decarboxylase) and Sam1p (S-adenosyl methionine synthetase) and a battery of respiratory genes sensitive to low heme levels are induced. Genes of the Aft1p-dependent iron regulon were induced specifically in the absence of Prx1p despite optimal mitochondrial Fe-S biogenesis, suggesting dysfunction of the mitochondria to the cytosol signaling pathway. Strikingly, requirement of Grx2p for these events places dithiolic Grx2 in the framework of iron metabolism.
Keywords: Glutathion, Iron Metabolism, Mitochondria, Oxidative Stress, Proteomics, Thiol, Yeast Metabolism, Glutaredoxin, Redox Proteomics, Thioredoxin
Introduction
Cysteine represents the most important redox-responsive amino acid within proteins, largely due to the wide range of oxidation states that sulfur can occupy. Thiols can be reversibly oxidized to sulfenic acid (-SOH), thiyl radicals (-S•), or nitrosothiols (-SNO); two thiol groups may form a disulfide bridge, which occurs in some proteins (PSSP), in oxidized glutathione (GSSG), or as a mixed disulfide in glutathionylated proteins (PSSG). These sulfur switches can provide an important and flexible means of reversibly controlling protein function (for a review, see Ref. 1). There is increasing evidence regarding the number of proteins whose function is regulated by the modification of specific Cys residues by H2O2, peroxinitrite or by a simple thiol disulfide exchange. Proteins regulated include transcription factors, enzyme activities, and components of the signal transduction machinery (2). In all cases, due to the chemical reactivity and regulatory logic, redox modifications of Cys are not spontaneously reduced when redox homeostasis is restored but are regenerated with the help of the redoxins (e.g. glutaredoxins (Grx)2 and thioredoxins (Trx)).
In this investigation, we have studied the yeast antioxidant defense enzymes Grx2p and Prx1p. Grx2p is a small protein belonging to the “thioredoxin fold” family, which, together with glutathione reductase and GSH, is thought to play an important role in redox regulation and antioxidant defense, allowing protein thiols to respond rapidly to changes in redox status (3). It has a dual localization in the cytosol and mitochondria due to alternative translation initiation from two in-frame AUG sites (4). Prx1p is a yeast mitochondrial peroxidase (5), whose mammalian counterpart, Prx3, has been reported to react with 90% of mitochondrial H2O2 (6). We have recently demonstrated that Grx2p together with GSH can regenerate the “peroxidatic” Cys of the active site in Prx1p (7).
Much is known about the functions of these “redoxins”; however, their exact action and the metabolic processes in which they play physiologically important roles remain to be completely elucidated. One way to assign functions to a protein is to identify other proteins that interact with it and have known activity. Any methodology that establishes a firm relationship either directly or indirectly between the protein of interest (a redoxin in our case) and any other protein(s) whose function is well known would help to throw light on the cellular role of the redoxin. The analysis of mutant proteomes by mass spectrometry (MS) has been suggested to provide important information for the identification of processes and pathways that are involved in establishing mutant phenotypes (8). Here we used a shotgun proteomic approach, consisting of searching within the redox proteome for proteins that are differentially modified when the cells do not express a particular redoxin. We have previously applied this method to select for redox-sensitive Cys-containing peptides or the “redox peptidome”(9). When a protein is detected as redox-modified in a mutant cell and not in the control, we conclude that the protein is modified as a consequence of the inflicted mutation. This strategy has allowed us to determine a kind of “redoxin functional interactome” without precluding subsequent confirmation of the existence of direct physical interaction.
Identification of redox changes in a protein does not necessarily imply that its function is affected correspondingly, but analysis of gene expression changes would provide complementary data on proteins not directly modified on their Cys residues but indirectly affected by postranslational modifications suffered by key proteins. We have carried out transcriptome expression profiling and have detected important and specific metabolic remodeling triggered by the absence of Grx2p, Prx1p, or both, including iron regulon, heme, amino acid, and cell wall biosynthesis. These findings show how deep the functions of the studied redoxins reach into the metabolic network, thus opening unexpected new doors to the knowledge of cellular adaptive redox mechanisms. Their relevance is enhanced by the fact that they occur in cells without apparent phenotypical symptoms of stress.
EXPERIMENTAL PROCEDURES
Yeast Strains
Saccharomyces cerevisiae strains CMML235 (WT) and MML44 (Δgrx2) were a kind gift from Prof. E. Herrero (University of Lleida, Spain) (10); strain Y13090 bearing a Δprx1 deletion was obtained from the Euroscarf collection; strain JR038 carrying a double mutation Δgrx2 and Δprx1 was obtained mating MATa and MATα haploid cells from strains MML44 and Y13090, followed by formation of diploids (MATa/α), meiosis induction, ascospore formation, and haploid selection by micromanipulation. After selection by growth in selective medium, genotype and phenotype were confirmed by PCR and Western blot, respectively. The strains used are described in Table 1.
TABLE 1.
Strains used in this study
The genotype and source/reference for each strain is included.
Determination of Protein Carbonylation, Glutathionylation, and ROS Detection
Proteins were derivatized with dinitrophenylhydrazine for carbonylation as described previously (11). For glutathionylation, protein was loaded in non-reducing buffer, and a specific antibody was used (Virogen). Protein concentrations were measured using Bradford reagent (Bio-Rad) (12) with BSA used as a standard. Following electrophoresis, proteins were transferred to nitrocellulose membranes and stained with Ponceau S (0.2%, w/v) in 5% (v/v) acetic acid to check for equivalent protein loading. Membranes were blocked and washed as described (11) except that primary antibody for carbonylation (anti-dinitrophenylhydrazine) was from Invitrogen and was used at 1:1500 dilution. Anti-GSH was diluted 1:1000. Anti-Dre2 was a kind gift from Dr. Laurence Vernis (CNRS/Institut Curie, Paris) and was used at a dilution of 1:2500. Chemiluminescent signal was detected using a LAS-3000 camera, and images were analyzed using Multi Gauge version 3.0 (Fujifilm, Japan). Intracellular ROS detection was measured using 5-carboxy-2′,7′-dichlorodihydrofluorescein diacetate according to the manufacturer's instructions (Invitrogen). Briefly 5 × 107 cells were resuspended in PBS containing 10 μm 5-carboxy-2′,7′-dichlorodihydrofluorescein diacetate. After a 30-min incubation, cells were washed and resuspended in growth medium, and fluorescence (excitation, 485 nm; emission, 525 nm) of 1 × 107 cells was measured.
Isolation of the Reversibly Oxidized Thiol “Redox Peptidome”
Cells were grown on YPD (1% (w/v) yeast extract, 2% (w/v) bactopeptone, 2% (w/v) dextrose) until they reached an A600 = 1–1.5. Cells were harvested by centrifugation at 7500 × g, and the pellet was washed twice with H2O and redissolved in lysis buffer (8 m urea, 100 mm NEM, 50 mm Tris-Cl, 2 mm EDTA, 0.1% (v/v) Triton X-100, and 2 mm PMSF, pH 8.0) containing washed sea sand. Cells were vortexed continually at 4 °C for 15 min, and samples were centrifuged at 15,000 × g for 10 min at 4 °C to obtain protein extract. Detection of proteins involved in reversible Cys modifications was carried out as described previously, except 250 μg of initial protein was tryptic digested (9). Three independent cultures were used for each strain.
LC-MS/MS Analysis
Samples were subjected to complete evaporation in a SpeedVac centrifuge and then were resuspended in 20 μl of 5% (v/v) acetonitrile, 2% (v/v) formic acid. 5 μl were used for the analysis in a Surveyor HPLC-LTQ Orbitrap XL (Thermo) instrument equipped with a nanospray source. Peptides were trapped and cleaned in an Agilent 300SB-C18 5 × 0.3-mm trap column (Agilent Technologies) at a flow rate of 10 μl/min of 5% (v/v) acetonitrile, 0.1% (v/v) formic acid for 15 min and then resolved in a Biobasic-18 100 × 0.075-mm column (Thermo) at a flow rate of 0.3 μl/min postsplitting for 60 min using an acetonitrile gradient, supplemented with 0.1% (v/v) formic acid, from 5 to 40%. The spray voltage and the capillary temperature at the mass spectrometer were set to 2.0 kV and 170 °C, respectively. Over the total analysis time, one full scan in FT mode at resolution setting of 30000 in the range 400–1500 m/z, followed by five collision-induced dissociation-activated MS/MS in dependent mode corresponding to the five most abundant masses, were acquired. Dynamic exclusion was set to on.
In total, we identified over 6000 cysteine-containing peptides for 203 distinct proteins involved in the redox proteome consistently between mutants and wild-type strains. Peptides that were matched to multiple proteins were assigned on the basis of top scoring protein in S. cerevisiae after performing BLAST analysis. There were a number of peptides that we would classify as false positives (those peptides identified without a cysteine; even after application of the filter, there were a little more than 2% of total peptides identified, and in many cases it was the same peptide across mutants and biological replicates). This indicates that either the peptide has been misidentified by the search engine or the peptide is in some way attached to the biotin-HPDP. Of the 203 proteins, 19 were identified using 24 distinct peptides that do not contain cysteines. The same raw data were searched in Mascot with identical criteria, and 7 of 24 of these peptides from 3 of 19 of these proteins were identified in both (i.e. their amino acid sequence coincided between search engines and suggests that they are true false positives and are attached in some way to avidin beads). Also, four of these proteins were identified in all strains and from independent cultures, suggesting that they are not an artifact.
Proteomic Data Analysis
Peptides were identified using version 27 of the software program Sequest to compare experimental tandem mass spectra with predicted fragmentation patterns of tryptic peptides generated from the protein data base for Saccharomyces downloaded from the National Center for Biotechnology Information nonredundant (NCBI nr) database on July 17, 2009 (20,690 sequences, 9,913,977 residues). A decoy data base, containing amino acid sequence-reversed analogs of each protein was appended to allow estimates of false discovery rates (13, 14). Default search parameters used were the following: enzyme, trypsin; maximum missed cleavages, 1; peptide mass tolerance, ±1.0 Da; fragment mass tolerance, ±0.6 Da; variable modifications, oxidation (M), destreak (C), and deamidation (N, Q). Peptide identifications were retained for Sequest results of XCorr ≥ 1.8 (z = 1), XCorr ≥ 2.5 (z = 2), or XCorr ≥ 3.5 (z = 3). These values yielded peptide false discovery rates of <5%. Information on the identified proteins was referenced from the Saccharomyces Genome Database. For inclusion in the list of proteins identified (supplemental Table S1), the protein had to be detected at least twice from two independent cultures of at least one yeast strain.
Expression Analysis
Yeast cells were grown to A600 = 1–1.5, cells were centrifuged and washed twice with deionized water. Expression analysis was performed in the laboratory of Dr. Joaquin Ariño (Universidad Autónoma Barcelona). Total RNA was purified using a RiboPure-Yeast kit (Ambion, Inc.), following the manufacturer's instructions. RNA quality was assessed by a gel electrophoresis RNA 6000 Nano Lab Chip with a 2100 Bioanalyzer (Agilent Technologies), and RNA quantification was performed by measuring absorbance at 260 nm in a Nanodrop Spectrophotometer (Thermo). Transcriptional analyses were performed using DNA microarrays containing PCR-amplified fragments from 6014 S. cerevisiae open reading frames in duplicate (15, 16). Fluorescent Cy3- and Cy5-labeled cDNA probes were prepared from 7 μg of purified total RNA by the indirect labeling method using a Chip Shot Indirect Labeling and Clean-Up System kit (Promega) and a CyDyeTM postlabeling reactive dye pack (GE Healthcare). Prehybridization, hybridization, and washes were carried out as recommended by the Institute for Genomic Research with minor modifications. For hybridizations of the DNA microarrays, both dried Cy3- and Cy5-labeled probes were resuspended in 53 μl of hybridization solution (50% (v/v) formamide, 5 × SSC, and 0.1% (w/v) SDS) and mixed. 5 μg of salmon sperm DNA were added to the mixture before denaturalization for 3 min at 95 °C. Hybridizations were carried out in an Array Booster hybridization station (Advalytix, Germany) for 16 h at 42 °C. A Scan Array GxPlus scanner (PerkinElmer Life Sciences) was used to obtain the Cy3 and Cy5 images at 10 μm resolution. The fluorescent intensity of the spots was measured and processed using GenePix Pro version 6.0 software (Molecular Devices Corp.). Software from the GEPAS server was used to merge duplicates. For each condition assayed, two independent experiments were performed, and dye swapping was carried out.
Other Methods
Protein extracts for enzyme activity assays were prepared as for analysis of the thiol redox peptidome, except that the buffer used was 50 mm Tris-Cl, 0.1% (v/v) Triton X-100, and 2 mm PMSF, pH 8.0. The aconitase assay and malate dehydrogenase assay were performed as described (17, 18). Free and protein-bound iron in protein extracts was determined using ferrozine (19), and ferric reductase activity in whole cells measured by the reduction of ferric to ferrous iron was determined as described elsewhere (20, 21). Data from immunoblots and enzymatic activities were determined at least in triplicate from three independent cultures and are expressed ± S.E. (*, differences between wild type and mutant strains that are significant at p < 0.05).
RESULTS
Phenotypic Analysis
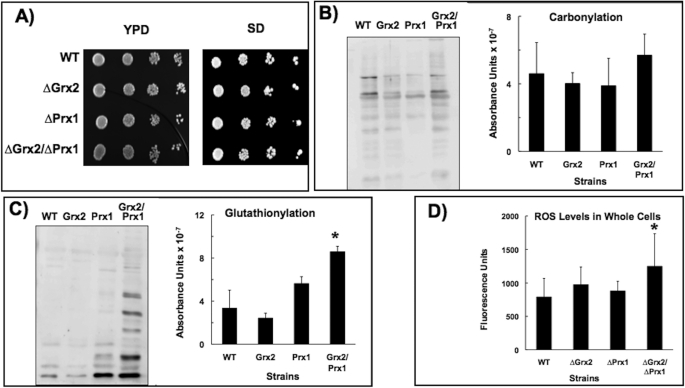
All four strains showed similar growth rates in complete YPD and selective SD media with glucose as the carbon source, indicating that a lack of one or both redoxins did not confer limitations to growth capability when the cells were not subject to exogenous stress conditions (Fig. 1A). Because the eliminated redoxins are thought to be involved in antioxidant defense and a basal level of ROS is produced endogenously under normal growth conditions (1), we checked for possible oxidative alterations. As a general indicator of oxidative damage, there appeared to be increases in carbonylation, but these were not significant (Fig. 1B). However, there were significant increases in glutathionylation in the double mutant (Fig. 1C), supporting the synergistic relationship between these two antioxidant proteins (7). A similar result was obtained from a general measure of intracellular ROS levels: no significant differences between wild type and single mutants, although there was a higher level of ROS in the double Δgrx2Δprx1 mutant (Fig. 1D). A marked decrease in protein thiol oxidation in mutants with an inactive GSH pathway has been reported (22), and we found a lower, although not significantly different glutathionylation level in the Δgrx2 mutant (Fig. 1C).
FIGURE 1.
Phenotypic analysis. A, 10-fold dilutions of exponentially growing yeast cells spotted onto glucose YPD or SD medium. Shown are carbonylation (B) and glutathionylation (C) levels in protein extracts as measured by chemiluminescence. 20 μg of total protein were loaded onto each lane in all cases. Representative sample immunoblots and quantitative analysis of three different experiments are shown. D, total levels of ROS in the cells. *, statistically significant (p < 0.05) relative to WT. Error bars, S.E.
Redox Proteomic Analysis
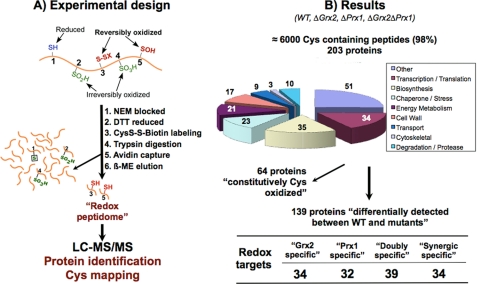
Lack of the antioxidant action of Prx1p could give rise to peroxide that would react with sensitive thiols in proteins, whereas a lack of Grx2p could compromise the recovery of reversibly oxidized cysteines through the glutathione pathway. To take a snapshot of the protein thiol status in the mutants, the comparative “redox peptidome” of all four strains was determined by a shotgun redox proteomics method we have recently developed (9) (Fig. 2). The method not only identifies the proteins with reversibly oxidized thiols but also determines the precise cysteine affected within each protein. The process of acquiring tandem MS is prone to undersampling (23), and technical replicates can show limited overlap with variations in protein identifications between runs (24). In redox proteomics, proteins that are modified to the greatest degree may be less abundant than those modified to a lesser degree and thus not detected. To circumvent under sampling and to increase reproducibility, three biological replicas for each experiment were always carried out, and reversibly oxidized cysteine-containing peptides were selected by sample prefractionation (Fig. 2A). These approaches greatly improve reproducibility between analyses (23).
FIGURE 2.
Redox proteomic approach and results. A, free thiols are initially blocked with NEM, and all reversibly oxidized thiols are reduced and subsequently labeled with biotin-HPDP. After tryptic digestion, biotin-labeled peptides are purified and analyzed by MS/MS. This method detects proteins with reversibly oxidized cysteine(s); reduced or irreversibly overoxidized cysteines are not detected. A protein that is not detected in WT but is reversibly oxidized in a mutant is expected to have its cysteine totally reduced in the WT (these proteins are marked with a plus sign in Table 2). A minor percentage of proteins were detected in WT but not in other mutant strain(s), probably due to irreversible overoxidation in the latter, because its antioxidant defenses are further diminished (these proteins are marked with a minus sign in Table 2). B, 203 proteins were consistently identified from the reversibly oxidized Cys-containing peptides. 64 “constitutively Cys-oxidized” proteins were detected in all four strains. A redox target is a protein whose thiol redox state changes differentially between the studied strains. “Grx2-specific” and “Prx1-specific” proteins are those proteins that change as a consequence of the absence of Grx2 and Prx1, respectively. A “doubly specific” protein is a protein specific for both redoxins and is detected with redox changes in both Δgrx2 and Δprx1 single mutants. “Synergic specific” proteins show redox changes only when both redoxins are absent, and are detected only in the double Δgrx2Δprx1 mutant.
Differences between proteins identified in both simple and double mutants may allow the identification of protein targets of these redoxins and also the redox-sensitive cysteines within those proteins. Supplemental Table S1 lists all proteins identified, the peptides used for identification, MH+ (Da), retention times, ions matched for each peptide, and the Sequest score. In total, we consistently identified over 6000 Cys-containing peptide ions for 203 distinct proteins involved in the redox proteome (Fig. 2B).
Sixty-four proteins were detected with reversibly oxidized Cys in independent replicates from all strains and were classified as “constitutively Cys-oxidized.” They included 10 enzymes of the glycolytic pathway to ethanol, meaning that the major metabolic pathway of energy production in the presence of glucose is thus not affected by the loss of Grx2p and/or Prx1p, and many of these proteins exist in vivo in a redox balance under normal conditions (1, 22).
139 proteins were consistently detected differentially between strains (Table 2). A protein is classified as “redoxin-specific” when its thiol redox state is specifically sensitive to the absence of that redoxin. According to the pattern of appearance among the four strains, there were 34 “Grx2-specific” and 32 “Prx1-specific” proteins. 39 proteins that were detected with redox changes in both Δgrx2 and Δprx1 single mutants are “doubly specific.” Finally, 34 proteins were “synergically specific” because they showed redox changes only when both redoxins were absent and were detected only in the double Δgrx2Δprx1 mutant. All important cell processes were represented although proteins related to the cell wall were not detected specifically in the single mutants.
TABLE 2.
Proteins with specifically modified cysteine(s) in the mutant strains
The abbreviated name of the proteins according to SGD or the ORF code are shown together with the position of the cysteine(s) residue(s) modified in the protein sequence. Criteria for inclusion of a protein in a particular group are explained under “Results” and in the legend to Fig. 2. *, the peptide detected does not contain cysteine. +, proteins detected only in the corresponding mutant (see also legend to Fig. 2); −, proteins are detected in all strains except in the corresponding mutant. Underlined names indicate that the pattern is the same in the double mutant.
| Grx2-specific |
Prx1-specific |
Doubly specific |
Synergic specific |
|||||
|---|---|---|---|---|---|---|---|---|
| Protein | Cys | Protein | Cys | Protein | Cys | Protein | Cys | |
| Biosynthesis | Aro4p (−) | 76, 244 | Aco1p (−) | 382 | Aah1p (+) | 142, 188, 252 | Erg2p (+) | 40 |
| Ham1p (+) | 123 | Asn2p (−) | 404, 431 | Adi1p (+) | 40 | Hom6p* (+) | ||
| Hem12p (+) | 26 | Bat1p (+) | 227, 361 | Adk1p (+) | 84 | Idi1p (−) | 58, 74 | |
| Lys9p (+) | 154, 340 | Fsh1p (−) | 39 | Bat2p (+) | 109 | |||
| Sam1p (+) | 91 | Gln1p (+) | 160 | Dys1p (+) | 208, 252 | |||
| Ura5p (+) | 86 | Lys4p (+) | 400/407/410 | Fcy1p (−) | 36, 71 | |||
| Ydl124wp (+) | 197 | Lys20p (−) | 202 | IIv3p (−) | 176, 258, 581 | |||
| Rib3p (−) | 34, 56, 133 | IIv5p (+) | 316 | |||||
| Trp5p (−) | 420 | IIv6p (+) | 270 | |||||
| Leu1p (+) | 63, 627 | |||||||
| Prs3p (+) | 273 | |||||||
| Transcription/Translation | Ala1p (+) | 532, 719 | Caf20p (−) | 82 | Pab1p (−) | 368 | Hyp2p* (−) | |
| Arc1p (−) | 266, 284 | Rnr2p (−) | 270 | Rpl18bp (−) | 121 | Rpl12bp (−) | 141 | |
| Rex2p (+) | 116 | Rpl23ap (+) | 25 | Rpl15bp (−) | 152 | Rps27ap* (−) | ||
| Rpc19p (+) | 138 | Rps17ap (−) | 35 | Rps0ap (−) | 162 | Ses1p (+) | 370, 400 | |
| Rps18bp (−) | 43, 113 | Rps1ap (−) | 69 | Vas1p (+) | 509, 758 | |||
| Rpl35ap (+) | 53 | Tif34p (−) | 67, 104 | |||||
| Sky1p (+) | 28 | Trm112p (+) | 16 | |||||
| Snu13p (+) | 91 | |||||||
| Tec1p (+) | 314 | |||||||
| Chaperone/Stress | Arf2p (+) | 159 | Dld3p (+) | 363 | Cct8p (+) | 252 | Cpr3p (−) | 137 |
| Ccs1p (+) | 159 | Dre2p (−) | 252 | Cpr5p (+) | 150 | Ykl069wp (−) | 91, 125 | |
| Hsp31p (+) | 138 | Hsp104p (+) | 209 | Sti1p (+) | 66 | |||
| Pdi1p (+) | 61/64, 406/409 | Ssb2p (−) | 20, 454 | |||||
| Yhb1p (+) | 328, 362 | |||||||
| Energy metabolism | Rpe1p (+) | 79 | Adh6p (−) | 101/103, 162 | Lsc1p (+) | 153 | Glo2p (−) | 18, 115, 127, 218 |
| Tkl1p (+) | 622 | Ald6p (+) | 133, 193/212 | Hrk1p (+) | 471 | |||
| Pgm2p* (−) | ||||||||
| Cell wall | Ccw14p (−) | 40/50/52 | Cwh41p (+) | 223 | ||||
| Guk1p (+) | 96 | Scw11p (+) | 323 | |||||
| Pmi40p (+) | 148, 291 | Mns1p (+) | 385 | |||||
| Psa1p (−) | 319 | Utr2p (+) | 67 | |||||
| Scw10p (+) | 176, 357 | |||||||
| Transport | Rad23p (+) | 37 | Prc1p (−) | 373/379, 409 | Pup2p (−) | 76, 221 | Doa1p (+) | 196 |
| Tfs1p (+) | 110/115 | Shp1p (−) | 88 | Ybr139wp (+) | 336 | |||
| Degradation/Proteases | Gre3p (+) | 20, 28 | Yrb1p (−) | 145 | Qcr2p (+) | 137 | Gda1p (+) | 87, 364 |
| Prd1p (+) | 478, 605 | Kin2p* (−) | ||||||
| Pep1p (+) | 896, 1096 | |||||||
| Other | Est2p* (+) | H+ATPase (−) | 19 | Acid phosphatase (−) | 276, 408/416 | Cin8p* (−) | ||
| Pho3p (−) | 263, 276, 408/416 | Pea2p* (−) | Pho84p (−) | 499 | Cpr1p (−) | 117 | ||
| Yor285wp (+) | 98 | Yar1p (+) | 44 | Pma1p (−) | 221, 409, 532 | Ddb1p (−) | 37/39, 87, 149 | |
| Ydl086wp (+) | 243 | Yjl217wp (−) | 170 | Pry2p (+) | 299 | Fas2p (+) | 1246 | |
| Yer079wp (−) | 59 | Ydr051cp (+) | 183 | Protoplast-secreted (−) | 80 | Hog1p (+) | 156 | |
| Ykl105cp (+) | 1023 | Ynl010wp (+) | 66 | Rhr2p (+) | 100 | orf 00954p (−) | 21/30/35/40 | |
| Ylr454wp* (+) | Unnamed* (−) | Yel047cp (+) | 196, 454 | Pyrophosphatase (+) | 83 | |||
| Ydr036p* (+) | Yir010wp* (+) | Yhr020wp (−) | 11 | |||||
| Yjr004cp (+) | 202 | Yjl218wp (−) | 178 | Ydr391cp (−) | 169 | |||
| Ygl137wp (−) | 160 | Pho8p (+) | 132, 150 | |||||
| Ydl037cp* (+) | ||||||||
| Yfr002wp* (−) | ||||||||
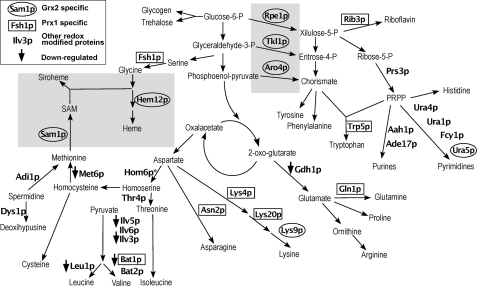
The biosynthesis of amino acids and nucleotides was where the greatest differences between proteins identified were detected. Fig. 3 shows 32 proteins detected in the various pathways with reversibly oxidized Cys. There are seven Grx2p-specific redox targets. Rpe1p (Cys79), Tkl1p (Cys622), and Aro4p (Cys76 and Cys244) play a key role linking the glycolytic metabolic flow to the biosynthesis of aromatic amino acids and nucleotides, and Hem12p (Cys26), Lys9p (Cys154 and Cys340), Sam1p (Cys91), and Ura5p (Cys86) are involved in heme, lysine, S-adenosylmethionine, and pyrimidine biosynthesis, respectively. Six other proteins are Prx1-specific redox targets. Fsh1p (Cys39), Asn2p (Cys404 and Cys431), Lys4p (Cys407), Gln1p (Cys160), Bat1p (Cys227 and Cys361), and Rib3p (Cys34, Cys56, and Cys133) are involved in glycine, asparagine, lysine, glutamine, isoleucine, and riboflavin biosynthesis, respectively. Rib3p is located in the cytoplasm but has also been reported to have an unrelated role in respiration in the mitochondrial intermembrane space (25).
FIGURE 3.
Integrated schematic diagram of 32 redox-modified proteins detected in amino acid and nucleotide biosynthesis. Gray shaded areas are pathways/enzymes preferentially discussed throughout. Of 32 modified proteins, six had reduced expression levels (↓). See the inset box for enzyme name symbols. “Other redox-modified proteins” are either “doubly specific” or “synergic specific” proteins. The function of the following proteins has been shown to be redox-sensitive in other studies (references included): Aro4p (31); Bat1p and Bat2p (27); Dys1p (29); Gln1p (28); Ilv3p, Leu1p, and Lys4p (30); Hem12p (32); Sam1p (33, 34); Rpe1p and Tkl1p (34). Proteins identified were as follows: Aah1p (adenine deaminase), Ade17p (Enzyme of “de novo” purine biosynthesis), Adi1p (acireductone dehydrogenase), Aro4p (3-deoxy-d-arabino-heptulosonate-7-phosphate synthase), Asn2p (glutamine-dependent asparagine synthetase), Bat1p (mitochondrial amino acid transferase), Bat2p (cytosolic branched-chain amino acid aminotransferase), Dys1p (deoxyhypusine synthase), Fcy1p (cytosine deaminase), Fsh1p (serine hydrolase 1), Gdh1p (NADP-dependent glutamate dehydrogenase 1), Gln1p (glutamine synthetase), Hem12p (uroporphyrinogen decarboxylase), Hom6p (homoserine dehydrogenase), Ilv3p (dihydroxydehydratase), Ilv5p (acetohydroxyreductoisomers), Ilv6p (regulatory subunit acetolactate synthase), Leu1p (isopropylmalate dehydrogenase), Lys4p (homoaconitase), Lys9p (saccharopine dehydrogenase), Lys20p (homocitrate synthase), Met6p (cobalamin-independent methionine synthase), Prs3p (5-phospho-ribosyl-1(α)-pyrophosphate synthetase), Rib3p (3,4-dihydroxy-2-butanone-4-phosphate synthase synthase), Rpe1p (d-ribulose-5-phosphate 3-epimerase), Sam1p (S-adenosylmethionine synthetase), Thr4p (threonine synthase), Tkl1p (transketolase), Trp5p (tryptophan synthase), Ura1p (dihydroorotate), Ura4p (dihydroorotase), and Ura5p (orotate phosphoribosyltransferase isozyme).
If these postranslational cysteine modifications had an effect on the functional performance of the proteins, then the metabolic flow through the pathways in which they participate would be altered (26). Although this would have to be demonstrated for each enzyme, a number of them have already been reported in the literature as redox-sensitive, reinforcing the validity of our thiol proteome screening results. For instance, the human BCATs, analogous to Bat1p and Bat2p, are inactivated by both S-nitrosation and S-glutathionylation, where activity in the latter can be recovered by glutaredoxin/glutathione reductase system (27). Gln1p Cys160 forms part of the regulatory citrate binding site (28), and we find this Cys as a specific Prx1p target. Similarly, in Dys1p, the Cys252 has been reported as essential for activity, and we have identified Cys208 as redox-sensitive (29). Ilv3p, Leu1p, and Lys4p are involved in Fe-S complexes (30). The equivalent of Aro4p Cys76 in Corynebacterium glutamicum is important for synthase activity (31). Hem12p, the enzyme that catalyzes the fifth step in heme biosynthesis, is oxidized at the residue Cys26, and the yeast protein is known to be thiol-dependent (32). The rat orthologue of Sam1p is thioltransferase-dependent (33), and we find the active site Cys91 reversibly oxidized in the Δgrx2 mutant. Rpe1p and Tkl1p belong to the non-oxidative part of the pentose phosphate pathway and possess H2O2-sensitive thiols. Moreover, the plant orthologues of Hem12p, Rpe1p, and Tkl1p have been detected as candidates for redox regulation (34).
Several redox protein targets play key metabolic roles; therefore, redox modulation of protein function, if validated, should have far reaching effects. To name just a few, Prs3p interacts with a number of other proteins, many involved in signaling pathways, and loss of the PRS3 gene is reported to disturb a wide variety of cell functions (35); Cys137 of a matrix-oriented component of the heme-containing cytochrome bc1 complex (Qcr2p) is a target of both redoxins; and similarly, the flavohemoglobin protein, nitric-oxide oxidoreductase (Yhb1p), involved in mitochondrial respiration, was again only detected in the absence of Grx2p with Cys328 and Cys362 reversibly oxidized and is reported to play a role in iron reduction (36). A group of cell wall-related proteins was detected with reversibly oxidized specific Cys but only by a synergic effect of the absence of both redoxins because they are only detected in the double mutant (Table 2). Interestingly, Erg2p, an enzyme that catalyzes a highly regulated step in ergosterol synthesis (37, 38), was detected in the same group with oxidized Cys40, a non-conserved Cys residue unique to yeast.
Transcriptome Analysis
As an alternative approach to validate functional changes in the identified proteins, we reasoned that if the posttranslational redox modifications detected do really constitute functional switches and modify metabolic and signaling flows as a primary effect, then gene transcription remodeling could take place as a secondary adaptive effect (26). To check this possibility, we undertook a parallel high throughput transcriptomic analysis. Comparison of mRNA levels between control and mutants confirmed that there is a profound metabolic remodeling as a consequence of the proteomic redox changes induced by the absence of Grx2p, Prx1p, or both.
Over 200 genes showed >2-fold up- or down-regulation in comparison with wild type as a result of loss of redoxins. Supplemental Table S2 lists the results of the expression analysis of mutants compared with wild type. A list of redoxin-related genes is also presented in Table 3. Only a small number of the proteins detected as reversibly oxidized by the proteomics analysis showed changes in their gene expression. This is an additional validation of our experimental approach because it indicates that the proteins are not detected because of an increase in transcription but rather because their redox state changes in the samples. Of the 32 redox-modified proteins selected for Fig. 3, only six had reduced expression levels in the mutants as compared with wild type, and the products of five of these genes are present in the mitochondria, ILV3, ILV5, LEU1, BAT1, and MET6. Each of these is detected in at least the Prx1p mutant and would point to adaptive mechanisms triggered by the absence of Prx1p.
TABLE 3.
Genes differentially expressed in the mutants
Genes differentially expressed in the Δgrx2, Δprx1 and Δgrx2/Δprx1 mutants relative to WT, classified by the mutant where they show expression changes and by cellular processes they are involved in. An underlined name indicates that the gene also changes in the double mutant. Genes in the last two columns are those that show changes only in the double mutant.
| Process | Δgrx2-specific genes |
Δprx1-specific genes |
Δgrx2/Δprx1-specific genes |
|||
|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | |
| Biosynthesis | BTN2, GAD1, TPK1 | ALD5, ARG1, BAT1, GDH1, ILV3, ILV5, INO2, LEU1, LEU4, MET13, OAC1, PUT1, SNZ1, YEH1 | CAR2, GCV2, LPP1 | BAR1, BAT1, BNA1, GDH1, INO2, LEU1, LEU4, MET13, OAC1, PUT1, RIB4 | MUP3 | MET6 |
| Transcription | NOP1, RTC3 | CIN5, NRG1 | ASG1, DED1, MNP1, MRM1, NOP1, PHO89, SOH1, TRM11, TUF1, VTS1 | CIN5, DAN1, GAT2, NRG1, RPL41A, SRB2 | PHD1 | |
| Iron regulon | FRE7 | FIT3,aFRE4,bICT1, IZH1 | ARN1, ARN2, CCC2, COT1, FET3, FET4, FET5, FIT1, FIT2, FIT3, FRE1, FTR1 | FRE4,bISU1 | ||
| Energy Metabolism | ATP2, GLK1, HXK1, HXT4, NDI1, PNC1, UGP1 | MAE1, MIG2, NCE103 | ACO1, ACO2, ATP12, CIT2, DLD3, GLK1, GSP2, HXK1, HXT4, IDH2, SIT1, TIS11 | NCE103 | ||
| Cell wall | GLC3, GSY1, GSY2, PGM2, SOL4, SPI1, TPS1, TPS2, TSL1, YGP1 | DAN3, MUM3, TIR 4 | AMS1, DAN1, DSE1, DSE4, GLC3, GSC2, ROX1, TIR1, TIR4 | AGA2, GAS2 | CBF2 | |
| Transport | COX5B, GYP7, TPO4 | AGP1, GAP1, YLR348C | AUS1, EGT2, PDR5, VID24 | AGP1, PDR12, STE6, TPO2, TPO3, TPO4, WSC4 | PIC2, SEO1 | |
| Degradation/Proteases | LAP4, PBI2, SNF8 | ATG3, LAP4 | CLB1 | |||
| Other | CMK2, FMP48, IRC9, ISF1, MSC1, PYK2, OPI10, STF2, YAK1, YBR056W, YER067W, YKR093W, YJL44W, YLR345W, YPC1, YPL014W | PAU1, PAU5, YBR012C, YCL021W, YCLX10C, YER074W, YGR035C, YIR042C, YKR075C, YLR348C, YLR349W, YNL024C, YOR225W, YPR157W | MATAα1, MAM33, MF(a)2, PYK2, SAG1, SDS23, SRP40, SSC1, STE3, YDL124W, YDL241W, YER067W, YER074W, YFR006W, YGR079W, YKL177W, YKR093W, YLR041W, YMR317W | MFA2, STE2, YBL029W, YCL021W, YCLX10C, YCR097WB, YCR102C, YGL041C, YIR035C, YLR132C, YLR257W, YNL024C, YPR157W, YRO2 | EMI2, RCN2, YLR194C | |
a FIT3 is down-regulated in Δgrx2 and up-regulated in Δprx1 and Δgrx2/Δprx1.
b FRE4 is down-regulated in all strains.
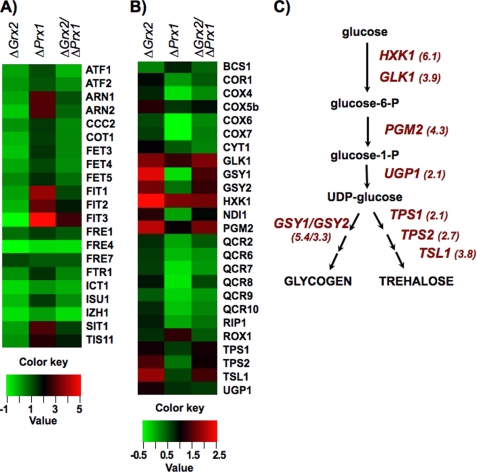
The transcriptomic analysis not only confirmed that the postranslational redox modifications have functional consequences but showed unexpected implications of Prx1p and Grx2p for iron homeostasis, amino acid biosynthesis, and storage carbohydrate pathways. Two remarkable findings could not have been detected from the proteomic analysis alone or from standard phenotypical assays. (i) The iron regulon is activated specifically in the absence of Prx1p but also requires the presence of Grx2p (Fig. 4A), and (ii) in the absence of Grx2p, there is an increase in expression of storage carbohydrate genes involved in glycogen and trehalose biosynthetic pathways: GSY1 and -2, UGP1, HXK1, GLK1, PGM2, TPS1 and -2, TSL1, and the gene coding for the respiratory complex NADH-ubiquinone reductase (NDI1) (Fig. 4, B and C). The genes for subunits of the other respiratory complexes, cytochrome bc1 (CYC1 and -7, CYT1, QCR2, and QCR6 to -9) and cytochrome oxidase (COX4, COX5B, and COX6 and -7) also showed an increase in Δgrx2 mutant. Up-regulation of storage carbohydrate genes and respiratory genes is reminiscent of adaptation to intermediate oxygen tension through heme-dependent signaling pathways and of changes that take place prior to diauxic shift (39, 40). These results suggest that redox changes inflicted on the proteome by the absence of Grx2p might cause the activation of the same transcription factors as those that govern both processes.
FIGURE 4.
Expression of “iron regulon” and storage carbohydrate biosynthesis genes in mutant strains. A, genes of the iron regulon under the control of Aft1p transcription factor; B, glycogen and trehalose biosynthesis and respiratory genes. In the absence of Prx1p and when Grx2p is present, there is an increase in genes involved in iron regulon response but not when both redoxins are absent, whereas in the absence of Grx2p, there is an increase in genes involved in glycogen and trehalose biosynthesis and in respiratory genes. -Fold change scales relative to wild type for both sets of genes are shown below (ratios in log2). C, the main intermediate metabolites from glucose to glycogen and trehalose are depicted; genes coding for enzymes of the pathways showing increased expression levels in the absence of Grx2p are named with the induction -fold relative to WT in parentheses.
Comparison of Iron Parameters
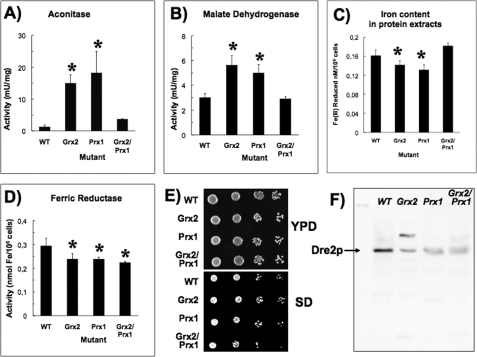
The finding of iron regulon activation in the absence of Prx1p prompted us to determine several indicators of the iron status in those cells. Analysis of the Fe-S mitochondrial aconitase is used as a measure of abnormalities in Fe-S cluster synthesis. Malate dehydrogenase is also a mitochondrial enzyme but lacks Fe-S clusters, and significant changes in absolute values would point to specific mitochondrial dysfunction (41); a recent study has indicated an interaction between malate dehydrogenase and cytochrome bc1 complex, pointing to a regulatory role in mitochondrial bioenergetics (42). There was a significant increase in aconitase and malate dehydrogenase activities in both Grx2p and Prx1p mutants (Fig. 5, A and B), a surprising result because the activation of the iron regulon usually coincides with iron depletion and with a loss of mitochondrial Fe-S cluster biogenesis (43). Our results indicate that dysfunction of Fe-S cluster biogenesis is not the cause of iron regulon activation and that disruption of the signaling pathway from mitochondrion to cytosolic Aft1p is triggered by other factor(s).
FIGURE 5.
Iron-dependent parameters in strains. A, aconitase activity; B, malate dehydrogenase activity. C, iron content in protein extracts. D, ferric reductase activity. Data in A–D are the average of three independent experiments; *, statistically significant relative to WT (p < 0.05). E, 10-fold dilutions of yeast cells under iron-limiting conditions (200 μm ferrozine included in YPD and SD glucose medium); the double mutant displays reduced growth rates when growing in selective medium (SD) with limited iron availability. F, immunoblot of anti-Dre2p; equal amounts of total protein were loaded on each lane; the arrow points to Dre2p monomer (∼50 kDa); a second band in the Δgrx2 extract may correspond to the Dre2p-Tah18p heterodimer (45). Error bars, S.E.
As a result of iron regulon activation, an increase in the iron content of protein extracts would be expected at least in the Δprx1 mutant. On the contrary, there was a slight although significant decrease in iron content in protein extracts from strains lacking Grx2p and Prx1p (Fig. 5C). This could be explained by low levels of iron in cytosolic proteins compensating for normal mitochondrial iron metabolism. Yeast iron uptake is heme-dependent, and one of the first steps is the reduction of Fe(III) to Fe(II) by heme-dependent ferric reductases. Although a crude measurement, this reduction has been used as an indication of the activity of these enzymes (20, 21). Our results indicate that in all mutant strains, there is a significant decrease in ferric reductase activity (Fig. 5D). Under iron starvation, the double mutants show a slightly diminished growth rate (Fig. 5E).
Activation of Aft1p under normal mitochondrial Fe-S center biogenesis indicates that the signaling pathway from mitochondria to the cytosol for iron homeostasis is dysfunctional. The Fe-S protein Dre2p has been reported as playing key roles in cytosolic Fe-S cluster assembly, and the Δdre2 mutant was deficient in cytosolic but not mitochondrial Fe-S clusters (44, 45). Dre2p was identified in our redox proteomics analysis as containing Cys252 reversibly oxidized in wild type and Δgrx2 strains but was not detected (overoxidized?) when Prx1p was absent. The transcriptomic analysis showed that mRNA levels decrease in all strains in comparison with wild type, and in Western blot using anti-Dre2p there is a decrease in abundance of the protein (Fig. 5F). Previously, it was found that Dre2p forms a 110-kDa complex with the apoptotic Tah18p in the cytoplasm that dissociates in response to H2O2, resulting in targeting Tah18 to the mitochondria with loss of mitochondrial integrity and cell death (45). It has been speculated that Dre2p is involved in iron binding and reduction in the cytoplasm that would require a source of electrons for reductase activity (44), and it would be tempting to suggest that Grx2p may provide this function. Nevertheless, Dre2p is one of the very few proteins where there are clear and consistent differences between mutants in gene expression, protein abundance, and redox state.
DISCUSSION
Experimental Strategy
Mitochondria are primarily known for their role in energy production, but this work was carried out in yeast growing on complete medium using glucose as an energy source; hence, respiration is not the primary source of energy production. This has allowed us to appreciate the roles of mitochondrial Prx1p and dually localized Grx2p on processes for which mitochondria are also essential: iron metabolism and amino acid biosynthesis. Under these conditions, the loss of both antioxidant enzymes has no significant effect on growth rate or irreversible protein damage within the cell. However, thanks to the combination of a redox proteomics with high throughput transcriptomic analysis and enzymatic assays, we have been able to detect latent metabolic remodeling responsible for the adaptation of cells to the absence of Grx2p or/and Prx1p. On one hand, the redox proteomics approach gives a real measurement of reversible redox postranslational modifications with the advantage that it selects for only those Cys-containing peptides of those proteins susceptible to redox modifications (9). On the other hand, we have determined the cellular effects of eliminating Grx2p, Prx1p, or both at the genomic level. The integration of both approaches has allowed us to establish a relationship between changes in gene expression patterns and primary changes at the protein level. This strategy provides valuable new insights into redox regulatory mechanisms, and further improvements with next generation quantitative proteomic methods could give the exact measure of the proportion of oxidized protein relative to the whole protein population.
Grx2p Control of Metabolic Energy Flow
Our results indicate that Grx2p could participate in modulating metabolic flow from energy production pathway toward aromatic amino acid and nucleotide biosynthesis as a function of the cellular redox state. Deviation of glycolytic flow toward storage carbohydrate biosynthesis by the absence of Grx2p could result from impairment of this control mechanism. The marked induction of glycogen and trehalose biosynthesis genes in the Δgrx2 mutant (Fig. 4, B and C) could be mediated by activation of the Msn2p-Msn4p transcription factor complex (40) through thiol redox changes (46). It would be of interest to check the redox state of this transcription factor in the Δgrx2 mutant in further studies to confirm this hypothesis, but it is beyond the scope of the present study because very low abundant proteins like this fall below the detection threshold of our redox proteomics method, which is a general limitation of redox proteomics (22).
Coherent with this hypothesis, Rpe1p, Tkl1p, and Aro4p, which play key roles to drive the metabolic flow from glycolysis to aromatic amino acid and nucleotide biosynthetic pathways (see Fig. 3), all suffer reversible thiol oxidation in the absence of Grx2p. We have not yet checked the relationship of reversible thiol oxidation of these key enzymes with their activities, but there are solid data in the literature that demonstrate their redox regulation. Rpe1p and Tkl1p orthologues have been identified as candidates for Trx redox regulation in spinach leaves by affinity capture using mutated monothiolic Trx (34) and could be substrates of Grx2p in yeast. Aro4p was detected with reversibly oxidized Cys after treatment of yeast cells with H2O2 (9), and oxidation of Cys76, as found in this study, inactivates the enzyme (31). TKL1 null mutant is auxotrophic for aromatic amino acids, and RPE1 and ARO4 null mutations induce glycogen accumulation (47).
Iron Regulon and Redoxins
Mitochondrial oxidative stress response due to the loss of Prx1p may be the cause of the iron regulon activation by impairment of mitochondria to cytosol signaling mechanisms. Genes belonging to the so-called iron regulon were markedly induced in the absence of Prx1p (Fig. 4A). An increase in its natural substrate H2O2 levels within the mitochondria would be expected in this mutant because this enzyme seems to account for the majority of peroxidase activity (6), and the level of glutathionylated proteins is slightly higher than in the WT (see Fig. 1C). However, ROS levels in these cells do not show significant differences between single mutants and the wild type (Fig. 1D). This parameter is a measure of total ROS within the cell and not specifically in mitochondria or cytoplasm. A small localized increase of H2O2 concentration cannot yet be discarded, but its measurement would require more sophisticated methodology and focused experimental approaches.
The activation of the iron regulon is known to take place under low iron conditions or upon disruption of mitochondrial Fe-S cluster biogenesis, by translocation of the iron-responsive transcription factor Aft1p monomer to the nucleus (30). Nucleo-cytoplasmic shuttling of Aft1 is inhibited under normal mitochondrial Fe-S cluster biosynthesis via a signaling pathway that includes the cytosolic monothiol glutaredoxins Grx3p and Grx4p and the ancillary proteins Fra1p and Fra2p (30, 48). The signal is exported through the mitochondrial inner membrane ATP-binding cassette transporter (Atm1p) in the presence of glutathione (30), but the specific mechanism of iron-dependent regulation of Aft1 localization by Fra2 and Grx3/4 is a key gap in our understanding of intracellular iron metabolism (48). In Δprx1, the Fra1/Fra2/Grx3/Grx4 pathway seems to be shut off, but surprisingly, mitochondrial Fe-S cluster biogenesis is not affected, as indicated by aconitase activity. The increased expression of TCA genes ACO1 (aconitase) and IDH2 (isocitrate dehydrogenase) and the Fe-S cluster-assembling protein ISU1 in this mutant is also coherent evidence. These results suggest that optimal Fe-S cluster biogenesis in the mitochondria is not a sufficient condition to export a signal molecule to the cytosol by the ATP-binding cassette transporter Atm1p and to retain the Aft1p transcription factor in the cytosol. Another factor(s) is missing or not functional in the Δprx1 mutant.
When both Prx1p and Grx2p are absent, induction of Aft1p-dependent genes is markedly attenuated or abolished (Fig. 4A), coinciding with a marked increase in the overall glutathionylation of proteins in the double mutant. It would be interesting to speculate whether Grx2p could be involved in cytosolic Fe-S maturation either directly or indirectly as a consequence of its effects on other proteins. Interestingly, heme biosynthesis could be diminished in the absence of Grx2p (see below), and it has been reported that inhibition of heme biosynthesis prevents transcription of iron uptake genes in yeast (49). Moreover, Grx2p is localized at three different compartments (cytosol, mitochondrial outer membrane, and matrix) (4, 50) and could be involved at several points along the mitochondria to cytosol signaling pathway, where GSH and other thiol disulfide proteins of the intermembrane space, like Erv1p, are also implicated (30).
The heme-regulated Qcr2p, subunit 2 of the mitochondrial ubiquinol cytochrome c reductase complex, and the dually localized Yhb1p have reversibly oxidized Cys in the mutants, although it is unknown how this modification affects the activity of the proteins. Indeed it has been suggested that Yhb1p may regulate iron reductase activity in plasma membrane of yeast cells (36), and this activity is slightly but significantly diminished in the mutants (see Fig. 5D).
Redox Control of Porphyrin Biosynthesis
Iron homeostasis may also be affected in the Δgrx2 mutant at the level of heme and siroheme biosynthesis because two key enzymes of the cytosolic part of these pathways, Hem12p and Sam1p, are oxidized at critical cysteine residues. Hem12p is known to be thiol-dependent and can be inactivated by sulfhydryl-specific reagents in yeast (32), although conserved Cys52 by itself is not essential for enzymatic activity (51). The oxidized residue in Δgrx2 cells, Cys26, is located in the sequence just after two successive proline residues and immediately preceding the conserved active site. A possible decrease in the activity of Hem12p caused by oxidation of Cys26 in the absence of Grx2p would slow down the rate of heme synthesis.
Redox regulation of Hem12p activity is worthy of further discussion because it may have implications in other organisms far from yeast. This enzyme is a constitutive protein; hence, postranslational modulation by thiol redox changes could provide a rapid means to control heme biosynthesis as a function of cytosolic redox state. Mutations in the human orthologue of HEM12 are responsible for the diseases hepatoerythropoietic porphyria and prophyria cutanea tarda, which shows the importance of this enzyme in porphyrin biosynthesis. The sequence of the peptide containing Cys26, VERPPCWIMR, is unique to this enzyme but only conserved in fungi (S. cerevisiae, Candida, and others), algae (Chlamydomonas, Synechococcus, and Prochlorococcus) plant (Zea mays), and some protobacteria, as determined by full pBLAST. In plants, chlorophyll biosynthesis has to be regulated by light/darkness fluctuations through the redox state of the chloroplast (52), and Hem12p orthologue (UROD) has been reported as a candidate for Trx-dependent redox control, although the affected Cys residue was not identified (34). One interesting extrapolation of our results suggests that UROD at Cys26 could be a site for this regulatory action in the chloroplast. The influence of Grx2p in tetrapirrole biosynthesis could go deeper because the divergent biosynthetic pathway to siroheme (and to chlorophyll in plants) has a downstream requirement for S-adenosylmethionine, and the redox-regulated enzyme responsible for its synthesis, Sam1p, is reversibly oxidized and presumably inactivated (33), in the Δgrx2 mutant.
Our findings show that in the absence of Grx2p, the cell responds by up-regulating respiratory genes, mimicking a situation of intermediate oxygen tension governed by Hap2/3/4/5p transcription factor (39). There is a strict correlation between oxygen tension and heme concentration (53), with heme serving as a secondary signal for oxygen. Moreover, when heme synthesis and oxygen tension are unbalanced, the intracellular heme concentration, not oxygen, controls the expression of respiratory genes (54). Heme-responsive transcription factor Hap1p is inactive under low heme concentration, and its target gene ROX1 is down-regulated (54). Low levels of its protein product, the aerobic repressor Rox1p, would alleviate expression of GSY1/2 (55) and CYC7 (56), coding for glycogen synthase and cytochrome c-iso-2, respectively, exactly what happens in the Δgrx2 mutant.
Conclusions
Redox proteomics coupled to transcriptomics analyses has provided insights on unprecedented regulatory mechanisms of metabolic flow and gene expression triggered by posttranslational modification of specific proteins in cells growing under optimal conditions. We have obtained a snapshot that may be indicative of disease-prone conditions where the loss of activity of one enzyme has no physiological or phenotypic trait, yet there are a number of metabolic routes that are affected. Redox regulation of metabolism through reversible thiol modifications was initially discovered in chloroplast as a response of Calvin cycle enzymes to light/darkness fluctuations, and the role of thioredoxins was evidenced therein. The concept has extended to regulation of a variety of processes from transcription factors to cell signaling in many organisms. Here we provide evidence that Cys-targeted redox regulation of certain metabolic pathways is universal and takes place for equivalent enzymes in non-photosynthetic organisms, although it is dependent on Grx instead of Trx. The absence of Grx2p in yeast changes the metabolism from active biosynthesis of amino acids, nucleotides, and porphyrins to conservative synthesis of storage carbohydrates. The mutant lacking mitochondrial Prx1p is also a model for dysfunction of mitochondria to cytosol communication that leads to induction of the Aft1p-dependent iron regulon without apparent disturbance of mitochondrial Fe-S biogenesis. The characteristics of the mitochondrially originated activation of Aft1p incorporate Grx2p into Fe-S and heme metabolism, joining other members of the Grx family, Grx3 to -7, and open new doors to our understanding of the interactions between mitochondria and cytosol.
Supplementary Material
Acknowledgments
We thank Lourdes Laura Muñoz for excellent technical assistance; L. Vernis for kindly providing anti-Dre2 antibody; and J. Ariño, A. Barceló, and X. Castells (Universidad Autónoma Barcelona) for expression analysis. Mass spectrometry was performed at the Proteomics Facility, SCAI, University of Córdoba, node 6 of the ProteoRed Consortium financed by Genoma España and ISCIII and part of the Andalusian Platform for Genomics, Proteomics, and Bioinformatics.
This work was supported by Andalusian Government Grant P06-CVI-01611 and Spanish Government Grants BFU2006-02990 and BFU2009-08004.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- Grx
- glutaredoxin(s)
- Trx
- thioredoxin(s)
- ROS
- reactive oxygen species.
REFERENCES
- 1. Jones D. P. (2008) Am. J. Physiol. Cell Physiol. 295, C849–C868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buchanan B. B., Balmer Y. (2005) Annu. Rev. Plant Biol. 56, 187–220 [DOI] [PubMed] [Google Scholar]
- 3. Gallogly M. M., Mieyal J. J. (2007) Curr. Opin. Pharmacol. 7, 381–391 [DOI] [PubMed] [Google Scholar]
- 4. Porras P., Padilla C. A., Krayl M., Voos W., Bárcena J. A. (2006) J. Biol. Chem. 281, 16551–16562 [DOI] [PubMed] [Google Scholar]
- 5. Pedrajas J. R., Miranda-Vizuete A., Javanmardy N., Gustafsson J. A., Spyrou G. (2000) J. Biol. Chem. 275, 16296–16301 [DOI] [PubMed] [Google Scholar]
- 6. Cox A. G., Winterbourn C. C., Hampton M. B. (2010) Biochem. J. 425, 313–325 [DOI] [PubMed] [Google Scholar]
- 7. Pedrajas J. R., Padilla C. A., McDonagh B., Bárcena J. A. (2010) Antioxid. Redox Signal. 13, 249–258 [DOI] [PubMed] [Google Scholar]
- 8. Gstaiger M., Aebersold R. (2009) Nat. Rev. Genet. 10, 617–627 [DOI] [PubMed] [Google Scholar]
- 9. McDonagh B., Ogueta S., Lasarte G., Padilla C. A., Bárcena J. A. (2009) J. Proteomics 72, 677–689 [DOI] [PubMed] [Google Scholar]
- 10. Rodríguez-Manzaneque M. T., Tamarit J., Bellí G., Ros J., Herrero E. (2002) Mol. Biol. Cell 13, 1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McDonagh B., Tyther R., Sheehan D. (2005) Aquat. Toxicol. 73, 315–326 [DOI] [PubMed] [Google Scholar]
- 12. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 13. Moore R. E., Young M. K., Lee T. D. (2002) J. Am. Soc. Mass Spectrom. 13, 378–386 [DOI] [PubMed] [Google Scholar]
- 14. Elias J. E., Gygi S. P. (2007) Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 15. Alberola T. M., García-Martínez J., Antúnez O., Viladevall L., Barceló A., Ariño J., Pérez-Ortín J. E. (2004) Int. Microbiol. 7, 199–206 [PubMed] [Google Scholar]
- 16. Viladevall L., Serrano R., Ruiz A., Domenech G., Giraldo J., Barceló A., Ariño J. (2004) J. Biol. Chem. 279, 43614–43624 [DOI] [PubMed] [Google Scholar]
- 17. Lushchak V., Semchyshyn H., Mandryk S., Lushchak O. (2005) Arch. Biochem. Biophys. 441, 35–40 [DOI] [PubMed] [Google Scholar]
- 18. Wallace M. A., Liou L. L., Martins J., Clement M. H., Bailey S., Longo V. D., Valentine J. S., Gralla E. B. (2004) J. Biol. Chem. 279, 32055–32062 [DOI] [PubMed] [Google Scholar]
- 19. Riemer J., Hoepken H. H., Czerwinska H., Robinson S. R., Dringen R. (2004) Anal. Biochem. 331, 370–375 [DOI] [PubMed] [Google Scholar]
- 20. Shakoury-Elizeh M., Protchenko O., Berger A., Cox J., Gable K., Dunn T. M., Prinz W. A., Bard M., Philpott C. C. (2010) J. Biol. Chem. 285, 14823–14833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dancis A., Klausner R. D., Hinnebusch A. G., Barriocanal J. G. (1990) Mol. Cell Biol. 10, 2294–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Le Moan N., Clement G., Le Maout S., Tacnet F., Toledano M. B. (2006) J. Biol. Chem. 281, 10420–10430 [DOI] [PubMed] [Google Scholar]
- 23. Nilsson T., Mann M., Aebersold R., Yates J. R., 3rd, Bairoch A., Bergeron J. J. (2010) Nat. Methods 7, 681–685 [DOI] [PubMed] [Google Scholar]
- 24. Malmström J., Lee H., Aebersold R. (2007) Curr. Opin. Biotechnol. 18, 378–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jin C., Barrientos A., Tzagoloff A. (2003) J. Biol. Chem. 278, 14698–14703 [DOI] [PubMed] [Google Scholar]
- 26. Klomsiri C., Karplus P. A., Poole L. B. (2011) Antioxid. Redox Signal. 14, 1065–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Coles S. J., Easton P., Sharrod H., Hutson S. M., Hancock J., Patel V. B., Conway M. E. (2009) Biochemistry 48, 645–656 [DOI] [PubMed] [Google Scholar]
- 28. He Y. X., Gui L., Liu Y. Z., Du Y., Zhou Y., Li P., Zhou C. Z. (2009) Proteins 76, 249–254 [DOI] [PubMed] [Google Scholar]
- 29. Abid R., Ueda K., Miyazaki M. (1997) Biol. Signals 6, 157–165 [DOI] [PubMed] [Google Scholar]
- 30. Lill R., Mühlenhoff U. (2008) Annu. Rev. Biochem. 77, 669–700 [DOI] [PubMed] [Google Scholar]
- 31. Helmstaedt K., Strittmatter A., Lipscomb W. N., Braus G. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9784–9789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Felix F., Brouillet N. (1990) Eur. J. Biochem. 188, 393–403 [DOI] [PubMed] [Google Scholar]
- 33. Martínez-Chantar M. L., Pajares M. A. (1996) FEBS Lett. 397, 293–297 [DOI] [PubMed] [Google Scholar]
- 34. Balmer Y., Koller A., del Val G., Manieri W., Schürmann P., Buchanan B. B. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Binley K. M., Radcliffe P. A., Trevethick J., Duffy K. A., Sudbery P. E. (1999) Yeast 15, 1459–1469 [DOI] [PubMed] [Google Scholar]
- 36. Lewinska A., Bartosz G. (2006) Redox Rep. 11, 231–239 [DOI] [PubMed] [Google Scholar]
- 37. Parks L. W., Smith S. J., Crowley J. H. (1995) Lipids 30, 227–230 [DOI] [PubMed] [Google Scholar]
- 38. Rahier A., Pierre S., Riveill G., Karst F. (2008) Biochem. J. 414, 247–259 [DOI] [PubMed] [Google Scholar]
- 39. Rintala E., Toivari M., Pitkänen J. P., Wiebe M. G., Ruohonen L., Penttilä M. (2009) BMC Genomics 10, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hazelwood L. A., Walsh M. C., Luttik M. A., Daran-Lapujade P., Pronk J. T., Daran J. M. (2009) Appl. Environ. Microbiol. 75, 6876–6885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Molina M. M., Bellí G., de la Torre M. A., Rodríguez-Manzaneque M. T., Herrero E. (2004) J. Biol. Chem. 279, 51923–51930 [DOI] [PubMed] [Google Scholar]
- 42. Wang Q., Yu L., Yu C. A. (2010) J. Biol. Chem. 285, 10408–10414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rutherford J. C., Ojeda L., Balk J., Mühlenhoff U., Lill R., Winge D. R. (2005) J. Biol. Chem. 280, 10135–10140 [DOI] [PubMed] [Google Scholar]
- 44. Zhang Y., Lyver E. R., Nakamaru-Ogiso E., Yoon H., Amutha B., Lee D. W., Bi E., Ohnishi T., Daldal F., Pain D., Dancis A. (2008) Mol. Cell Biol. 28, 5569–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vernis L., Facca C., Delagoutte E., Soler N., Chanet R., Guiard B., Faye G., Baldacci G. (2009) PLoS One 4, e4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boisnard S., Lagniel G., Garmendia-Torres C., Molin M., Boy-Marcotte E., Jacquet M., Toledano M. B., Labarre J., Chédin S. (2009) Eukaryot. Cell 8, 1429–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilson W. A., Wang Z., Roach P. J. (2002) Mol. Cell Proteomics 1, 232–242 [DOI] [PubMed] [Google Scholar]
- 48. Li H., Mapolelo D. T., Dingra N. N., Naik S. G., Lees N. S., Hoffman B. M., Riggs-Gelasco P. J., Huynh B. H., Johnson M. K., Outten C. E. (2009) Biochemistry 48, 9569–9581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Crisp R. J., Pollington A., Galea C., Jaron S., Yamaguchi-Iwai Y., Kaplan J. (2003) J. Biol. Chem. 278, 45499–45506 [DOI] [PubMed] [Google Scholar]
- 50. Porras P., McDonagh B., Pedrajas J. R., Bárcena J. A., Padilla C. A. (2010) Biochim. Biophys. Acta 1804, 839–845 [DOI] [PubMed] [Google Scholar]
- 51. Di Flumeri C., Acheson N. H., Keng T. (1997) Can. J. Microbiol. 43, 792–795 [DOI] [PubMed] [Google Scholar]
- 52. Stenbaek A., Jensen P. E. (2010) Phytochemistry 71, 853–859 [DOI] [PubMed] [Google Scholar]
- 53. Zitomer R. S., Lowry C. V. (1992) Microbiol. Rev. 56, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang L., Hach A. (1999) Cell Mol. Life Sci. 56, 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Unnikrishnan I., Miller S., Meinke M., LaPorte D. C. (2003) J. Biol. Chem. 278, 26450–26457 [DOI] [PubMed] [Google Scholar]
- 56. Lowry C. V., Zitomer R. S. (1988) Mol. Cell Biol. 8, 4651–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.