Abstract
Hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) is a component of the ESCRT-0 protein complex that captures ubiquitylated cargo proteins and sorts them to the lysosomal pathway. Although Hrs acts as a key transporter for ubiquitin-dependent endosomal sorting, we previously reported that Hrs is also involved in ubiquitin-independent endosomal sorting of interleukin-2 receptor β (IL-2Rβ). Here, we show direct interactions between bacterially expressed Hrs and interleukin-4 receptor α (IL-4Rα), indicating that their binding is not required for ubiquitylation of the receptors, similar to the case for IL-2Rβ. Examinations of the Hrs binding regions of the receptors reveal that a hydrophobic amino acid cluster in both IL-2Rβ and IL-4Rα is essential for the binding. Whereas the wild-type receptors are delivered to LAMP1-positive late endosomes, mutant receptors lacking the hydrophobic amino acid cluster are sorted to lysobisphosphatidic acid-positive late endosomes rather than LAMP1-positive late endosomes. We also show that the degradation of these mutant receptors is attenuated. Accordingly, Hrs functions during ubiquitin-independent endosomal sorting of the receptors by recognizing the hydrophobic amino acid cluster. These findings suggest the existence of a group of cargo proteins that have this hydrophobic amino acid cluster as a ubiquitin-independent sorting signal.
Keywords: Cell Surface Receptor, Cytokine, Endosomes, Interleukin, Membrane Trafficking, Ubiquitin, Endosomal Sorting
Introduction
Endosomal sorting of cell surface receptors plays a key role in the destiny of the receptors, as some receptors in sorting endosomes are recycled back to the plasma membrane, whereas others are delivered to the lysosome for degradation and attenuation of receptor-mediated signal transduction. During endosomal sorting, ubiquitylation of the receptors serves as a sorting signal (1–3). Internalized receptors are pinched off from the plasma membrane into vesicles and then caught by the endosomal sorting complex required for transport (ESCRT)2 protein complexes ESCRT-I and ESCRT-II at the late endosome stage. These ESCRT complexes, which consist of class E vacuolar protein sorting (Vps) proteins, recognize ubiquitylated cargo or receptor proteins via subunits that interact directly with the ubiquitylated proteins (4–6). An ESCRT-I complex recruits an ESCRT-II complex on the endosomal membrane, and the recruited ESCRT-II complex stimulates the assembly of an ESCRT-III complex, which acts as the membrane abscission machinery for the budding membrane to form a multivesicular body (MVB) (7–11). Mature MVBs fuse with the lysosome, thereby releasing the receptor-containing vesicles into the hydrolytic lumen for degradation.
Hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) and signal transducing adaptor molecule (STAM) were identified as phosphotyrosine proteins after stimulation with hepatocyte growth factor (12) and interleukin-2 (IL-2) (13), respectively. Hrs (Vps27 in yeast) is associated with STAM (Hse1 in yeast) (14), and both proteins contain a ubiquitin-interacting motif (UIM) domain that binds to ubiquitylated cargo proteins (15–18). Because Hrs and STAM complexes recruit ESCRT-I during the process for endosomal sorting of ubiquitylated cargo or receptor proteins, these complexes are sometimes referred to as ESCRT-0 (19). Mutations of the UIM domain of Hrs abrogate its ability to bind to ubiquitylated proteins (20) and consequently prevent ubiquitylated cargo proteins from being sorted into the vacuolar lumen (21). Accordingly, Hrs appears to function as a critical transporter in the early stage of ubiquitin-dependent endosomal sorting.
IL-2 is a cytokine that is mainly produced by T cells and promotes T cell growth, differentiation, and activation of various types of hematopoietic cells through its interactions with IL-2 receptor (IL-2R) complexes. Functional IL-2R complexes are composed of α, β, and γc chains or β and γc chains, indicating that the β and γc chains are indispensable for the formation of functional IL-2R complexes (22). We previously found a direct interaction between bacterially expressed IL-2Rβ and Hrs, indicating that this binding is independent of ubiquitin (23). Furthermore, an IL-2Rβ mutant lacking the Hrs binding region exhibited impaired endosomal sorting to LAMP1-positive late endosomes, and the degradation rate of the IL-2Rβ mutant was diminished compared with that of wild-type IL-2Rβ (23). These findings led us to propose the following two hypotheses; 1) there is a group of cytokine receptors that interact with Hrs in a ubiquitin-independent manner, because it does not follow that only IL-2Rβ interacts with Hrs in this manner, and 2) there is a motif that interacts with Hrs in the cytoplasmic region of such receptors. In the present study, we tried to identify a ubiquitin-independent Hrs binding motif present in cytokine receptors.
EXPERIMENTAL PROCEDURES
Plasmids
Expression vectors containing HA-tagged wild-type Hrs (pKU-HrsHA) and its derivative mutants HrsHAd257–277 (dUIM) and HrsHAd428–466 were used (23). pME18s-IL-4Rα, which contains a human IL-4Rα cDNA, was kindly provided by S. Watanabe (The University of Tokyo, Tokyo, Japan). An expression vector for FLAG-tagged IL-4Rα (FLAG-IL4Rα) was generated by inserting the FLAG epitope (DYKDHDIDYKDDDDK) between amino acid positions Met-90 and Asp-91 in the IL-4Rα coding region of pME18s-IL4Rα using PCR. A series of IL-4Rα mutants were generated by PCR-based site-directed mutagenesis using FLAG-IL-4Rα as a template. FLAG-IL-4Rαd379, FLAG-IL-4Rαd399, FLAG-IL-4Rαd435, FLAG-IL-4Rαd400–418, FLAG-IL-4Rαd400–436, FLAG-IL-4RαmH, and FLAG-IL-4RαmA were IL-4Rα mutants with C-terminal truncations at positions 379, 399, and 435, deleted regions at positions 400–418 and 400–436, or mutations at positions 410–415 (410LFLDLL/AAADAA415) and 372–380 (372EEEEVEEE/AAAAVAAA380), respectively. Expression vectors containing FLAG-tagged wild-type IL-2Rβ (FLAG-IL-2Rβ) and its derivative mutants FLAG-IL-2Rβd268–348 and FLAG-IL-2Rβd349–410 were used (23). Additional IL-2Rβ mutants were created by PCR using FLAG-IL-2Rβ as a template. FLAG-IL-2RβmH1, FLAG-IL-2RβmH2, FLAG-IL-2RβmH3, FLAG-IL-2RβmA, and FLAG-IL-2RβmY were IL-2Rβ mutants with mutations at positions 336-338 (336LLL/AAA338), 365–369 (365FFFHL/AAAHA369), 407–411 (407PLQPL/AAQAA411), 389–394 (389EEDPDE/AAAPAA394) and 364–367 (364YFFF/AFFF367), respectively. pSRB5 is a human IL-2Rβ expression vector (24). Non-tagged pME18s-IL-4RαmH and pSRβmH2 were also constructed by PCR using plasmids pME18s-IL4Rα and pSRβ5, respectively, as templates. pMXs-IL-4Rα, pMXs-IL-2Rα, and pMXs-IL-2Rγc were constructed by insertion of human IL-4Rα, IL-2Rα, and IL-2Rγc cDNAs, respectively, into a Moloney murine leukemia virus-derived vector, pMXs. pMXs-IL-2Rα-β269–551 and pMXs-IL-2Rα-β365–369 were generated by inserting the cytoplasmic tail (residues 269–551) and hydrophobic amino acid cluster (residues 365–369), respectively, of human IL-2Rβ at the C terminus of IL-2Rα. pMXsβ constructed by the insertion of human IL-2Rβ into pMXs was used (23). pMXsβmHP2 and pMXs-IL-4RαmH were created by PCR-based site-directed mutagenesis using pMXsβ and pMXs-IL-4Rα, respectively, as templates. pcDNA3-HA-ubiquitin was generously provided by K. Miyazono (The University of Tokyo, Tokyo). All constructs were sequenced with an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems) to verify the amino acid changes.
Cell Lines
HEK293T cells and mouse embryonic fibroblast (MEF) cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and antibiotics. MEFβ, MEFβ-mH2, MEF-IL4Rα, and MEF-IL4Rα-mH were stable cell clones expressing wild-type IL-2Rβ, IL-2RβmH2 mutant, wild-type IL-4Rα, and IL-4RαmH mutant, respectively. To introduce these genes into MEF cells, we used a pMXs retrovirus vector system (kindly provided by T. Kitamura, The University of Tokyo) and obtained single clones by the limiting dilution method. The mouse IL-3-dependent pro-B cell line BAF-B03 was maintained in RPMI 1640 medium supplemented with 10% FBS, 10% conditioned medium derived from WEHI-3 cell line cultures (as a source of IL-3), 50 μm 2-mercaptoethanol, and antibiotics. BAFβ, BAFβ-mH2, BAF-IL-4Rα, and BAF-IL-4Rα-mH were stable cell clones expressing wild-type IL-2Rβ, IL-2RβmH2 mutant, wild-type IL-4Rα, and IL-4RαmH mutant, respectively. Each gene and a hygromycin-resistance gene were co-introduced by electroporation into BAF-B03 cells, and positive clones were selected by their hygromycin resistance and the limiting dilution method.
Flow Cytometry
Cell surface marker expressions were examined by flow cytometry. Cells (1 × 106) were incubated with 10% FBS in phosphate-buffered saline (PBS) for 30 min on ice and then incubated with an anti-IL-2Rβ monoclonal antibody (TU11) (25) or anti-IL-4Rα monoclonal antibody (MAB230) (R&D Systems) for 30 min on ice. After three washes with PBS, the cells were incubated with a FITC-conjugated secondary antibody (MP Biomedicals) for 30 min on ice. After washing, the cells were fixed with 1% paraformaldehyde in PBS before analysis using a FACSCalibur (BD Biosciences).
Immunoprecipitation and Immunoblotting
Immunoprecipitation and immunoblotting were carried out as described previously (14). Briefly, HEK293T cells (1 × 106) were transfected with 3 μg of each vector using a calcium phosphate precipitation method and then lysed with Nonidet P-40 cell extraction buffer (1% Nonidet P-40, 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm Na3VO4, 2.5 mm sodium pyrophosphate, 1 mm β-glycerol phosphate, and 1 mm aprotinin). After preclearing, the lysates were immunoprecipitated with various antibodies immobilized on Protein A-Sepharose beads (GE Healthcare) at 4 °C for 1 h. The immunoprecipitates were extensively washed with a buffer (1% Nonidet P-40, 20 mm Tris-HCl, pH 7.5, 500 mm NaCl, 1 mm EDTA, 1 mm Na3VO4, 2.5 mm sodium pyrophosphate, 1 mm β-glycerol phosphate), boiled in SDS sample buffer, separated by SDS-PAGE, and transferred onto Immobilon-P membranes (Millipore). After blocking with 5% skim milk and 0.1% Tween 20 in Tris-buffered saline, the membranes were incubated with various primary antibodies at room temperature for 1 h, washed, and incubated with horseradish-peroxidase-conjugated secondary antibodies (Cell Signaling Technology) at room temperature for 1 h. After a thorough washing, the positive signals were visualized using the ImmobilonTM Western substrate (Millipore). The primary antibodies utilized were: rabbit anti-HA, anti-IL-2Rβ (C20), anti-IL-4Rα (C20), rabbit anti-STAT5 (C17), and rabbit anti-phospho-STAT6 (Y641) antibodies (Santa Cruz Biotechnology, Inc.), rabbit anti-STAT6 antibody and mouse anti-phosphotyrosine antibody (PY100) (Cell Signaling Technology), mouse anti-FLAG monoclonal antibody (M2) (Sigma), rabbit anti-phospho-STAT5 antibody (EPITOMICS), and rat anti-Hrs monoclonal antibody (Imos-1) (14).
Immunofluorescence Microscopy and Immunostaining
Cells grown on coverslips were fixed with 3% paraformaldehyde in PBS for 15 min at room temperature, permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature or iced methanol for 5 min on ice, and incubated with 10% FBS in PBS for 30 min to block nonspecific antibody binding. For immunostaining, the cells were incubated with various primary antibodies at 30 °C for 1 h, washed 3 times with PBS, and incubated with appropriate secondary antibodies (anti-rabbit, anti-mouse, and anti-rat IgG antibodies conjugated with Alexa 488, Alexa 594, and FITC, respectively (Molecular Probes)) at 30 °C for 1 h. The coverslips were washed 5 times with PBS and mounted on glass slides with glycerol containing 0.1% p-phenylenediamine. Fluorescence images were captured using a confocal microscope (TCS SP2 (Leica) or LSM EXCITER (Carl Zeiss)). The primary antibodies utilized were: rabbit anti-IL-2Rβ antibody (C20), rabbit anti-IL-4Rα antibody (C20), mouse anti-IL-2Rβ antibody (TU11), mouse anti-IL-4Rα antibody (MAB230), rabbit anti-Hrs antibody (14), rat anti-mouse LAMP1 monoclonal antibody (1D4B), and rat anti-mouse transferrin receptor monoclonal antibody (R17217.1.4) (Santa Cruz Biotechnology Inc.); mouse anti-IL-2Rα antibody (H-31) (26), rat anti-IL-2Rγc antibody (TUGh4) (27), and mouse anti-lysobisphosphatidic acid (LBPA) monoclonal antibody (generously provided by T. Kobayashi, RIKEN Advanced Science Institute, Saitama, Japan).
GST Pulldown Assay
A His-tagged Hrs (His-Hrs) expression vector was constructed by insertion of wild-type Hrs into pCOLD® (TaKaRa). GST-tagged IL-2Rβ (GST-IL-2Rβcy) and IL-4Rα (GST-IL-4Rαcy) expression vectors were generated by ligating the cytoplasmic tails of IL-2Rβ (amino acids 296–551) and IL-4Rα (amino acids 257–825), respectively, into pGEX-4T-1 (GE Healthcare). GST-IL-2RβcymH2 and GST-IL-4RαcymH were created by PCR using GST-IL-2Rβcy and GST-IL-4Rαcy, respectively, as templates. The BL21 strain of Escherichia coli was used to express the proteins, and the His-Hrs and GST fusion proteins were purified using nickel-nitrilotriacetic acid-agarose (Qiagen) and glutathione-Sepharose 4B beads (GE Healthcare), respectively. To investigate the interactions between Hrs and IL-2Rβ or IL-4Rα in vitro, glutathione-Sepharose 4B beads containing the immobilized GST fusion proteins were incubated with His-Hrs in a buffer (1% Nonidet P-40, 50 mm Hepes, pH 7.4, 150 mm NaCl, 0.1 mm EDTA, 1 mm PMSF) at 30 °C for 15 min. The beads were washed 3 times with washing buffer (1% Nonidet P-40, 50 mm Hepes, pH 7.4, 500 mm NaCl, 0.1 mm EDTA), and the bound proteins were analyzed by immunoblotting with an anti-His monoclonal antibody (Cell Signaling Technology).
Internalization and Degradation Assay
Anti-IL-2Rβ antibody (TU-11) and anti-IL-4Rα antibody (MAB230) were radiolabeled with Na125I (MP Biomedicals) by the chloramine-T method. Recombinant IL-2 (Shionogi Co.) and recombinant IL-4 (PeproTech) were radiolabeled with Na125I using a modified iodogen method (28). The specific activities of 125I-TU-11, 125I-MAB230, 125I-IL-2, and 125I-IL-4 were 4.8 × 106, 4.7 × 106, 2.0 × 106, and 9.5 × 105 dpm/pmol, respectively. For internalization assays, cells (2 × 106) were incubated with PBS, 10% FBS containing an 125I-labeled antibody or ligand at 0 °C for 15 min. The cells were then washed 3 times with PBS, resuspended in RPMI/10% FBS, and incubated at 37 °C in a 5% CO2 incubator for specified times. After centrifugation of the cell suspensions, the culture supernatants were collected, and the cell pellets were treated with chilled glycine buffer (200 mm glycine, pH 2.2, 150 mm NaCl) on ice for 10 min. The radioactivities of the culture supernatant fractions, acid-removable glycine buffer fractions, and acid-unremovable cell precipitates were counted. For degradation assays, cells (2 × 106) were incubated with PBS, 10% FBS containing an 125I-labeled antibody or ligand at 0 °C for 15 min and washed with a cross-linking buffer (PBS containing 1 mm MgCl2, pH 8.3). To cross-link cell surface receptors with the 125I-labeled components, the cells were incubated with 300 μm dithiobis(sulfosuccinimidylpropionate) (DTSSP; Thermo Fisher Scientific, Inc.) in the cross-linking buffer on ice for 10 min. The cells were then washed twice with PBS containing 50 mm Tris-HCl, pH 7.4, resuspended in RPMI, 10% FBS, and incubated at 37 °C in a 5% CO2 incubator for the specified times. After centrifugation of the cell suspensions, the radioactivities of the culture supernatants and cell pellets were counted. The culture supernatants were further treated with 10% trichloroacetic acid (TCA) and incubated at 4 °C overnight. The radioactivities of the TCA-soluble fractions were counted.
Colocalization Image Analysis
Quantification of colocalization images captured with an LSM EXCITER confocal microscope (Carl Zeiss) was carried out using the ZEN2009 software (Carl Zeiss). Confocal images of single cells were captured randomly (carefully avoiding signal saturation), and each image was redistributed into 0–255 intensity levels. The fluorescence signals of more than 100 intensity levels per pixel were counted (29). The primary antibodies utilized were: rabbit anti-IL-2Rβ antibody (C20), rabbit anti-IL-4Rα antibody (C20), rat anti-mouse LAMP1 monoclonal antibody (1D4B), and mouse anti-LBPA monoclonal antibody.
RESULTS
UIM-deleted Hrs Interacts with IL-4Rα
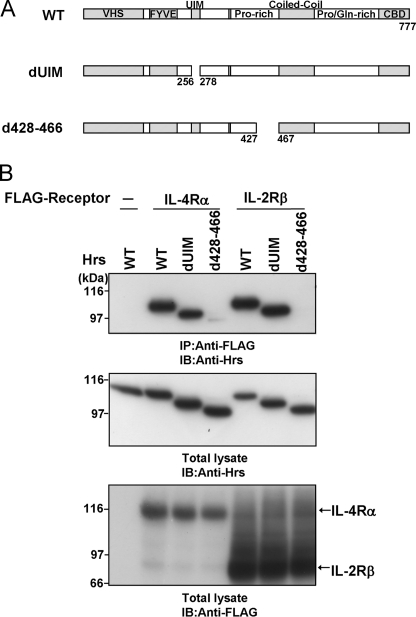
We previously found that IL-2Rβ interacts with Hrs in a ubiquitin-independent manner (23). Thus, analyses of the cytoplasmic regions of both IL-2Rβ and another cytokine receptor that interacts with Hrs in the same manner will contribute to the identification of an Hrs binding motif in the ubiquitin-independent Hrs binding regions of the receptors. The receptors for IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21 utilize the IL-2Rγc chain as an essential subunit (14, 22), suggesting the possibility that these receptors may have similar biological and biochemical characters for endosomal sorting. Therefore, we selected the IL-4 receptor α chain (IL-4Rα) and examined this possibility. The UIM domain of Hrs is necessary for ubiquitin binding and sorting of ubiquitylated cargo proteins into the MVB pathway (16, 20, 21). We previously reported that a UIM-deleted Hrs mutant can bind to IL-2Rβ (23). Accordingly, to examine whether the UIM-deleted mutant can bind to IL-4Rα, we performed coimmunoprecipitation assays. Human embryonic kidney-derived (HEK) 293T cells were transiently transfected with FLAG-tagged IL-4Rα or IL-2Rβ and Hrs expression vectors (Fig. 1A). Lysates of the HEK293T cells were immunoprecipitated with an anti-FLAG antibody and immunoblotted with an anti-Hrs antibody. Similar to IL-2Rβ, wild-type Hrs and the UIM-deleted mutant were clearly coimmunoprecipitated with IL-4Rα (Fig. 1B). We also previously reported that amino acids 428–466 of Hrs are a key domain for its ubiquitin-independent binding to IL-2Rβ (23). The association of Hrs lacking amino acids 428–466 with IL-4Rα was drastically reduced, similar to the case for IL-2Rβ (Fig. 1B). These results suggest that the manner of IL-4Rα binding to Hrs is similar to that of IL-2Rβ.
FIGURE 1.
The UIM domain of Hrs is dispensable for the interaction between Hrs and IL-4Rα. A, structures of wild-type Hrs and its mutants are shown. The Vps27-Hrs-STAM (VHS), Fab1-YGL023-Vps27-EEA1 (FYVE), ubiquitin interacting motif (UIM), proline (Pro)-rich region, coiled-coil, proline/glutamine (Pro/Gln)-rich region, and clathrin binding domain (CBD) are indicated. B, HEK293T cells (1 × 106) were cotransfected with 3 μg of wild-type FLAG-IL-2Rβ, wild-type FLAG-IL-4Rα, or empty vector and 3 μg of wild-type Hrs or its mutants. Aliquots (400 μg) of the cell lysates were immunoprecipitated with an anti-FLAG monoclonal antibody and immunoblotted with an anti-Hrs monoclonal antibody (top panel). The expression levels of Hrs and FLAG-tagged receptors in total lysates of the transfected HEK293T cells were examined by immunoblotting with an anti-Hrs monoclonal antibody and anti-FLAG monoclonal antibody, respectively. Total lysate, aliquots (10 μg) of the lysates were immunoblotted with an anti-Hrs antibody (middle panel) or anti-FLAG monoclonal antibody (lower panel). IP, immunoprecipitation; IB, immunoblotting.
Hrs Binding Motif in IL-4Rα and IL-2Rβ
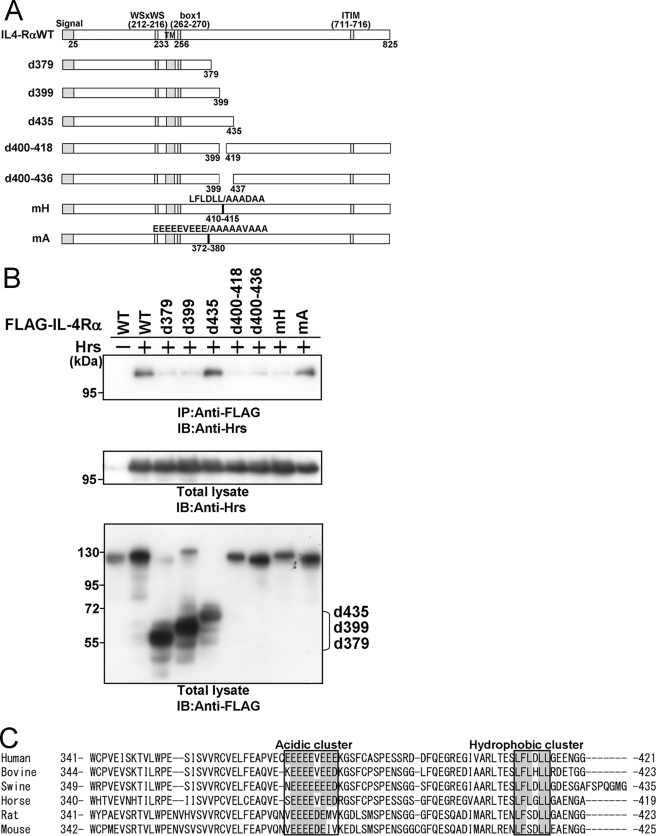
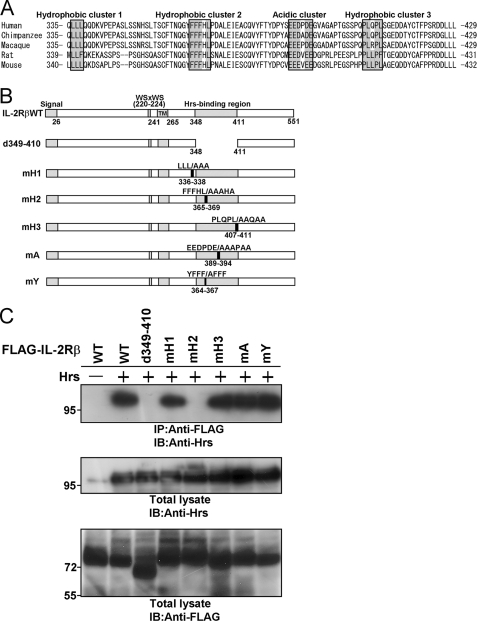
To define the region of IL-4Rα required for its interaction with Hrs, we constructed IL-4Rα mutants truncated at amino acids 379, 399, and 435 (Fig. 2A) and investigated their binding abilities toward Hrs by coimmunoprecipitation analyses (Fig. 2B). Hrs binding was clearly detected in the lysates of HEK293T cells transfected with the IL-4Rα mutant comprising amino acids 1–435 (d435) but was markedly reduced after coimmunoprecipitation with the mutants comprising amino acids 1–379 (d379) and 1–399 (d399). These observations suggested that amino acids 400–435 include an important region for Hrs binding. Further experiments revealed that deletion mutants lacking amino acids 400–418 (d400–418) and 400–436 (d400–436) (Fig. 2A) were hardly able to bind to Hrs (Fig. 2B). These findings suggest the possibility that there is an amino acid cluster or motif for Hrs binding within amino acids 400–418. We then aligned the amino acid sequences of human, bovine, swine, horse, rat, and mouse IL-4Rα and found a hydrophobic amino acid cluster within amino acids 400–418 (Fig. 2C). In addition, we detected an acidic cluster (amino acids 372–380) (Fig. 2C) that is often identified as a binding motif between proteins during endosomal sorting (1). We constructed an mH mutant in which the hydrophobic amino acids were substituted with alanine within residues 410–415 and an mA mutant in which the acidic amino acids were substituted with alanine within residues 372–380. Although significant binding was observed between Hrs and the mA mutant, an association of the mH mutant with Hrs was barely detectable (Fig. 2B). Accordingly, we aligned the amino acid sequences of human and other mammalian IL-2Rβ proteins and found three hydrophobic clusters and one acidic amino acid cluster (Fig. 3A). We then substituted the hydrophobic amino acids in the three hydrophobic amino acid clusters in IL-2Rβ with alanine and generated the mH1, mH2, and mH3 mutants with mutations located at residues 336–338, 365–369, and 407–411, respectively (Fig. 3B). In addition, we substituted the acidic amino acids of the Hrs binding region in IL-2Rβ with alanine and generated the mA mutant with mutations located at residues 389–394 (Fig. 3B). The deletion mutant lacking residues 349–410 was previously reported as an IL-2Rβ mutant lacking Hrs binding ability (23). The mH2 and d349–410 mutants were completely unable to bind to Hrs, although the mH1, mH3, and mA mutants strongly bound to Hrs (Fig. 3C). Thus, one of the three hydrophobic amino acid clusters is involved in the interactions between these proteins, suggesting that a steric structure beside the cluster may play an important part in Hrs binding. We further generated an mY mutant in which tyrosine residue 364 was substituted with alanine, as the tyrosine at residue 364 is located at the N-terminal side of amino acids 365–369 (mutated area in the mH2 mutant) and corresponds to a tyrosine-based signal (YXXϕ (ϕ indicates a residue with bulky hydrophobic side chains)) involved in endosomal sorting (Fig. 3, A and B). The mY mutant as well as wild-type IL-2Rβ bound to Hrs (Fig. 3C). These findings indicate that tyrosine residue 364 and the acidic amino acid cluster are not essential for Hrs binding.
FIGURE 2.
A hydrophobic amino acid cluster in the cytoplasmic tail of IL-4Rα is required for Hrs binding. A, structures of wild-type IL-4Rα and its mutants are shown. The signal sequence, WSXWS (tryptophan, serine, any amino acid, tryptophan, serine) motif, transmembrane region (TM), box1 motif, and immunoreceptor tyrosine-based inhibitory motif (ITIM) are indicated. B, HEK293T cells (1 × 106) were cotransfected with 3 μg of FLAG-tagged wild-type IL-4Rα or its mutants and 3 μg of wild-type Hrs or empty vector. Aliquots (400 μg) of the cell lysates were immunoprecipitated with an anti-FLAG monoclonal antibody and immunoblotted with an anti-Hrs monoclonal antibody (top panel). The expression levels of Hrs and IL-4Rα were examined by immunoblotting with an anti-Hrs monoclonal antibody and anti-FLAG monoclonal antibody, respectively. Total lysate, aliquots (10 μg) of the lysates were immunoblotted with an anti-Hrs monoclonal antibody (middle panel) or anti-FLAG monoclonal antibody (lower panel). IP, immunoprecipitation; IB, immunoblotting. C, multiple alignment of the Hrs binding regions of human, bovine, swine, horse, rat, and mouse IL-4Rα is shown. The regions defined as acidic or hydrophobic clusters are boxed.
FIGURE 3.
A hydrophobic amino acid cluster in the cytoplasmic tail of IL-2Rβ is required for Hrs binding. A, multiple alignment of the Hrs binding regions of human, chimpanzee, macaque, rat, and mouse IL-2Rβ is shown. The regions defined as acidic and hydrophobic clusters are boxed. B, shown are structures of wild-type IL-2Rβ and its mutants. The signal sequence, WSXWS motif, transmembrane region (TM), and Hrs binding region are indicated. C, HEK293T cells (1 × 106) were cotransfected with 3 μg of FLAG-tagged wild-type IL-2Rβ or its mutants and 3 μg of wild-type Hrs or empty vector. Aliquots (400 μg) of the cell lysates were immunoprecipitated with an anti-FLAG monoclonal antibody and immunoblotted with an anti-Hrs monoclonal antibody (top panel). The expression levels of Hrs and IL-2Rβ were examined by immunoblotting with an anti-Hrs monoclonal antibody and anti-FLAG monoclonal antibody, respectively. Total lysate, aliquots (10 μg) of the lysates were immunoblotted with an anti-Hrs monoclonal antibody (middle panel) or anti-FLAG monoclonal antibody (lower panel). IP, immunoprecipitation; IB, immunoblotting.
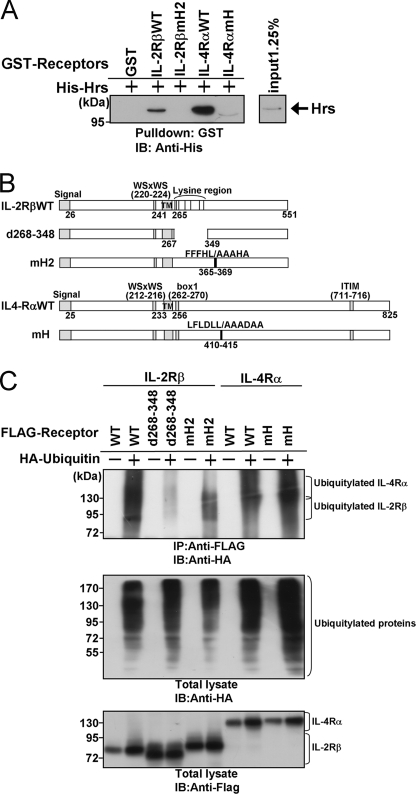
The Hydrophobic Amino Acid Cluster Is Involved in Direct Interactions between Hrs and IL-4Rα or IL-2Rβ
We previously identified a direct interaction between bacterially expressed IL-2Rβ and Hrs in GST pulldown assays (23). To explore whether the hydrophobic amino acid cluster is involved in the direct association with Hrs, we prepared purified protein extracts from E. coli expressing His-tagged Hrs, GST-tagged IL-2Rβ, and GST-tagged IL-2Rβ with the hydrophobic amino acid cluster substituted with alanine (IL-2RβmH2) or GST-tagged IL-4Rα or GST-tagged IL-4Rα with the hydrophobic amino acid cluster substituted with alanine (IL-4RαmH). The GST-tagged fusion proteins were immobilized on glutathione-Sepharose beads, and each fusion protein was incubated with His-tagged Hrs. The beads were washed, and bound material was immunoblotted with an anti-His antibody. Wild-type IL-2Rβ and IL-4Rα associated with His-tagged Hrs, whereas IL-2RβmH2, IL-4RαmH, and GST alone did not (Fig. 4A). These results suggested that the hydrophobic amino acid clusters in IL-2Rβ and IL-4Rα are involved in their Hrs binding abilities in a ubiquitylation-independent manner. Next, we examined whether the hydrophobic amino acid cluster affects the ubiquitylation of the receptors. HEK293T cells were transfected with HA-tagged ubiquitin and wild-type or mutant receptors as shown in Fig. 4C. Although the IL-2Rβ mutant lacking residues 268–348, in which all the lysine residues of the cytoplasmic tail are located, was hardly ubiquitylated, IL-2RβmH2 and IL-4RαmH were similarly ubiquitylated to the wild-type receptors (Fig. 4, B and C). These observations indicated that the ubiquitylation of the receptors is not affected by the issue of whether Hrs interacts with the receptors through the hydrophobic amino acid cluster.
FIGURE 4.
Hrs directly associates with the cytokine receptors IL-2Rβ and IL-4Rα by recognizing the hydrophobic amino acid cluster in a ubiquitin-independent manner. A, glutathione-Sepharose beads containing immobilized GST, GST-fused cytoplasmic tail fragment of IL-2Rβ (GST-IL-2RβcyWT or GST-IL-2RβcymH2), or GST-fused cytoplasmic tail fragment of IL-4Rα (GST-IL-4RαcyWT or GST-IL-4RαcymH) were incubated with His-tagged Hrs. The bound proteins were separated by SDS-PAGE and analyzed by immunoblotting with an anti-His tag antibody. The input control of His-Hrs is shown in the right panel. B, structures of the IL-2Rβ and IL-4Rα mutants used in the receptor ubiquitylation assays are shown. C, HEK293T cells were cotransfected with 2 μg of HA-ubiquitin or empty vector and 2 μg of FLAG-IL-2Rβ, FLAG-IL-2Rβd268–348, FLAG-IL-2RβmH2, FLAG-IL-4Rα, or FLAG-IL-4RαmH. Aliquots (200 μg) of the cell lysates were immunoprecipitated with an anti-FLAG monoclonal antibody and immunoblotted with an anti-HA monoclonal antibody (top panel). The expression levels of the receptors (IL-2Rβ and IL-4Rα) and ubiquitylated total proteins were examined by immunoblotting with an anti-FLAG monoclonal antibody and anti-HA monoclonal antibody, respectively. Total lysate, aliquots (10 μg) of the lysates were immunoblotted with an anti-HA antibody (middle panel) or anti-FLAG antibody (lower panel). IP, immunoprecipitation; IB, immunoblotting.
The Hydrophobic Amino Acid Clusters of IL-2Rβ and IL-4Rα Are Required for Endosomal Sorting to Late Endosomes
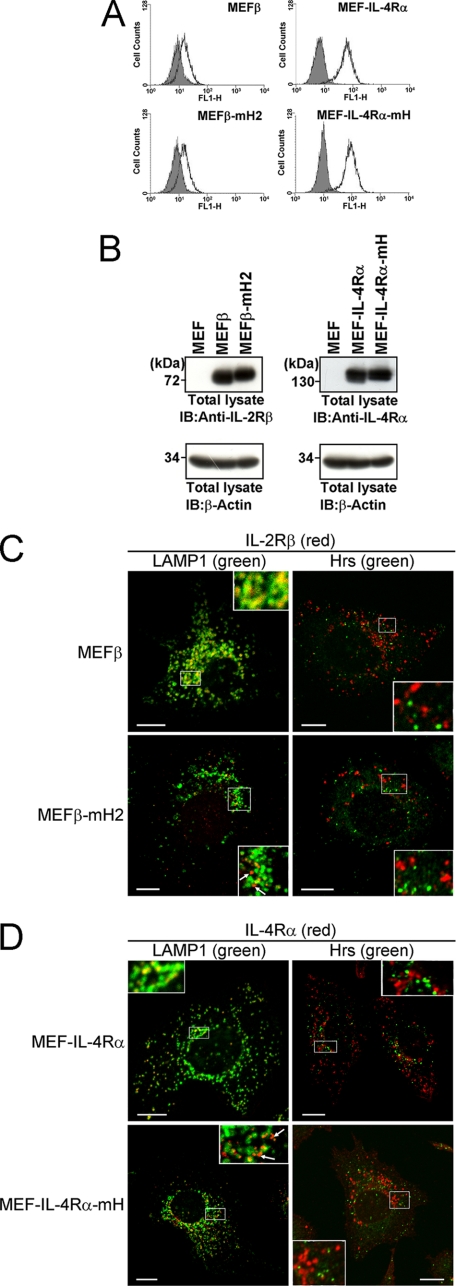
We previously examined the endocytic intracellular localization of IL-2Rβ by confocal microscopy using an anti-IL-2Rβ antibody together with antibodies against the early endosome marker EEA1 (or Hrs) or late endosome/lysosome marker LAMP1. The experiments revealed that IL-2Rβ is localized to LAMP1-positive compartments and rarely found in EEA1-positive and Hrs-positive compartments under steady-state conditions (23). In addition, a kinetics study on IL-2Rβ endosomal sorting revealed that IL-2Rβ is delivered to LAMP1-positive compartments through Hrs-positive compartments (23). Therefore, we investigated the effects of the hydrophobic amino acid cluster on the localizations of IL-2Rβ and IL-4Rα. cDNAs encoding the IL-2Rβ mutant with the hydrophobic amino acid cluster substituted with alanine and wild-type IL-2Rβ were introduced into a MEF cell line to generate MEFβ-mH2 and MEFβ cells, respectively. MEF-IL-4Rα-mH and MEF-IL-4Rα cells were similarly established as MEF cells expressing IL-4RαmH and IL-4Rα, respectively. FACS and immunoblotting analyses indicated that there were no significant differences between the amounts of the wild-type and mutant receptors on the cell surfaces or in the intracellular expressions of the transfectants (Fig. 5, A and B). Although wild-type IL-2Rβ in MEFβ cells and IL-4Rα in MEF-IL-4Rα cells were localized to LAMP1-positive compartments, the mutant IL-2Rβ in MEFβ-mH2 cells and mutant IL-4Rα in MEF-IL-4Rα-mH cells were localized to punctate structures in the cytoplasm rather than to LAMP1-positive compartments (Fig. 5,C and D). Taken together with the data shown in Fig. 4A, these findings suggest that the hydrophobic amino acid clusters involved in the interactions between Hrs and the receptors play a key role in the sorting to LAMP1-positive compartments.
FIGURE 5.
Effects of the hydrophobic amino acid clusters in IL-2Rβ and IL-4Rα on their late-endosomal localizations. A, the IL-2Rβ and IL-4Rα expression levels on the surface of MEF transfectants were examined by flow cytometry. MEFβ, MEFβ-mH2, MEF-IL-4Rα, and MEF-IL-4Rα-mH cells were incubated with an anti-IL-2Rβ monoclonal antibody (TU11) or anti-IL-4Rα monoclonal antibody (MAB230) followed by a FITC-conjugated secondary antibody. B, the expression levels of IL-2Rβ and IL-4Rα in the indicated cells were examined by immunoblotting. Aliquots (15 μg) of the total lysates were immunoblotted with an anti-IL-2Rβ antibody (C20) or anti-IL-4Rα antibody (C20) (upper panels) or with an anti-β-actin antibody (lower panels). C and D, fluorescence images were observed using a confocal laser microscope. The indicated cells were grown on coverslips, fixed, and double-labeled with an anti-IL-2Rβ antibody (TU11) or anti-IL-4Rα antibody (MAB230) and an anti-LAMP1 monoclonal antibody or anti-Hrs antibody. Subsequently, the cells were incubated with fluorescently labeled secondary antibodies. Fairly large amounts of IL-2RβmH2 and IL-4RαmH are not sorted to LAMP1-positive compartments (arrows). Scale bars, 10 μm. IB, immunoblotting.
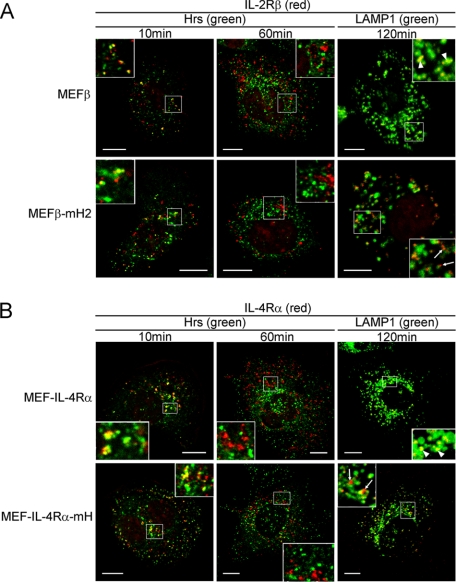
In a previous kinetics study on IL-2Rβ endosomal sorting, IL-2Rβd349–410 lacking amino acids 349–410, which include the hydrophobic amino acid cluster (residues 365–369), was observed in Hrs-positive compartments at 10 min after the receptor internalization and had departed from these compartments at 30 min (23). However, the sorting of IL-2Rβd349–410 to LAMP1-positive compartments was largely impaired (23). Therefore, we investigated whether IL-4RαmH and IL-2RβmH2 are delivered to the punctate structures shown in Fig. 5, C and D, through Hrs-positive compartments. MEFβ-mH2 and MEFβ cells were incubated at 0 °C to suppress the receptor internalization and treated with the anti-IL-2Rβ antibody TU11. The receptors were covalently linked to TU11 by incubation with DTSSP as a chemical cross-linker. Next, the cells were incubated at 37 °C for the indicated times and analyzed by confocal microscopy. IL-2Rβ and IL-2RβmH2 were observed in Hrs-positive compartments at 10 min and had departed from these compartments at 60 min (Fig. 6A). At 120 min, a large proportion of IL-2Rβ had been delivered to LAMP1-positive compartments, whereas IL-2RβmH2 had been partially delivered to LAMP1-positive compartments (Fig. 6A). Similarly, IL-4Rα was delivered to LAMP1-positive compartments through Hrs-positive compartments, whereas blockade of IL-4RαmH transportation to the LAMP1-positive compartments was observed (Fig. 6B). These findings indicate that the hydrophobic amino acid clusters in these receptors play a key role in endosomal sorting of the receptors from early to late endosomes.
FIGURE 6.
Kinetics of the endosomal localizations of IL-2Rβ and IL-4Rα. A and B, the indicated MEF transfectants were grown on coverslips, and the cell surface receptors were bound by an anti-IL-2Rβ antibody (TU11) or anti-IL-4Rα antibody (MAB230) at 0 °C followed by treatment with the chemical cross-linker DTSSP. The cells were cultured at 37 °C, fixed at the indicated times, and incubated with an anti-Hrs antibody or anti-LAMP1 monoclonal antibody. Fluorescence labeling was carried out for IL-2Rβ (red), IL-4Rα (red), Hrs (green), and LAMP1 (green). Large proportions of IL-2Rβ and IL-4Rα are delivered to LAMP1-positive compartments (arrowheads). In contrast, fairly large amounts of IL-2RβmH2 and IL-4RαmH are not delivered to LAMP1-positive compartments (arrows). Scale bars, 10 μm.
Subsequently, we examined whether the hydrophobic amino acid cluster introduced into a recycling receptor serves as the sorting signal to LAMP1-positive endosomes. We then constructed chimeric receptors of IL-2Rα, which is constitutively internalized and rapidly recycled back to the plasma membrane (30). The hydrophobic amino acid cluster (residues 365–369) or the cytoplasmic tail (residues 269–551) of IL-2Rβ was inserted at the C terminus of IL-2Rα (supplemental Fig. S1A). MEF cells transiently expressing wild-type IL-2Rα, the chimeric receptor including the residues 365–369 (IL-2Rα-β365–369), or the residues 269–551 (IL-2Rα-β269–551) were incubated at 0 °C and treated with the anti-IL-2Rα antibody H-31. Next, the cells were incubated at 37 °C for 120 min and analyzed by confocal microscopy. Although a large part of wild-type IL-2Rα was observed on the plasma membrane, IL-2Rα-β269–551, including the cytoplasmic tail of IL-2Rβ, was detected in LAMP1-positive compartments (supplemental Fig. S1B). On the other hand, IL-2Rα-β365–369, including the hydrophobic amino acid cluster of IL-2Rβ as well as wild-type IL-2Rα, was found on the plasma membrane, indicating that the other cytoplasmic region along with the hydrophobic amino acid cluster is also needed for the function as the sorting signal.
Involvement of the Hydrophobic Amino Acid Clusters in the Degradation of IL-2Rβ and IL-4Rα in BAF-B03 Transfectants
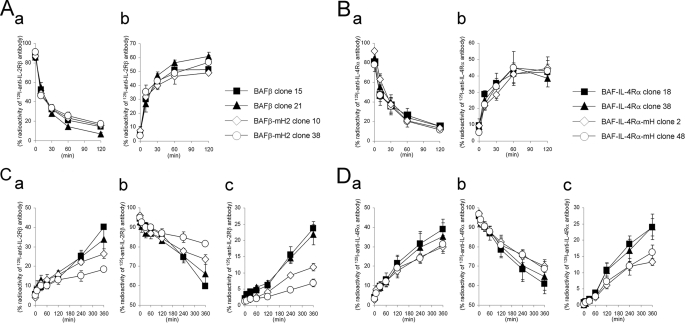
To analyze the internalization and degradation of IL-2RβmH2 and IL-4RαmH lacking the hydrophobic amino acid cluster required for Hrs binding, we used IL-2Rβ-deficient BAF-B03 cells expressing mouse IL-2Rα and IL-2Rγc. The mouse pro-B cell line BAF-B03 is an IL-3-dependent cell line, and its transfectants with human cytokine receptor genes (IL-2Rβ and IL-4Rα) have been used to analyze cytokine signal transduction (31, 32). cDNAs encoding the IL-2Rβ mutant with the hydrophobic amino acid cluster substituted with alanine and wild-type IL-2Rβ were introduced into BAF-B03 cells to generate BAFβ-mHP2 and BAFβ cells, respectively. BAF-IL-4Rα-mH and BAF-IL-4Rα cells were similarly established as BAF-B03 cells expressing IL-4RαmH and IL-4Rα, respectively. Using FACS analyses, we selected two types of cell clones; one expressing a higher amount of each receptor and the other expressing a lower amount of each receptor on the cell surface (supplemental Fig. S2, A and B). First, we examined the kinetics of the receptor internalization in the transfectants. BAFβ clone 15, BAFβ clone 21, BAFβ-mH2 clone 10, and BAFβ-mHP2 clone 38 cells were incubated with 125I-anti-IL-2Rβ antibody (TU11). TU11 does not block receptor assembly among the IL-2Rα, -β and, -γc chains (24). In addition, TU11 neither stimulates nor inhibits IL-2R-mediated cell growth (33). To evaluate the receptor internalization, the cells were collected at the indicated times (Fig. 7A). Treatment of the cells with 200 mm glycine buffer, pH 2.2, was used to distinguish the cell surface-bound 125I-TU11 from the intracellular 125I-TU11. The radioactivity of the cell surface-bound acid-removable 125I-TU11 decreased rapidly (Fig. 7Aa) accompanied by a rapid increase in the radioactivity of intracellular acid-unremovable 125I-TU11 (Fig. 7Ab), indicating that IL-2Rβ is constitutively internalized in the absence of the ligand, as previously reported (23). The kinetics of 125I-TU11 internalization in BAFβ-mHP2 cells was similar to that in BAFβ cells, and the different levels of receptor expression in these clones had no effect on the kinetics (Fig. 7A). Similar to BAFβ cells, BAF-IL-4Rα clone 18, BAF-IL-4Rα clone 38, BAF-IL-4Rα-mH clone 2, and BAF-IL-4Rα-mH clone 48 cells were incubated with an 125I-anti-IL-4Rα antibody, which blocks IL-4 binding to IL-4Rα, and then analyzed for the internalization of IL-4Rα. The kinetics of 125I-conjugated anti-IL-4Rα antibody internalization in BAF-IL-4Rα-mH cells was similar to that in BAF-IL-4Rα cells (Fig. 7B), and the kinetics of the blocking-type anti-IL-4Rα antibody was similar to that of TU11 (Fig. 7, A and B). Subsequently, we evaluated the receptor degradation based on the amount of radioactivity released into the culture supernatants by the cells. The transfectants expressing IL-2Rβ and IL-4Rα were incubated with the 125I-TU11 and 125I-anti-IL-4Rα antibodies, respectively, and the receptors were covalently linked with the 125I-TU11 or 125I-anti-IL-4Rα antibodies using the chemical cross-linker DTSSP before the culture supernatants were collected at the indicated times (Fig. 7, C and D). The radioactivities in the culture supernatants increased (Fig. 7, Ca and Da), and the increases were quantitatively correlated with decreases in the cell-bound radioactivity (Fig. 7, Cb and Db). The radioactivity in the culture supernatants of BAFβ-mHP2 clones was apparently lower than that of BAFβ clones (Fig. 7Ca), and the radioactivity in the culture supernatants of BAF-IL-4Rα-mH clones was apparently lower than that of BAF-IL-4Rα clones (Fig. 7Da). Furthermore, we extracted the degraded short peptides from the culture supernatants by TCA precipitation and found lower amounts of degraded short peptides in the culture supernatants of BAFβ-mHP2 and BAF-IL-4Rα-mH cells compared with those in the culture supernatants of BAFβ and BAF-IL-4Rα cells, respectively (Fig. 7, Cc and Dc). Taken together, these findings indicate that the degradation rates of the mutant receptors with the hydrophobic amino acid clusters substituted with alanine are lower than those of the wild-type receptors.
FIGURE 7.
Internalization and degradation of IL-2Rβ and IL-4Rα in BAF-B03 transfectants. A and B, shown is internalization of IL-2Rβ and IL-4Rα in the transfectants. BAF transfectants were incubated with 125I-anti-IL-2Rβ antibody (TU11) or 125I-anti-IL-4Rα antibody (MAB230) at 0 °C. The cells were then cultured at 37 °C and harvested at the indicated times. The radioactivities of the cell surface-bound acid-removable fractions (a) and intracellular acid-unremovable fractions (b) were counted. C and D, degradation of IL-2Rβ and IL-4Rα in the transfectants is shown. BAF transfectants were incubated with 125I-anti-IL-2Rβ antibody or 125I-anti-IL-4Rα antibody at 0 °C followed by treatment with the chemical cross-linker DTSSP. The cells were then cultured at 37 °C and harvested at the indicated times. The radioactivities of the culture supernatants (a), cell precipitate fractions (b), and TCA-soluble fractions of the culture supernatants (c) were counted. The values represent the means ± S.E. of three separate experiments.
As shown in Fig. 6, confocal microscopy analyses using MEFβ-mH2, MEFβ, MEF-IL-4Rα-mH, and MEF-IL-4Rα cells revealed that the sorting of IL-2RβmH2 and IL-4RαmH to LAMP1-positive compartments was partially impaired. Therefore, we used the MEF transfectants to investigate the receptor internalization and degradation. The kinetics of 125I-TU11 internalization in MEFβ-mHP2 cells was the same as that in MEFβ cells (supplemental Fig. S3A). In addition, the kinetics of 125I-anti-IL-4Rα antibody internalization in MEF-IL-4Rα-mH cells was the same as that in MEF-IL-4Rα cells (supplemental Fig. S3B). Subsequently, however, the radioactivities in the culture supernatants of MEFβ-mHP2 and MEF-IL-4Rα-mH cells were apparently lower than those in the culture supernatants of MEFβ and MEF-IL-4Rα cells, respectively (supplemental Fig. S3, C and D), and the amounts of the degraded short peptides in the culture supernatants of MEFβ-mHP2 and MEF-IL-4Rα-mH cells were lower than those in the culture supernatants of MEFβ and MEF-IL-4Rα cells, respectively (supplemental Fig. S3, Cc and Dc). Therefore, the lower amounts of the degraded short peptides observed for the mutant receptors may reflect a partial sorting of the mutant receptors to LAMP1-positive compartments.
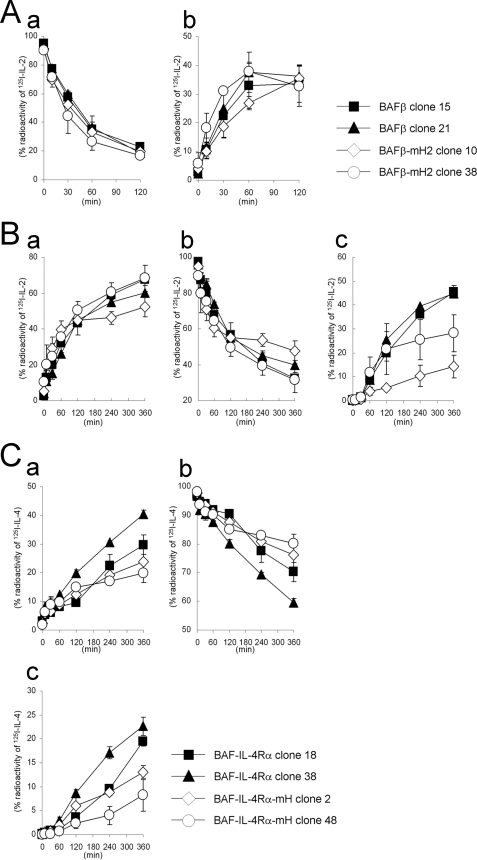
Next, we examined the kinetics of the ligand degradation in the BAF-B03 transfectants. The transfectants expressing IL-2Rβ were incubated with 125I-IL-2 and analyzed for IL-2 internalization. The radioactivity of the cell surface-bound acid-removable 125I-IL-2 decreased rapidly (Fig. 8Aa) accompanied by a rapid increase in the radioactivity of intracellular acid-unremovable 125I-IL-2 (Fig. 8Ab). The kinetics of 125I-IL-2 internalization in BAFβ-mH2 cells was similar to that in BAFβ cells (Fig. 8A). Subsequently, we evaluated the 125I-IL-2 degradation based on the amount of radioactivity released into the culture supernatants by the cells. The radioactivity in the culture supernatants increased rapidly (Fig. 8Ba), and the increase was quantitatively correlated with a decrease in the cell-bound radioactivity (Fig. 8Bb). The radioactivity in the culture supernatants of BAFβ-mHP2 clones was similar to that of BAFβ clones (Fig. 8Ba), but the amounts of the degraded short peptides obtained by TCA precipitation in the culture supernatants of BAFβ-mHP2 cells were apparently lower than those in the culture supernatants of BAFβ cells (Fig. 8Bc). We also tried to examine the kinetics of 125I-IL-4 degradation in the BAF-B03 transfectants. However, we found that a small amount of 125I-IL-4 was dissociated from the receptors during the indicated incubation times. Therefore, we used a chemical cross-linker to link 125I-IL-4 and the receptor. In degradation analyses, the radioactivity in the culture supernatants of BAF-IL-4Rα-mH cells was lower than that in the culture supernatants of BAF-IL-4Rα cells (Fig. 8C, a and b). Furthermore, the amounts of the degraded short peptides in the culture supernatants of BAF-IL-4Rα-mH cells were apparently lower than those in the culture supernatants of BAF-IL-4Rα cells (Fig. 8Cc). The degradation patterns of the antibodies against the receptors in the transfectants were similar to those of the ligands, suggesting that the antibodies were delivered by the same endosomal sorting route as the ligands. Therefore, these findings support the possibility that the hydrophobic amino acid clusters of the receptors are required for their accurate transport to the lysosome for degradation.
FIGURE 8.
Internalization and degradation of 125I-IL-2 and 125I-IL-4 in BAF-B03 transfectants. A, internalization of 125I-IL-2 in the transfectants is shown. BAF transfectants were incubated with 350 pm 125I-IL-2 at 0 °C. The cells were then cultured at 37 °C and harvested at the indicated times. The radioactivities of the cell surface-bound acid-removable fractions (a) and intracellular acid-unremovable fractions (b) were counted. B, degradation of 125I-IL-2 in the transfectants is shown. BAF transfectants were incubated with 350 pm 125I-IL-2 and then cultured for the indicated times. The radioactivities of the culture supernatants (a), cell precipitate fractions (b), and TCA-soluble fractions of the culture supernatants (c) were counted. C, degradation of 125I-IL-4 in the transfectants is shown. BAF transfectants were incubated with 250 pm 125I-IL-4, treated with the cross-linker DTSSP, and then cultured for the indicated times. The radioactivities of the culture supernatants (a), cell precipitate fractions (b), and TCA-soluble fractions of the culture supernatants (c) were counted. The values represent the means ± S.E. of three separate experiments.
On the other hand, the kinetics on the tyrosine phosphorylations of STAT5 and STAT6, downstream molecules of IL-2R and IL-4R, respectively, were similar between the wild-type and mutant receptor expressing cells (supplemental Fig. S4, A and B). The kinetics on the tyrosine phosphorylations of IL-2Rβ and IL-4Rα were also similar between the wild-type and mutant receptor expressing cells (supplemental Fig. S4, A and B). The difference of the receptor degradation pattern between the wild-type and mutant IL-2Rβ (or IL-4Rα) became marked after 120 min (Fig. 7), whereas the tyrosine phosphorylations of the receptors and the STAT molecules by the ligand stimulations were observed within 120 min. Thus, the kinetics on the tyrosine phosphorylations of these molecules may not be influenced by the differences between the wild-type and mutant receptor degradations.
Because functional IL-2R and IL-4R are composed of a combination of each the receptor subunit and IL-2Rγc, we examined whether IL-2Rγc is colocalized with each the subunit using a confocal microscope. Similar to the case of wild-type IL-2Rβ in MEFβ cells, IL-2RβmH2 expressed in MEFβ-mH2 was colocalized with IL-2Rγc under steady-state conditions in the presence of IL-2 (supplemental Fig. S5A). The degree of the colocalization between IL-2Rγc and IL-4RαmH in MEF-IL-4Rα-mH cells was also similar to that of the colocalization between IL-2Rγc and wild-type IL-4Rα in MEF-IL-4Rα cells in the presence of IL-4 (supplemental Fig. S5B). At 40 min after the ligand stimulation, wild-type IL-2Rβ had been departed from transferrin receptor-positive early endosomes as described previously (23). We then tried to examine the colocalization between IL-2Rβ and IL-2Rγc departed from early endosomes and here found that wild and the mutant IL-2Rβ were colocalized with IL-2Rγc at 40 min after IL-2 stimulation (supplemental Fig. S5C). These observations suggested that IL-2Rγc and its receptor counterpart stably exist in the same compartments during the course of the endosomal sorting.
The Mutant Receptors Lacking the Hydrophobic Amino Acid Cluster Are Localized to LBPA-positive Compartments Rather than LAMP1-positive Compartments
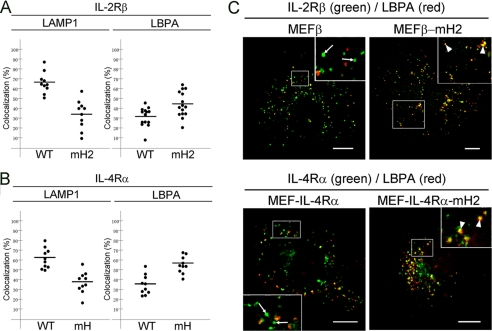
The data shown in Fig. 5, C and D, indicated that although the wild-type receptors were localized to LAMP1-positive compartments, the receptors lacking the hydrophobic amino acid cluster were found as punctuate structures that differed from the LAMP1-positive compartments. Previously, we reported that an IL-2Rβ mutant lacking amino acids 349–410, which includes the hydrophobic amino acid cluster, is partially mis-sorted to transferrin receptor-positive recycling endosomes, resulting in interference with its transport to LAMP1-positive compartments (23). Similar to the result of IL-2Rβ mutant lacking amino acids 349–410, the receptors lacking the hydrophobic amino acid cluster were also partially mis-sorted to transferrin receptor-positive recycling endosomes in the kinetics study of these receptors (supplemental Fig. S6). However, the localization of the mutant at recycling endosomes was hardly detectable under steady-state conditions. Thus, we further searched for the subcellular localizations of IL-2RβmH2 and IL-4RαmH under steady-state conditions. LBPA is a component of luminal vesicles in late endosomes and a marker of MVBs (34, 35). Therefore, using the MEF transfectant MEFβ-mH2, MEFβ, MEF-IL-4Rα-mH, and MEF-IL-4Rα cells, we tried to compare the ratios of mutant receptors localized to LAMP1-positive compartments with those localized to LBPA-positive compartments. Consistent with the results shown in Fig. 5, wild-type IL-2Rβ expressed in MEFβ cells was colocalized with 66% of LAMP1-positive compartments, whereas IL-2RβmH2 lacking the hydrophobic amino acid cluster expressed in MEFβ-mH2 cells was colocalized with 34% of LAMP1-positive compartments (Fig. 9A). In contrast, wild-type IL-2Rβ was colocalized with 32% of LBPA-positive compartments, whereas IL-2RβmH2 was colocalized with 44% of LBPA-positive compartments (Fig. 9, A and C). Similar results were obtained for the localizations of IL-4Rα in MEF-IL-4Rα cells and IL-4RαmH in MEF-IL-4Rα-mH cells. Wild-type IL-4Rα and IL-4RαmH lacking the hydrophobic amino acid cluster were colocalized with 62 and 38% of LAMP1-positive compartments, respectively (Fig. 9B), whereas wild-type IL-4Rα and IL-4RαmH were colocalized with 35 and 56% of LBPA-positive compartments, respectively (Fig. 9, B and C). These findings suggest that some of the mutant receptors lacking the hydrophobic amino acid cluster are localized to LBPA-positive compartments rather than LAMP1-positive compartments and that the transport of the mutant receptors to LAMP1-positive compartments is partially impaired, resulting in accumulation of the mutant receptors in LBPA-positive compartments.
FIGURE 9.
Localizations of IL-2Rβ and IL-4Rα mutants lacking the hydrophobic amino acid cluster to LBPA-positive compartments. MEF transfectants were grown on coverslips, fixed, and double-labeled with an anti-IL-2Rβ antibody (C20) or anti-IL-4Rα antibody (C20) and an anti-LAMP1 monoclonal antibody or anti-LBPA monoclonal antibody. Fluorescence images of the MEF transfectants (n ≥ 10 cells) were captured using a confocal laser microscope. The percentages of the IL-2Rβ-positive (A) and IL-4Rα-positive (B) pixel areas that were colocalized with LAMP1 (left) and LBPA (right) were analyzed. C, the IL-2Rβ-positive and IL-4Rα-positive pixel areas that were colocalized with LBPA (arrowheads) and not with LBPA (arrows) are indicated. Fluorescence labeling was carried out for IL-2Rβ (green), IL-4Rα (green), and LBPA (red). Scale bars, 10 μm.
DISCUSSION
The Hydrophobic Amino Acid Cluster Is a Ubiquitin-independent Endosomal Sorting Signal
The present results indicate that the hydrophobic amino acid cluster is an Hrs binding motif on the basis of our analyses of two cytokine receptors and functions as an endosomal sorting signal for the receptors. There are currently two major endosomal sorting signal sequences, namely YXXϕ and (D/E)XXXL(L/I), which are the consensus motifs for tyrosine-based and dileucine-based signals, respectively (1). Both sequences are recognized by the adaptor protein (AP) complexes AP-1, AP-2, AP-3, and AP-4. YXXϕ signal sequences are widely found in molecules not only at the plasma membrane (36, 37) but also at the trans-Golgi network (38) and lysosome (39, 40), suggesting multiple roles in endosomal sorting. YXXϕ signal sequences, one of which is located in the cytoplasmic region of transferrin receptor (41), play an essential role for the internalization of membrane proteins. Another type of tyrosine-based signal sequence (NPXY) is often found in membrane molecules such as low density lipoprotein receptor, insulin receptor, and epidermal growth factor receptor and is involved in the internalization but not other endosomal sorting events of a subset of type I integral membrane proteins (42). (D/E)XXXL(L/I) signal sequences are found in type I, type II, and multispanning transmembrane proteins, which are widely distributed from the plasma membrane to late endosomes, lysosomes, and specialized antigen-processing compartments, similar to YXXϕ signal sequences. For example, the (D/E)XXXL(L/I) signal sequence in the CD3-γ chain mediates its rapid internalization and lysosomal targeting (43), and internalization of glucose transporter 8 is mediated by the interaction of (D/E)XXXL(L/I) signal sequence with AP2 complex (44), indicating that (D/E)XXXL(L/I) signal sequence plays a essential role for the internalization in the membrane proteins. In contrast, the hydrophobic amino acid clusters found in this study were involved in the interaction with Hrs and not needed for the internalization of the receptors. Another type of dileucine-based signal sequence (DXXLL) is present in molecules at the plasma membrane, trans-Golgi network, and endosomes and is recognized by another family of adaptor proteins known as Golgi-localized γ-ear-containing Arf-binding proteins (GGAs). The DXXLL sequence interacts with the VHS domain of GGAs (45, 46). On the other hand, the VHS domains of both Hrs and STAM bind to ubiquitin but not the DXXLL sequence (45, 47). Regarding other signal sequences, acidic amino acid clusters are present in some transmembrane proteins and are considered to play roles in their retrieval from endosomes to the trans-Golgi network (1). The hydrophobic amino acid cluster identified in this study is unique in the following points; (i) the cluster is involved in ubiquitin-independent sorting (ii) the cluster is recognized by Hrs, a known sorting component of the ubiquitin-dependent machinery ESCRT-0, and (iii) the cluster comprises FFFHL in IL-2Rβ and LFLDLL in IL-4Rα, indicating that the amino acid sequence of the cluster may vary a great deal. Thus, we speculate that Hrs may recognize the hydrophobic amino acid clusters in various receptors with broad specificity. In this regard, our analyses to identify a key domain (amino acids 428–466) in Hrs that influences the receptor binding are important. Although there is no motif involved in the protein-protein interaction, we focused on a hydrophobic amino acid cluster comprising amino acids 453–457 (LLELL) in the C-terminal half of Hrs. We generated an Hrs mutant with the hydrophobic amino acids (residues 453–457) with alanine and examined the binding between this Hrs mutant and IL-2Rβ or IL-4Rα. Only slight reductions in the associations of the Hrs mutant with the receptors were observed (data not shown). Accordingly, extensive analyses, including solution of the crystallographic structure, will be needed to explore this region (amino acids 428–466) of Hrs. Recently, the complete crystal structure of human ESCRT-0 core complex was clarified (48). Analyses of the crystal structure revealed that the coiled-coil domains of Hrs and STAM form an antiparallel two-stranded coiled-coil. The coiled-coil domain of Hrs consists of four α-helix strands, α1 (amino acids 406–429), α2 (amino acids 432–436), α3-N (amino acids 437–453), and α3-C (amino acids 470–498). Because the hydrophobic amino acid cluster binding region (residues 428–466) of Hrs includes the hinge region between α1 and α2, the area around the hinge region may be a candidate site that contributes to the interaction.
On the other hand, what are the biological features of the receptors that are recognized by Hrs in a ubiquitin-independent manner? The epidermal growth factor receptor is localized on the cell surface membrane in the absence of ligand stimulation, and receptor internalization and transport to the MVB pathway are initiated following receptor ubiquitylation after ligand stimulation (49–52). In contrast, IL-2Rβ is constitutively internalized and delivered to the lysosomal pathway in the absence of its ligand (23). We found that IL-4Rα as well as IL-2Rβ was localized to LAMP1-positive compartments without ligand stimulation, suggesting constitutive internalization and endosomal sorting of IL-4Rα. Consequently, we speculate that the ubiquitin-independent binding targets of Hrs may be certain kinds of receptors that have the properties of constitutive internalization and sorting to lysosomes.
In conclusion, ESCRT complexes including ESCRT-0, which consists of Hrs and STAM, serve as the transport machinery for ubiquitylated cargo proteins. Therefore, it is noteworthy that Hrs associates with cytokine receptors in a ubiquitin-independent manner and is involved in their transport during endosomal sorting. In the present study we found that Hrs recognized a hydrophobic amino acid cluster in two cytokine receptors and played a role in the precise delivery of the receptors to late endosomes. These findings suggest the existence of a group of cargo proteins that are independent of ubiquitylation for endosomal sorting and have the hydrophobic amino acid cluster as an endosomal sorting signal motif.
Supplementary Material
Acknowledgments
We thank Dr. T. Kobayashi (RIKEN Advanced Science Institute, Saitama, Japan) for providing the anti-LBPA antibody, Dr. K. Miyazono (The University of Tokyo) for providing the pcDNA-HA-ubiquitin, and Dr. T. Kitamura (The University of Tokyo) for providing the pMXs. We also thank the Instrumental Analysis Research Center for Human and Environmental Sciences at Shinshu University for technical assistance with the DNA sequencing, flow cytometry analyses, and confocal microscopy.
This work was supported in part by a Grant-in-aid for Scientific Research C21590530 (to T. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- ESCRT
- endosomal sorting complex required for transport
- GGA
- Golgi-localized, γ-ear-containing, Arf-binding protein
- IL-2Rβ
- IL-2 receptor β chain
- IL-4Rα
- IL-4 receptor α chain
- Hrs
- hepatocyte growth factor-regulated tyrosine kinase substrate
- STAM
- signal transducing adaptor molecule
- MVB
- multivesicular body
- UIM
- ubiquitin interacting motif
- Vps
- vacuolar protein sorting
- DTSSP
- dithiobis(sulfosuccinimidylpropionate)
- LBPA
- lysobisphosphatidic acid
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1. Bonifacino J. S., Traub L. M. (2003) Annu. Rev. Biochem. 72, 395–447 [DOI] [PubMed] [Google Scholar]
- 2. Di Fiore P. P., Polo S., Hofmann K. (2003) Nat. Rev. Mol. Cell. Biol. 4, 491–497 [DOI] [PubMed] [Google Scholar]
- 3. Hicke L., Schubert H. L., Hill C. P. (2005) Nat. Rev. Mol. Cell. Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 4. Katzmann D. J., Babst M., Emr S. D. (2001) Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 5. Alam S. L., Sun J., Payne M., Welch B. D., Blake B. K., Davis D. R., Meyer H. H., Emr S. D., Sundquist W. I. (2004) EMBO J. 23, 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Slagsvold T., Aasland R., Hirano S., Bache K. G., Raiborg C., Trambaiolo D., Wakatsuki S., Stenmark H. (2005) J. Biol. Chem. 280, 19600–19606 [DOI] [PubMed] [Google Scholar]
- 7. Hanson P. I., Roth R., Lin Y., Heuser J. E. (2008) J. Cell Biol. 180, 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lata S., Schoehn G., Jain A., Pires R., Piehler J., Gottlinger H. G., Weissenhorn W. (2008) Science 321, 1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teis D., Saksena S., Emr S. D. (2008) Dev. Cell 15, 578–589 [DOI] [PubMed] [Google Scholar]
- 10. Saksena S., Wahlman J., Teis D., Johnson A. E., Emr S. D. (2009) Cell 136, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. (2009) Nature 458, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Komada M., Kitamura N. (1995) Mol. Cell. Biol. 15, 6213–6221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takeshita T., Arita T., Asao H., Tanaka N., Higuchi M., Kuroda H., Kaneko K., Munakata H., Endo Y., Fujita T., Sugamura K. (1996) Biochem. Biophys. Res. Commun. 225, 1035–1039 [DOI] [PubMed] [Google Scholar]
- 14. Asao H., Sasaki Y., Arita T., Tanaka N., Endo K., Kasai H., Takeshita T., Endo Y., Fujita T., Sugamura K. (1997) J. Biol. Chem. 272, 32785–32791 [DOI] [PubMed] [Google Scholar]
- 15. Lloyd T. E., Atkinson R., Wu M. N., Zhou Y., Pennetta G., Bellen H. J. (2002) Cell 108, 261–269 [DOI] [PubMed] [Google Scholar]
- 16. Polo S., Sigismund S., Faretta M., Guidi M., Capua M. R., Bossi G., Chen H., De Camilli P., Di Fiore P. P. (2002) Nature 416, 451–455 [DOI] [PubMed] [Google Scholar]
- 17. Raiborg C., Bache K. G., Gillooly D. J., Madshus I. H., Stang E., Stenmark H. (2002) Nat. Cell Biol. 4, 394–398 [DOI] [PubMed] [Google Scholar]
- 18. Mizuno E., Kawahata K., Kato M., Kitamura N., Komada M. (2003) Mol. Biol. Cell 14, 3675–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raiborg C., Stenmark H. (2009) Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 20. Bishop N., Horman A., Woodman P. (2002) J. Cell Biol. 157, 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shih S. C., Katzmann D. J., Schnell J. D., Sutanto M., Emr S. D., Hicke L. (2002) Nat. Cell Biol. 4, 389–393 [DOI] [PubMed] [Google Scholar]
- 22. Sugamura K., Asao H., Kondo M., Tanaka N., Ishii N., Ohbo K., Nakamura M., Takeshita T. (1996) Annu. Rev. Immunol. 14, 179–205 [DOI] [PubMed] [Google Scholar]
- 23. Yamashita Y., Kojima K., Tsukahara T., Agawa H., Yamada K., Amano Y., Kurotori N., Tanaka N., Sugamura K., Takeshita T. (2008) J. Cell Sci. 121, 1727–1738 [DOI] [PubMed] [Google Scholar]
- 24. Takeshita T., Asao H., Ohtani K., Ishii N., Kumaki S., Tanaka N., Munakata H., Nakamura M., Sugamura K. (1992) Science 257, 379–382 [DOI] [PubMed] [Google Scholar]
- 25. Suzuki J., Takeshita T., Ohbo K., Asao H., Tada K., Sugamura K. (1989) Int. Immunol. 1, 373–377 [DOI] [PubMed] [Google Scholar]
- 26. Tanaka Y., Tozawa H., Hayami M., Sugamura K., Hinuma Y. (1985) Microbiol. Immunol. 29, 959–972 [DOI] [PubMed] [Google Scholar]
- 27. Ishii N., Takeshita T., Kimura Y., Tada K., Kondo M., Nakamura M., Sugamura K. (1994) Int. Immunol. 6, 1273–1277 [DOI] [PubMed] [Google Scholar]
- 28. Fraker P. J., Speck J. C., Jr. (1978) Biochem. Biophys. Res. Commun. 80, 849–857 [DOI] [PubMed] [Google Scholar]
- 29. Gillooly D. J., Raiborg C., Stenmark H. (2003) Histochem. Cell Biol. 120, 445–453 [DOI] [PubMed] [Google Scholar]
- 30. Radhakrishna H., Donaldson J. G. (1997) J. Cell Biol. 139, 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taniguchi T., Minami Y. (1993) Cell 73, 5–8 [DOI] [PubMed] [Google Scholar]
- 32. Harada N., Yang G., Miyajima A., Howard M. (1992) J. Biol. Chem. 267, 22752–22758 [PubMed] [Google Scholar]
- 33. Ohbo K., Takeshita T., Asao H., Kurahayashi Y., Tada K., Mori H., Hatakeyama M., Taniguchi T., Sugamura K. (1991) J. Immunol. Methods 142, 61–72 [DOI] [PubMed] [Google Scholar]
- 34. Kobayashi T., Stang E., Fang K. S., de Moerloose P., Parton R. G., Gruenberg J. (1998) Nature 392, 193–197 [DOI] [PubMed] [Google Scholar]
- 35. Matsuo H., Chevallier J., Mayran N., Le Blanc I., Ferguson C., Fauré J., Blanc N. S., Matile S., Dubochet J., Sadoul R., Parton R. G., Vilbois F., Gruenberg J. (2004) Science 303, 531–534 [DOI] [PubMed] [Google Scholar]
- 36. Canfield W. M., Johnson K. F., Ye R. D., Gregory W., Kornfeld S. (1991) J. Biol. Chem. 266, 5682–5688 [PubMed] [Google Scholar]
- 37. Jadot M., Canfield W. M., Gregory W., Kornfeld S. (1992) J. Biol. Chem. 267, 11069–11077 [PubMed] [Google Scholar]
- 38. Humphrey J. S., Peters P. J., Yuan L. C., Bonifacino J. S. (1993) J. Cell Biol. 120, 1123–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Williams M. A., Fukuda M. (1990) J. Cell Biol. 111, 955–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harter C., Mellman I. (1992) J. Cell Biol. 117, 311–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marks M. S., Woodruff L., Ohno H., Bonifacino J. S. (1996) J. Cell Biol. 135, 341–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen W. J., Goldstein J. L., Brown M. S. (1990) J. Biol. Chem. 265, 3116–3123 [PubMed] [Google Scholar]
- 43. Letourneur F., Klausner R. D. (1992) Cell 69, 1143–1157 [DOI] [PubMed] [Google Scholar]
- 44. Schmidt U., Briese S., Leicht K., Schürmann A., Joost H. G., Al-Hasani H. (2006) J. Cell Sci. 119, 2321–2331 [DOI] [PubMed] [Google Scholar]
- 45. Puertollano R., Aguilar R. C., Gorshkova I., Crouch R. J., Bonifacino J. S. (2001) Science 292, 1712–1716 [DOI] [PubMed] [Google Scholar]
- 46. Zhu Y., Doray B., Poussu A., Lehto V. P., Kornfeld S. (2001) Science 292, 1716–1718 [DOI] [PubMed] [Google Scholar]
- 47. Ren X., Hurley J. H. (2010) EMBO J. 29, 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ren X., Kloer D. P., Kim Y. C., Ghirlando R., Saidi L. F., Hummer G., Hurley J. H. (2009) Structure 17, 406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Joazeiro C. A., Wing S. S., Huang H., Leverson J. D., Hunter T., Liu Y. C. (1999) Science 286, 309–312 [DOI] [PubMed] [Google Scholar]
- 50. Levkowitz G., Waterman H., Ettenberg S. A., Katz M., Tsygankov A. Y., Alroy I., Lavi S., Iwai K., Reiss Y., Ciechanover A., Lipkowitz S., Yarden Y. (1999) Mol. Cell 4, 1029–1040 [DOI] [PubMed] [Google Scholar]
- 51. Soubeyran P., Kowanetz K., Szymkiewicz I., Langdon W. Y., Dikic I. (2002) Nature 416, 183–187 [DOI] [PubMed] [Google Scholar]
- 52. Petrelli A., Gilestro G. F., Lanzardo S., Comoglio P. M., Migone N., Giordano S. (2002) Nature 416, 187–190 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.