Abstract
Clostridium sordellii lethal toxin and Clostridium novyi α-toxin, which are virulence factors involved in the toxic shock and gas gangrene syndromes, are members of the family of clostridial glucosylating toxins. The toxins inactivate Rho/Ras proteins by glucosylation or attachment of GlcNAc (α-toxin). Here, we studied the activation of the autoproteolytic processing of the toxins by inositol hexakisphosphate (InsP6) and compared it with the processing of Clostridium difficile toxin B. In the presence of low concentrations of InsP6 (<1 μm), toxin fragments consisting of the N-terminal glucosyltransferase (or GlcNAc-transferase) domains and the cysteine protease domains (CPDs) of C. sordellii lethal toxin, C. novyi α-toxin, and C. difficile toxin B were autocatalytically processed. The cleavage sites of lethal toxin (Leu-543) and α-toxin (Leu-548) and the catalytic cysteine residues (Cys-698 of lethal toxin and Cys-707 of α-toxin) were identified. Affinity of the CPDs for binding InsP6 was determined by isothermal titration calorimetry. In contrast to full-length toxin B and α-toxin, autocatalytic cleavage and InsP6 binding of full-length lethal toxin depended on low pH (pH 5) conditions. The data indicate that C. sordellii lethal toxin and C. novyi α-toxin are InsP6-dependently processed. However, full-length lethal toxin, but not its short toxin fragments consisting of the glucosyltransferase domain and the CPD, requires a pH-sensitive conformational change to allow binding of InsP6 and subsequent processing of the toxin.
Keywords: Bacterial Toxins, Cysteine Protease, Inositol Phosphates, Protein Processing, Protein Structure, Isothermal Titration Calorimetry
Introduction
Various pathogens of the genus Clostridium produce highly potent glucosylating toxins, clostridial glucosylating toxins (CGTs),3 which act on host target cells by modification and inactivation of Rho and Ras GTPases (1–5). This group of toxins, comprising Clostridium difficile toxins A and B, Clostridium novyi α-toxin, and Clostridium sordellii lethal and hemorrhagic toxins, are major virulence factors (6). Whereas C. difficile toxins A and B cause antibiotic-associated diarrhea and pseudomembranous colitis (7), C. sordellii lethal toxin is implicated in toxic shock syndrome (e.g. after medically induced abortion) (8) and, like C. novyi α-toxin, plays a pathogenic role in gas gangrene syndrome (9, 10).
CGTs are single-chain proteins subdivided into at least four functional domains (11). Binding of the toxins to cell-surface receptors is mediated by a C-terminal region including a series of repetitive oligopeptides (12). After uptake of the toxins by clathrin-dependent endocytosis (13), a central region harboring a pattern of hydrophobic amino acids is suggested to mediate pore formation and translocation from endosomes into the cytosol (12, 14). The glucosyltransferase domain is located at the N-terminal end of the toxins (15). As shown for toxins A and B, this domain is released into the cytosol by autocatalytic cleavage induced by an adjacent cysteine protease domain (CPD) (16–19).
Previous studies revealed that autocatalytic processing of CGTs depends on inositol hexakisphosphate (InsP6) (17, 18). It was shown for C. difficile toxins A and B that InsP6 can bind to the intrinsic CPD (20, 21). Binding of InsP6 causes conformational changes in the catalytic center of the CPD, resulting in activation of the protease, cleavage of the toxin molecule between the glucosyltransferase domain and the CPD, and release of the glucosyltransferase into the cytosol (21–23).
This study deals with the biochemical characterization of the InsP6-induced autocatalytic cleavage of C. sordellii lethal toxin and C. novyi α-toxin. For this purpose, exclusively recombinant toxins were used. Although no major differences could be observed in the binding affinity of InsP6 for the CPDs of all CGTs, significant differences between CGTs were observed in the InsP6-induced autocatalytic processing of the holotoxins. In particular, we report on the activation of the InsP6-induced autoproteolytic cleavage of C. sordellii lethal toxin, which suggests differences in the modular organization and function of the toxin domains of the various members of the CGT family.
EXPERIMENTAL PROCEDURES
Cultivation of Mammalian and Bacterial Cells
Vero cells were grown at 37 °C with 5% CO2 in DMEM (Biochrom AG, Berlin, Germany) supplemented with 10% FCS, 1% nonessential amino acids (PAA Laboratories GmbH, Pasching, Austria), 4 mm penicillin/streptomycin (PAN Biotech GmbH, Aidenbach, Germany), and 1% sodium pyruvate (Biochrom AG). Bacillus megaterium (strain WH320; MoBiTec, Göttingen, Germany) bacteria were cultivated in LB medium at 37 °C.
Constructs, Cloning, and Mutagenesis
For cloning of C. novyi α-toxin into the B. megaterium expression vector pHIS1522 (MoBiTec), the gene was amplified by PCR from genomic DNA (C. novyi strain 6018) in two parts. First, the upstream sequence of an intrinsic SpeI restriction site at bp 2965 was amplified with oligonucleotides introducing a 5′-BsrGI restriction site, and second, the downstream sequence of the SpeI restriction site was amplified by omitting the stop codon and by introducing a 3′-KpnI restriction site. Subsequent ligation of the two DNA fragments into BsrGI/KpnI-digested pHIS1522 resulted in α-toxin-His6. C. difficile toxin B and C. sordellii lethal toxin were cloned into the same expression vector essentially as described previously (13, 24). For the generation of His-tagged constructs of α-toxin, lethal toxin, and toxin B comprising only the glucosyltransferase domain (toxin B-(1–546)), the CPD, or the glucosyltransferase domain (GD) and the CPD (GD-CPD) in pHIS1522, the corresponding sequences were amplified with oligonucleotides introducing a 5′-BsrGI and a 3′-KpnI restriction site and using the full-length pHIS1522 constructs as template. The ligation of BsrGI/KpnI-digested pHIS1522 vector and PCR products was then performed. Eventually, mutations were introduced by site-directed mutagenesis (QuikChange, Stratagene, La Jolla, CA).
Expression and Purification of Recombinant Proteins
Full-length versions or only parts of toxin B, lethal toxin, and α-toxin that were cloned into the pHIS1522 vector were recombinantly expressed in B. megaterium and purified as His-tagged proteins by nickel affinity chromatography. Briefly, B. megaterium protoplasts were first transformed with plasmids according to the manufacturer's instructions (MoBiTec). The cells were then grown at 37 °C in LB medium until A600 nm = 0.3. Subsequently, protein expression was induced by the addition of 5% xylose and by further incubation at 37 °C for 3–6 h or at 29 °C overnight. The cells were then harvested by centrifugation, and the pellet was suspended in lysis buffer (20 mm Tris-HCl (pH 8.0), 300 mm NaCl, 20 mm imidazole, 10% glycerol, and 500 μm EDTA) supplemented with Complete protease inhibitor mixture (Roche Applied Science). The bacteria were lysed using a Microfluidizer (Microfluidics, Newton, MA) at 15000 p.s.i., and cell debris was removed by centrifugation at 164,000 × g for 1 h at 4 °C. The supernatant was then passed through a pre-equilibrated nickel-charged HisTrap column (GE Healthcare). The column was washed with buffer containing 20 mm Tris-HCl (pH 8.0), 300 mm NaCl, 50 mm imidazole, and 10% glycerol, and bound His-tagged proteins were eluted with 20 mm Tris-HCl (pH 8.0), 300 mm NaCl, 500 mm imidazole, and 10% glycerol. In addition, the eluted proteins were further purified by size-exclusion chromatography on a Superdex 200 column (GE Healthcare) equilibrated with buffer containing 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 10% glycerol. Rac1, Ha-Ras, and RhoA were cloned as GST-tagged versions and purified from Escherichia coli BL21(DE3) essentially as described previously (25).
Intoxication of Cultured Vero Cells
Cultured Vero cells were seeded in 24-well plates and cultivated in DMEM to form semiconfluent monolayers prior to washing with PBS and intoxication by the addition of toxins in DMEM at the indicated concentrations. Upon onset of intoxication characteristics (cell rounding), images were taken using an Axiovert 25 inverted microscope (Carl Zeiss, Jena, Germany).
In Vitro Glucosylation Assay
At the indicated concentrations, full-length toxins or toxin fragments were incubated in glucosylation buffer (50 mm Hepes (pH 7.5), 100 mm KCl, 2 mm MgCl2, 1 mm MnCl2, and 100 μg/ml bovine serum albumin) in the presence of 10 μm UDP-[14C]glucose or UDP-[14C]GlcNAc (α-toxin) and 5 μm recombinant Rac1, Ha-Ras, or RhoA for 20–45 min at 37 °C. The reaction was stopped by the addition of Laemmli buffer and boiling for 5 min at 95 °C. Radiolabeled proteins were separated by SDS-PAGE and visualized by autoradiography using a PhosphorImager (GE Healthcare).
In Vitro Cleavage Assay
The in vitro cleavage assay was performed with 2 μg of full-length toxins or toxin fragments either in neutral cleavage buffer (100 mm Tris-HCl (pH 7.4)) or under acidic conditions (100 mm sodium acetate (pH 5.0)). Cleavage was initiated by the addition of the indicated concentrations of InsP6, and the reaction was incubated at 37 °C for the indicated time points. The reaction was stopped by the addition of Laemmli buffer and boiling for 5 min at 95 °C. Cleavage products were visualized as Coomassie Blue-stained protein bands after SDS-PAGE.
Filter Binding Assay with Radiolabeled InsP6
Full-length toxins or toxin fragments (1 μm) were incubated with a mixture of 4.8 μm InsP6 and 0.2 μm [3H]InsP6 in binding buffer containing 100 mm Tris-HCl (pH 7.4) or 100 mm sodium acetate (pH 5.0), each supplemented with 1 mg/ml bovine serum albumin, for 10 min at 4 °C. The reactions were then terminated by rapid vacuum filtration through a prewetted 0.45-μm nitrocellulose membrane (Whatman), followed by washing with 3 ml of binding buffer to separate free and bound [3H]InsP6. Membrane-bound radioactivity was determined by liquid scintillation counting.
Isothermal Titration Calorimetry
The Kd values of InsP6 binding to the CPDs of toxin B, lethal toxin, and α-toxin were determined by isothermal titration calorimetry (ITC200, GE Healthcare) at 25 °C under neutral pH conditions in buffer containing 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 10% glycerol or under acidic conditions in buffer containing 20 mm sodium acetate (pH 5), 150 mm NaCl, and 10% glycerol. The reaction cell was filled with 50–100 μm protein, and InsP6 was injected at 10-fold higher concentrations. As a control, InsP6 was injected alone into the cell filled with buffer only. Data were evaluated with the manufacturer's software.
RESULTS
CPDs of C. sordellii Lethal Toxin and C. novyi α-Toxin Are Responsible for Autocatalytic Processing
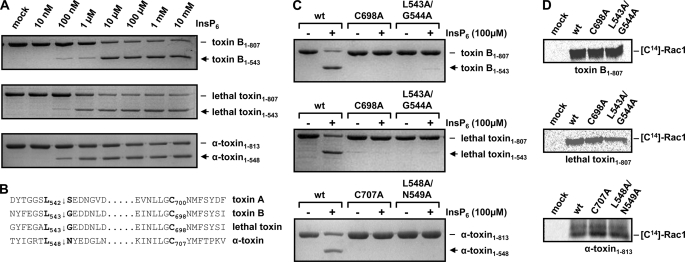
Studies by Pruitt et al. (21) delimited the CPD of toxin A to amino acids 543–809 (corresponding to amino acids 544–807 in toxin B). Sequence comparison of toxins A and B with lethal toxin and α-toxin revealed putative CPDs of lethal toxin and α-toxin at residues 544–807 and 549–813, respectively. To study the autocatalytic cleavage of the toxins, we therefore expressed toxin fragments consisting of the GDs and the putative CPDs from various CGTs (toxin B-(1–807), lethal toxin-(1–807), and α-toxin-(1–813)). An in vitro cleavage assay was then performed by incubation of increasing concentrations of InsP6 with the respective GD-CPD fragments of toxins. As shown in Fig. 1A, InsP6-induced autocatalytic processing was detected with each toxin fragment at concentrations of 0.1–1 μm InsP6. Thus, autocatalytic processing of all CGT members occurred independently of any domains that are located beyond the intrinsic CPD.
FIGURE 1.
Comparative analysis of CGT fragments comprising the GD and the putative CPD. A, InsP6-induced in vitro cleavage of CGT fragments comprising the GD and the adjacent CPD. The toxin fragments were expressed as recombinant proteins in B. megaterium and purified. Processing of CGT fragments (2 μg) was induced with increasing concentrations of InsP6 as indicated and incubation for 1 h at 37 °C. B, alignment of primary sequences of the CGTs representing the putative autocatalytic cleavage site (↓) and the conserved catalytic cysteine residue of the cysteine protease homology region (shown in boldface). C, the amino acids highlighted in B were substituted in each CGT fragment with alanine and analyzed for autocatalytic processing in the presence or absence of 100 μm InsP6 and incubated for 1 h at 37 °C. D, in vitro glucosylation of Rac1 with CGT fragments and mutants shown in C in the presence of radiolabeled UDP-glucose or UDP-GlcNAc (α-toxin), respectively. Samples were subjected to SDS-PAGE, and modified Rac1 was visualized by autoradiography.
Recently, the cleavage sites of the CPDs in toxins A and B, as well as amino acids that are crucial for the activity of the protease, were characterized (17, 19, 21). To identify the cleavage sites and the catalytically active cysteine residues of lethal toxin and α-toxin, a sequence comparison of all CGTs was performed. The multiple alignments of CGT sequences surrounding the cleavage sites and the catalytically active cysteine residues of toxins A and B is depicted in Fig. 1B. This analysis revealed the cleavage sites Leu-543 and Leu-548 for lethal toxin and α-toxin, respectively. Furthermore, by sequence comparison, we identified Cys-698 and Cys-707 as catalytic cysteine residues of the CPDs of lethal toxin and α-toxin, respectively. The in silico data were confirmed by mutagenesis studies. A change of the proposed catalytic cysteine residues of the three toxin fragments (C698A-toxin B-(1–807), C698A-lethal toxin-(1–807), and C707A-α-toxin-(1–813)) to alanine inhibited autocatalytic cleavage in the presence of 100 μm InsP6. Moreover, we corroborated the proposed cleavage sites by mutation. Again, all cleavage site mutants (L543A/G543A-toxin B-(1–807), L543A/G543A-lethal toxin-(1–807), and L548A/N549A-α-toxin-(1–813)) were stable toward autocatalytic cleavage. In vitro glucosylation of Rac1 in the presence of radiolabeled UDP-glucose (or UDP-GlcNAc for α-toxin) was performed with each GD-CPD mutant, showing that the recombinant proteins were correctly folded, because Rac1 was modified in comparable levels by the mutants compared with wild-type GD-CPD fragments (Fig. 1D).
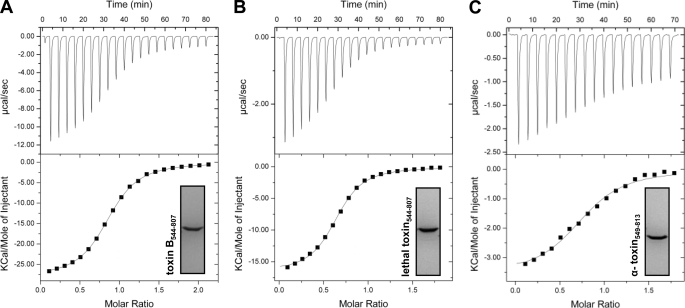
A prerequisite of the autoproteolytic cleavage of the CGTs is the binding of InsP6 to the CPD. We compared the binding affinity of InsP6 for the CPDs by isothermal titration calorimetry at pH 7.4 (Fig. 2, A–C). The studies were performed with the recombinant CPDs. Titrations were carried out twice and yielded Kd values of 4.4 and 5.1 μm for toxin B-(544–807), 2.1 and 2.8 μm for lethal toxin-(544–807), and 6.7 and 8.9 μm for α-toxin-(549–813). These results indicated that InsP6 has a similar affinity for binding to the CPD of each CGT. The stoichiometries of the complex were 0.81 and 0.85 for toxin B-(544–807), 0.64 and 0.84 for lethal toxin-(544–807), and 0.74 and 0.77 for α-toxin-(549–813), indicating that InsP6 binds to all CPDs of the toxins at a ratio of 1:1.
FIGURE 2.
Binding affinity of InsP6 for the CGT CPDs determined by isothermal calorimetry. The respective CPDs of the CGTs were expressed as recombinant proteins in B. megaterium and purified prior to analysis of the binding of InsP6 at pH 7.4 by isothermal calorimetry. A, toxin B-(544–807); B, lethal toxin-(544–807); C, α-toxin-(549–813).
Full-length C. sordellii Lethal Toxin Differs in Its Requirement for InsP6-induced Autocatalytic Processing
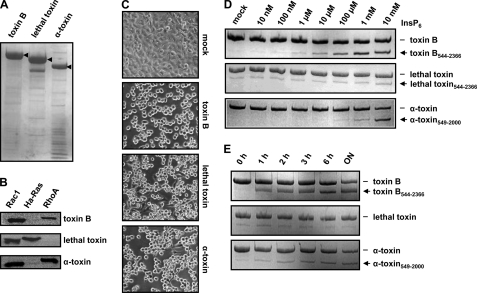
For biochemical characterization of the autocatalytic cleavage of the full-length CGTs, we expressed the proteins in B. megaterium. Fig. 3A shows the SDS-PAGE of the purified recombinant proteins. To test the glucosyltransferase activity of the toxins, in vitro glucosylation assays were performed with Rac1, Ha-Ras, and RhoA in the presence of radiolabeled UDP-[14C]glucose (or UDP-[14C]GlcNAc for α-toxin) as a donor substrate. As expected, incorporation of radiolabeled glucose (or GlcNAc) was detected only in Rac1 and RhoA with toxin B or α-toxin and only in Rac1 and Ha-Ras with lethal toxin (Fig. 3B), confirming same substrate specificity as described for the native toxins (3, 26). Moreover, the addition of recombinant CGTs to cultured Vero cells induced phenotypic alterations in cell morphology (e.g. cell rounding) that are typical for cytotoxicity (Fig. 3C). This finding implies the proper folding of domains of the recombinant CGTs implicated in the uptake of the proteins into endosomal compartments and the release of the GD into the cytosol.
FIGURE 3.
Functional characterization of recombinant CGTs. A, C. difficile toxin B, C. sordellii lethal toxin, and C. novyi α-toxin were purified from B. megaterium and subjected to SDS-PAGE, and separated proteins were stained with Coomassie Blue. The bands indicated by arrowheads represent the respective purified toxins. B, in vitro glucosylation of Rac1, Ha-Ras, and RhoA with recombinant CGTs. C, intoxication of Vero cells with recombinant CGTs. Images were obtained after overnight (control (mock)), 1-h (100 pm toxin B), 6-h (1 nm lethal toxin), and overnight (1 nm α-toxin) treatment. D, InsP6-induced in vitro cleavage of recombinant CGTs. Processing of CGTs (2 μg) was induced with increasing concentrations of InsP6 as indicated and incubation for 1 h at 37 °C. E, processing of CGTs (2 μg) was induced with 100 μm InsP6, followed by an incubation period as indicated at 37 °C.
Next, the autocatalytic processing of the recombinant full-length CGTs was studied in in vitro cleavage assays at pH 7.4 with increasing concentrations of InsP6. Surprisingly, significant autocatalytic cleavage of CGTs at physiological concentrations of InsP6 (≤100 μm) was observed only for toxin B and α-toxin (Fig. 3D). In contrast, lethal toxin exhibited much less autocatalytic processing at up to 10 mm InsP6. Also the time course of InsP6 (100 μm)-induced autocatalysis revealed that full-length lethal toxin was much less processed (even after overnight incubation) under the conditions used (Fig. 3E). Obviously, in contrast to CGT fragments consisting of the GD and CPD, full-length CGTs differed in their susceptibility to InsP6-induced autocatalytic processing.
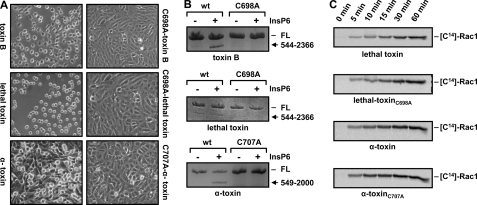
A Functional CPD Is a Prerequisite for Intoxication of Vero Cells by Lethal Toxin
Because full-length lethal toxin showed no autocatalytic cleavage upon incubation with InsP6 under in vitro conditions, we aimed to determine the biological significance of autocatalytic processing by performing intoxication assays on cultured Vero cells with lethal toxin carrying an inactive CPD (C698A-lethal toxin). Accordingly, CPD-inactive toxin mutants were also synthesized for toxin B (C698A-toxin B) and α-toxin (C707A-α-toxin). Interestingly, a lack of cytotoxicity compared with wild-type toxins was observed not only for the CPD-inactive toxin B and α-toxin mutants but also for the CPD mutant of lethal toxin (Fig. 4A). Therefore, CPD-dependent processing represents an essential step in complete intoxication not only by toxin B and α-toxin but also by lethal toxin. The inability of the CPD-inactive toxin mutants to undergo autocatalytic processing was confirmed in vitro by incubation with InsP6 (Fig. 4B). In addition, to exclude the possibility that the lethal toxin and α-toxin mutants lack cytotoxicity due to general misfolding, a time-dependent in vitro glucosylation assay with recombinant Rac1 was performed. Similar kcat values for the wild-type and mutant lethal toxins (912.6 ± 37.1 h−1 (S.D.) versus 1098 ± 84.9 h−1) and α-toxins (120.1 ± 29.8 h−1 (S.D.) versus 187.8 ± 54.6 h−1) were calculated from three time points in the linear range (5, 10, and 15 min), confirming that the CPD-inactive mutants are fully active in glucosyltransferase or GlcNAc-transferase activity (Fig. 4C). The importance of the autocatalytic cleavage in the intoxication process was demonstrated for toxins A and B previously (17, 27).
FIGURE 4.
Intoxication of cultured Vero cells with CPD-inactive toxin mutants. A, cultured Vero cells were incubated with wild-type toxins (100 pm toxin B, 1 nm lethal toxin, and 100 pm α-toxin) or CPD-inactive versions (100 pm C698A-toxin B, 1 nm C698A-lethal toxin, and 100 pm C707A-α-toxin), and images were obtained after 1 h (toxin B/C698A-toxin B) and 6 h (lethal toxin/C698A-lethal toxin and α-toxin/C707A-α-toxin). B, in vitro cleavage assay. The wild-type and mutant toxins (2 μg each) shown in A were incubated for 1 h at 37 °C with 100 μm (toxin B/C698A-toxin B and lethal toxin/C698A-lethal toxin) or 1 mm (α-toxin/C707A-α-toxin) InsP6, followed by SDS-PAGE and visualization of proteins by Coomassie Blue staining. FL, full-length. C, time-dependent in vitro glucosylation of Rac1. 10 nm lethal toxin/C698A-lethal toxin or 10 nm α-toxin/C707A-α-toxin was incubated with 5 μm Rac1 in the presence of radiolabeled UDP-glucose or UDP-GlcNAc (α-toxin), respectively. Samples were taken at the indicated time points and subjected to SDS-PAGE, and modified Rac1 was visualized by autoradiography.
InsP6-induced Autocatalytic Processing of Full-length Lethal Toxin Requires Acidic pH
Because autocatalytic processing of full-length lethal toxin was not induced by incubation with InsP6, we wondered whether steric hindrance may prevent binding of InsP6 to the CPD. Therefore, we studied the direct interaction of InsP6 with the full-length CGT. Because isothermal titration calorimetry is hampered by the large size of full-length toxins (note that very large amounts of toxins would be necessary), we employed a filter binding assay. To this end, recombinant holotoxins were preincubated with radiolabeled [3H]InsP6 at neutral pH, and non-bound InsP6 was removed by filtration through nitrocellulose filters. These binding studies revealed significant binding of InsP6 to full-length variants of toxin B and α-toxin but not lethal toxin (Fig. 5A).
FIGURE 5.
Influence of pH in binding of InsP6 to lethal toxin for induction of autocatalytic processing. A, filter binding assay. 1 μm recombinant CGT protein was incubated with [3H]InsP6 for 10 min at 4 °C. Thereafter, the mixture was loaded onto nitrocellulose filters, and after extensive washing, bound radioactivity was measured by liquid scintillation counting. The GD of toxin B (toxin B-(1–546)) was included in the assay as a negative control. B, InsP6-induced in vitro cleavage of lethal toxin at low pH. Processing of full-length toxin and GD-CPD fragments of lethal toxin was induced at pH 5 with increasing concentrations of InsP6 as indicated and incubation for 1 h at 37 °C. C, the filter binding assay (in the presence of [3H]InsP6) shown in A was repeated with lethal toxin and toxin B-(1–546) (negative control) at pH 5. D, the filter binding assay shown in A was performed with full-length toxin B and full-length lethal toxin under various pH conditions as indicated. The highest InsP6-binding values for each toxin were set to 100%. E, filter binding assay in the presence of [3H]InsP6 with lethal toxin and toxin B at pH 7.4 after lowering the pH of the mixture to pH 5 and after incubation of the toxins at pH 5 for 10 min, followed by readjustment from pH 5 to pH 7.4. Bound radioactivity is given as a percent of maximal binding (100%). F, isothermal calorimetry of the CPD of lethal toxin and InsP6 at pH 5. Error bars in A and C–E represent S.D. (n ≥ 3).
CGTs are clathrin-dependently endocytosed to reach low pH endosomes (13). It is widely accepted that the low pH triggers conformational changes within CGTs, thereby enabling the insertion of the toxins into endosomal membranes (28). We speculated that conformational changes induced by low pH may be a prerequisite for InsP6 binding to the CPD of lethal toxin. Therefore, we studied the autocatalytic cleavage of full-length lethal toxin with increasing concentrations of InsP6 at pH 5 and compared the results with the processing of the short toxin fragments (GD-CPD), which consisted of the GD and CPD. Interestingly, full-length lethal toxin and GD-CPD fragments were processed equally at physiological concentrations of InsP6 (10–100 μm) at low pH (Fig. 5B).
Apparently, low pH-induced rearrangements within full-length lethal toxin enabled binding of InsP6, activation of the CPD, and subsequent autocatalytic cleavage. Accordingly, a filter binding assay performed with InsP6 and full-length lethal toxin at pH 5 revealed binding of InsP6 under low pH conditions (Fig. 5C). Direct comparison of InsP6 binding of lethal toxin with that of toxin B showed that the latter toxin exhibited maximal binding at pH 7.4 and reduced binding at pH 5 (Fig. 5D). We next asked whether the binding of InsP6 to full-length lethal toxin is reversibly controlled by the pH of the incubation medium. For this purpose, lethal toxin was first incubated in buffer at pH 7.4. The samples were then acidified to pH 5, and thereafter, the mixture was again brought to pH 7.4, which was followed by the binding assay with radiolabeled InsP6. As shown in Fig. 5E, InsP6 did not bind to full-length lethal toxin after readjustment of the medium from acidic pH to pH 7.4. In contrast, toxin B, which was tested in parallel, partially bound InsP6 after preincubation in acidic buffer and readjustment to neutral pH (Fig. 5E). Moreover, we determined the influence of acidification on binding of InsP6 to the CPD fragment of lethal toxin by isothermal titration calorimetry. This study revealed that, at low pH (pH 5), the CPD of lethal toxin exhibited an ∼10-fold reduced affinity for InsP6 (Kd = 33.0 and 40.4 μm) compared with results obtained at neutral pH (Figs. 2C and 5F). These data indicate that the low pH shift facilitated binding of InsP6 only to full-length lethal toxin but not to the GD-CPD fragment, supporting the notion that the holotoxin undergoes a major conformational change at low pH, which allows InsP6 binding.
DISCUSSION
Here, we studied the InsP6-dependent autocatalytic processing of three CGTs. Initial studies with short fragments of the toxins covering the glucosyltransferase (GlcNAc-transferase of α-toxin) domain and the CPD revealed that all toxins needed InsP6 for cleavage. This is not surprising because the CPDs of lethal toxin and α-toxin are 78 and 34% identical to the CPD of toxin B, and the proposed catalytic triad (toxin B Asp-587/His-653/Cys-698) and the InsP6-binding sites are highly conserved among the toxins. Accordingly, we determined very similar Kd values for the binding of InsP6 to the toxin protease domains. Moreover, the concentration range of InsP6 needed for cleavage was similar, indicating that the mechanism of InsP6-induced activation of the protease activity is similar among all these toxins. In addition, we predicted and confirmed the cleavage sites of all three toxins by mutagenesis studies, indicating that the toxins are split after a conserved leucine residue.
Whereas the short fragments covering the glucosyltransferase (GlcNAc-transferase of α-toxin) domains and CPDs of the toxins exhibited similar properties with respect to InsP6 binding and activation, full-length toxins differed significantly. Although full-length toxin B and α-toxin exhibited InsP6 sensitivity at 1 and 100 μm, respectively, lethal toxin was much more resistant against autocatalytic cleavage even at high concentrations of InsP6 (≥1 mm). Moreover, we were not able to detect significant binding of InsP6 to lethal toxin in filter binding assays, whereas toxin B and α-toxin exhibited InsP6 binding under these conditions. This discrepancy was not caused by misfolding of recombinant B. megaterium-expressed proteins because glucosyltransferase activity and cytotoxicity were confirmed, indicating proper folding of the proteins.
To study the impact of toxin processing on cytotoxicity of full-length toxins, we changed the catalytic cysteine residues of the CPD to alanine. This resulted in inhibition of cytotoxicity of all three toxins, including lethal toxin, which was largely resistant to in vitro processing even in the presence of a high concentration of InsP6. These findings indicate that also processing of lethal toxin is essential for full cytotoxicity.
As CGTs translocate from acidic endosomal compartments into the cytosol (11, 28, 29), we wondered whether incubation at low pH might affect the susceptibility of the toxins to InsP6. We observed that lowering the pH strongly increased the autocatalytic cleavage of full-length lethal toxin in the presence of InsP6. In line with this finding, we observed that the binding of InsP6 to full-length lethal toxin was largely increased at low pH. This effect was most likely caused by conformational changes in the overall structure of lethal toxin and not only by charge effects at the binding site. A decrease in the pH should rather reduce the binding of the negatively charged InsP6 to the toxin. This was confirmed in a direct comparison of the InsP6 binding of full-length lethal toxin with that of full-length toxin B at different pH values. Whereas the binding of InsP6 to lethal toxin increased at low pH, the reverse was true for toxin B.
The notion that, at low pH, the binding of InsP6 to its binding pocket at the cysteine protease is reduced was confirmed by isothermal titration calorimetry with the isolated protease domain of lethal toxin, showing an ∼10-fold reduced binding affinity for InsP6. Thus, these findings indicate that the pH shift causes major conformational changes in the overall structure of lethal toxin, resulting in exposure and/or structural changes in the CPD, allowing InsP6 binding.
Major pH-dependent structural changes in lethal toxin have been reported before. Ballard and co-workers (30) showed that the cytotoxicity of C. sordellii lethal toxin is increased in a pH-dependent manner. A short pH pulse strongly increased the velocity of cytotoxic effects. They showed major conformational changes in lethal toxin by fluorescence methods at low pH, which were reversible (30). These data are in agreement with our studies, indicating that, at low pH, a major conformational change in the toxin exposes the InsP6-binding site and allows binding of InsP6 and subsequent autocatalytic processing. As processing is important for activity, low pH exposure of the toxin results in an increase in cytotoxicity.
More recently, it was proposed that lethal toxin forms high molecular complexes, which dissociate at low pH and reassociate after an increase in pH (31). We suggest that these studies performed with native lethal toxin purified from C. sordellii might be biased by pH-dependent impurities, which could affect complex formation. We used recombinant toxins expressed in B. megaterium. By gel filtration of recombinant full-length lethal toxin, we could confirm a monodisperse preparation of monomers. Therefore, formation of multimeric toxin complexes most likely did not play a major role in our studies. However, we also observed reversibility of the pH-dependent effect. After readjustment of the low pH to pH 7.4, the binding of InsP6 and the processing of the toxin were again inhibited. This finding indicates that the proposed pH-dependent conformational change, which allows InsP6 binding, is largely reversible.
Recent studies solved the crystal structures of the GDs of C. difficile toxin B (32) and C. novyi α-toxin and C. sordellii lethal toxin (33); the CPD of toxin A (21); and parts of the C-terminal repetitive oligopeptide domain (34, 35). However, the structure of the holotoxin is still enigmatic. Related CPD structures are available not only for toxin A but also for Vibrio cholerae MARTX toxin. Notably, both CPDs share conserved lysine residues, which are involved in InsP6 binding. However, the conformation of the bound allosteric activator is largely different in the CPDs of toxin A and MARTX (21, 22, 36). Because the InsP6-binding properties of the isolated CPDs of toxin B, α-toxin, and lethal toxin are very similar, we do not suggest that major structural differences exist in the CPDs of these toxins. Therefore, structural features located distantly to the CPD may affect the conformation and binding of InsP6 to the CPD in full-length lethal toxin. A recent study with negative stain electron microscopy revealed a model of holotoxins A and B and proposed conformational changes occurring at the low pH of endosomes (37). In this model, the large extension of the C-terminal domain is remarkable, not even excluding interaction with N-terminal structures of the toxins. Especially the C-terminal polypeptide repeat domain, which has an extended solenoid-like structure, differs between the various CGTs. Therefore, it remains to be studied whether the C-terminal domain is involved in the different susceptibility of holotoxins to InsP6.
What is the physiological reason for the unique, low pH-dependent InsP6-binding properties of lethal toxin? One might speculate that C. sordellii has adopted to environments that frequently exhibit high concentrations of extracellular InsP6. The disintegration of cells in wounds and injured tissues (frequent sites of C. sordellii infection) would allow cytosolic InsP6 to reach extracellular compartments. Unique structural characteristics of lethal toxin under neutral pH conditions thereby prevent activation of the CPD by InsP6 binding prior to cell entry. Such prerequisites might not be necessary for C. difficile toxin B, for example, because phytase enzymes that hydrolyze extracellular luminal InsP6 efficiently are present in the small intestine (38). After endocytosis, however, all CGTs are exposed to endosomal acidification, resulting in partial unfolding and/or activation of domains involved in membrane insertion and delivery of the enzyme portions across the endosomal membrane. Eventually, the CPD is accessible for InsP6-driven activation, leading to the release of the glucosyltransferase into the cytosol. However, one has to keep in mind that activation of the CPD and release of the GD should not occur before entering the cytosol. Otherwise, the delivery of the biologically active part is altered. Therefore, it makes sense that the affinity of toxin B for InsP6 is reduced at low pH (e.g. which is observed in endosomes).
Acknowledgments
We greatly appreciate the excellent technical assistance of Otilia Wunderlich and Sven Hornei.
This work was supported by Deutsche Forschungsgemeinschaft Grant AK6/16-3.
- CGT
- clostridial glucosylating toxin
- CPD
- cysteine protease domain
- InsP6
- inositol hexakisphosphate
- GD
- glucosyltransferase domain.
REFERENCES
- 1. Just I., Selzer J., Wilm M., von Eichel-Streiber C., Mann M., Aktories K. (1995) Nature 375, 500–503 [DOI] [PubMed] [Google Scholar]
- 2. Selzer J., Hofmann F., Rex G., Wilm M., Mann M., Just I., Aktories K. (1996) J. Biol. Chem. 271, 25173–25177 [DOI] [PubMed] [Google Scholar]
- 3. Just I., Selzer J., Hofmann F., Green G. A., Aktories K. (1996) J. Biol. Chem. 271, 10149–10153 [DOI] [PubMed] [Google Scholar]
- 4. Just I., Gerhard R. (2004) Rev. Physiol. Biochem. Pharmacol. 152, 23–47 [DOI] [PubMed] [Google Scholar]
- 5. Voth D. E., Ballard J. D. (2005) Clin. Microbiol. Rev. 18, 247–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boriello S. P., Aktories K. (2005) in Topley & Wilson's Microbiology & Microbial Infections (Boriello S. P., Murray P. R., Funke G. eds) pp. 1089–1136, Hodder Arnold, Ltd., London [Google Scholar]
- 7. Kelly C. P., LaMont J. T. (2008) N. Engl. J. Med. 359, 1932–1940 [DOI] [PubMed] [Google Scholar]
- 8. Ho C. S., Bhatnagar J., Cohen A. L., Hacker J. K., Zane S. B., Reagan S., Fischer M., Shieh W. J., Guarner J., Ahmad S., Zaki S. R., McDonald L. C. (2009) Am. J. Obstet. Gynecol. 201, 459–467 [DOI] [PubMed] [Google Scholar]
- 9. Cohen A. L., Bhatnagar J., Reagan S., Zane S. B., D'Angeli M. A., Fischer M., Killgore G., Kwan-Gett T. S., Blossom D. B., Shieh W. J., Guarner J., Jernigan J., Duchin J. S., Zaki S. R., McDonald L. C. (2007) Obstet. Gynecol. 110, 1027–1033 [DOI] [PubMed] [Google Scholar]
- 10. Tsokos M., Schalinski S., Paulsen F., Sperhake J. P., Püschel K., Sobottka I. (2008) Int. J. Legal Med. 122, 35–41 [DOI] [PubMed] [Google Scholar]
- 11. Jank T., Aktories K. (2008) Trends Microbiol. 16, 222–229 [DOI] [PubMed] [Google Scholar]
- 12. von Eichel-Streiber C., Sauerborn M., Kuramitsu H. K. (1992) J. Bacteriol. 174, 6707–6710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Papatheodorou P., Zamboglou C., Genisyuerek S., Guttenberg G., Aktories K. (2010) PLoS ONE 5, e10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Genisyuerek S., Papatheodorou P., Guttenberg G., Schubert R., Benz R., Aktories K. (2011) Mol. Microbiol. 79, 1643–1654 [DOI] [PubMed] [Google Scholar]
- 15. Hofmann F., Busch C., Prepens U., Just I., Aktories K. (1997) J. Biol. Chem. 272, 11074–11078 [DOI] [PubMed] [Google Scholar]
- 16. Pfeifer G., Schirmer J., Leemhuis J., Busch C., Meyer D. K., Aktories K., Barth H. (2003) J. Biol. Chem. 278, 44535–44541 [DOI] [PubMed] [Google Scholar]
- 17. Egerer M., Giesemann T., Jank T., Satchell K. J., Aktories K. (2007) J. Biol. Chem. 282, 25314–25321 [DOI] [PubMed] [Google Scholar]
- 18. Reineke J., Tenzer S., Rupnik M., Koschinski A., Hasselmayer O., Schrattenholz A., Schild H., von Eichel-Streiber C. (2007) Nature 446, 415–419 [DOI] [PubMed] [Google Scholar]
- 19. Rupnik M., Pabst S., Rupnik M., von Eichel-Streiber C., Urlaub H., Söling H. D. (2005) Microbiology 151, 199–208 [DOI] [PubMed] [Google Scholar]
- 20. Egerer M., Giesemann T., Herrmann C., Aktories K. (2009) J. Biol. Chem. 284, 3389–3395 [DOI] [PubMed] [Google Scholar]
- 21. Pruitt R. N., Chagot B., Cover M., Chazin W. J., Spiller B., Lacy D. B. (2009) J. Biol. Chem. 284, 21934–21940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Egerer M., Satchell K. J. (2010) PLoS Pathog. 6, e1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prochazkova K., Satchell K. J. (2008) J. Biol. Chem. 283, 23656–23664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang G., Zhou B., Wang J., He X., Sun X., Nie W., Tzipori S., Feng H. (2008) BMC Microbiol. 8, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Just I., Selzer J., von Eichel-Streiber C., Aktories K. (1995) J. Clin. Invest. 95, 1026–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Popoff M. R., Chaves-Olarte E., Lemichez E., von Eichel-Streiber C., Thelestam M., Chardin P., Cussac D., Antonny B., Chavrier P., Flatau G., Giry M., de Gunzburg J., Boquet P. (1996) J. Biol. Chem. 271, 10217–10224 [DOI] [PubMed] [Google Scholar]
- 27. Kreimeyer I., Euler F., Marckscheffel A., Tatge H., Pich A., Olling A., Schwarz J., Just I., Gerhard R. (2011) Naunyn-Schmiedeberg's Arch. Pharmacol. 383, 253–262 [DOI] [PubMed] [Google Scholar]
- 28. Barth H., Pfeifer G., Hofmann F., Maier E., Benz R., Aktories K. (2001) J. Biol. Chem. 276, 10670–10676 [DOI] [PubMed] [Google Scholar]
- 29. Qa'Dan M., Spyres L. M., Ballard J. D. (2000) Infect. Immun. 68, 2470–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qa'Dan M., Spyres L. M., Ballard J. D. (2001) Infect. Immun. 69, 5487–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Voth D. E., Qa'Dan M., Hamm E. E., Pelfrey J. M., Ballard J. D. (2004) Infect. Immun. 72, 3366–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reinert D. J., Jank T., Aktories K., Schulz G. E. (2005) J. Mol. Biol. 351, 973–981 [DOI] [PubMed] [Google Scholar]
- 33. Ziegler M. O., Jank T., Aktories K., Schulz G. E. (2008) J. Mol. Biol. 377, 1346–1356 [DOI] [PubMed] [Google Scholar]
- 34. Ho J. G., Greco A., Rupnik M., Ng K. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18373–18378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Greco A., Ho J. G., Lin S. J., Palcic M. M., Rupnik M., Ng K. K. (2006) Nat. Struct. Mol. Biol. 13, 460–461 [DOI] [PubMed] [Google Scholar]
- 36. Prochazkova K., Shuvalova L. A., Minasov G., Voburka Z., Anderson W. F., Satchell K. J. (2009) J. Biol. Chem. 284, 26557–26568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pruitt R. N., Chambers M. G., Ng K. K., Ohi M. D., Lacy D. B. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 13467–13472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iqbal T. H., Lewis K. O., Cooper B. T. (1994) Gut 35, 1233–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]