Abstract
Membrane-bound receptors induce biochemical signals to remodel the actin cytoskeleton and mediate cell motility. In association with receptor tyrosine kinases, several downstream mitogen-induced kinases facilitate cell migration. Here, we show a role for c-Jun N-terminal kinase 2 (JNK2) in promoting mammary cancer cell migration through inhibition of epidermal growth factor substrate 8 (EPS8) expression, a key regulator of EGF receptor (R) signaling and trafficking. Using jnk2−/− mice, we found that EPS8 expression is higher in polyoma middle T antigen (PyVMT)jnk2−/− mammary tumors and jnk2−/− mammary glands compared with the respective jnk2+/+ controls. The inverse relationship between the jnk2 and eps8 expression was also associated with cancer progression in that patients with basal-type breast tumors expressing high jnk2 and low eps8 experienced poor disease-free survival. In mammary tumor cell lines, the absence of jnk2 greatly reduces cell migration that is rescued by EPS8 knockdown. Subsequent studies show that JNK2 enhances formation of the EPS8-Abi-1-Sos-1 complex to augment EGFR activation of Akt and ERK, whereas the absence of JNK2 promotes ESP8/RN-Tre association to inhibit endocytotic trafficking of the EGFR. Together, these studies unveil a critical role for JNK2 and EPS8 in receptor tyrosine kinase signaling and trafficking to convey distinctly different effects on cell migration.
Keywords: Cell Motility, Cell Surface Receptor, Endosomes, Receptor Endocytosis, Serine/Threonine Protein Kinase, EPS8, JNK, EGFR
Introduction
Eukaryotic cell migration is essential for embryonic development and tissue homeostasis. In cancer, tumor progression is associated with cell extravasation into the surrounding tissue and vasculature. Up-regulation of motility-associated genes in tumors increases sensitivity to micro-environmental cues and leads to an invasive phenotype. Because the survival rate of metastatic breast cancer remains dismal, proteins involved in cancer cell migration are promising prognostic markers or targets for treatment of metastatic disease.
Cell migration commences in response to chemotactic cues that lead to transient cell polarization and accumulation of signaling complexes at the leading edge of the cell. This response is facilitated by the intrinsic vesicular trafficking machinery (1) and results in actin polymerization to form lamellipodia at the advancing end (2–4).
At the molecular level, ligand-activated RTKs2 induce Ras and subsequently Rac activity to initiate actin reorganization. Active Rac is imperative for lamellipodia and filopodia formation at the leading edge of a migrating cell (5, 6). Appropriate localization and activation of cell signaling pathways are achieved through an intricate interplay of adaptor proteins (Grb2, Shc, etc.), guanine nucleotide exchange factors (GEF), including Son of Sevenless-1 (Sos-1), and scaffold proteins such as Abl interaction-1 (Abi-1) (1, 7). Sos-1, having a dual GEF property, activates membrane-associated Ras. Other upstream kinases such as phosphoinositide 3-kinase (PI3K), Raf, and Rac then induce Akt, ERK, and JNK.
The literature supports an evolutionarily conserved role for JNK in cell movement. There is evidence for the involvement of JNK in forming F-actin stress fibers, altering cell shape, maintaining planar cell polarity, inducing migration during dorsal closure in Drosophila (8), and mediating neural tube and eyelid closure in developing mouse embryos (9, 10). JNKs phosphorylate transcription factors, including c-Jun, ATF2, and other proteins. JNK signaling is associated with proliferation, differentiation, and apoptosis in an isoform-, stimulus-, and tissue-specific manner (11). In higher organisms, the JNK family is represented by three jnk genes (jnk1, jnk2, and jnk3), resulting in 10 isoforms with both common and discrete functions. Huang et al. (12) previously reported that JNK1 induces phosphorylation of paxillin, a focal adhesion adaptor protein, to maintain labile adhesions required for rapid cell migration. No such mechanism is known for JNK2, the isoform most commonly activated by EGF-induced mitogenic responses (13, 14). There is evidence that JNK2 is up-regulated in cancers, including glioblastomas and skin carcinoma (15–17), but the role of JNK2 in breast cancer remains elusive.
New evidence indicates that abnormal vesicular trafficking alters the localization and amplitude of cell surface growth factor receptors and integrins to alter cell proliferation, viability, and migration. In fact, derailed endocytosis has been proposed as a new hallmark of cancer as it provides a mechanism for sustaining (recycling via endosomes) or attenuating (degrading in the lysosome) mitogenic signals (4, 18). This study focuses on a novel role for JNK2 in promoting mammary tumor cell migration by facilitating EGFR trafficking via EPS8.
EPS8 is a multifunctional adaptor protein that acts as an EGFR substrate to integrate signals from Ras and PI3K to Rac (7, 19). EPS8 regulates receptor trafficking by binding with RN-Tre to prevent Rab5-mediated EGFR internalization (20). Moreover, EPS8 interacts with the actin cytoskeleton to facilitate Rac localization and subsequent cell migration (21–23). RTK stimulation of cell migration depends upon the interplay between Abi-1 and Grb2 for Sos-1 binding into an EPS8-containing complex (21, 24, 25). The binding of Abi-1 to EPS8 enables the second GEF function of Sos-1 to enhance the Rac and JNK activity (21, 25, 26).
Here, we used a variety of biochemical and microscopy techniques to show that JNK2 alters mammary tumor cell migration. JNK2 controls cell migration and EGFR endocytosis by regulating the expression and localization of EPS8. To extend these findings to clinical breast cancer, we also found an association between high jnk2 expression and poor disease-free survival (DFS) of patients with basal-type breast tumors. Importantly, the low eps8 expression in this group of basal-type tumors stratified patients with shorter DFS. These findings suggest that the interactions between JNK2 and EPS8 may play a role in disease progression.
EXPERIMENTAL PROCEDURES
Materials
All materials and chemicals were purchased from Sigma unless otherwise noted. The JNK inhibitor (TAT-JIP) was purchased from Calbiochem. Human fibronectin (BD Biosciences) was used for migration assays. JNK2 (D2) and Grb2 (C23) antibodies were obtained from Santa Cruz Biotechnology. pAkt (Ser-473), pJNK, and pERK (Thr-202/Tyr-204) antibodies were purchased from Cell Signaling; EGFR antibodies were purchased from Millipore, and JNK1/2 and EPS8 were from BD Biosciences.
Tumor Data Base Analyses
To estimate the clinical relevance of jnk2 expression in breast cancer, patient data on breast tumors from publicly available sets were used (GSE2990, GSE6532, and GSE9185). These sets are from the same institute and same platform (Affy U133) and were normalized together, and the median was centered before further analysis. Subtypes were stratified based on gene signature (27–29).
Cell Culture
Mouse mammary tumor cell lines were created from PyVMTjnk2+/+ and PyVMTjnk2−/− tumors (30). Cells were maintained in DMEM/F-12 media (Cellgro, Mediatech) with 10% FBS (Gemini Bio-Products), 10 μg/ml insulin (Humulin R), 5 ng/ml EGF (PeproTech), and antibiotics.
Serum-free medium (SFM) consisted of DMEM/F-12 medium containing 10 mm HEPES (pH 7.4), transferrin (2 μg/ml), human fibronectin (2 μg/ml), 1× trace elements (BIOSOURCE), and antibiotics. The 4T1.2 cell line was maintained in α-minimal essential medium (Invitrogen) plus 10% FBS and antibiotics. For growth factor stimulation experiments, cells were washed twice with warm phosphate-buffered saline (PBS). Medium was replaced with SFM. Sixteen to 20 h later, cells were stimulated with FBS or EGF (as indicated). For experiments using small molecule inhibitors, cells were preincubated with TAT-JIP for 30 min prior to incubation with FBS stimulation plus TAT-JIP.
Immunoblotting
Cells were lysed using either RIPA buffer (50 mm Tris-HCl (pH 7.4), 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA) or EB buffer (0.05% Nonidet P-40, 20 mm Tris-HCl (pH 7.6), 0.25 m NaCl, 3 mm EDTA). For tumors, a portion of snap-frozen tissue was homogenized in EB buffer. Lysates were separated using SDS-PAGE and transferred to nitrocellulose. Proteins were visualized using a STORM 860 PhosphorImager (GE Healthcare).
Immunocytochemistry
Cells were plated overnight in chamber slides with fibronectin. Cells were rinsed with PBS and then cultured in SFM for 6–10 h prior to treatment with FBS-containing medium. At the time of harvest, cells were fixed. Slides were blocked with 10% normal goat serum, and primary antibody was diluted in 10% normal goat serum. Slides were then washed and incubated with secondary antibody (Molecular Probes). Finally, slides were mounted with Vectashield DAPI. Fluorescent signal was detected using a CCD camera mounted on a Nikon Diaphot 300 inverted microscope.
Cell Migration
Cell migration was measured using a modified Boyden chamber (BD Biosciences). Cells were placed in the upper compartment of the Transwell chamber. Lower chambers were filled with 1% FBS-containing medium. Cells were allowed to migrate for 8 h. Migrated cells were fixed and stained with 0.5% crystal violet. Four randomly chosen fields were quantified (at 10× objective), or crystal violet was dissolved and the absorbance read. Where indicated, cells were pretreated for 30 min with TAT-JIP prior to migration assays.
For scratch wound assays, a confluent monolayer was scraped in a straight line to create a “scratch” using a sterile p200 pipette tip. Cells were washed and then cultured in 1% FBS-containing media. To obtain the same field during the image acquisition, markings were placed on the plate bottom with an ultrafine tip marker. Changes in the open wound widths were measured by comparing the distance measured at the time of wounding versus the time of harvest.
EGF Internalization
Cells were plated in chamber slides. After serum starvation, slides were kept on ice for 10 min to stall EGFR activity. 100 ng/ml FITC-EGF or Texas Red-EGF (Molecular Probes) was added to the cells. At each time point, acid stripping of membrane-bound ligand was performed using ice-cold 0.2 mm acetic acid, 0.5 mm NaCl (pH 2.5) followed by neutralization in ice-cold Hanks' buffered saline solution (Cellgro, Mediatech).
RN-Tre Overexpression and Co-immunoprecipitation
PyVMT jnk2−/− cells were transiently transfected using Lipofectamine 2000 (Invitrogen) and 2 μg of eGFP RN-Tre or control eGFP plasmids. After 36 h, cells were serum-starved overnight. Cells were then treated with 20 ng/ml EGF for 1 h, and the media were aspirated off and incubated with a cross-linker, namely 2.5 mm dithiobis[succinimidyl propionate] (Pierce). Cells were washed and then lysed (0.5% Nonidet P-40, 20 mm Tris-HCl (pH 8), 150 mm NaCl, 1 mm phenylmethanesulfonyl fluoride, and Miniprotease Inhibitor Mixture (Roche Applied Science)). 300 μg of total protein was immunoprecipitated using protein A/G Plus-agarose beads (Santa Cruz Biotechnology) and anti-EPS8 at 4 °C overnight. Beads were washed with lysis buffer and boiled with SDS sample buffer before resolving by SDS-PAGE.
One day after transfection, a scratch wound was created. Cells were cultured for 12 h, and then images were captured. Transfected cells were then lysed and immunoblotted to determine RN-Tre expression.
Lentiviral Transduction
The lentiviral GFP-tagged nonsilencing control (gipz), shjnk2, or sheps8 plasmids (Open Biosystems) along with three constructs P1, P2, and P3 plasmids (encoding for viral proteins Gag, Pol, and Env) were transfected at a 1:2 (target gene versus viral coat proteins) into Phoenix cells using Lipofectamine 2000. 48 h post-transfection, the supernatant from transfected Phoenix cells was collected, filtered, and mixed with Polybrene. Transduced cells were infected with the virus-containing supernatant and then placed in media supplemented with puromycin.
Real Time PCR
Total RNA was isolated from PyVMTjnk2+/+ and PyVMTjnk2−/− cells. First-strand complementary cDNA was made using Superscript II reverse transcriptase and oligo(dT) primer (Invitrogen). PCR products of jnk1α1, jnk1β1, jnk2α1, jnk2β1, jnk3α1, and jnk3β1 were amplified using the primers described previously (68). Real time PCR was performed on a Stratagene Mx3005P thermocycler using the Brilliant SYBR Green PCR master mix (Stratagene).
Statistical Analysis
Non-patient- based results are presented as mean ± S.E. Results were evaluated using analysis of variance followed by nonparametric, post hoc Student's t test to identify differences between groups. Differences between means yielding p values of ≤0.05 were considered statistically significant.
RESULTS
High JNK2 Expression Is Correlated with Poor Survival in Basal-type Breast Tumors
Breast cancer is a heterogeneous disease with certain subtypes demonstrating more frequent metastasis. Hence, we explored the potential clinical relevance of JNK2 expression in human breast cancer subtypes. To do so, we evaluated if the jnk2 (mapk9) expression level in tumors correlated with the clinical outcome of 469 breast cancer patients using all tumor grades and subtypes. Fig. 1A shows that when all subtypes of breast cancers are included in the analysis, high jnk2 (mapk9) expression showed a trend toward poorer DFS, but it was not statistically significant. Subgroup analysis was then performed to compare jnk2 expression and DFS in luminal A, luminal B, or basal-type breast tumors individually. For luminal A or B tumors, grouping tumors into high or low jnk2 expression did not show differences in DFS (data not shown). This may be due, in part, to the longer survival observed in these patient groups because median DFS was not reached in these data sets. In contrast, high jnk2 expression was associated with poorer DFS in patients with basal-type tumors (Fig. 1B, p < 0.018), which are more frequently metastatic than hormone receptor-positive luminal tumors (31, 32). These data suggest that high jnk2 expression in basal-type breast tumors may negatively influence patient survival. Our observation was further supported by another recent publication (33). Together, these retrospective and correlative studies provided justification to more mechanistically evaluate the role of jnk2 in mammary tumor models.
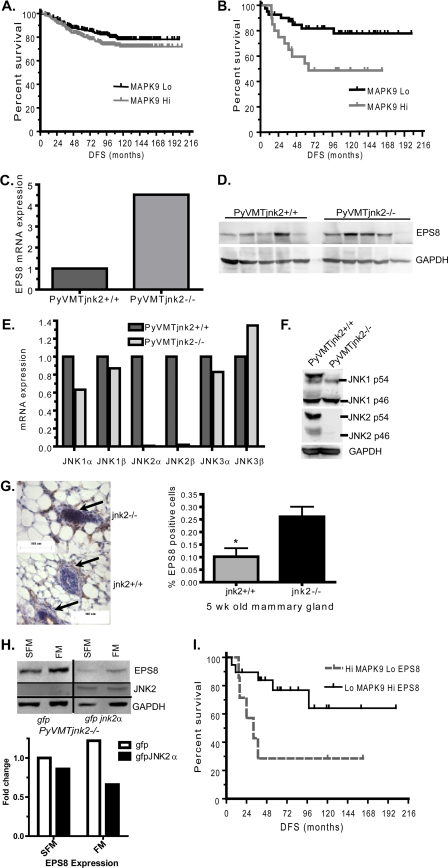
FIGURE 1.
Loss of jnk2 increases EPS8 expression. A, human tumor data base was used to evaluate if jnk2 (mapk9) expression level is correlated with patient DFS using all tumor subtypes. B, correlation of jnk2 expression and DFS was performed in basal-type breast tumors. C, PyVMTjnk2+/+ and PyVMTjnk2−/− mouse mammary tumors. cDNA was reverse-transcribed from each tumor, as described under “Experimental Procedures.” Quantitative PCRs were performed using eps8 and gapdh. D, PyVMTjnk2+/+ and PyVMTjnk2−/− mammary tumors were snap-frozen and homogenized in lysis buffer and analyzed by Western blot using EPS8 primary antibody. Each lane represents an individual tumor. E, RNA was isolated from PyVMTjnk2+/+ and PyVMTjnk2−/− cell lines and reverse-transcribed to cDNA. Quantitative PCRs were performed using jnk1, jnk2, jnk3, and gapdh (as housekeeping gene control) primers. F, PyVMTjnk2+/+ and PyVMTjnk2−/− cell lines were lysed. Membranes were probed using JNK1, JNK2, or GAPDH primary antibodies. G, EPS8 expression in 5-week-old mammary glands; 5-μm sections of fixed, paraffin-embedded tissues were probed with EPS8 antibody. Ductal mammary epithelial cells are designated by arrows. Increased positive staining (brown) was evidenced in jnk2−/− mammary glands (*, p = 0.0168). I, PyVMT/jnk2−/− cells expressing GFP or GFP-JNK2 were cultured in SFM or full medium (FM) for 48 h after which cells were lysed and EPS8 expression determined by Western blot. GAPDH expression served as the loading control. H, basal type breast tumors were evaluated for jnk2 (mapk9) and eps8 expression levels and correlated for DFS were evaluated.
JNK2 Modulates EPS8 Expression in Mammary Tissue
The effects of systemic jnk2 knock-out were evaluated using BALB/c mice expressing the PyVMT transgene. Transgene expression of PyVMT induces PI3K and MAPK pathways in mammary cells in a fashion similar to RTK overexpression (34). Tumor progression in the PyVMT model closely reflects human breast cancer progression (35). Specifically, the early stages resemble ductal carcinoma in situ where estrogen and progesterone receptors are expressed. As tumors progress to late stage, ErbB2 expression becomes elevated, indicating a dependence upon RTK signaling. This model is also one of the few spontaneous mammary tumor models known to metastasize (34).
For our purposes, late stage tumors were harvested to compare gene expression differences in mammary tumors. Microarray analysis showed EPS8 mRNA was elevated 3.9–7.8-fold in PyVMTjnk2−/− mammary tumors compared with their wild type counterparts (data not shown). Real time PCR with EPS8 primers confirmed over 4-fold increase in EPS8 mRNA expression in the PyVMTjnk2−/− tumors compared with controls (Fig. 1C). Consistent with increased mRNA levels, EPS8 protein levels were higher on average when analyzing several individual tumors from each genotype (Fig. 1D).
To characterize the functional implications for the regulation of EPS8 expression by JNK2, cell lines derived from PyVMTjnk2+/+ and PyVMTjnk2−/− tumors were used (30). First, we examined if deletion of jnk2 induced a compensatory increase in JNK1 or JNK3 expression. Quantitative PCR analysis of jnk1 and jnk3 mRNA reflected similar expression levels between the cells lines (Fig. 1E). As anticipated, jnk2α1 and jnk2β1 (normalized to GAPDH) expressions were 50- and 150-fold higher, respectively, in the PyVMTjnk2+/+ cells compared with the PyVMTjnk2−/− cells. These expression patterns were confirmed at the protein level where both cell lines expressed similar levels of JNK1 α1/2 (p54) and JNK1 β2 (p46) proteins. Finally, JNK2 isoforms are only detected in PyVMTjnk2+/+ lysates (Fig. 1F).
To determine whether regulation of eps8 expression by jnk2 was specific to mammary tumor cells, we next assessed if EPS8 expression was also elevated in noncancerous jnk2−/− mammary epithelial cells. Mammary glands were harvested from 5-week-old virgin female mice. Paraffin-embedded tissue was immunostained. Positively staining epithelial cells were scored and compared with the total number of mammary epithelial cells. EPS8 expression was observed in luminal and myoepithelial cells (shown as brown stain, Fig. 1G). Positively staining cells were observed more frequently in jnk2−/− mammary epithelial cells (Fig. 1G, indicated by arrows) compared with jnk2+/+ controls. These data show that the effect of jnk2 on EPS8 expression occurs in both normal and PyVMT-transformed mammary cells.
Previous work has reported that EPS8 expression is induced by FBS in myocytes (36). Therefore, we evaluated whether FBS exposure would alter EPS8 expression in our model and whether expression of JNK2 would affect this response. To this end, PyVMTjnk2−/− gfp and PyVMTjnk2−/− gfp jnk2α subconfluent cells (30) were exposed to SFM or full medium or 48 h. When normalized to the GAPDH loading controls, serum starvation led to only 20% less EPS8 in the GFP-JNK2α-expressing cells compared with the GFP controls (Fig. 1H). In contrast, serum treatment resulted in an even larger expression difference. In the absence of jnk2, EPS8 expression increased, and JNK2α expression led to further reduction in EPS8 with serum exposure. These data suggest that the inhibition of EPS8 expression by JNK2 is growth factor-dependent.
Given the inverse relationship that we observed between jnk2 and eps8 expression in the PyVMT mouse mammary tumors, we used the same human basal-type tumor data base from Fig. 1B to inquire if jnk2 and eps8 expression would further stratify this patient group for DFS. Although eps8 expression alone was not associated with any differences in DFS (data not shown), tumors with high mapk9 (jnk2) and low eps8 expression were associated with shorter DFS compared with the low mapk9/high eps8-expressing tumors (Fig. 1I, p = 0.031). These data led us to hypothesize that the ability of jnk2 to suppress eps8 expression may negatively affect patient outcome. Basal-type breast tumors typically overexpress EGFR, which may further implicate an important function of EPS8 in this breast tumor group.
Absence of JNK2 Inhibits Cell Migration
EPS8 plays a role in actin remodeling and EGFR signaling. Thus, the migration potential of PyVMTjnk2+/+ or PyVMT/jnk2−/− cells was examined using a Transwell assay. PyVMT/jnk2+/+ cells migrate five times more than the PyVMT/jnk2−/− cells (observe untreated groups in Fig. 2A and representative pictures). To delineate the contributions of the other JNK isoforms, cells were preincubated with TAT-JIP, a pan-JNK inhibitor (37, 38). Migration of PyVMTjnk2+/+ cells was reduced with increasing TAT-JIP concentrations (Fig. 2A). At 10 μm TAT-JIP, migration was maximally inhibited by 80%. In contrast, PyVMTjnk2−/− cell migration remained unaltered by TAT-JIP (Fig. 2A), suggesting that the JNK2 isoforms play a key role in serum-induced cell migration. Although a higher concentration was required, a similar effect of TAT-JIP was observed in migration of the basal-type, human breast cancer cell line MDA MB 231 (Fig. 2B).
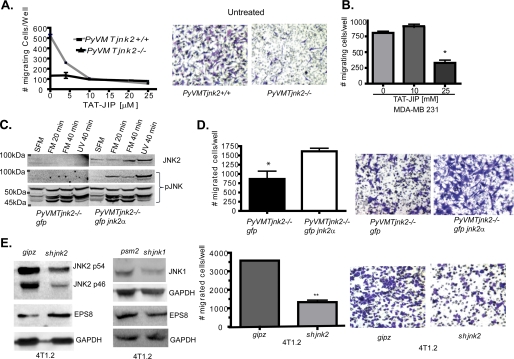
FIGURE 2.
JNK2 mediates cancer cell migration. A, PyVMTjnk2+/+ and PyVMTjnk2−/− cells were plated in the upper chamber of 8-μm inserts. 1% FBS-containing media was added as a chemoattractant. Cell migration was measured after 8 h. Four fields were counted per well. A representation of at least five independent experiments is shown. Representative pictures are shown of migratory, stained cells. B, MDA-MB 231 human breast cancer cells were treated with TAT-JIP at indicated concentrations, and cell migration was assayed 8 h later as described under “Experimental Procedures.” C, PyVMTjnk2−/− cells expressing GFP and GFP-JNK2α were treated with full medium (FM) containing 10% FBS, EGF, and insulin for 20 or 40 min or UV light (10 J/m2). Membranes were probed as indicated using phospho-JNK and JNK2 antibodies. D, GFP or GFP JNK2α-expressing PyVMTjnk2−/− cells were assayed for cell migration, as above. Four individual fields were counted per insert (*, p = 0.0302, two-sided Student's t test). Representative pictures are shown of migratory stained cells. E, 4T1.2 mouse mammary tumor cells were transduced with shjnk2, shjnk1, or psm2 and gipz (nonsilencing vectors) expressing lentivirus, as indicated. Cells were evaluated for JNK1, JNK2, and EPS8 expression by Western blot analysis. Expression was quantified and normalized to individual GAPDH loading controls and compared with gipz-expressing cells. Subsequently, gipz- and shjnk2-expressing 4T1.2 cells were plated in a modified Boyden chamber. 5% FBS was used as a chemoattractant in the lower chamber. After 8 h, cells remaining on the lower side of the filter were stained with crystal violet and counted (**, p = 0.002, two-sided Student's t test). Representative pictures are shown of migratory stained cells.
To further confirm a role for JNK2 in mammary cell migration, the PyVMTjnk2−/− cells expressing GFP-JNK2α were compared with the GFP-expressing control cells. We first evaluated if JNK signaling differs in response to serum or UV treatment (a well known JNK stimulant). Typically, testing isoform-specific responses is difficult due to the similar sizes of the various JNK isoforms, as well as the identical homology in the dually phosphorylated region recognized by the phosphor-specific antibody. In our model, expression of GFP-JNK2α fusion protein (∼95 kDa) in jnk2-deficient cell allowed us to specifically address its phosphorylation relative to the other isoforms.
Fig. 2C shows that GFP-JNK2α is phosphorylated in response to either serum or UV treatment. A 49-kDa JNK protein is also phosphorylated by both treatment types, whereas the 46-kDa JNK protein was increased more by FBS than UV light. Finally, a 54-kDa protein shows no change in phosphorylation with either treatment. These data show that JNK2 and another JNK1 or JNK3 isoform respond to serum treatment. Importantly, expression of JNK2α does not alter the phosphorylation of the other JNK isoforms in response to either serum or UV light. To address whether JNK2 enhances cell migration in response to growth factor exposure, Boyden chamber-based migration assays were performed. GFP-JNK2α expression rescued motility of the PyVMTjnk2−/− cells (Fig. 2D, p = 0.032) in response to serum stimulation. These observations were further validated using a wound-based assay (data not shown). Collectively, these data show that JNK2 phosphorylation is increased in response to serum, and a large portion of cell migration is mediated solely by JNK2α.
Breast cancer is a heterogeneous disease with distinct subtypes. Among the subtypes, the basal-type tumors are known to be the most metastatic. Thus, we evaluated a well characterized basal type, 4T1.2 murine mammary cancer cells (39), and transduced them with a short hairpin RNA (shRNA) targeting jnk2. shjnk2 expression resulted in an 88% reduction in JNK2 protein. Moreover, JNK2 knockdown led to a concomitant increase in endogenous EPS8 protein (Fig. 2E), although knockdown of JNK1 did not affect EPS8 expression. In agreement with our previous observation, reduction of JNK2 decreased cell migration 3-fold compared with the gipz controls (Fig. 2D). Again, these observations strongly support a central role for JNK2 in positively modulating breast cancer cell migration.
JNK2 Regulates EPS8 Localization
EPS8 can interact with several critical signaling molecules within the cell. Although the outcomes or predominance of all those interactions are not known, previous studies have elucidated functions for EPS8 in the context of RTK signaling. EPS8, when bound to Abi-1 and Sos-1, transmits EGFR-induced signals from Ras to Rac. Because localization of activated Rac at the migration front is critical, we tested if JNK2 participates in the EPS8-Abi-1-Sos-1 complex or directly interacts with any of these proteins. Failure to detect any endogenous interactions led us to propose an alternative mechanism by which JNK2 could indirectly affect EPS8 function in RTK signaling and cell migration.
EPS8 is necessary for correct localization of Rac GTPase to the sites of F-actin remodeling at the leading edge of a migrating cell (21, 22). Earlier studies detected EPS8 at membrane ruffles in growth factor-stimulated epithelial cells (40, 41), and these ruffles were completely abrogated in cells lacking any eps8 (7, 22, 26) suggesting a role for EPS8 in facilitating the dynamic membrane activity observed during migration.
These reports led us to determine whether JNK2 influences EPS8 localization or the formation of membrane ruffles in the presence or absence of JNK2. Using immunocytochemistry, we observed classic membrane ruffles, indicative of advancing lamellipodia in response to serum or EGF, only in the PyVMTjnk2+/+ cells (Fig. 3A). In these cells EPS8 is mainly localized at membrane ruffles. In contrast, membrane ruffling is negligible in PyVMTjnk2−/− cells, and EPS8 is restricted to “focal adhesion”-like structures (Fig. 3B). Similarly, reduction in JNK2 expression coincided with the absence of EPS8 at membrane ruffles in the 4T1.2 cells (supplemental Fig. 1, A and B). These data show that JNK2 is necessary for EPS8 localization at membrane ruffles and subsequent membrane ruffle formation. Accurate localization of EPS8 in the leading edge ruffles may poise cells for an effective migratory response.
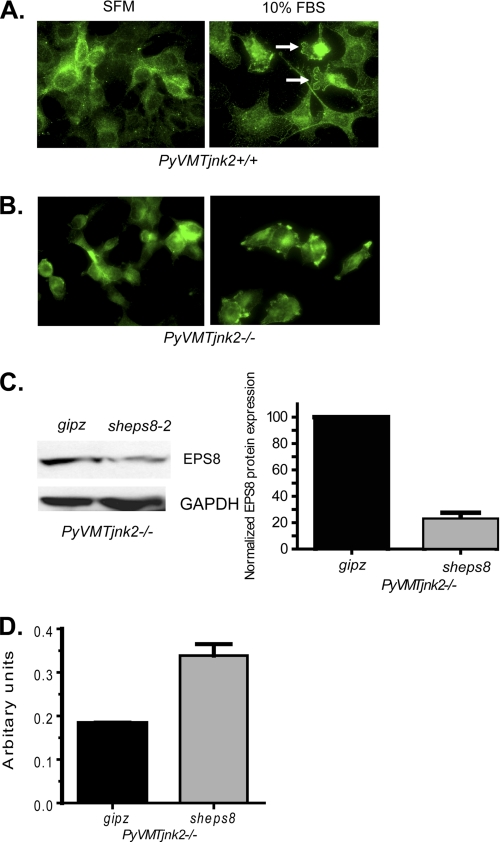
FIGURE 3.
ESP8 localization is JNK2-dependent and sheps8 expression in PyVMTjnk2−/− cells increases cell migration. PyVMTjnk2+/+ (A) and PyVMTjnk2−/− (B) cells suspended in SFM were plated onto fibronectin-coated chamber slides. Cells remained in SFM or were treated with 10% FBS- and EGF-containing media. 60 min later, cells were fixed. EPS8 primary antibody (green) was used for cytochemical analysis. White arrows designate membrane ruffles. C, PyVMT jnk2−/− mammary tumor cells were transduced with sheps8 or gipz nonsilencing vector expressing lentivirus. After puromycin selection, EPS8 expression was measured using Western blot analysis. EPS8 bands were quantified, and values were normalized to individual GAPDH-loading controls and then compared with gipz-expressing cells. D, PyVMT jnk2−/− gipz- and sheps8-expressing cells were plated in the upper well of a modified Boyden chamber using 1% FBS as a chemoattractant. At 8 h, cells in the lower chamber were stained with crystal violet. Crystal violet was then dissolved and absorbance of each sample measured.
EPS8 Inhibits Cell Migration in the Absence of JNK2
EPS8 mediates RTK response and actin dynamics, and our data show that JNK2 facilitates EPS8 localization in the membrane ruffles. We then questioned if there is a threshold of EPS8 that is detrimental to migration. In other words, is the down-regulation of eps8 expression by JNK2 critical for cell migration? To address this question, stable expression of sheps8 in PyVMTjnk2−/− cells decreased the otherwise elevated levels of EPS8 protein by ∼80% (Fig. 3C). Indeed, reduction of EPS8 increased migration of jnk2-deficient cells (Fig. 3D). These data indicate that the ability of JNK2 to regulate EPS8 expression has functional consequences on cancer cell migration.
JNK2 Abrogates EPS8 Binding to RN-Tre to Restore Wound Repair
Our data show that the localization and effect of EPS8 on cell migration differs in a JNK2-dependent fashion. Despite the existing information on the various complexes in which EPS8 engages, the conditions that favor one signaling complex over the other are not well understood. In the canonical pathway, EPS8 is tethered to Sos-1 to activate Rac signaling when associated with Abi-1. Measurement of cellular EPS8 and Abi-1 revealed a 1:3 ratio (22). In a jnk2-deficient scenario, the overabundance of EPS8 may alter this balance, allowing excess EPS8 to engage a new partner, namely RN-Tre (42), via the same binding site. RN-Tre binding to EPS8 reduces EGF response by inhibiting Rab5-mediated EGFR internalization and trafficking (20). The net biological outcome would be decreased cell migration, as we consistently observed.
To test if EGF stimulation alters binding between RN-Tre and EPS8, PyVMTjnk2−/− cells were transfected with GFP-RN-Tre and then serum-starved. Fig. 4A shows that EPS8 and RN-Tre interact in the absence of EGF. After EGF exposure, the interaction between EPS8 and RN-Tre binding is diminished, consistent with the competitive binding that occurs between Abi-1 and RN-Tre for EPS8. These data support that less RN-Tre binding allows EPS8 to interact with the Abi and Sos-1 to transduce EGFR signaling (7).
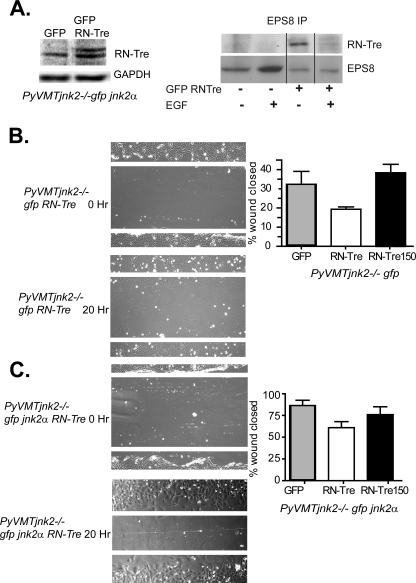
FIGURE 4.
Expression of JNK2 abrogates EPS8-RN-Tre interactions and restores wound repair. A, PyVMTjnk2−/− cells were transfected with eGFP or eGFP RN-Tre and then serum-starved overnight. Cells were harvested 1 h after EGF treatment, and samples were immunoprecipitated with anti-EPS8. RN-Tre binding was detected using Western blot analysis. PyVMTjnk2−/−gfp (B) and PyVMTjnk2−/−gfp jnk2 (C) cells were plated in 6-well dishes. The next day, cells were transiently transfected with eGFP, RN-Tre, or RN-Tre150 expression plasmids. 24 h after transfection, a scratch wound was created in each sample, and culture media were replaced with 1% FBS-containing media. Wound closure was measured at time 0 and at 20 h post-wounding.
To then evaluate how RN-Tre influences cell migration, GFP-RN-Tre and a mutant GFP-RN-Tre 150 (lacking GAP activity at residue 150) were overexpressed in PyVMTjnk2−/−gfp or PyVMTjnk2−/−gfpjnk2α cells. Wound assays were performed to measure the influence of RN-Tre on cell migration. Representative images, depicted in Fig. 4B, show stronger inhibition of wound closure in the PyVMTjnk2−/−gfp cells with RN-Tre overexpression. In contrast, wound closure was less restricted when RN-Tre was overexpressed in the PyVMTjnk2−/−gfp jnk2α cells (Fig. 4C). These data further confirm that EPS8 binding to Abi-1 or RN-Tre is dependent on protein expression ratios. An increase in RN-Tre-EPS8 complexes usurps EPS8-Abi1-Sos-1 signaling and cell migration in the same way as higher EPS8 expression.
Absence of JNK2 Impairs EGF Internalization
Both RTK activity and appropriate localization are necessary for cells to polarize and then initiate migration through localized actin remodeling (43, 44). Internalization of activated receptors allows ligand recycling and compartmentalization of signaling modules (45, 46). Thus, “site-specific” enrichment of migratory signals occurs via endocytosis to facilitate directional chemotactic cell movement (47, 48).
At this point, it was still unclear how EPS8 regulates RTK signaling and cell migration in the context of JNK2. Previous publications have shown that the EPS8-Abi-1-Sos-1-PI3K complex promotes RTK signaling (7). Alternatively, when EPS8 binds to RN-Tre (via the same Src homology 3 domain that binds Abi-1), it inhibits Rab5-mediated EGFR internalization (20, 49).
To shed light on whether JNK2 affects EGFR signaling, we first compared EGFR trafficking in PyVMTjnk2+/+ and PyVMTjnk2−/− cells. EGFR internalization was visualized using pulsed FITC-EGF treatment. Mild acid stripping removes membrane-bound EGF, allowing clearer visualization of internalized ligand. The left panel of Fig. 5A shows membrane-bound FITC-EGF prior to acid stripping for EGF detection at the beginning of the assay. After 15 min, FITC-EGF was dispersed throughout the cytoplasm in the PyVMTjnk2+/+ cells (Fig. 5A, right panel). In contrast, green punctates, depicting internalized EGF/EGFR complexes, were rare in the jnk2-deficient cells (even though the basal EPS8 expression is higher). This observation suggests that jnk2 deficiency in cells expressing high EPS8 favors the alternative pathway where EPS8 binds to RN-Tre, an interaction known to inhibit EGFR internalization. If this is indeed the case, then reducing EPS8 in PyVMTjnk2−/− cells will increase receptor internalization and signaling.
FIGURE 5.
Loss of JNK2 impairs EGFR internalization. A, cells were treated with 100 ng/ml FITC-EGF at 37 °C for 15 min. Media were aspirated, and membrane-bound ligand was acid-stripped with 0.01% acetic acid solution. FITC-EGF uptake was captured live using ImagePro 6.1 software and color overlay (EGF = green, ×100). Data are representative of two independent experiments. B, conditions were similar to A, except PyVMTjnk2−/−gipz and PyVMTjnk2−/−sheps8 cells were treated with Texas Red-EGF. Acid stripping removed unbound EGF (EGF = red, ×100). C, subconfluent cell lines were lysed and later probed with EGFR antibody. GAPDH was used as the internal loading control. D, subconfluent PyVMTjnk2−/−gipz and PyVMTjnk2−/−sheps8 cells were serum-starved overnight and then stimulated with 1% FBS for 0, 0.5, 1, 2, and 4 h. Cell lysates were probed using primary antibodies, as indicated. GAPDH was used to evaluate equal loading among samples. E, model diagram. In the leading edge of a migrating cell, activated EGFR is internalized into endosomes via active Rab5 GTPase. EPS8, when complexed with Abi-1 and Sos-1, transmits signals from EGFR to activate Rac and JNK, which facilitate actin remodeling and migration. JNK2 inhibits eps8 expression such that low levels of EPS8 bind to Abi-1 and Sos-1. With JNK2 deficiency, eps8 transcription is up-regulated, and the Abi-1/Sos-1 pool becomes rate-limiting. Excess EPS8 binds to RN-Tre. EPS8 binding activates RN-Tre, a GTPase-activating enzyme for Rab5, resulting in Rab5 inactivation and reduced EGFR internalization.
To test this possibility, the PyVMTjnk2−/−sheps8 cells were assayed using a Texas Red-EGF pulse. Red EGF is abundant in the cytoplasm of PyVMTjnk2−/−sheps8 cells compared with their controls (Fig. 5B), strongly supporting our hypothesis that lowering EPS8 in the jnk2−/− cells restores canonical RTK internalization and signaling. We also evaluated EGFR to determine whether expression differences influenced EGF internalization. PyVMTjnk2+/+ and PyVMTjnk2−/− cells showed similar levels of EGFR, demonstrating that only internalization of EGFR is affected by JNK2 and EPS8 (Fig. 5C).
Analysis of fluorescently labeled EGF in Fig. 4, A and B, showed EGF (and perhaps EGFR) in the nucleus of the PyVMTjnk2−/− cells. EGFR nuclear transport has been previously reported so we further evaluated its localization by fractionating cell lysates. The supplemental Fig. 2 compares EGFR cytoplasmic and nuclear fractions in PyVMTjnk2+/+ and PyVMTjnk2−/− with or without EGF treatment. Although small amounts of EGFR were observed in nuclear extracts of the PyVMT/jnk2−/− cells, EGF exposure had no effect on EGFR abundance in either compartment. We then compared PyVMTjnk2−/−gfp and PyVMTjnk2−/− gfp jnk2α cells. Again, EGF treatment did not affect EGFR distribution (supplemental Fig. 2), but PyVMTjnk2−/−gfp cells had higher nuclear EGFR levels. Together, these data indicate that JNK2 may be important in retaining EGFR in the cytoplasm independently of EGF. However, with regard to interpreting the nuclear location of FITC-EGF or Texas Red-EGF in Fig. 4B, it seems unlikely that the fluorescent signals represent EGF-EGFR complexes.
Immunocytochemical analyses were also performed to visualize EGFR localization after EGF exposure. An increase in internalized EGFR was observed throughout the cytoskeleton and lamellipodia within 30 min of EGF stimulation in the PyVMTjnk2+/+ cells (supplemental Fig. 3). In contrast, the jnk2-deficient cells showed weak fluorescence and starkly reduced cytoplasmic EGFR. EGFR appeared to co-localize with the actin cytoskeleton, as reported by others both in vitro and in vivo (50). Taken together, our data support that JNK2 affects EPS8 localization at membrane ruffles as well as EGFR trafficking in response to ligand binding.
EPS8 Knockdown Sensitizes Cells to EGF Stimulation in the Absence of JNK2
After EGFR internalization, ligand-bound receptor complexes continue to signal within endosomes (48). To understand how less EPS8 in jnk2-deficient cells migrate more successfully, we examined EGFR signaling using phosphorylation of ERK, Akt, and JNK as downstream readouts of activated EGFR. Each kinase family has an established role in cell migration (51–53). EGF treatment of PyVMTjnk2−/−sheps8 cells increased phosphorylation of Akt and ERK compared with the PyVMTjnk2−/−gfp or PyVMTjnk2−/− GFPjnk2α cells (Fig. 5D). In a similar fashion, EGF-induced phosphorylation of the JNK1/3 isoforms was also higher in the PyVMTjnk2−/−sheps8 cells compared with PyVMTjnk2−/−gipz-expressing cells. These differences coincide with increased EGFR internalization and a higher migration phenotype observed in the sheps8-expressing jnk2 knock-out cells. Considering our data and the existing view that EPS8 enhances EGFR signaling (42, 54, 55), we conclude that JNK2 is critical for EGFR internalization and recruitment of EPS8 to complexes that propagate signals to ERK, Akt, and JNK.
DISCUSSION
Tumor cell migration is essential for tissue invasion and metastasis. Up-regulation of motility-associated genes allows cancer cells to develop a metastatic phenotype. Indeed, Yao et al. (56) recently reported low frequency amplification of EPS8, in association with H2AFJ and KRAS in the same region, in high grade breast tumors. EPS8 has also been implicated in Src-mediated transformation (57). Migration machinery in cancer cells provides a strategic target for treatment of the metastatic disease. To this end, we report a novel role for JNK2 and the multifunctional scaffold protein EPS8 in mammary cancer cell migration. Human tumor data provide further support that high jnk2 expression may contribute to the poorer survival observed in patients with basal-type tumors. This relationship was further strengthened in a combined analysis of high jnk2- and low eps8-expressing tumors. Our cell line data provide important insight into how JNK2 expression influences EPS8 abundance and function in EGFR-expressing mammary tumor cells. Together, these data support the concept that EPS8 function is influenced by the abundance of other regulatory proteins.
Interestingly, JNK is also a convergence point for Rac and FAK (58, 59), supporting its role in metastasis (60). Our tools, including biochemical inhibitors of JNK and various genetic knock-out and knockdown models, enabled us to isolate the role of JNK2 in mammary cancer cells. We determined that JNK2 is the key isoform that drives cell migration in breast cancer cells via a novel pathway involving EPS8.
EPS8 possesses several key properties through which JNK2 influences EGFR signaling and cell migration. Cellular interactions involving EPS8 also include association with Dishevelled IRSp53 and Shc (61). A well studied function of EPS8 is its ability to transmit and diversify RTK signaling. This property gains clinical relevance in cancer both in facilitating tumor growth and metastasis. Elevated levels of EPS8 have been reported in lung, thyroid, pituitary, colon, pancreatic, and breast tumors (42, 54, 55, 56, 62, 63). However, with respect to cell migration, most of the data are correlative or inconclusive. In fact, overexpression of EPS8 failed to increase cell migration in pituitary tumors (55). We found that jnk2 expression level showed a stronger association with patient survival in breast cancer, although eps8 expression alone did not discriminate DFS when tumor subtypes were grouped or when basal type alone was evaluated. Our studies of the role of EPS8 in growth factor signaling using the invasive PyVMT model, along with other basal-type cell lines, underlines its potential importance in human disease.
In our studies, the reduction or loss of JNK2 expression increased EPS8 levels and decreased cell migration. Decreased EPS8 expression in PyVMTjnk2−/− cells enhanced cell migration. We observed this phenotype in both PyVMT and 4T1.2 cells, raising the question of why, despite higher levels of EPS8, the JNK2-deficient cells migrate more poorly. There are two possible explanations for the reduced migratory response. First, EPS8 is essential for transmitting growth factor signals from Ras to Rac, and JNK is a downstream effector. In the absence of JNK2, Rac activity and actin remodeling are impaired. We observed a JNK2 dependence of actin polymerization and lamellipodia formation. Also, EPS8 was not detected at ruffles in the absence of JNK2. The second possibility is that EPS8 acts directly on actin fibers. Several in vitro and in vivo studies showed that the effector domain of EPS8 has a high affinity for actin filament barbed ends. EPS8 functions as a capping protein in the presence of Abi-1 (22). Localized actin polymerization on the fast growing barbed end allows protrusion of the cell membrane, but higher levels of EPS8 inhibit actin filament growth (65).
Disanza et al. (66) reported that EPS8 and Abi-1 exist in cells at a 1:3 ratio. Its relative abundance may be a critical determinant in the participation of EPS8 in various protein complexes. JNK2 influences EPS8 expression and localization, perhaps allowing it to facilitate EGFR signaling by binding with Abi-1-Sos-1 or to inhibit EGFR trafficking by binding with RN-Tre (7, 20). The EPS8-RN-Tre complex inhibits EGFR internalization by reducing Rab5 activity (67). We observed that PyVMTjnk2+/+ cells internalize EGF more efficiently than their jnk2−/− counterparts. Considering these findings, we propose that in the absence of JNK2, higher levels of EPS8 interact with RN-Tre, which inhibits RTK internalization and cell migration (Fig. 5E).
Strength and duration of RTK responses are dependent upon endocytosis and degradation. Receptor endocytosis directs it to various cellular compartments (48). Activation of endosome-associated EGFR supports cell survival by stimulating the PI3K-Akt and ERK pathways. Recently, it was shown that depletion of a Rab5 GEF inhibited EGFR signaling and proliferation in A549 cells (64). This indicates that EGFR internalization is essential for its signaling in these cancer cells. Our model reinforces that increased ERK and Akt activity occurs when RTK signals are driven through the EPS8-Sos-1-Abi-1 complex.
We predicted that jnk2-deficient cells expressing more EPS8 have higher ratios of EPS8-RN-Tre complexes. RN-Tre overexpression inhibited migration more strongly in jnk2-deficient cells. Thus, if JNK2 is essential for EGFR endocytosis then presumably the EGFR-EPS8-RN-Tre complex is targeted to endocytic vesicles.
Overall, our model suggests that the consequence of low jnk2 expression is dictated by the balance between the EPS8-Abi-1-Sos-1 complex that promotes migration and the EPS8-RN-Tre complex that inhibits receptor internalization and the signaling necessary for cell motility. Given that high jnk2 expression was associated with poorer DFS in human breast tumors, we propose that JNK2 conveys important RTK-related signals in breast tumors. This may also extend to EGFR binding partners like ErbB2, but the sample size was too low to detect a statistical difference in the ErbB2-type tumors. We show for the first time that JNK2 enhances growth factor-induced cell migration by regulating RTK trafficking and modulating EPS8 levels. These findings underline the critical role of JNK2 in RTK signaling and migration and stresses its importance in cancer.
Supplementary Material
Acknowledgments
We thank Dr. Robin Anderson for generously providing the 4T1.2 cells. Drs. DeFiori and Scita generously donated the expression plasmids and RN-Tre antibody. We also thank Gayathri J. Kollessery for valuable technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant CA 100238 (to C. L. V. D. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- RTK
- receptor tyrosine kinase
- EGFR
- EGF receptor
- SFM
- serum-free medium
- DFS
- disease-free survival
- GDF
- guanine nucleotide exchange factor
- eGFP
- enhanced GFP.
REFERENCES
- 1. Ridley A. J. (2001) J. Cell Sci. 114, 2713–2722 [DOI] [PubMed] [Google Scholar]
- 2. Sorkin A. (2001) Biochem. Soc. Trans. 29, 480–484 [DOI] [PubMed] [Google Scholar]
- 3. Yamazaki T., Zaal K., Hailey D., Presley J., Lippincott-Schwartz J., Samelson L. E. (2002) J. Cell Sci. 115, 1791–1802 [DOI] [PubMed] [Google Scholar]
- 4. Disanza A., Frittoli E., Palamidessi A., Scita G. (2009) Mol. Oncol. 3, 280–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003) Science 302, 1704–1709 [DOI] [PubMed] [Google Scholar]
- 6. Cory G. O., Ridley A. J. (2002) Nature 418, 732–733 [DOI] [PubMed] [Google Scholar]
- 7. Scita G., Nordstrom J., Carbone R., Tenca P., Giardina G., Gutkind S., Bjarnegård M., Betsholtz C., Di Fiore P. P. (1999) Nature 401, 290–293 [DOI] [PubMed] [Google Scholar]
- 8. Xia Y., Karin M. (2004) Trends Cell Biol. 14, 94–101 [DOI] [PubMed] [Google Scholar]
- 9. Weston C. R., Wong A., Hall J. P., Goad M. E., Flavell R. A., Davis R. J. (2003) Genes Dev. 17, 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sabapathy K., Jochum W., Hochedlinger K., Chang L., Karin M., Wagner E. F. (1999) Mech. Dev. 89, 115–124 [DOI] [PubMed] [Google Scholar]
- 11. Bogoyevitch M. A. (2006) BioEssays 28, 923–934 [DOI] [PubMed] [Google Scholar]
- 12. Huang C., Rajfur Z., Borchers C., Schaller M. D., Jacobson K. (2003) Nature 424, 219–223 [DOI] [PubMed] [Google Scholar]
- 13. Antonyak M. A., Moscatello D. K., Wong A. J. (1998) J. Biol. Chem. 273, 2817–2822 [DOI] [PubMed] [Google Scholar]
- 14. Bost F., McKay R., Bost M., Potapova O., Dean N. M., Mercola D. (1999) Mol. Cell. Biol. 19, 1938–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsuiki H., Tnani M., Okamoto I., Kenyon L. C., Emlet D. R., Holgado-Madruga M., Lanham I. S., Joynes C. J., Vo K. T., Wong A. J. (2003) Cancer Res. 63, 250–255 [PubMed] [Google Scholar]
- 16. Chen N., Nomura M., She Q. B., Ma W. Y., Bode A. M., Wang L., Flavell R. A., Dong Z. (2001) Cancer Res. 61, 3908–3912 [PubMed] [Google Scholar]
- 17. Tong C., Yin Z., Song Z., Dockendorff A., Huang C., Mariadason J., Flavell R. A., Davis R. J., Augenlicht L. H., Yang W. (2007) Am. J. Pathol. 171, 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mosesson Y., Mills G. B., Yarden Y. (2008) Nat. Rev. Cancer 8, 835–850 [DOI] [PubMed] [Google Scholar]
- 19. Fazioli F., Minichiello L., Matoska V., Castagnino P., Miki T., Wong W. T., Di Fiore P. P. (1993) EMBO J. 12, 3799–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lanzetti L., Rybin V., Malabarba M. G., Christoforidis S., Scita G., Zerial M., Di Fiore P. P. (2000) Nature 408, 374–377 [DOI] [PubMed] [Google Scholar]
- 21. Scita G., Tenca P., Areces L. B., Tocchetti A., Frittoli E., Giardina G., Ponzanelli I., Sini P., Innocenti M., Di Fiore P. P. (2001) J. Cell Biol. 154, 1031–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Disanza A., Carlier M. F., Stradal T. E., Didry D., Frittoli E., Confalonieri S., Croce A., Wehland J., Di Fiore P. P., Scita G. (2004) Nat. Cell Biol. 6, 1180–1188 [DOI] [PubMed] [Google Scholar]
- 23. Croce A., Cassata G., Disanza A., Gagliani M. C., Tacchetti C., Malabarba M. G., Carlier M. F., Scita G., Baumeister R., Di Fiore P. P. (2004) Nat. Cell Biol. 6, 1173–1179 [DOI] [PubMed] [Google Scholar]
- 24. Yang S. S., Van Aelst L., Bar-Sagi D. (1995) J. Biol. Chem. 270, 18212–18215 [DOI] [PubMed] [Google Scholar]
- 25. Innocenti M., Tenca P., Frittoli E., Faretta M., Tocchetti A., Di Fiore P. P., Scita G. (2002) J. Cell Biol. 156, 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Innocenti M., Frittoli E., Ponzanelli I., Falck J. R., Brachmann S. M., Di Fiore P. P., Scita G. (2003) J. Cell Biol. 160, 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu Z., Fan C., Oh D. S., Marron J. S., He X., Qaqish B. F., Livasy C., Carey L. A., Reynolds E., Dressler L., Nobel A., Parker J., Ewend M. G., Sawyer L. R., Wu J., Liu Y., Nanda R., Tretiakova M., Ruiz Orrico A., Dreher D., Palazzo J. P., Perreard L., Nelson E., Mone M., Hansen H., Mullins M., Quackenbush J. F., Ellis M. J., Olopade O. I., Bernard P. S., Perou C. M. (2006) BMC Genomics 7, 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loi S., Haibe-Kains B., Desmedt C., Lallemand F., Tutt A. M., Gillet C., Ellis P., Harris A., Bergh J., Foekens J. A., Klijn J. G., Larsimont D., Buyse M., Bontempi G., Delorenzi M., Piccart M. J., Sotiriou C. (2007) J. Clin. Oncol. 25, 1239–1246 [DOI] [PubMed] [Google Scholar]
- 29. Loi S., Haibe-Kains B., Desmedt C., Wirapati P., Lallemand F., Tutt A. M., Gillet C., Ellis P., Ryder K., Reid J. F., Daidone M. G., Pierotti M. A., Berns E. M., Jansen M. P., Foekens J. A., Delorenzi M., Bontempi G., Piccart M. J., Sotiriou C. (2008) BMC Genomics 9, 239–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen P., O'Neal J. F., Ebelt N. D., Cantrell M. A., Mitra S., Nasrazadani A., Vandenbroek T. L., Heasley L. E., Van Den Berg C. L. (2010) PLoS ONE 5, e10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perou C. M., Sørlie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Rees C. A., Pollack J. R., Ross D. T., Johnsen H., Akslen L. A., Fluge O., Pergamenschikov A., Williams C., Zhu S. X., Lønning P. E., Børresen-Dale A. L., Brown P. O., Botstein D. (2000) Nature 406, 747–752 [DOI] [PubMed] [Google Scholar]
- 32. Prat A., Parker J. S., Karginova O., Fan C., Livasy C., Herschkowitz J. I., He X., Perou C. M. (2010) Breast Cancer Res. 12, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang X., Chao L., Li X., Ma G., Chen L., Zang Y., Zhou G. (2010) Hum. Pathol. 41, 401–406 [DOI] [PubMed] [Google Scholar]
- 34. Webster M. A., Hutchinson J. N., Rauh M. J., Muthuswamy S. K., Anton M., Tortorice C. G., Cardiff R. D., Graham F. L., Hassell J. A., Muller W. J. (1998) Mol. Cell. Biol. 18, 2344–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin E. Y., Jones J. G., Li P., Zhu L., Whitney K. D., Muller W. J., Pollard J. W. (2003) Am. J. Pathol. 163, 2113–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gallo R., Provenzano C., Carbone R., Di Fiore P. P., Castellani L., Falcone G., Alemà S. (1997) Oncogene 15, 1929–1936 [DOI] [PubMed] [Google Scholar]
- 37. Barr R. K., Kendrick T. S., Bogoyevitch M. A. (2002) J. Biol. Chem. 277, 10987–10997 [DOI] [PubMed] [Google Scholar]
- 38. Barr R. K., Bogoyevitch M. A. (2001) Int. J. Biochem. Cell Biol. 33, 1047–1063 [DOI] [PubMed] [Google Scholar]
- 39. Lelekakis M., Moseley J. M., Martin T. J., Hards D., Williams E., Ho P., Lowen D., Javni J., Miller F. R., Slavin J., Anderson R. L. (1999) Clin. Exp. Metastasis 17, 163–170 [DOI] [PubMed] [Google Scholar]
- 40. Goicoechea S., Arneman D., Disanza A., Garcia-Mata R., Scita G., Otey C. A. (2006) J. Cell Sci. 119, 3316–3324 [DOI] [PubMed] [Google Scholar]
- 41. Offenhäuser N., Borgonovo A., Disanza A., Romano P., Ponzanelli I., Iannolo G., Di Fiore P. P., Scita G. (2004) Mol. Biol. Cell 15, 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matoskova B., Wong W. T., Salcini A. E., Pelicci P. G., Di Fiore P. P. (1995) Mol. Cell. Biol. 15, 3805–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones M. C., Caswell P. T., Norman J. C. (2006) Curr. Opin. Cell Biol. 18, 549–557 [DOI] [PubMed] [Google Scholar]
- 44. Kawada K., Upadhyay G., Ferandon S., Janarthanan S., Hall M., Vilardaga J. P., Yajnik V. (2009) Mol. Cell. Biol. 29, 4508–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Orth J. D., McNiven M. A. (2006) Cancer Res. 66, 11094–11096 [DOI] [PubMed] [Google Scholar]
- 46. Sorkin A., Goh L. K. (2008) Exp. Cell Res. 314, 3093–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Palamidessi A., Frittoli E., Garré M., Faretta M., Mione M., Testa I., Diaspro A., Lanzetti L., Scita G., Di Fiore P. P. (2008) Cell 134, 135–147 [DOI] [PubMed] [Google Scholar]
- 48. Gould G. W., Lippincott-Schwartz J. (2009) Nat. Rev. Mol. Cell Biol. 10, 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Matòsková B., Wong W. T., Nomura N., Robbins K. C., Di Fiore P. P. (1996) Oncogene 12, 2679–2688 [PubMed] [Google Scholar]
- 50. Tang J., Gross D. J. (2003) Biochem. Biophys. Res. Commun. 312, 930–936 [DOI] [PubMed] [Google Scholar]
- 51. Joslin E. J., Opresko L. K., Wells A., Wiley H. S., Lauffenburger D. A. (2007) J. Cell Sci. 120, 3688–3699 [DOI] [PubMed] [Google Scholar]
- 52. Huang C. Y., Fong Y. C., Lee C. Y., Chen M. Y., Tsai H. C., Hsu H. C., Tang C. H. (2009) Biochem. Pharmacol. 77, 794–803 [DOI] [PubMed] [Google Scholar]
- 53. Teranishi F., Takahashi N., Gao N., Akamo Y., Takeyama H., Manabe T., Okamoto T. (2009) Cancer Sci. 100, 770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maa M. C., Lee J. C., Chen Y. J., Chen Y. J., Lee Y. C., Wang S. T., Huang C. C., Chow N. H., Leu T. H. (2007) J. Biol. Chem. 282, 19399–19409 [DOI] [PubMed] [Google Scholar]
- 55. Xu M., Shorts-Cary L., Knox A. J., Kleinsmidt-DeMasters B., Lillehei K., Wierman M. E. (2009) Endocrinology 150, 2064–2071 [DOI] [PubMed] [Google Scholar]
- 56. Yao J., Weremowicz S., Feng B., Gentleman R. C., Marks J. R., Gelman R., Brennan C., Polyak K. (2006) Cancer Res. 66, 4065–4078 [DOI] [PubMed] [Google Scholar]
- 57. Liu P. S., Jong T. H., Maa M. C., Leu T. H. (2010) Oncogene 29, 3977–3989 [DOI] [PubMed] [Google Scholar]
- 58. Almeida E. A., Iliæ D., Han Q., Hauck C. R., Jin F., Kawakatsu H., Schlaepfer D. D., Damsky C. H. (2000) J. Cell Biol. 149, 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shintani Y., Wheelock M. J., Johnson K. R. (2006) Mol. Biol. Cell. 17, 2963–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Behrens A., Jochum W., Sibilia M., Wagner E. F. (2000) Oncogene 19, 2657–2663 [DOI] [PubMed] [Google Scholar]
- 61. Inobe M., Katsube K., Miyagoe Y., Nabeshima Y., Takeda S. (1999) Biochem. Biophys. Res. Commun. 266, 216–221 [DOI] [PubMed] [Google Scholar]
- 62. Welsch T., Endlich K., Giese T., Büchler M. W., Schmidt J. (2007) Cancer Lett. 255, 205–218 [DOI] [PubMed] [Google Scholar]
- 63. Griffith O. L., Melck A., Jones S. J., Wiseman S. M. (2006) J. Clin. Oncol. 24, 5043–5051 [DOI] [PubMed] [Google Scholar]
- 64. Tomshine J. C., Severson S. R., Wigle D. A., Sun Z., Beleford D. A., Shridhar V., Horazdovsky B. F. (2009) J. Biol. Chem. 284, 26331–26339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mouneimne G., Soon L., DesMarais V., Sidani M., Song X., Yip S. C., Ghosh M., Eddy R., Backer J. M., Condeelis J. (2004) J. Cell Biol. 166, 697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Disanza A., Mantoani S., Hertzog M., Gerboth S., Frittoli E., Steffen A., Berhoerster K., Kreienkamp H. J., Milanesi F., Di Fiore P. P., Ciliberto A., Stradal T. E., Scita G. (2006) Nat. Cell Biol. 8, 1337–1347 [DOI] [PubMed] [Google Scholar]
- 67. Lanzetti L., Palamidessi A., Areces L., Scita G., Di Fiore P. P. (2004) Nature 429, 309–314 [DOI] [PubMed] [Google Scholar]
- 68. Dreskin S. C., Thomas G. W., Dale S. N., Heasley L. E. (2001) J. Immunol. 166, 5646–5653 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.