Abstract
Snail, a zinc finger-containing transcriptional regulator, migrates into the nucleus where it controls gene expression. We demonstrated previously that importin β1 directly recognizes the zinc finger domain of Snail and transports it into the nucleus. Here, using in vitro and in vivo assays, we show that importin α, an adaptor protein for importin β1, negatively regulates the nuclear import of Snail mediated by importin β1. In vitro binding assays indicated that importin α interacted with the zinc finger domain of Snail to compete with the binding of importin β1 and that Snail did not form a ternary complex with importin α/importin β1. Overexpression of importin α in A549 cells reduced the endogenous Snail protein level, which was restored by inhibitors of the proteasome and glycogen synthase kinase 3β. Furthermore, knockdown of importin α by siRNA treatment increased the endogenous Snail protein level in several cancer cell lines. This study provides a novel regulatory mechanism of the nuclear protein import process by importin α and gives an implication to control Snail activity by inhibiting its nuclear localization.
Keywords: Protein Degradation, Protein Stability, Protein Targeting, Protein Translocation, Tumor, Snail, Importin, Nuclear Transport
Introduction
The epithelial mesenchymal transition (EMT)2 functions as a developmental switch that changes the morphology and behavior of epithelial cells to the highly motile mesenchymal phenotype. The hallmark of EMT is a decrease in E-cadherin expression (1). E-cadherin is a cell-cell adhesion molecule that forms stable epithelial adherent junctions and maintains the epithelial phenotype. Down-regulation of E-cadherin is mediated by several transcription factors such as Snail, Slug, ZEB, or Twist which repress E-cadherin transcription via interaction with the E-cadherin promoter (2). The loss of epithelial characteristics and the acquisition of migratory properties in EMT have recently been proposed to be the initial step of tumor metastasis. Indeed, the E-cadherin expression level is often inversely correlated to tumor grade (3). Although the loss of E-cadherin is one of the important markers of EMT, it is noteworthy that additional genes should be regulated to diminish the epithelial characteristics and to provoke the mesenchymal transition (4).
Snail is one of the prominent transcriptional regulators that contributes to EMT by suppressing E-cadherin expression (5, 6) and participates in EMT concerning the mesoderm and neural crest formation during embryonic development. In fact, Snail knockout mice represent early embryonic lethality (7). Emerging evidence indicates that Snail is also a key factor in the tumor progression and metastasis in melanoma, bladder, colorectal, and pancreatic carcinomas (5, 6, 8, 9). In these tumors, the down-regulation of E-cadherin was observed to correlate with the high expression of Snail. Signaling pathways such as TGFβ and Wnt regulate Snail gene expression at the transcriptional level (10).
On the other hand, it has been shown that Snail is a fragile protein and that phosphorylation of Snail is critical for its posttranslational regulation, which induces the degradation of the protein by the ubiquitin-proteasome system (11). In addition, the subcellular localization of Snail is regulated by serine phosphorylation, which induces its nuclear export by exportin1 (XPO1/CRM1) (12). Glycogen synthase kinase 3β and p21-activated kinase (PAK1) have been identified as kinases for Snail (11, 13). It has been shown that serine phosphorylation induces Snail localization in the cytoplasm, where it is rapidly degraded by the proteasome pathway (11, 14), whereas overexpression of small C-terminal domain phosphatase dephosphorylates Snail to enhance its activity (15). Furthermore, it has been reported that loss of the zinc transporter LIV1 causes the cytoplasmic localization of Snail in zebrafish (16). Taken together, the control of the nucleocytoplasmic localization of Snail is crucial for its activity.
The nuclear envelope restricts nuclear access of cellular molecules. Although the nuclear pore complex present in the nuclear envelope allows small molecules (40–60 kDa) to diffuse passively, an active transport system is required for large macromolecules to cross. Specific transport factors called importins and exportins facilitate translocation of macromolecules through the nuclear pore complex. Importin β family members play a central role in nucleocytoplasmic protein transport and recognize a broad class of nuclear localization signals (NLS) directly or indirectly to transport macromolecules into the nucleus through the nuclear pore complex via transient interactions with phenylalanine-glycine (FG) repeat-containing nucleoporins (17, 18). For classical basic-type NLS-containing proteins, importin β1 uses importin α family members as NLS receptors and forms a ternary complex, cargo/importin α/importin β1, to translocate through the nuclear pore complex into the nucleus. After the translocation, the import complex is dissociated in the nucleus by the binding of GTP-bound form of Ran (RanGTP) to importin β family members (19). Then, importins are exported from the nucleus to the cytoplasm for the next round of transport.
We have demonstrated previously that importin β1 mediates the nuclear import of Snail through direct binding with its zinc finger domains (20). On the other hand, it has recently been shown that other importin β family members, importin 7 and transportin, are also involved in the nuclear transport of Snail (21). Although it has been reported that the nuclear export of Snail is controlled in a phosphorylation-dependent manner, the regulation of its nuclear import remains to be elucidated.
In this study, we show that importin α, an adaptor protein between karyophilic proteins and importin β1, functions as an inhibitor of the nuclear import of Snail, indicating a novel regulatory mechanism of nuclear protein transport by importin α. Furthermore, we demonstrated that the suppression of Snail nuclear accumulation by importin α leads to a decrease in its cellular protein level through degradation by the proteasome system. This has implications for the prevention of tumor cell invasion by inhibiting Snail nuclear localization.
EXPERIMENTAL PROCEDURES
Cell Culture
Cells were maintained in Dulbecco's modified Eagle's medium (Sigma) (for HeLa, A172, A549, and A2058), L-15 medium (Gibco) (for MDA-MB 231, MDA-MB 435, SW480, and SW620) or RPMI 1640 (Gibco) (for DLD1) supplemented with 10% fetal bovine serum. For microinjection, HeLa cells were seeded on the day before injection at 1.5 × 105 cells on a coverslip in 35-mm dishes. For the in vitro transport assay, 1 × 106 HeLa cells were grown on eight-well slide glass (ICN) in 100-mm dishes for 24–48 h before the assay.
Antibodies
Antibodies used in this study were anti-Snail (Cell Signaling Technology), anti-GST (Santa Cruz Biotechnology), anti-GAPDH (Zymed Laboratories, Inc.), anti-GFP (Invitrogen), anti-importin α1 (Santa Cruz Biotechnology), anti-importin α5 (Abnova), anti-E-cadherin, anti-importin β1, and anti-Ran (BD Biosciences).
Plasmids and Recombinant Proteins
cDNA of full-length human Snail was inserted into pGEX-GFP (22) to produce the GST-Snail-GFP fusion protein in bacteria. The Escherichia coli BL21 (DE3) strain containing the Snail expression vector was cultured at 18 °C for 16–20 h in the presence of 0.1 mm IPTG, and GST fusion proteins were purified by glutathione-Sepharose (GE Healthcare) according to the manufacturer's instructions. GST-Snail-GFP was applied to a MonoQ column and eluted with a 0.1–0.6 m NaCl gradient using the AKTA system (GE Healthcare). HA-importin β1, FLAG-importin αs, Ran, NTF2, and GST-SV40 NLS-GFP were prepared as described previously (22).
Microinjection and in Vitro Transport Assay
Purified GST-Snail-GFP proteins (3 mg/ml) was microinjected into HeLa cytoplasm with FLAG-importin α5 (4 mg/ml) or BSA (4 mg/ml). Alexa Fluor 568-labeled IgG (Molecular Probes) was coinjected as an injection marker. After 30 min at 37 °C, the cells were fixed with 3.7% formaldehyde. In vitro transport assays using digitonin-permeabilized HeLa cells were performed as described previously (22).
Transfection, Western Blotting, and RT-PCR
pEGFP or pEGFP-importin α5 was transfected into A549 cells using Effecten (Qiagen). After 24 h, the cells were further treated with or without MG132 (10 μm) and LiCl (20 mm) for 6 h. They were then fixed by 3.7% formaldehyde, permeabilized with 0.5% Triton X-100, and blocked with 5% skim milk in PBS. The subcellular localization of endogenous Snail was detected with anti-Snail antibody, followed by Alexa Fluor 568-labeled goat anti-mouse IgG (Molecular Probes).
For Western blotting, transfected cells were concentrated by G418 treatment (900 μg/ml) for 5 days and then lysed in SDS sample buffer. MG132 (10 μm) and LiCl (20 mm) were added to the medium for 6 h before protein extraction. Whole cell extracts were subjected to SDS-PAGE and transferred to PVDF membranes. The membranes were incubated with antibodies against Snail, GAPDH, GFP, E-cadherin, importin β1, or Ran, followed by HRP-conjugated secondary antibody. Signals were detected by enhanced chemiluminescence (GE Healthcare).
For RT-PCR, total RNA was extracted from transfected cells by TRIzol reagent (Invitrogen). Snail (35 cycles) and GAPDH (30 cycles) were amplified using specific primers: Snail sense, 5′-gagctgcaggactctaatccagag-3′; and antisense 5′-agcctggagatccttggcctcag-3′. GAPDH sense, 5′-atggggaaggtgaaggtcggagtc-3′; and antisense 5′-tggaggccatgtgggccatgaggtc-3′.
siRNA Treatment
siRNA duplexes were reverse-transfected to HeLa or SW480 cells using Lipofectamine RNAiMAX (Invitrogen) for 72 h. siRNAs were synthesized by NIPPON GENE: si-control sense, 5′-ggcuacguccaggagcgcatt-3′; and antisense, 5′-ugcgcuccuggacguagcctt-3′. si-imp α1 sense, 5′-gcagauucuuccuaccuuatt-3′; and antisense 5′-uaagguaggaagaaucugctt-3′. si-imp α5 sense, 5′-ggaagcugcuuucaaaagatt-3′; and antisense 5′-ucuuuugaaagcagcuucctt-3′.
Binding Assays
Proteins were incubated with glutathione-Sepharose or anti-FLAG-agarose (Sigma) in transport buffer (22) containing 0.1% Triton X-100 at 4 °C for 2 h. After extensive washing, SDS sample buffer was added to the beads, and the samples were subjected to SDS-PAGE. Bound proteins were detected by Coomassie Brilliant Blue staining or Western blotting using the indicated antibodies.
RESULTS
Importin α Inhibits Nuclear Import of Snail in Vitro and in Vivo
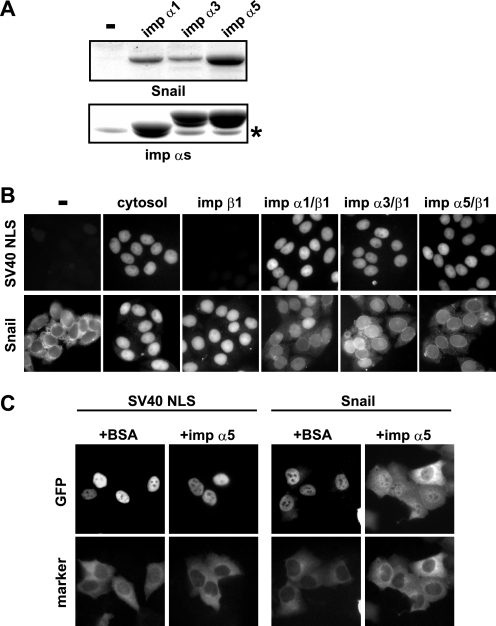
It has been shown that importin β1 directly recognizes and transports Snail into the nucleus (20). Because importin β1-dependent nuclear transport usually uses importin α as an adaptor, we analyzed whether importin α is also involved in the nuclear import of Snail. We found that three importin α family members, importin α1/Rch1, importin α3/Qip1, and importin α5/NPI-1, directly bound to Snail in vitro (Fig. 1A), suggesting the possibility that, in addition to the importin β1-mediated pathway, the classical importin α/β1 transport machinery also plays some role in Snail nuclear import. To test this possibility, we performed a digitonin-permeabilized cell-free transport assay using recombinant proteins. Unexpectedly, the presence of an equal molar amount of importin α suppressed the nuclear accumulation of Snail (Fig. 1B). Furthermore, microinjected Snail protein was retained in the cytoplasm when importin α was coinjected (Fig. 1C), although nuclear import of GST-SV40 NLS-GFP, a well known classical NLS-containing control substrate, was not affected by the coinjection of importin α (Fig. 1C). These results indicate that importin α can act as an inhibitor of the nuclear import of Snail.
FIGURE 1.
Importin α inhibits nuclear import of Snail in vitro and in vivo. A, three importin αs showed direct binding activity on the Snail protein. GST-Snail-GFP was incubated with FLAG-importin α in the presence of anti-FLAG-agarose. The asterisk indicates the position of the heavy chain of the anti-FLAG antibody. B, in vitro nuclear transport assay. Purified cargo protein, importin β1, Ran, and NTF2 with or without equivalent molar amounts of importin αs were applied to digitonin-permeabilized HeLa cells and incubated at 30 °C for 30 min. C, importin α inhibited the nuclear import of microinjected Snail. GFP fusion proteins were microinjected into HeLa cytoplasm with BSA or importin α5 and incubated at 37 °C for 30 min.
Importin α Competes with the Binding of Importin β1 to Snail
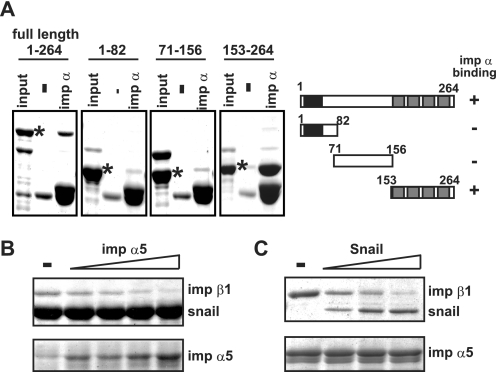
To clarify the inhibitory effect of importin α on Snail nuclear import, first, the importin α-binding region of Snail was determined using full-length and three truncated mutants of Snail. As shown in Fig. 2A, the Snail C-terminal fragment (153–264), which contains four zinc finger domains, was directly recognized by importin α5. Previous studies showed that the zinc finger domains of Snail act as an NLS and that several importin β family members directly recognize this same domain to transport it into the nucleus (20, 21). It is therefore likely that importin α competes with the binding of importin β1 to Snail. Indeed, the addition of importin α5 disrupted the Snail/importin β1 complex in a dose-dependent manner (Fig. 2B).
FIGURE 2.
Importin α competes with the binding of importin β1 to Snail. A, determining the importin α-binding region of Snail. Full-length and three truncated mutant Snail proteins were incubated with FLAG-importin α5 in the presence of anti-FLAG-agarose. The asterisk indicates the position of recombinant Snail protein. B, competition assay. GST-Snail-GFP prebound to glutathione-Sepharose was incubated with importin β1 in the presence of increasing amounts of importin α5 (0.5, 1, 2, and 5 molar excess). C, Snail does not form a ternary complex. The importin α/β1 complex prebound to anti-FLAG-agarose was incubated with increasing amounts of GST-Snail-GFP (1, 3, and 9 molar excess).
It has been shown that for some karyophilic proteins a ternary complex of importin α/β1/cargo is non-functional for nuclear import (23, 24). In addition, it is known that importin α dimerized with importin β1 increases the binding affinity to cargo protein (25, 26). Thus, we tested whether the importin α/β1 heterodimer forms a complex with Snail to suppress its nuclear import. When the preformed importin α5-β1 complex was incubated with increasing amounts of Snail, the complex dissociated and the importin α5-Snail complex was formed (Fig. 2C). This indicates that importin α disrupts the Snail-importin β1 transport complex and does not form a ternary complex comprising importin α-β1-Snail.
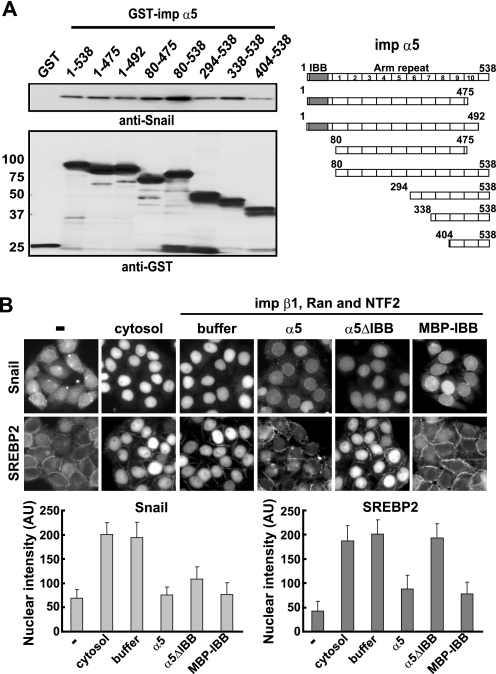
The importin α family consists of two functional domains, a short N-terminal importin β binding (IBB) domain and a large NLS binding domain comprising 10 Armadillo (Arm) repeats. Because the Snail binding region in importin α would overlap with the importin β binding site, the Snail binding region of importin α was determined using various deletion mutants of importin α5. Interestingly, Snail bound to a broad region of importin α5, including the IBB domain (Fig. 3A). To address the effects of the IBB domain of importin α on Snail nuclear localization, in vitro transport assays using importin α mutants were performed. SREBP2 is a well characterized substrate for the importin β1-dependent nuclear import pathway (27, 28). Although importin α does not associate directly with SREBP2, importin α disrupts the SREBP2-importin β1 complex by binding to importin β1 through the IBB domain of importin α (27). As shown in Fig. 3B, an importin α5 ΔIBB mutant suppressed the nuclear import of Snail, whereas it failed to inhibit SREBP2 nuclear accumulation. On the other hand, maltose binding protein (MBP)-IBB significantly inhibited the nuclear accumulation of both Snail and SREBP2 (Fig. 3B), consistent with the result that Snail bound to broad regions of importin α5 (Fig. 3A). However, the Arm repeat region showed the most efficient binding activity (Fig. 3A). Thus, the Snail nuclear import is inhibited in part by the IBB domain but mainly by Arm repeats in importin α, indicating that the inhibitory effect of importin α on Snail nuclear import is distinct from that on SREBP2 import.
FIGURE 3.
Arm repeat of importin α contributes to inhibition of Snail nuclear import. A, the Snail-binding region of importin α5. Purified full-length and truncated GST-importin α5 were incubated with Snail-GFP at 4 °C for 2h in the presence of glutathione-Sepharose. After extensive washing, bound proteins were detected by anti-Snail and anti-GST antibodies. B, the IBB domain of importin α5 is dispensable in the inhibition of Snail nuclear import. In vitro transport assays were performed using GST-Snail-GFP or GST-GFP-SREBP2 as a substrate with the indicated recombinant proteins at 30 °C for 30 min. The nuclear intensity of Snail or SREBP2 was quantitated and represented as mean ± S.D. across three experiments. AU, arbitrary unit.
Overexpression of Importin α Affects Snail Protein Levels
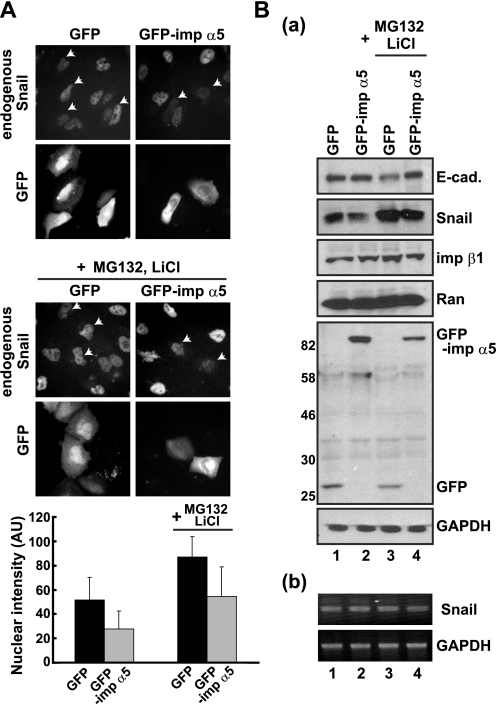
To confirm that importin α actually inhibits the nuclear import of endogenous Snail in living cells, we overexpressed importin α5 and detected the subcellular localization of endogenous Snail. When A549 cells were transfected with GFP-importin α5, the fluorescence intensity of Snail was greatly decreased compared with that of GFP-expressing control cells (Fig. 4A). It has been reported that Snail is a fragile protein that is rapidly degraded by the proteasome system (11). In A549 cells, we found that the half-life of endogenous Snail was ∼50 min and that treatment with MG132 and LiCl, a proteasome inhibitor and a GSK3β inhibitor, respectively, effectively stabilized the Snail protein (supplemental Fig. S1). These results raised the idea that inhibition of Snail nuclear import by importin α enhances its degradation in the cytoplasm by the proteasome.
FIGURE 4.
Importin α overexpression reduces endogenous Snail protein. A, the effects of importin α5 expression on Snail subcellular localization. GFP or GFP-importin α5 was transfected into A549 cells for 24 h, and then MG132 (10 μm) and LiCl (20 mm) were added for a further 6 h. The subcellular localization of endogenous Snail protein was detected by indirect immunofluorescence using anti-Snail antibody. The arrows indicate transfected cells. The nuclear intensity of endogenous Snail was quantitated and represented as mean ± S.D. across three experiments. AU, arbitrary unit. B, transfected cells were selected by G418 for 4 days, and the whole cell extracts were analyzed by Western blotting using the indicated antibodies (a). RT-PCR (b). Cells were transfected and treated with or without MG132 and LiCl as described in Fig. 3B, a. Total RNAs were extracted and subjected to RT-PCR using Snail- or GAPDH-specific primers.
To test this, we treated transfected cells with MG132 and LiCl to prevent the degradation of Snail. We found that inhibition of proteasome function increased the nuclear fluorescence intensity of Snail (Fig. 4A). These results suggest that importin α can negatively regulate Snail nuclear import to facilitate its degradation in the cytoplasm, whereas a proportion of the Snail proteins escape from the importin α-mediated suppression of nuclear import and degradation by the proteasome to enter the nucleus.
Next, we concentrated the transfected cells and analyzed protein levels. Consistent with the results in Fig. 4A, although Snail protein levels were decreased in GFP-importin α5 expressing cells, MG132 and LiCl treatment restored them (Fig. 4B, a). Furthermore, we confirmed that the mRNA level of Snail was not affected under these conditions (Fig. 4B, b) and that the protein levels of importin β1 and Ran were not altered by GFP-importin α5 overexpression (B, a). From these findings, we conclude that the function of Snail is negatively regulated by importin α-mediated suppression of its nuclear import as well as by proteasome-dependent degradation in living cells.
Knockdown of Importin α Increased Snail Protein Levels
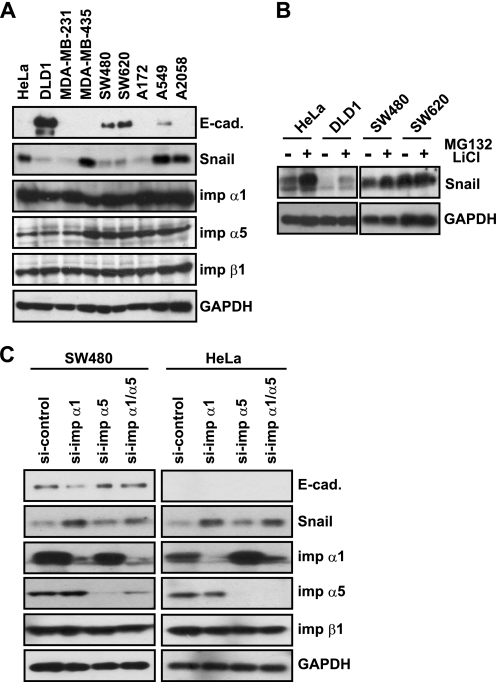
To address the biological significance of the negative regulation of Snail nuclear import by importin α, we analyzed the protein expression level in several human cancer cell lines. As shown in Fig. 5A, low Snail expression correlated with a high E-cadherin protein level in DLD1, SW480, and SW620 cells; however, no correlation between importin α and the Snail protein level was found. MG132 and LiCl treatment increased the Snail protein level in HeLa and SW480 cells (Fig. 5B), indicating that Snail stability was regulated by the proteasome system in these cells. It is likely that decreased importin α expression would stabilize the Snail protein. We introduced a siRNA duplex into these cells to knock down importin α1 or importin α5. Fig. 5C indicated that knockdown of importin α1 increased the Snail protein in HeLa and SW480 cells and decreased E-cadherin expression in SW480 cells. Knockdown of importin α5 showed a weaker effect than that of importin α1 on the Snail protein level (Fig. 5C). These results suggested that the protein level of importin α affects Snail protein stability, and, thus, importin α controls both Snail nuclear import and its stability in living cells.
FIGURE 5.
Knockdown of importin α increases endogenous Snail protein in cancer cells. A, Western blotting of human cancer cell lines. Whole cell extract of each cell lines were subjected to Western blotting using indicated antibodies. B, effects of MG132 and LiCl on endogenous Snail protein. Cells were treated with or without MG132 (10 μm) and LiCl (20 mm) for 6 h. Whole cell extracts were subjected to Western blotting using anti-Snail and anti-GAPDH antibodies. C, knockdown of importin αs by siRNA. Control siRNA, siRNA for importin α1, or for importin α5 was transfected to HeLa or SW480 cells for 72 h. The expressions of E-cadherin, Snail, importin α1, importin α5, importin β1, and GAPDH were detected by Western blotting.
DISCUSSION
Proper subcellular localization of proteins is essential for their function. Snail is a transcriptional regulator and should be transported into the nucleus to function. In this study, we demonstrated the inhibitory effects of importin α on Snail nuclear import in vitro and in vivo. Our findings have two major implications for the regulatory mechanisms of nuclear protein transport and for Snail activity in EMT and tumor metastasis.
Importin α is an adaptor for importin β1 in the protein nuclear import pathway. In addition to its adaptor function, it directly recognizes and transports CAMKIV (29) and HIV1 Vpr (30) into the nucleus without using importin β1. In contrast, it has been shown that importin α sometimes inhibits the nuclear import of proteins. For example, PTHrP and TRF1 form a complex with importin α/β1, but this ternary complex is not active and might bind to cytoplasmic components, leading to their cytoplasmic retention (23, 24). The formation of similar non-active complexes containing importin α was reported in importin 7/importin β1-mediated histone H1 nuclear transport (31). Furthermore, although the effects of importin α on nuclear import are not clear, the zinc finger-(like) proteins EKLF (32) and DMRT1 (33) were recognized by both importin α and importin β. Thus, the inhibition of Snail nuclear import may be one of the characteristic features of importin α. This implies that importin α-mediated control of nuclear protein import might be more common than expected, although its biological significance is still unclear in most cases.
Importin α/β-mediated nuclear import is thought to be widely used in cells. For most cargo proteins containing the classical NLS, the Arm repeat region of importin α recognizes their NLS, and the IBB domain of importin α associates with the C-terminal half of importin β1 to generate a ternary complex. Why does Snail-bound importin α fail to associate with importin β1 to form a ternary complex? Using importin α5 deletion mutants, we found that broad regions of importin α5 associated with Snail but that the Arm repeat was predominant in the recognition of Snail (Fig. 3A), consistent with a previous report showing that the zinc finger-type transcription factor, SP1, has a similar binding property (34). Furthermore, the N-terminal half of importin β1, which contains the Ran binding domain, was mapped as the Snail-binding region (supplemental Fig. S2). Mingot et al. (21) identified that the basic residues are important for Snail nuclear localization and that these residues are distributed among the three zinc finger domains rather than in a single cluster. The characteristic binding of Snail to importin α and importin β1 may be due to such non-consensus NLS of Snail, and the ability of importin α to bind to Snail across a broad region may interfere spatially with ternary complex formation. Crystallographic studies of the Snail/importin complexes would provide answers as to why importin α disrupts the Snail-importin β1 complex and why Snail does not form a ternary complex with importin α/β1.
A recent study revealed that, in addition to importin β1, additional import factors, such as importin 7 and transportin, contribute to the nuclear import of Snail and that both import factors recognized the zinc finger domain of Snail (21), suggesting that importin α may inhibit their transport pathway. However, this study also showed that the importin α/β1 system worked on the nuclear accumulation of Snail (21). This discrepancy with our data may arise because of the different constructs used: GST-Snail-GFP in our study and GST-GFP-Snail in that of Mingot et al. (21). To test this, we prepared GST-GFP-Snail and used it in our experimental conditions. We found that GST-GFP-Snail also associated directly with the importin αs or β1 in vitro and that, similar to GST-Snail-GFP, its nuclear import was inhibited by the presence of importin α (supplemental Fig. S3). Further studies will be required to resolve this difference.
The EMT-related genes are thought to be critical for tumor metastasis. Recent studies have indicated that high levels of Snail expression are closely correlated with low E-cadherin expression in several tumors (5, 6, 8). Consistent with these studies, knockdown of Snail affects tumor growth and invasiveness (35). However, E-cadherin and Snail mRNA have been detected in several types of tumors (12, 36). It is likely that highly expressed importin α would suppress nuclear import of Snail to facilitate its degradation, resulting in the down-regulation of Snail function and high expression of E-cadherin without a decrease in Snail mRNA levels. Consistently, Snail mRNA levels were not affected in importin α-overexpressing cells (Fig. 4B, b).
Furthermore, the expression level of importin α1 affected the Snail protein level in living cells (Fig. 5C). Although the sensitivity to importin α might vary with tumor cells, some cells would be affected by importin α, resulting in low Snail protein and high E-cadherin expression. Thus, it is possible that importin α contributes to a poor metastatic potential of tumors that express high levels of Snail mRNA. Collectively, elucidating the precise mechanism of inhibition of Snail nuclear import by importin α will suggest novel approaches for the suppression of tumor metastasis.
Supplementary Material
Acknowledgment
We thank Dr. Y. Yasuda for providing the GFP-importin α5 expression vector.
This work was supported in part by a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and JST, CREST.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- EMT
- epithelial mesenchymal transition
- NLS
- nuclear localization signal
- IBB
- importin β binding
- Arm
- Armadillo
- SREBP2
- sterol regulatory element-binding protein 2.
REFERENCES
- 1. Thiery J. P. (2002) Nat. Rev. Cancer 2, 442–454 [DOI] [PubMed] [Google Scholar]
- 2. Peinado H., Olmeda D., Cano A. (2007) Nat. Rev. Cancer 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 3. Cowin P., Rowlands T. M., Hatsell S. J. (2005) Curr. Opin. Cell Biol. 17, 499–508 [DOI] [PubMed] [Google Scholar]
- 4. Nieto M. A. (2009) Int. J. Dev. Biol. 53, 1541–1547 [DOI] [PubMed] [Google Scholar]
- 5. Cano A., Pérez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F., Nieto M. A. (2000) Nat. Cell Biol. 2, 76–83 [DOI] [PubMed] [Google Scholar]
- 6. Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., García de Herreros A. (2000) Nat. Cell Biol. 2, 84–89 [DOI] [PubMed] [Google Scholar]
- 7. Carver E. A., Jiang R., Lan Y., Oram K. F., Gridley T. (2001) Mol. Cell. Biol. 21, 8184–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poser I., Domínguez D., de Herreros A. G., Varnai A., Buettner R., Bosserhoff A. K. (2001) J. Biol. Chem. 276, 24661–24666 [DOI] [PubMed] [Google Scholar]
- 9. De Craene B., Gilbert B., Stove C., Bruyneel E., van Roy F., Berx G. (2005) Cancer Res. 65, 6237–6244 [DOI] [PubMed] [Google Scholar]
- 10. Barrallo-Gimeno A., Nieto M. A. (2005) Development 132, 3151–3161 [DOI] [PubMed] [Google Scholar]
- 11. Zhou B. P., Deng J., Xia W., Xu J., Li Y. M., Gunduz M., Hung M. C. (2004) Nat. Cell. Biol. 6, 931–940 [DOI] [PubMed] [Google Scholar]
- 12. Domínguez D., Montserrat-Sentís B., Virgós-Soler A., Guaita S., Grueso J., Porta M., Puig I., Baulida J., Francí C., García de Herreros A. (2003) Mol. Cell. Biol. 23, 5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang Z., Rayala S., Nguyen D., Vadlamudi R. K., Chen S., Kumar R. (2005) Cancer Res. 65, 3179–3184 [DOI] [PubMed] [Google Scholar]
- 14. Ko H., Kim H. S., Kim N. H., Lee S. H., Kim K. H., Hong S. H., Yook J. I. (2007) Cells Tissues Organs 185, 66–72 [DOI] [PubMed] [Google Scholar]
- 15. Wu Y., Evers B. M., Zhou B. P. (2009) J. Biol. Chem. 284, 640–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamashita S., Miyagi C., Fukada T., Kagara N., Che Y. S., Hirano T. (2004) Nature 429, 298–302 [DOI] [PubMed] [Google Scholar]
- 17. Stewart M. (2007) Nat. Rev. Mol. Cell Biol. 8, 195–208 [DOI] [PubMed] [Google Scholar]
- 18. Forwood J. K., Lange A., Zachariae U., Marfori M., Preast C., Grubmüller H., Stewart M., Corbett A. H., Kobe B. (2010) Structure 18, 1171–1183 [DOI] [PubMed] [Google Scholar]
- 19. Lee S. J., Matsuura Y., Liu S. M., Stewart M. (2005) Nature 435, 693–696 [DOI] [PubMed] [Google Scholar]
- 20. Yamasaki H., Sekimoto T., Ohkubo T., Douchi T., Nagata Y., Ozawa M., Yoneda Y. (2005) Genes Cells 10, 455–464 [DOI] [PubMed] [Google Scholar]
- 21. Mingot J. M., Vega S., Maestro B., Sanz J. M., Nieto M. A. (2009) J. Cell Sci. 122, 1452–1460 [DOI] [PubMed] [Google Scholar]
- 22. Sekimoto T., Fukumoto M., Yoneda Y. (2004) EMBO J. 23, 1934–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lam M. H., Briggs L. J., Hu W., Martin T. J., Gillespie M. T., Jans D. A. (1999) J. Biol. Chem. 274, 7391–7398 [DOI] [PubMed] [Google Scholar]
- 24. Forwood J. K., Jans D. A. (2002) Biochemistry 41, 9333–9340 [DOI] [PubMed] [Google Scholar]
- 25. Catimel B., Teh T., Fontes M. R., Jennings I. G., Jans D. A., Howlett G. J., Nice E. C., Kobe B. (2001) J. Biol. Chem. 276, 34189–34198 [DOI] [PubMed] [Google Scholar]
- 26. Harreman M. T., Hodel M. R., Fanara P., Hodel A. E., Corbett A. H. (2003) J. Biol. Chem. 278, 5854–5863 [DOI] [PubMed] [Google Scholar]
- 27. Nagoshi E., Imamoto N., Sato R., Yoneda Y. (1999) Mol. Biol. Cell 10, 2221–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee S. J., Sekimoto T., Yamashita E., Nagoshi E., Nakagawa A., Imamoto N., Yoshimura M., Sakai H., Chong K. T., Tsukihara T., Yoneda Y. (2003) Science 302, 1571–1575 [DOI] [PubMed] [Google Scholar]
- 29. Kotera I., Sekimoto T., Miyamoto Y., Saiwaki T., Nagoshi E., Sakagami H., Kondo H., Yoneda Y. (2005) EMBO J. 24, 942–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nitahara-Kasahara Y., Kamata M., Yamamoto T., Zhang X., Miyamoto Y., Muneta K., Iijima S., Yoneda Y., Tsunetsugu-Yokota Y., Aida Y. (2007) J. Virol. 81, 5284–5293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jäkel S., Albig W., Kutay U., Bischoff F. R., Schwamborn K., Doenecke D., Görlich D. (1999) EMBO J. 18, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Quadrini K. J., Bieker J. J. (2002) J. Biol. Chem. 277, 32243–32252 [DOI] [PubMed] [Google Scholar]
- 33. Ying M., Chen B., Tian Y., Hou Y., Li Q., Shang X., Sun J., Cheng H., Zhou R. (2007) Biochim. Biophys. Acta 1773, 804–813 [DOI] [PubMed] [Google Scholar]
- 34. Ito T., Kitamura H., Uwatoko C., Azumano M., Itoh K., Kuwahara J. (2010) Biochem. Biophys. Res. Commun. 403, 161–166 [DOI] [PubMed] [Google Scholar]
- 35. Olmeda D., Jordá M., Peinado H., Fabra A., Cano A. (2007) Oncogene 26, 1862–1874 [DOI] [PubMed] [Google Scholar]
- 36. Hajra K. M., Chen D. Y., Fearon E. R. (2002) Cancer Res. 62, 1613–1618 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.