Abstract
Expression of the Arabidopsis CGS1 gene, encoding the first committed enzyme of methionine biosynthesis, is feedback-regulated in response to S-adenosyl-l-methionine (AdoMet) at the mRNA level. This regulation is first preceded by temporal arrest of CGS1 translation elongation at the Ser-94 codon. AdoMet is specifically required for this translation arrest, although the mechanism by which AdoMet acts with the CGS1 nascent peptide remained elusive. We report here that the nascent peptide of CGS1 is induced to form a compact conformation within the exit tunnel of the arrested ribosome in an AdoMet-dependent manner. Cysteine residues introduced into CGS1 nascent peptide showed reduced ability to react with polyethyleneglycol maleimide in the presence of AdoMet, consistent with a shift into the ribosomal exit tunnel. Methylation protection and UV cross-link assays of 28 S rRNA revealed that induced compaction of nascent peptide is associated with specific changes in methylation protection and UV cross-link patterns in the exit tunnel wall. A 14-residue stretch of amino acid sequence, termed the MTO1 region, has been shown to act in cis for CGS1 translation arrest and mRNA degradation. This regulation is lost in the presence of mto1 mutations, which cause single amino acid alterations within MTO1. In this study, both the induced peptide compaction and exit tunnel change were found to be disrupted by mto1 mutations. These results suggest that the MTO1 region participates in the AdoMet-induced arrest of CGS1 translation by mediating changes of the nascent peptide and the exit tunnel wall.
Keywords: Gene Regulation, Peptide Conformation, Plant, Ribosome Function, Translation Regulation, Nascent Peptide, Translation Arrest
Introduction
The CGS1 gene encodes cystathionine γ-synthase (EC 2.5.1.48) (1), which catalyzes the first committed step of methionine biosynthesis in higher plants (2). Expression of Arabidopsis thaliana CGS1 (gene ID At3g01120) is negatively feedback-regulated by mRNA degradation in response to S-adenosyl-l-methionine (AdoMet),4 a direct metabolite of methionine (3, 4). mto1 mutants, which overaccumulate soluble methionine (5), are deficient in this post-transcriptional regulation and overaccumulate CGS1 mRNA (3). mto1 mutants bear single amino acid sequence alterations within a short stretch of amino acid sequence, termed the MTO1 region, 77RRNCSNIGVAQIVA90, encoded within the first exon of CGS1. The amino acid sequence of the MTO1 region is highly conserved among multicellular plants (3, 6). Stable and transient expression experiments using CGS1 exon 1-reporter fusions demonstrated that the exon 1 coding region is necessary and sufficient for the feedback regulation (3, 7). These experiments also revealed that the MTO1 amino acid sequence is involved in this regulation by acting in cis (3, 6, 7).
Post-transcriptional regulation of CGS1 occurs during translation (8) and is reproducible in a cell-free translation system of wheat germ extract (WGE) (4). Studies using WGE revealed that temporal translation elongation arrest occurs prior to mRNA degradation at the Ser-94 codon located immediately downstream of the MTO1 region. The ribosome is stalled at the translocation step, and peptidyl-tRNASer occupies the A-site of the stalled ribosome (9).
During translation of proteins, new peptide bonds are formed in the ribosomal large subunit at the peptidyltransferase (PTase) center, and the nascent peptide passes through the ribosomal exit tunnel that penetrates the large subunit. The ribosomal exit tunnel is ∼100 Å in length, whereas the diameter is 10–20 Å in both prokaryotes and eukaryotes (10–15). In several prokaryotic systems, including Escherichia coli tnaC gene arrest in response to elevated tryptophan levels (16, 17), arrest in Bacillus subtilis ermC leader translation for erythromycin resistance (18), and E. coli secM translation arrest for regulation of the secretory pathway (19), the arrest of translation involves interaction between nascent peptide and ribosomal components, including the PTase center and exit tunnel (18, 20–28). In addition, formation of a compact structure of the SecM nascent peptide is crucial for its translation arrest (29). In eukaryotes, cryo-electron microscopy studies of the upstream open reading frame of human cytomegalovirus UL4 gene (30) and that of fungal CPA1 gene, termed the arginine attenuator peptide (AAP) (31), showed specific contacts between nascent peptide and the exit tunnel wall of the stalled ribosome (32). These studies led to the concept that the exit tunnel is not only a simple path for nascent polypeptides but that interactions between nascent peptides and the tunnel wall can also regulate translational activity of ribosome itself.
In the CGS1 system of A. thaliana, translation elongation is arrested immediately downstream of a 14-amino acid MTO1 region. As the ribosomal exit tunnel normally holds 30–40 amino acids of nascent peptide during translation (33, 34), this implies that the MTO1 amino acid sequence resides inside the ribosomal exit tunnel when translation arrest occurs at Ser-94. The effect of AdoMet is temporal and readily reversible (9). Among the possibilities that could explain this effect, we hypothesized that a change of the nascent peptide conformation itself and/or that of the ribosomal exit tunnel triggers the translation arrest.
It is possible to gain insight on the topological exposure of specific amino acids in a translated protein by using the pegylation assay (35). The large molecule of polyethyleneglycol maleimide (PEG-MAL) efficiently reacts with exposed or accessible cysteine residues, leading to an increase in peptide mass that can be monitored by SDS-PAGE. This assay can also be applied to nascent peptides in the ribosomal exit tunnel. PEG-MAL efficiently reacts with cysteine residues located outside the translating ribosome, whereas this efficiency is reduced if the target cysteine is located inside the ribosomal exit tunnel and remains relatively inaccessible. A change in pegylation efficiency may therefore be interpreted as reflecting conformational changes of the nascent peptide in the exit tunnel (36–38).
In this study, we have applied the pegylation assay to test the hypothesis that AdoMet induces a change of CGS1 nascent peptide conformation inside the ribosomal exit tunnel during translation arrest. We show here that CGS1 nascent peptide arrested at Ser-94, which includes the MTO1 sequence, forms a compact conformation in response to AdoMet. Further analyses using methylation protection and UV cross-link of the 28 S rRNA showed that this compaction is associated with specific changes in methylation protection and UV cross-link patterns in the ribosomal exit tunnel.
EXPERIMENTAL PROCEDURES
Chemicals
AdoMet (iodide salt, A4377) and PEG-MAL were purchased from Sigma. Dimethyl sulfate (DMS) and 1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate were purchased from Wako Pure Chemicals (Osaka, Japan), and kethoxal was from MP Biomedicals (Irvine, CA). Other chemicals were obtained as described previously (9).
Plasmids
Plasmid pYK00 carries the M8:Ex1(WT) DNA in the pSP64 poly(A) vector (Promega, Madison, WI). To construct pYK00, the Ex1(WT) coding region was amplified by PCR from plasmid pMN1(WT), which carries GST:Ex1(WT) DNA (9), using primers Koz-Met8f and TP2 (supplemental Table S1), and the amplified fragment was digested with SalI and BamHI and inserted between the SalI and BamHI sites of the pSP64 poly(A) vector.
Plasmids pNO1 and pNO2 carry the M8:ND5(WT) and M8:ND5(mto1-1) DNA in the pSP64 poly(A) vector, respectively. To generate these plasmids, the XbaI-BamHI fragment containing Ex1(WT) coding region of pYK00 was replaced with the XbaI-BamHI fragment of ND5(WT) or ND5(mto1-1), respectively (6).
Amino acid substitution mutagenesis and introduction of mto1 mutation alleles for plasmids containing M8:ND5 DNA were accomplished by the overlap extension PCR method (39, 40) using the primers shown in supplemental Table S1. Mutated ND5 DNA fragments were recloned into pYK00.
Plasmids pNO35 and pNO36 carry three consecutive Strep-tag II sequences (IBA, Göttingen, Germany) fused in-frame at the N terminus of the CGS1 exon 1 coding region in the pSP64 poly(A) vector and harbor wild-type and mto1-1 mutant MTO1 sequences, respectively. To construct these plasmids, a double-stranded DNA fragment containing two Strep-tag II and an SpeI site was first generated by annealing the 5′-phosphorylated oligonucleotides strep-A′f and strep-A′r (supplemental Table S1). This fragment was then inserted between the SalI and XbaI sites of pSP64 poly(A) vectors that contained either CGS1 Ex1(WT) or CGS1 Ex1(mto1-1) between the XbaI and SacI sites. The resulting plasmids were subsequently digested with SpeI and XbaI, and an additional Strep-tag II was inserted using a DNA fragment generated by annealing the 5′-phosphorylated oligonucleotides strep-B′f and strep-B′r (supplemental Table S1). In all constructs, the integrity of PCR-amplified regions was confirmed by sequence analysis.
In Vitro Transcription
DNA templates in the pSP64 poly(A) vector were linearized with EcoRI and purified as described previously (4). Templates for nonstop RNAs were prepared by amplifying corresponding regions from pNO1 through pNO36 by PCR using forward primer SP65′fP and reverse primer S94r (supplemental Table S1) with PrimeSTAR® DNA polymerase (Takara Bio, Ohtsu, Japan). The amplified fragments were purified as described previously (9). In vitro transcription in the presence of a cap analog m7G[5′]ppp[5′]GTP (Epicenter Technologies, Madison, WI) was carried out as described previously (4).
Pegylation Assay
WGE used for in vitro translation was prepared as described previously (41). M8:ND5 RNA or nonstop RNA (5 pmol) was translated in a 50-μl WGE reaction mixture (20 μl of WGE, 36 mm Hepes-KOH (pH 7.6), 10 mm creatine phosphate, 50 μg ml−1 creatine kinase, 5 mm DTT, 2.1 mm Mg(OAc)2, 53 mm KOAc, 0.4 mm spermidine, 1.2 mm ATP, 0.1 mm GTP, 0.08 mm amino acids mixture minus methionine, 0.1 μm methionine, 0.3 μm [35S]methionine (37 TBq mmol−1, American Radiolabeled Chemicals, St. Louis), 0.8 units μl−1 RNase inhibitor (Promega)) with or without AdoMet for 30 min at 25 °C. For pegylation assays, an input mRNA amount of 5 pmol was used to increase the population of translating mRNA carrying a single ribosome (42).
Pegylation assays were carried out as described previously (37) with minor modifications. In short, 50 μl of completed in vitro translation reaction was loaded on a 50-μl sucrose cushion (37) and centrifuged in a TLA100.3 rotor (Beckman Coulter, Brea, CA) at 76,000 rpm for 20 min at 4 °C to obtain RNCs. The RNC mixture was resuspended in 25 μl of reaction buffer (37), and the pegylation reaction was started by adding 25 μl of 2 mm PEG-MAL in the same buffer. The reaction was carried out at 4 °C for 30 min. Preliminary experiments showed that the pegylation reaction reached an almost plateau level at 30 min (supplemental Fig. S1). AdoMet was included in the sucrose cushion and reaction buffer as appropriate. Reactions were terminated by adding DTT to a final concentration of 100 mm (36). For translation-only reactions, PEG-MAL was inactivated with DTT before adding to the reaction mixture. Samples were treated with 0.5 mg ml−1 RNase A for 15 min at 25 °C, heated for 10 min at 70 °C, separated on a NuPAGE 4–12% BisTris gel and Mes running buffer (Invitrogen), dried, and visualized using a FLA-7000 image analyzer (Fuji Photo Film, Tokyo, Japan), and band intensities were measured using MultiGauge software (Fuji Photo Film). Pegylation efficiency for translation products arrested at Ser-94 was calculated by dividing the band intensity of the pegylated product (23 kDa) by the sum of unpegylated (10 + 11 kDa) and pegylated products.
Chemical Modification and UV Cross-link Analyses
Methylation protection and UV cross-link assays were performed essentially as described previously (21, 43). Strep3:Ex1(S94-ns) nonstop RNA (80 pmol) was translated in WGE for 30 min in an 800-μl reaction mixture containing 1 mm AdoMet. Monosome RNCs were prepared as described previously (44) with slight modifications. In short, ice-cooled 800-μl reaction mixtures were centrifuged for 15 h at 23,600 rpm, 4 °C, in an SW40 Ti rotor (Beckman Coulter, Brea, CA) on a 10–50% sucrose gradient in buffer S1 (44) containing 1 mm AdoMet. The monosome fraction was collected using an ISCO 520 density gradient system at 4 °C and purified through a Strep-Tactin-Sepharose column (IBA) to obtain an ∼300-μl RNC preparation in buffer S2 (44).
For the methylation protection assay, 3 μl of DMS (1:6 dilution in ethanol) was added to 75 μl of RNC sample and incubated for 10 min at 25 °C. Reactions were stopped by the addition of 37.5 μl of stop solution (1 m Hepes, 10% (v/v) 2-mercaptoethanol). Total RNA was then extracted using Isogen LS (Nippon Gene, Tokyo, Japan) and subjected to primer extension analysis using Thermoscript RNase H− reverse transcriptase (Invitrogen) and 32P-labeled oligonucleotide primers (supplemental Table S2) for 60 min at 58 °C. Chemical modification experiments using 1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate and kethoxal were carried out as described previously (45) except that the reactions were carried out at 25 °C. Separation of cDNA products on a 6% polyacrylamide gel containing 7 m urea was performed as described previously (4), after which signals were visualized using a FLA-7000 image analyzer, and band intensities were measured using MultiGauge software.
For UV cross-link, 75 μl of RNC sample placed in an open-cap 1.5-ml microcentrifuge tube was irradiated with UV light (312 nm) for 10 min on ice using a SpectroLinker XL-1000 UV cross-linker (Spectronics Corp., New York). Samples were then treated with proteinase K (Wako Pure Chemical Industries) for 15 min at 37 °C before RNA extraction using Isogen LS. Primer extension analysis and quantification of the band intensities were carried out as described for the methylation protection assays.
RESULTS
Pegylation Assay of CGS1 Nascent Peptide
To test whether the CGS1 nascent peptide changes conformation inside the ribosomal exit tunnel, the pegylation assay (36, 37) was applied in a WGE cell-free translation system that enables post-transcriptional regulation of CGS1 to be reproduced (4, 9). Application of the pegylation assay to the CGS1 system requires a single cysteine residue on the nascent peptide, such that if the nascent peptide of the RNC changes conformation in response to AdoMet, the accessibility to PEG-MAL to this cysteine residue will be changed.
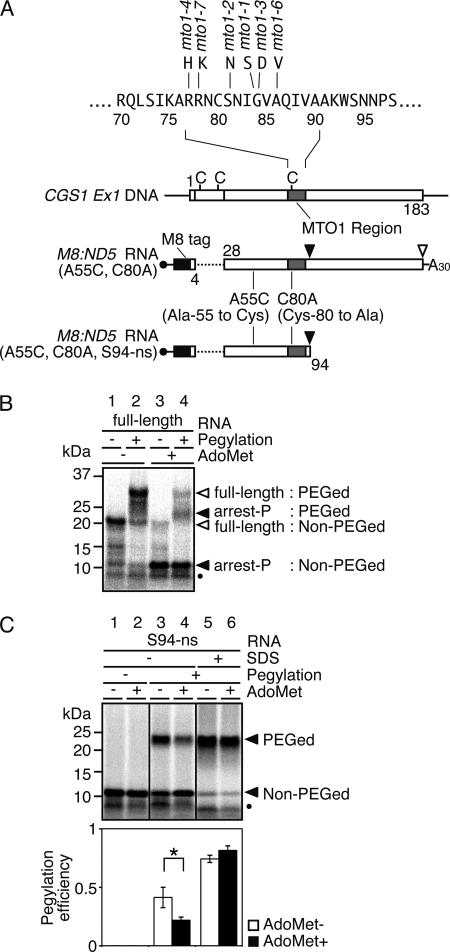
As the ribosomal exit tunnel holds 30–40 amino acids (33, 34), it was expected that a cysteine residue located between 30 and 40 amino acids upstream of the arrest site (Ser-94) would be near the outlet of the exit tunnel and that the pegylation efficiency for this residue would change prominently depending on the conformation of the nascent peptide. Wild-type CGS1 exon 1 coding sequence up to the Ser-94 codon (codon numbers of CGS1 relative to first methionine of native CGS1 are used hereafter, irrespective of N-terminal tag sequences added) carries three cysteine residues (Fig. 1A). None of these endogenous cysteine residues are located 30–40 amino acids upstream of the arrest site, and thus application of the pegylation assays required a cysteine-scanning approach to introduce cysteine into this upstream region. To provide an mRNA that encoded only one cysteine after cysteine-scanning substitution, a modified CGS1 gene was first constructed from which the three endogenous cysteine codons were eliminated. An ND5 deletion, which carries a deletion between the 5th and 27th codons and is capable of post-transcriptional regulation of CGS1 (6), was used to eliminate two of the three cysteine codons (Cys-8 and Cys-22). The remaining Cys-80 codon resided within the MTO1 region and was substituted to alanine (C80A). Although substitution of most amino acid residues within the MTO1 region renders the CGS1 mRNA insensitive to the regulation (the mto1 phenotype) (6), the C80A substitution is tolerated (supplemental Fig. S2). An eight consecutive methionine tag (M8 tag) was next fused in-frame to the N terminus that enabled nascent peptide products to be detected by [35S]methionine labeling. A single cysteine was then introduced at 40 amino acids upstream of the arrest site by substituting Ala-55 to cysteine (A55C) to generate the M8:ND5(A55C, C80A) construct (Fig. 1A).
FIGURE 1.
Pegylation assay of CGS1 nascent peptide. A, schematic representation of CGS1 exon 1 DNA (CGS1 Ex1 DNA), the “full-length” CGS1 exon 1 RNA construct M8:ND5(A55C, C80A), and the nonstop RNA construct M8:ND5(A55C, C80A, S94-ns). Positions of endogenous cysteine codons are marked with vertical bars and labeled with C. Gray boxes indicate the MTO1 region, and the black box indicates the M8 tag sequence. Amino acid sequence around the MTO1 region and mto1 mutant alleles are shown above the top construct. Positions of introduced amino acid substitutions are indicated. B, pegylation of translation products carrying a single cysteine residue at the 55th position in the “full-length” CGS1 exon 1 RNA. M8:ND5(A55C, C80A) RNA was translated for 30 min in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 1 mm AdoMet. Translation products were labeled with [35S]methionine. RNCs were isolated by ultracentrifugation and subjected to pegylation reaction (Pegylation +; lanes 2 and 4) or translation-only reaction (Pegylation −; lanes 1 and 3). Samples were separated by SDS-PAGE followed by detection of the radioactive signals. Positions of 32-kDa pegylated full-length (full-length: PEGed), 20-kDa unpegylated (full-length: Non-PEGed), 23-kDa pegylated (arrest-P: PEGed), and 11-kDa unpegylated (arrest-P: Non-PEGed) products are indicated. Unpegylated arrested peptide (10 kDa) produced by a secondary arrested ribosome is marked with a black dot. A representative result of triplicate experiments is shown. C, pegylation of translation arrest products carrying a single cysteine residue at the 55th position in the Ser-94 nonstop RNA. Upper panel, M8:ND5(A55C, C80A, S94-ns) RNA was translated for 30 min in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of 1 mm AdoMet. Translation products were labeled with [35S]methionine. RNCs were isolated by ultracentrifugation and subjected to pegylation reaction (lanes 3–6) or translation-only reaction (Pegylation −; lanes 1 and 2). In lanes 5 and 6, RNCs were treated with 1% SDS for 20 min at 25 °C prior to the pegylation reaction. Samples were separated by SDS-PAGE followed by detection of the radioactive signals. Positions of the 23-kDa pegylated (PEGed) and 11-kDa unpegylated (Non-PEGed) products are indicated. Unpegylated arrested peptide (10 kDa) produced by a secondary arrested ribosome is marked with a black dot. A representative result of triplicate experiments is shown. Lower panel, radioactive signals of the pegylated and unpegylated bands were quantified, and the pegylation efficiency was calculated (see “Experimental Procedures”). Average ± S.D. of at least three independent experiments are shown. Significant difference in pegylation efficiency between the presence and absence of AdoMet is indicated by an asterisk (p < 0.05 by t test).
Peptidyl-tRNASer accumulates the highest at 30 min after the start of the translation reaction in the presence of AdoMet (9). After translating M8:ND5(A55C, C80A) RNA in WGE for 30 min, RNCs were collected by ultracentrifugation, and nascent peptides were examined in a gel-shift assay using SDS-PAGE. When M8:ND5(A55C, C80A) RNA was translated in the absence of AdoMet, a major band of ∼20 kDa was observed representing the fully translated product (Fig. 1B, lane 1). A minor band was also observed at ∼11 kDa, which became the major band when RNA was translated in the presence of AdoMet (Fig. 1B, lane 3) and represented the product temporally arrested at Ser-94. An additional band was also present at ∼10 kDa, which is due to a second ribosome stacked directly behind the initial arrested ribosome (42). When RNCs were subjected to a pegylation reaction following translation, the 20-kDa band corresponding to the fully translated product shifted to ∼32 kDa (Fig. 1B, lanes 1 and 2) and the 11-kDa temporally arrested product shifted to ∼23 kDa (Fig. 1B, lanes 3 and 4). The 10-kDa band corresponding to the second ribosome would also contain the introduced Cys-55 and therefore can be pegylated. Because the pegylated products of the 10- and 11-kDa peptides cannot be dissolved in our SDS-PAGE system, this 10-kDa band was also included in the pegylation efficiency calculation.
The pegylation efficiency for the arrested product was calculated as 0.20 ± 0.01 (n = 3); however, it is not possible to establish whether this represents an AdoMet-induced conformational change because translation continues in the absence of AdoMet and comparison cannot be made with the same product. To provide robust comparison of nascent peptide conformation in the ribosomal exit tunnel according to the presence or absence of AdoMet and to prevent further state change due to possible resumption of translation (9), the construct was truncated at the 3′ end to provide an RNA without a stop codon that terminated at Ser-94. Use of this nonstop RNA, M8:ND5(A55C, C80A, S94-ns) (Fig. 1A), provides an RNC that contains the nascent peptide arrested at Ser-94 irrespective of AdoMet conditions, enabling the influence of AdoMet on the nascent peptide to be tested.
When M8:ND5(A55C, C80A, S94-ns) nonstop RNA was translated in the absence of AdoMet, arrested bands of 10 and 11 kDa and their pegylated products at 23 kDa were observed (Fig. 1C, lanes 3 and 4). The 23-kDa band was not observed when the pegylation reaction was omitted (Fig. 1C, lanes 1 and 2). The relative amount of 23-kDa pegylated product greatly increased when RNCs were disrupted by SDS prior to the pegylation assay (Fig. 1C, lanes 5 and 6), confirming that the pegylation efficiency reflects the state of the nascent peptide within the intact RNC and changes according to Cys-55 accessibility in the A55C construct. When the M8:ND5(A55C, C80A, S94-ns) RNA was translated in the presence of 1 mm AdoMet, a lower relative amount of pegylated product was observed (Fig. 1C, lane 4), suggesting that accessibility of PEG-MAL to the Cys-55 residue was reduced compared with that in the absence of AdoMet. This difference was not observed when RNCs were disrupted with SDS (Fig. 1C, lanes 5 and 6), indicating that the reduced accessibility induced by AdoMet occurred in the context of an intact RNC.
CGS1 Nascent Peptide Takes a Compact Conformation Inside the Ribosomal Exit Tunnel in Response to AdoMet
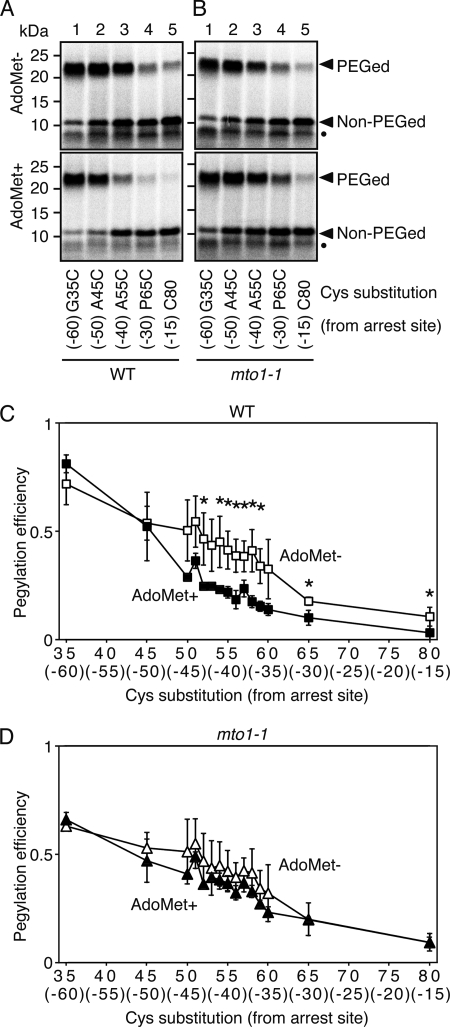
The above results suggested that the Cys-55 residue was located outside the ribosomal exit tunnel in the absence of AdoMet and inside the tunnel in the presence of AdoMet. If this interpretation is correct, it was predicted that pegylation efficiency would be higher when cysteine substitution was carried out at residues located a greater distance from the arrest site, whereas pegylation efficiency would be reduced when residues located closer to the arrest site were substituted. It was also predicted that in both cases the difference between the presence and absence of AdoMet would be diminished. To test this, mRNAs were constructed that carried cysteine substitutions at Gly-35 (G35C; 60 amino acids from the arrest site), Ala-45 (A45C; 50 amino acids from arrest site), and Pro-65 (P65C; 30 amino acids from arrest site). The original Cys-80 (C80) carried by the MTO1 sequence, located 15 amino acids from arrest site, was also used.
When M8:ND5(G35C, C80A, S94-ns) and M8:ND5(A45C, C80A, S94-ns) RNAs were subjected to the pegylation assay in the absence of AdoMet, increased pegylation efficiencies relative to that for the A55C construct were observed (Fig. 2A, upper panel, lanes 1–3). However, pegylation efficiencies of M8:ND5(P65C, C80A, S94-ns) and M8:ND5(C80, S94-ns) were reduced compared with that of A55C (Fig. 2A, upper panel, lanes 3–5). These results confirmed that a decrease in pegylation efficiency reflects a shift of the cysteine residue into the ribosomal exit tunnel. It should be noted that pegylation efficiency of the G35C construct was as high as that in the SDS-treated RNCs (Fig. 1C), suggesting that Cys-35 was located well outside the ribosomal exit tunnel.
FIGURE 2.
Cysteine-scanning and pegylation assay of CGS1 nascent peptide. A and B, pegylation assay of wild-type MTO1 (A) and mto1-1 mutant (B) versions of M8:ND5(G35C, C80A, S94-ns) (lane 1), M8:ND5(A45C, C80A, S94-ns) (lane 2), M8:ND5(A55C, C80A, S94-ns) (lane 3), M8:ND5(P65C, C80A, S94-ns) (lane 4), and M8:ND5(S94-ns) (lane 5) RNAs in the absence (upper panel) and presence (lower panel) of 1 mm AdoMet was carried out as described for Fig. 1C. Representative results of triplicate experiments are shown. Positions of the 23-kDa pegylated (PEGed) and 11-kDa unpegylated (Non-PEGed) products are indicated. Unpegylated arrested peptide (10 kDa) produced by a secondary arrested ribosome is marked with a black dot. The single cysteine position is indicated below each lane, and the distance of this cysteine from the arrest site (Ser-94) is indicated in parentheses. C and D, pegylation experiments were carried out using a series of cysteine substitution constructs of wild-type MTO1 (C) and mto1-1 mutant (D) versions. The radioactive signals of the pegylated and unpegylated bands were quantified. Pegylation efficiencies were calculated as in Fig. 1C, and average ± S.D. of at least three independent experiments are shown. Significant difference in pegylation efficiency between the absence and presence of AdoMet are marked with asterisks (p < 0.05 by t test).
When the same reactions were carried out in the presence of AdoMet, both M8:ND5(G35C, C80A, S94-ns) and M8:ND5(A45C, C80A, S94-ns) retained the increased pegylation efficiency (Fig. 2A, lower panel, lanes 1 and 2), which was equivalent to that in the absence of AdoMet (Fig. 2, A, upper panel, and C) and consistent with these residues remaining outside the exit tunnel irrespective of AdoMet conditions. A decrease in pegylation efficiency for M8:ND5(P65C, C80A, S94-ns) and M8:ND5(C80, S94-ns) was observed in the presence of AdoMet (Fig. 2, A, lower panel, lanes 4 and 5, and C), consistent with reduced accessibility of residues located inside the ribosomal tunnel according to AdoMet. More detailed cysteine scanning was also carried out for each amino acid between codons Ser-50 (45 amino acids from the arrest site) and Gly-60 (35 amino acids from the arrest site) codons (Fig. 2C, supplemental Fig. S3). Each construct carrying a cysteine substitution between Cys-50 and Cys-60 showed decreased pegylation in response to AdoMet. These results are consistent with AdoMet inducing a compaction of the CGS1 nascent peptide inside the ribosomal exit tunnel. Hereafter, the Ser-58 to cysteine substitution construct (S58C, 37 amino acids from the arrest site) was used for subsequent pegylation experiments.
Compaction of CGS1 Nascent Peptide Is Correlated with Regulatory Function of the MTO1 Region
The MTO1 region within CGS1 nascent peptide functions in cis in an AdoMet-dependent manner to mediate post-transcriptional regulation of CGS1 (3, 4, 6, 9). In the above experiments, compaction of the CGS1 nascent peptide was also shown to occur in an AdoMet-dependent manner. To assess whether nascent peptide compaction is dependent on the MTO1 sequence, pegylation experiments using mto1 mutations were performed. mto1 mutations, which harbor single amino acid alterations in the MTO1 region of CGS1 (Fig. 1A), have been shown to abolish post-transcriptional regulation of CGS1 (3, 4, 6, 9).
Cysteine-scanning experiment was performed using M8:ND5(C80A, S94-ns) constructs containing the mto1-1 mutation (Fig. 2B and supplemental Fig. S3). The results showed that reduction of pegylation efficiency observed with the wild-type MTO1 constructs (Fig. 2C) was greatly diminished (Fig. 2D), suggesting that the mto1-1 mutation inhibits AdoMet-induced compaction of CGS1 nascent peptide.
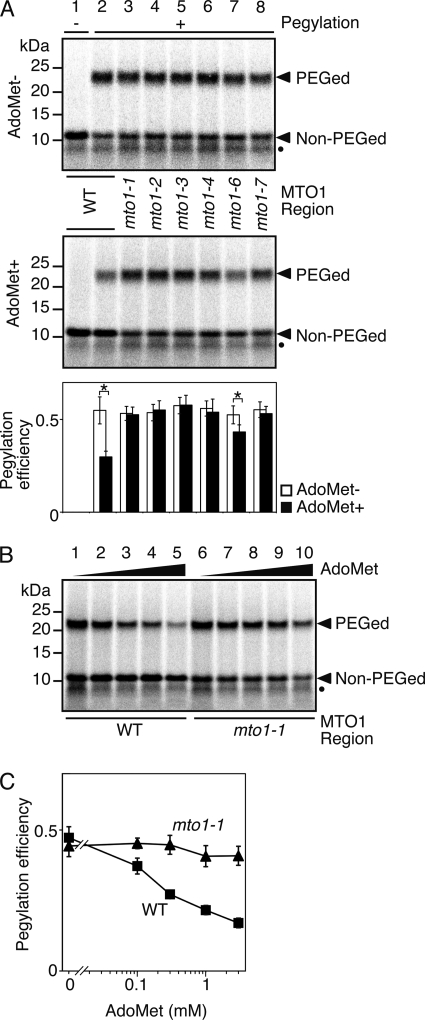
To further assess the importance of the MTO1 region, M8:ND5(S58C, C80A, S94-ns) constructs harboring different mto1 mutations were used for pegylation assay. All the mto1 mutant constructs showed enhanced pegylation in the presence of AdoMet compared with that for the wild-type construct (Fig. 3A, middle panel). The pegylation efficiency of mto1 mutant constructs in the presence of AdoMet was similar to that without AdoMet, indicating AdoMet-induced compaction of nascent peptides is impaired in the presence of an mto1 mutation. It is intriguing to note that the construct containing mto1-6, which is a weak mto1 allele (6), showed significantly lower pegylation efficiency in the presence of AdoMet than in its absence (Fig. 3A, lower panel; p < 0.05 by t test). Based on these results, we concluded that the wild-type CGS1 nascent peptide forms a compact conformation within the RNC in response to AdoMet, whereas the mto1 mutant peptide is impaired in this response.
FIGURE 3.
Correlation between regulatory function of the MTO1 region and pegylation efficiency. A, effects of different alleles of mto1 mutations were tested. Pegylation assays of wild-type MTO1 (lane 2), mto1-1 (lane 3), mto1-2 (lane 4), mto1-3 (lane 5), mto1-4 (lane 6), mto1-6 (lane 7), and mto1-7 (lane 8) mutant versions of M8:ND5(S58C, C80A, S94-ns) RNA in the absence (upper panel) and presence (middle panel) of 1 mm AdoMet (representative result of quadruplicate experiments) and quantification of radioactive signals (lower panel) were carried out as described for Fig. 1C. A translation-only reaction using wild-type MTO1 version is shown in lane 1. Significant difference in pegylation efficiency between the presence and absence of AdoMet are indicated by asterisks (p < 0.05 by t test). Positions of the 23-kDa pegylated (PEGed) and 11-kDa unpegylated (Non-PEGed) products are indicated. Unpegylated arrested peptide (10 kDa) produced by a secondary arrested ribosome is marked with a black dot. B, pegylation assay at different AdoMet concentrations was carried out as in Fig. 1C using wild-type MTO1 (lanes 1–5) and mto1-1 mutant (lanes 6–10) versions of M8:ND5(S58C, C80A, S94-ns) RNAs. The samples were translated for 30 min with 0 (lanes 1 and 6), 0.1 mm (lanes 2 and 7), 0.3 mm (lanes 3 and 8), 1 mm (lanes 4 and 9), and 3 mm (lanes 5 and 10) AdoMet. The 23-kDa pegylated (PEGed) and 11-kDa unpegylated (Non-PEGed) products are indicated. A representative result of triplicate experiments is shown. Unpegylated arrested peptide (10 kDa) produced by a secondary arrested ribosome is marked with a black dot. C, radioactive signals in B were quantified for the wild-type MTO1 (B, lanes 1–5) and mto1-1 mutant (B, lanes 6–10) versions. The pegylation efficiency was calculated as in Fig. 1C, and average ± S.D. of three independent experiments are shown.
To confirm that AdoMet-induced compaction was indeed lost by mto1 mutations, additional pegylation assays were performed in the presence of different AdoMet concentrations. AdoMet-induced post-transcriptional regulation of CGS1 is more evidently observed as AdoMet concentration is increased (4), whereas changes in AdoMet concentration does not affect expression of CGS1 harboring the mto1-1 mutation (4). A clear negative correlation was observed between AdoMet concentration and pegylation efficiency for the wild-type MTO1 construct, whereas pegylation efficiency of the mto1-1 construct was not appreciably altered by a change in AdoMet concentration (Fig. 3, B and C). The data confirmed that AdoMet-induced compaction of CGS1 nascent peptide was indeed lost by mto1-1 mutation and suggests a tight link between MTO1 function in translation arrest and compaction of the nascent peptide in the ribosomal exit tunnel.
Compaction of CGS1 Nascent Peptide Is Associated with Changes in Methylation Protection and UV Cross-link Patterns in 28 S rRNA
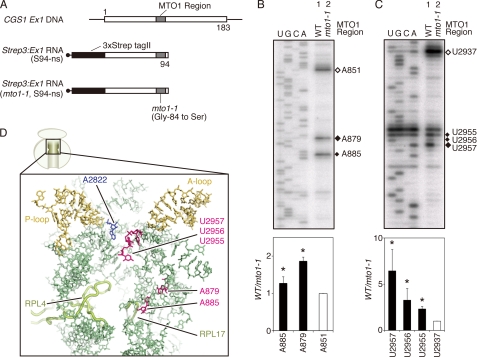
As described above, AdoMet induced CGS1 nascent peptide compaction within the ribosomal exit tunnel. To determine whether the peptide compaction was also associated with alteration of the exit tunnel, footprinting analysis of 28 S rRNA in the arrested RNC was carried out using DMS chemical modification and UV cross-link experiments. DMS methylates rRNA at specific nucleotides according to accessibility, whereas UV cross-link treatment results in bond formation between nearby molecules. Both of these treatments generate sites that inhibit primer extension reactions, resulting in a specific pattern of stop products depending on the molecular environment of rRNA residues such as accessibility of DMS or distance from nearby molecules (21, 43, 46). For the 28 S rRNA analysis, nonstop RNA similar to that for pegylation experiments was used, except that no cysteine substitution was introduced and the N terminus contained three consecutive Strep-tag II sequence (Strep3) fused in-frame to facilitate isolation of highly pure RNCs (Fig. 4A).
FIGURE 4.
Methylation protection and UV cross-link experiments of 28 S rRNA. A, schematic representation of Strep3:Ex1(S94-ns) and Strep3:Ex1(mto1-1, S94-ns) RNAs. Gray boxes indicate the MTO1 region, and the black boxes indicate the three consecutive Strep-tag II sequence. B, upper panel, methylation protection assay of arrested ribosome. Wild-type MTO1 (lane 1) and mto1-1 (lane 2) versions of Strep3:Ex1(S94-ns) RNAs were translated in WGE for 30 min in the presence of 1 mm AdoMet. RNCs were subjected to methylation reaction using DMS. Total RNA was extracted and used for primer extension analysis using 32P-labeled oligonucleotides complementary to wheat 28 S rRNA. Primer extension signals were detected after separation on a sequence gel. Filled diamonds mark the nucleotides in which a methylation difference was detected between lane 1 (WT) and lane 2 (mto1-1), and an open diamond marks the position of A851 that was taken as no difference between the two lanes. Sequence ladders were synthesized using the same oligonucleotides as primer and are shown in the rRNA sequence. A representative result of triplicate experiments is shown. Lower panel, radioactive signals of the bands marked in the upper panel were quantified. Relative methylation levels were calculated as lane 1 (WT)/lane 2 (mto1-1) and normalized with that for A851. Average ± S.D. of three independent experiments are shown. Significant difference in methylation between lanes 1 (WT) and lane 2 (mto1-1) are indicated by asterisks (p < 0.05 by t test). C, upper panel, UV cross-link assay of arrested ribosome. Wild-type MTO1 (lane 1) and mto1-1 (lane 2) versions of Strep3:Ex1(S94-ns) RNAs were translated in WGE as in B. RNCs were irradiated with UV, and primer extension analysis was carried out as in B. Filled diamonds mark the nucleotides in which a difference in UV cross-link was detected between lanes 1 (WT) and lane 2 (mto1-1), and an open diamond marks the position of U2937 that was taken as no difference between the two lanes. Sequence ladders were synthesized as in B. A representative result of triplicate experiments is shown. Lower panel, radioactive signals of the bands marked in the upper panel were quantified. Relative levels of UV cross-link were calculated as lane 1 (WT)/lane 2 (mto1-1) and normalized with that for U2937. Average ± S.D. of three independent experiments are shown. Significant difference in UV cross-link levels between lane 1 (WT) and lane 2 (mto1-1) are indicated by asterisks (p < 0.05 by t test). D, wheat 60 S subunit showing components in the upper half region of the exit tunnel, including A2822 (EcA2451) in the PTase center (blue) and ribosomal proteins L4 and L17 that constitutes the constriction region (light green). Nucleotides A879, A885, U2955, U2956, and U2957 are shown in magenta; the A-loop and P-loop are marked in ocher; and other 28 S rRNA nucleotides are shown in pale green. Protein Data Bank codes 3IZ9 and 3IZR for Triticum aestivum 28 S rRNA and ribosomal proteins (14, 54), and PyMol software are used.
Primer extension analysis of DMS-treated 28 S rRNA isolated from reactions containing AdoMet and an mRNA with the wild-type MTO1 region revealed increased methylation at nucleotides A879 (U744 in E. coli 23 S rRNA; EcU744) and A885 (EcA750) compared with that of reactions containing an mto1 mutant construct (Fig. 4B). More than 90% of the entire 28 S rRNA sequence was analyzed in this methylation protection assay, and these were the only two differences that could be detected (supplemental Fig. S4). These nucleotides are known to constitute the wall of the exit tunnel and are included in the stem-loop structure located adjacent to ribosomal protein L17, which is one of the components of the constriction region in the exit tunnel (Fig. 4D) (11).
In the UV cross-link experiments, 28 S rRNA extracted from reactions containing AdoMet and an mRNA with the wild-type MTO1 region showed enhanced signals at U2955, U2956, and U2957 (EcU2584, EcU2585, and EcU2586, respectively) compared with that for reactions containing an mto1 mutant construct (Fig. 4C). Of more than 90% of the entire 28 S rRNA sequence analyzed, these were the only differences detectable using the UV cross-link assay (supplemental Fig. S5). The U2955, U2956, and U2957 nucleotides are located within the PTase loop in domain V of large subunit rRNA (Fig. 4D) (11). The results suggest that the MTO1 region mediates these changes in the ribosomal exit tunnel in response to AdoMet.
DISCUSSION
In this study, we have demonstrated that the nascent peptide of the CGS1 gene is induced to adopt a compact conformation within an arrested eukaryotic ribosome according to AdoMet, the effector molecule for feedback regulation of CGS1 expression. In vitro analyses using a plant cell-free translation system was combined with a pegylation assay to assess changes in nascent peptide accessibility. These assays showed that cysteine residues located at around Cys-55 (40 amino acids from the arrest site) became less accessible in an AdoMet-dependent manner, consistent with conformational change of the CGS1 nascent peptide inside the exit tunnel. This conformational change was measured as change in the signal intensities of pegylated translation arrest product (23 kDa) relative to that of the main (11 kDa) and weaker (10 kDa) unpegylated products. The weaker 10-kDa arrest product is due to a second ribosome stacked directly behind the primarily stalled ribosome (42). Because a ribosome covers about 30 nucleotides of mRNA (47), the 10-kDa peptide is expected to also carry the cysteine residue at a position about 10 residues closer to the PTase center compared with that for the 11-kDa peptide. Although it is possible this calculation may obscure the exact final pegylation efficiency values, the intensity of the 10-kDa band relative to 11 kDa without pegylation reaction (Fig. 1C) is only ∼20%, and the pegylation efficiency of the 10-kDa peptide is expected to be no larger than the 11-kDa one. Thus, the data presented here support the conclusion that CGS1 nascent peptide takes a conformational change with reduced PEG-MAL accessibility in response to AdoMet and that this conformational change is inhibited by mto1 mutations.
The MTO1 region is a 14-amino acid sequence located between codons 77 and 90 of the CGS1 nascent peptide. This region is highly conserved in higher plants and has been shown to act in mediating translation arrest and post-transcriptional regulation of CGS1 in response to AdoMet (4, 6, 9). Amino acid residues essential for MTO1 activity have been identified by characterization of phenotypic plant mutants and mutational analysis using recombinant constructs (4, 7). A change in amino acid sequence at these residues, termed mto1 mutations, causes loss of function of the MTO1 region and normal translation of CGS1 even in the presence of AdoMet (4, 9, 48). As shown above, the introduction of mto1 mutations into constructs used for the pegylation assays resulted in AdoMet-induced compaction of CGS1 peptide to be greatly diminished. In this analysis, only mutations known to functionally disrupt MTO1 function caused loss of the AdoMet-induced compaction, whereas sequence changes within the MTO1 region that do not affect its function (C80A) (supplemental Fig. S2) showed the compaction response. A mutation that has a less detrimental effect on MTO1 function (mto1-6, A86V) (6) exhibited partial reduction of pegylation efficiency in response to AdoMet (Fig. 3). These results show that CGS1 nascent peptide compaction induced by AdoMet is correlated with functional activity of the MTO1 region, rather than simply any amino acid sequence alteration in this region. This correlation likely represents a dependence on MTO1 function, although experiments with full-length mRNA constructs would be required to demonstrate this definitively. As shown in Fig. 1B, the pegylation efficiency of arrested product for full-length mRNA (0.20 ± 0.01) was not significantly different (p < 0.05, t test) from that for the nonstop construct used here (0.22 ± 0.03, see Fig. 1C), suggesting that translation arrest of the nonstop RNA in the presence of AdoMet reflects AdoMet-induced translation arrest of CGS1. However, it is not possible to examine compaction of the arrested product using full-length mRNA as comparative analysis cannot be performed in the absence of AdoMet (Fig. 1B).
Methylation protection and UV cross-link analysis of 28 S rRNA from purified RNCs revealed specific changes in footprinting patterns in parallel to the compaction state of the CGS1 nascent peptide. Both types of treatment have been used to detect changes in the molecular environment of rRNA residues (21, 43, 46). The methylation protection analysis used DMS, which creates primer extension stop sites by modifying accessible adenine, cytosine, or guanine nucleotides (45), whereas UV cross-link creates primer extension stop sites due to formation of new bonds between nearby molecules (46). The nucleotides A879 and A885 showed a difference in DMS accessibility according to nascent peptide compaction state (Fig. 4B), whereas the UV cross-link treatment revealed changes at U2955, U2956, and U2957 (Fig. 4C). Over 90% of the entire 28 S rRNA, including that covering the exit tunnel region (11, 14), was explored in this analysis, and these were the only differences that were of statistical significance. In addition, no nucleotide differences were identified in the upper tunnel region using 1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate and kethoxal chemical modification analyses (data not shown) (45). A879 and A885 are among the large subunit rRNA residues that constitute the wall of the exit tunnel, whereas U2955, U2956, and U2957 reside in the PTase loop of large subunit rRNA (Fig. 4D) (11). These observations suggest that, when AdoMet-induced compaction of CGS1 nascent peptide occurs, either the stalled ribosome also adopts a specific conformational change in the tunnel wall or relative positioning of nascent peptide and the tunnel wall is altered.
In eukaryotes, compaction of a nascent peptide inside the ribosomal exit tunnel has been reported for transmembrane proteins (37, 38, 49) and fungal AAP (32), and it was suggested that a preferable zone for secondary structure formation is present near the PTase center (32, 37, 49). Because the MTO1 sequence exists at 5–18 amino acids from the PTase center when translation is arrested at Ser-94, the MTO1 sequence would reside in the region of the exit tunnel thought to be preferable for secondary structure formation. Although it is not yet possible to clarify whether nascent peptide compaction occurs either prior to or immediately following translation arrest, these findings strongly suggest that the MTO1 region functions in nascent peptide-mediated translation elongation arrest of CGS1 mRNA by directing changes of both the nascent peptide and ribosomal exit tunnel.
How then could AdoMet-induced compaction of CGS1 nascent peptide be mediated by the internal MTO1 region? Because AdoMet serves as a major methyl donor in most biological methyl transfer reactions, one possibility is that AdoMet-mediated methylation of either the body of translating ribosome or the nascent peptide in the ribosomal exit tunnel induces nascent peptide compaction. However, this is unlikely as S-adenosyl-l-homocysteine, a competitive inhibitor of AdoMet-mediated methyl transfer reactions (50), does not affect the AdoMet-dependent down-regulation in CGS1 (4). Two alternative scenarios can also be envisaged for the mechanism of AdoMet induction of nascent peptide compaction within the exit tunnel. The nascent peptide may directly interact with AdoMet that results in a change of folding state (scenario 1), or it is also possible that AdoMet interacts with the translating ribosome, and the signal is transduced to the nascent peptide either via the exit tunnel wall or via both the PTC and the tunnel wall (scenario 2).
Although data are not yet available to fully assess the above possible scenarios, recent reports on translation arrest and compaction of nascent peptide deserve discussion. Compaction of nascent peptide upon translation arrest accompanied by interaction between specific residues in the nascent peptide and the exit tunnel wall has been suggested in the E. coli SecM system (27, 29). Local contacts with the tunnel wall also appear to be a feature of the E. coli TnaC arrest sequence, although this sequence seems to take a relatively extended conformation (26).
Prokaryotic translation elongation inhibitors erythromycin, carbomycin A, and sparsomycin have also been shown to interact with the wall of exit tunnel (51–53). It is plausible that AdoMet can also access and act within the exit tunnel, enabling AdoMet to interact directly with the MTO1 peptide that mediates the nascent peptide compaction of CGS1. Together with the fact that the MTO1 region, 77RRNCSNIGVAQIVA90, contains several amino acids whose side chains can be involved in hydrogen bond formation, this would argue for scenario 1.
The accessibility of DMS changed in the stalled ribosome containing compacted CGS1 nascent peptide at A879 and A885 (Fig. 4B). These nucleotides correspond to E. coli 23 S rRNA residues EcU744 and EcA750, which are adjacent to EcA749-EcA753 that occupy the narrowest region of the exit tunnel (10, 14, 54). The importance of this rRNA region for translation arrest has been suggested by genetic analysis of the nascent peptide-mediated translation arrest of secM and tnaC. Mutations at or near the EcA751 residue inhibit ribosome stalling in both systems (20, 21, 24), whereas EcA749-EcA753 residues are suggested to make close contacts with the TnaC and SecM nascent peptides upon translation arrest (21, 26, 55). Our methylation protection result therefore raises the possibility that nascent peptide-mediated translation arrest in CGS1, tnaC, and secM utilize homologous regions of eukaryotic and prokaryotic ribosomes, respectively. U2955, U2956, and U2957, which showed a difference in UV cross-link patterns (Fig. 4C), correspond to EcU2584, EcU2585, and EcU2586, respectively (14, 54). This region of rRNA is known to contribute to the PTase reaction and binding of the 3′ end of peptidyl-tRNA to the P-site (56–59). EcU2585 and EcU2586 are suggested to interact with Pro-24 and Asp-21 of the TnaC nascent peptide, respectively (26), and mutation of the neighboring EcG2583 and EcU2584 have been shown to reduce ribosome stalling efficiency in the TnaC system (60). The structure of the fungal AAP-stalled wheat ribosome revealed compaction of the nascent peptide by a turn of the α-helix (32). This compaction also involved interaction with the tunnel wall at A886 (EcA751) and U2956 (EcU2585) residues. These reports suggest involvement of both the U2955–U2957 region and the vicinity of A885 in AdoMet-induced translation arrest of CGS1 and support an argument for scenario 2. Secondary structure prediction of the CGS1 amino acid sequence up to Ser-94 using PSIPRED indicated a potential α-helix at 5–11 amino acids from Ser-94, which would also be located in the upper region of the ribosomal exit tunnel. Based on the recent reports of peptide secondary structure inside the exit tunnel (15, 32, 37, 38, 49), it is interesting to speculate that formation of an α-helix structure could play a role in CGS1 compaction, although experimental determination of the nascent peptide CGS1 structure in the presence of AdoMet is required to correctly examine this possibility.
In a previous report, we demonstrated that the anticodon end of peptidyl-tRNA resides at the A-site of the ribosome that is arrested at the translocation step and not at the PTase reaction step (9). In SecM, peptidyl-tRNAGly-165 containing an arrest motif at its C terminus resides in the P-site, and prolyl-tRNAPro-166 in the A-site functions as a crucial factor for interfering with the PTase reaction (23). CGS1 post-transcriptional regulation shares the importance for peptide compaction with that of secM; however, there are clear differences with regard to the ribosomal site of peptidyl-tRNA occupation (A-site versus P-site) and the arrested step (ribosomal translocation versus PTase reaction). The CGS1 system described here thus implies that multiple regulatory mechanisms and modes of action exist in higher organisms for nascent peptide-mediated translation arrest.
Supplementary Material
Acknowledgments
We are grateful to Drs. Yukako Chiba and Junpei Takano for critical reading of the manuscript and valuable discussion. We also thank Saeko Yasokawa for skillful technical assistance and Kumi Fujiwara and Naoe Konno for general assistance. We used the Radioisotope Laboratory of the Graduate School of Agriculture, Hokkaido University.
This work was supported in part by Grant-in-aid for Scientific Research (B) 20370016 (to S. N.), Grant-in-aid for JSPS Fellows 3032 (to N. O.) from the Japan Society for the Promotion of Science, Grant-in-aid for Scientific Research on Innovative Areas 22119006 (to S. N.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by Core Research for Evolutional Science and Technology PJ34085001 (to H. O.) from the Japan Science and Technology Agency.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S5, and Tables S1 and S2.
This article was selected as a Paper of the Week.
- AdoMet
- S-adenosyl-l-methionine
- PTase
- peptidyltransferase
- AAP
- arginine attenuator peptide
- PEG-MAL
- polyethyleneglycol-maleimide
- DMS
- dimethyl sulfate
- RNC
- ribosome-nascent chain complex
- M8
- an eight consecutive methionine tag
- WGE
- wheat germ extract
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Kim J., Leustek T. (1996) Plant Mol. Biol. 32, 1117–1124 [DOI] [PubMed] [Google Scholar]
- 2. Matthews B. F. (1999) in Plant Amino Acids: Biochemistry and Biotechnology (Singh B. K. ed) pp. 205–225, Marcel Dekker, Inc., New York [Google Scholar]
- 3. Chiba Y., Ishikawa M., Kijima F., Tyson R. H., Kim J., Yamamoto A., Nambara E., Leustek T., Wallsgrove R. M., Naito S. (1999) Science 286, 1371–1374 [DOI] [PubMed] [Google Scholar]
- 4. Chiba Y., Sakurai R., Yoshino M., Ominato K., Ishikawa M., Onouchi H., Naito S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10225–10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Inaba K., Fujiwara T., Hayashi H., Chino M., Komeda Y., Naito S. (1994) Plant Physiol. 104, 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ominato K., Akita H., Suzuki A., Kijima F., Yoshino T., Yoshino M., Chiba Y., Onouchi H., Naito S. (2002) J. Biol. Chem. 277, 36380–36386 [DOI] [PubMed] [Google Scholar]
- 7. Suzuki A., Shirata Y., Ishida H., Chiba Y., Onouchi H., Naito S. (2001) Plant Cell Physiol. 42, 1174–1180 [DOI] [PubMed] [Google Scholar]
- 8. Lambein I., Chiba Y., Onouchi H., Naito S. (2003) Plant Cell Physiol. 44, 893–900 [DOI] [PubMed] [Google Scholar]
- 9. Onouchi H., Nagami Y., Haraguchi Y., Nakamoto M., Nishimura Y., Sakurai R., Nagao N., Kawasaki D., Kadokura Y., Naito S. (2005) Genes Dev. 19, 1799–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A. (2000) Science 289, 905–920 [DOI] [PubMed] [Google Scholar]
- 11. Nissen P., Hansen J., Ban N., Moore P. B., Steitz T. A. (2000) Science 289, 920–930 [DOI] [PubMed] [Google Scholar]
- 12. Menetret J. F., Neuhof A., Morgan D. G., Plath K., Radermacher M., Rapoport T. A., Akey C. W. (2000) Mol. Cell 6, 1219–1232 [DOI] [PubMed] [Google Scholar]
- 13. Morgan D. G., Ménétret J. F., Radermacher M., Neuhof A., Akey I. V., Rapoport T. A., Akey C. W. (2000) J. Mol. Biol. 301, 301–321 [DOI] [PubMed] [Google Scholar]
- 14. Armache J. P., Jarasch A., Anger A. M., Villa E., Becker T., Bhushan S., Jossinet F., Habeck M., Dindar G., Franckenberg S., Marquez V., Mielke T., Thomm M., Berninghausen O., Beatrix B., Söding J., Westhof E., Wilson D. N., Beckmann R. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 19748–19753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bhushan S., Gartmann M., Halic M., Armache J. P., Jarasch A., Mielke T., Berninghausen O., Wilson D. N., Beckmann R. (2010) Nat. Struct. Mol. Biol. 17, 313–317 [DOI] [PubMed] [Google Scholar]
- 16. Gong F., Yanofsky C. (2001) J. Biol. Chem. 276, 1974–1983 [DOI] [PubMed] [Google Scholar]
- 17. Gong F., Ito K., Nakamura Y., Yanofsky C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8997–9001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vazquez-Laslop N., Thum C., Mankin A. S. (2008) Mol. Cell 30, 190–202 [DOI] [PubMed] [Google Scholar]
- 19. Nakatogawa H., Ito K. (2001) Mol. Cell 7, 185–192 [DOI] [PubMed] [Google Scholar]
- 20. Nakatogawa H., Ito K. (2002) Cell 108, 629–636 [DOI] [PubMed] [Google Scholar]
- 21. Cruz-Vera L. R., Rajagopal S., Squires C., Yanofsky C. (2005) Mol. Cell 19, 333–343 [DOI] [PubMed] [Google Scholar]
- 22. Gong F., Yanofsky C. (2002) Science 297, 1864–1867 [DOI] [PubMed] [Google Scholar]
- 23. Muto H., Nakatogawa H., Ito K. (2006) Mol. Cell 22, 545–552 [DOI] [PubMed] [Google Scholar]
- 24. Cruz-Vera L. R., New A., Squires C., Yanofsky C. (2007) J. Bacteriol. 189, 3140–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cruz-Vera L. R., Yanofsky C. (2008) J. Bacteriol. 190, 4791–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seidelt B., Innis C. A., Wilson D. N., Gartmann M., Armache J. P., Villa E., Trabuco L. G., Becker T., Mielke T., Schulten K., Steitz T. A., Beckmann R. (2009) Science 326, 1412–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yap M. N., Bernstein H. D. (2009) Mol. Cell 34, 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vázquez-Laslop N., Ramu H., Klepacki D., Kannan K., Mankin A. S. (2010) EMBO J. 29, 3108–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Woolhead C. A., Johnson A. E., Bernstein H. D. (2006) Mol. Cell 22, 587–598 [DOI] [PubMed] [Google Scholar]
- 30. Janzen D. M., Frolova L., Geballe A. P. (2002) Mol. Cell. Biol. 22, 8562–8570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spevak C. C., Ivanov I. P., Sachs M. S. (2010) J. Biol. Chem. 285, 40933–40942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bhushan S., Meyer H., Starosta A. L., Becker T., Mielke T., Berninghausen O., Sattler M., Wilson D. N., Beckmann R. (2010) Mol. Cell 40, 138–146 [DOI] [PubMed] [Google Scholar]
- 33. Malkin L. I., Rich A. (1967) J. Mol. Biol. 26, 329–346 [DOI] [PubMed] [Google Scholar]
- 34. Blobel G., Sabatini D. D. (1970) J. Cell Biol. 45, 130–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu J., Deutsch C. (2001) Biochemistry 40, 13288–13301 [DOI] [PubMed] [Google Scholar]
- 36. Lu J., Deutsch C. (2005) Biochemistry 44, 8230–8243 [DOI] [PubMed] [Google Scholar]
- 37. Lu J., Deutsch C. (2005) Nat. Struct. Mol. Biol. 12, 1123–1129 [DOI] [PubMed] [Google Scholar]
- 38. Tu L. W., Deutsch C. (2010) J. Mol. Biol. 396, 1346–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 40. Pogulis R. J., Vallejo A. N., Pease L. R. (1996) Methods Mol. Biol. 57, 167–176 [DOI] [PubMed] [Google Scholar]
- 41. Erickson A. H., Blobel G. (1983) Methods Enzymol. 96, 38–50 [DOI] [PubMed] [Google Scholar]
- 42. Haraguchi Y., Kadokura Y., Nakamoto M., Onouchi H., Naito S. (2008) Plant Cell Physiol. 49, 314–323 [DOI] [PubMed] [Google Scholar]
- 43. Cruz-Vera L. R., Gong M., Yanofsky C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3598–3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schaffitzel C., Ban N. (2007) J. Struct. Biol. 159, 302–310 [DOI] [PubMed] [Google Scholar]
- 45. Moazed D., Noller H. F. (1986) Cell 47, 985–994 [DOI] [PubMed] [Google Scholar]
- 46. Noah J. W., Shapkina T. G., Nanda K., Huggins W., Wollenzien P. (2003) Biochemistry 42, 14386–14396 [DOI] [PubMed] [Google Scholar]
- 47. Wolin S. L., Walter P. (1988) EMBO J. 7, 3559–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Onouchi H., Haraguchi Y., Nakamoto M., Kawasaki D., Nagami-Yamashita Y., Murota K., Kezuka-Hosomi A., Chiba Y., Naito S. (2008) Plant Cell Physiol. 49, 549–556 [DOI] [PubMed] [Google Scholar]
- 49. Woolhead C. A., McCormick P. J., Johnson A. E. (2004) Cell 116, 725–736 [DOI] [PubMed] [Google Scholar]
- 50. Ueland P. M. (1982) Pharmacol. Rev. 34, 223–253 [PubMed] [Google Scholar]
- 51. Hansen J. L., Ippolito J. A., Ban N., Nissen P., Moore P. B., Steitz T. A. (2002) Mol. Cell 10, 117–128 [DOI] [PubMed] [Google Scholar]
- 52. Hansen J. L., Moore P. B., Steitz T. A. (2003) J. Mol. Biol. 330, 1061–1075 [DOI] [PubMed] [Google Scholar]
- 53. Tu D., Blaha G., Moore P. B., Steitz T. A. (2005) Cell 121, 257–270 [DOI] [PubMed] [Google Scholar]
- 54. Cannone J. J., Subramanian S., Schnare M. N., Collett J. R., D'Souza L. M., Du Y., Feng B., Lin N., Madabusi L. V., Müller K. M., Pande N., Shang Z., Yu N., Gutell R. R. (2002) BMC Bioinformatics 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mitra K., Schaffitzel C., Fabiola F., Chapman M. S., Ban N., Frank J. (2006) Mol. Cell 22, 533–543 [DOI] [PubMed] [Google Scholar]
- 56. Moazed D., Noller H. F. (1989) Cell 57, 585–597 [DOI] [PubMed] [Google Scholar]
- 57. Porse B. T., Garrett R. A. (1995) J. Mol. Biol. 249, 1–10 [DOI] [PubMed] [Google Scholar]
- 58. Schmeing T. M., Huang K. S., Kitchen D. E., Strobel S. A., Steitz T. A. (2005) Mol. Cell 20, 437–448 [DOI] [PubMed] [Google Scholar]
- 59. Schmeing T. M., Huang K. S., Strobel S. A., Steitz T. A. (2005) Nature 438, 520–524 [DOI] [PubMed] [Google Scholar]
- 60. Yang R., Cruz-Vera L. R., Yanofsky C. (2009) J. Bacteriol. 191, 3445–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.