Abstract
The Notch pathway is a conserved cell-to-cell signaling mechanism that mediates cell fate decisions in metazoans. Canonical signaling results in changes in gene expression, which is regulated by the nuclear effector of the pathway CSL (CBF1/RBP-J, Su(H), Lag-1). CSL is a DNA binding protein that functions as either a repressor or an activator of transcription, depending upon whether it is complexed by transcriptional corepressor or coactivator proteins, respectively. In stark contrast to CSL-coactivator complexes, e.g. the transcriptionally active CSL-Notch-Mastermind ternary complex, the structure and function of CSL-corepressor complexes are poorly understood. The corepressor MINT (Msx2-interacting nuclear target protein) has been shown in vivo to antagonize Notch signaling and shown in vitro to biochemically interact with CSL; however, the molecular details of this interaction are only partially defined. Here, we provide a quantitative thermodynamic binding analysis of CSL-MINT complexes. Using isothermal titration calorimetry, we demonstrate that MINT forms a high affinity complex with CSL, and we also delineate the domains of MINT and CSL that are necessary and sufficient for complex formation. Moreover, we show in cultured cells that this region of MINT can inhibit Notch signaling in transcriptional reporter assays. Taken together, our results provide functional insights into how CSL is converted from a repressor to an activator of transcription.
Keywords: Calorimetry, Corepressor Transcription, Crystal Structure, Notch Pathway, Protein-protein Interactions, Signal Transduction, Transcription Factors
Introduction
Notch refers to an evolutionary conserved signal transduction pathway that occurs between neighboring cells (1). The central components of the pathway are the receptor Notch, the ligand DSL (Delta, Serrate, Lag-2), and the nuclear effector CSL (CBF1/RBP-J, Su(H), Lag-1). Genetic ablation of the central components results in embryonic lethality, emphasizing the role of Notch during embryonic development (2–6); tissue specific ablation of Notch signaling results in severe phenotypes during hematopoiesis (7), organogenesis (8), and angiogenesis (9). Correspondingly, mutations in the central components underlie the pathogenesis of many human diseases, including certain types of cancer (10), cardiovascular defects (11), and congenital syndromes (12). At present, much effort has been devoted to developing reagents that block Notch signaling for therapeutic purposes (13); however, it has also been suggested that the identification of reagents that promote Notch signaling may have clinical applications for diseases that are characterized by insufficient signaling (14).
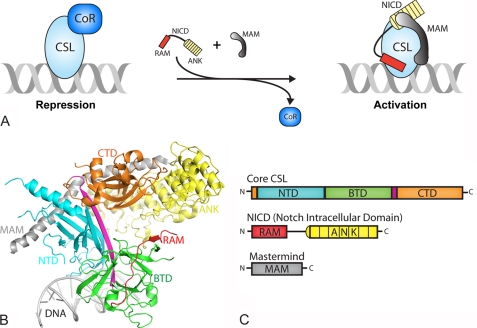
Extracellular interactions between the receptor Notch and the ligand DSL activate signaling, which results in proteolytic cleavage of Notch and release of its intracellular domain (NICD)3 from the cell membrane (1). NICD localizes to the nucleus where it binds CSL and the transcriptional coactivator Mastermind (MAM), forming a transcriptionally active ternary complex and leading to the expression of genes that are responsive to Notch signals. Our group and others have determined the x-ray structures of CSL and CSL-NICD-MAM ternary complexes bound to DNA, and quantitatively characterized the assembly of these complexes using a bevy of biochemical and biophysical approaches (15). As shown below (see Fig. 1), the structure of core CSL is composed of three domains: N-terminal domain (NTD), β-trefoil domain (BTD), and C-terminal domain (CTD), in which its BTD and CTD interact with the RAM (RBP-J-associated molecule) and ankyrin repeats (ANK) domains of NICD, respectively. MAM forms an elongated bent helix, which interacts with a continuous groove formed by ANK-CTD and the NTD of CSL.
FIGURE 1.
Overview of Notch transcription complexes. A, current model in the field for how transcription is regulated in the Notch pathway. In the absence of a Notch signal, the nuclear effector of the pathway, CSL, specifically binds DNA elements within the promoter and/or enhancer regions of target genes and interacts with transcriptional corepressor proteins in order to repress transcription. Upon pathway activation, the Notch receptor is cleaved at the plasma membrane and its intracellular domain translocates to the nucleus where it interacts with CSL through its RAM and ANK domains. NICD is thought to displace or outcompete corepressors for CSL binding. The CSL-NICD binary complex recruits the transcriptional coactivator MAM, which forms the transcriptionally active CSL-NICD-MAM ternary complex, a necessary step for up-regulating transcription from Notch target genes. B, X-ray structure of the CSL-NICD-MAM ternary complex bound to a cognate DNA (30, 33). CSL is composed of three domains: NTD, BTD, and CTD, which are colored cyan, green, and orange, respectively. A β-strand that makes hydrogen-bonding interactions with all three domains, integrating the three domains into one overall fold, is colored magenta. The NTD and BTD interact with the DNA (gray). NICD interacts with the BTD and CTD through its RAM (red) and ANK (yellow) domains, respectively. MAM, colored gray, forms a bent elongated helix that binds a continuous groove formed by the CTD-ANK interface and the NTD of CSL. C, domain schematics for CSL, NICD, and MAM, coloring is the same as in B.
In the absence of a Notch signal, it is thought that CSL functions as a repressor by interacting with transcriptional corepressor proteins, such as MINT (Msx2-interacting nuclear target protein), which is also known as SHARP (SMRT/HDAC-1-associated repressor protein) in humans (16, 17). Corepressors are components of large multiprotein histone deacetylase-containing complexes, which serve to link CSL to the repressive histone deacetylase machinery in the nucleus and localize deacetylase activity at Notch target genes (18). The competitive and mutually exclusive binding of corepressors or NICD to CSL have lead to current models in the field that suggest, upon pathway activation, NotchIC binding to CSL displaces corepressors from CSL (19–21), leading to a sequence of events that ultimately activate transcription (Fig. 1A). Although the interactions of NICD with CSL have been well characterized (15), the interactions of corepressors with CSL and the molecular basis of NICD and corepressor competition for CSL are not well understood.
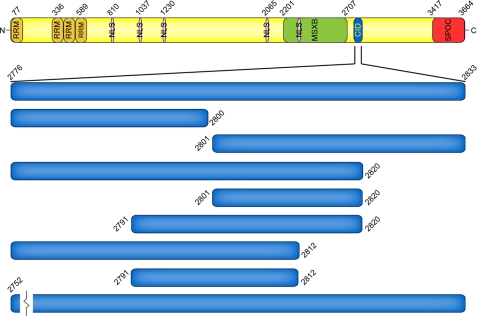
MINT is a large multidomain protein that functions as a transcriptional corepressor for multiple transcription factors (Fig. 2) (22). MINT null mice are embryonic lethal; however, fetal liver cells isolated from MINT (−/−) mice display defects in both B and T cell development (16, 23). The interaction between MINT and CSL was first identified in yeast two-hybrid screens, and subsequent biochemical studies mapped a region in MINT, of ∼14 amino acids, that is necessary for binding CSL (16, 17). As similarly observed for other corepressors, MINT competes with NICD for binding to CSL (16). MINT also was shown in cell culture studies to repress transcription from reporters that are responsive to activated forms of Notch (17). In addition to its interaction with CSL, MINT functions as a binding platform for other transcriptional corepressors, including C-terminal binding protein and C-terminal binding protein interacting protein (24), Ski-interacting protein (17), ETO (eight-twenty-one) (25), and silencing mediator for retinoid and thyroid (26).
FIGURE 2.
Domain organization of the transcriptional corepressor MINT. MINT is a multidomain 3,664-residue protein composed of N- and C-terminal RNA recognition motifs (RRM) and Spen paralog and ortholog C-terminal domain (SPOC) domains, respectively. MINT also contains several nuclear localization sequences (NLS), a region that interacts with the transcription factor Msx2 (MSXB), and a CSL-interacting domain (CID). The constructs that were utilized in the herein described CSL-MINT binding study are expanded below the CSL-interacting domain.
To quantitatively characterize the interactions of MINT with CSL and increase our understanding of how CSL functions as a transcriptional repressor, we performed a thorough biophysical analysis of CSL-MINT complexes using circular dichroism and isothermal titration calorimetry. Our binding data demonstrates that MINT forms a high affinity interaction with CSL, comparable with the interaction of NICD with CSL. We also show that MINT binds the BTD and CTD of CSL, similar to NICD; however, the relative affinities of MINT and NICD for these two domains of CSL are strikingly different. In addition, we demonstrate that the region of MINT defined in our binding studies to interact with CSL is a random coil in solution. Nonetheless, this region of MINT is a potent inhibitor of transcription from a Notch-responsive reporter in cells. Taken together, our results provide molecular insights into how MINT and NICD compete for binding to CSL, which is important for understanding how CSL is converted from a repressor to an activator of transcription from Notch target genes.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Protein Purification
The cloning, expression, and purification of Mus musculus core CSL (RBP-J), residues 53–474, BTD (203–363), and the RAM (1744–1771), and RAMANK (1744–2113) domains of Notch1 were described previously (27). Constructs that correspond to either the CTD (residues 367–474) or BTD-CTD (residues 203–474) of CSL were cloned into pGEX6P-1 and purified similarly to core and BTD CSL proteins. Briefly, following overexpression in bacteria, GST-CTD and GST-BTD-CTD fusion proteins were isolated via glutathione-Sepharose affinity chromatography; the fusion protein was cleaved with PreScission Protease, and CTD or BTD-CTD was purified to homogeneity using a combination of ion exchange and size exclusion chromatography.
All MINT constructs were cloned into a modified pET28b(+) vector that contains an N-terminal His6 tag and SMT3 (suppressor of Mif2 temperature-sensitive mutant 3) (28), resulting in His-SMT3-MINT fusion proteins. MINT constructs were transformed into bacteria, BL21(DE3) Tuner, and grown/overexpressed using autoinduction medium and methods (29). Bacteria were harvested by centrifugation, frozen, and lysed using a Niro Saovi Panda high pressure homogenizer. Bacterial lysates were cleared by centrifugation and incubated overnight in batch mode with nickel-nitrilotriacetic acid affinity resin. The lysate-bead slurry was poured into an empty gravity column and washed with buffer, and the fusion protein was eluted with 0.5 m imidazole, following the manufacturer's protocol. His-SMT3-MINT fusion proteins were cleaved using the protease Ulp-1, which leaves only an additional N-terminal serine residue attached to the MINT peptide. For MINT constructs that start with residues 2776 or 2752, serine is the native residue at these positions; however, for MINT constructs that encode different starting residues (e.g. 2791 or 2801), these peptides will have an additional non-native serine residue at their N termini. The MINT peptides were further purified using ion exchange and size exclusion chromatography. In some cases, fusion proteins were not cleaved but were purified to homogeneity in a similar manner to isolated MINT peptides. CSL and MINT peptides were flash-frozen in liquid nitrogen and stored at −80 °C.
Isothermal Titration Calorimetry
Isothermal titration calorimetry (ITC) experiments were performed using a MicroCal VP-ITC microcalorimeter. All standard experiments were carried out at 25 °C in a buffer consisting of 50 mm sodium phosphate, pH 6.5, and 150 mm NaCl. CSL and MINT peptides were degassed and buffer-matched using size exclusion chromatography. A typical experiment consisted of 10 μm CSL in the cell and 100 μm MINT in the syringe. For experiments that contained CSL bound to DNA, an oligomeric 19-mer duplex corresponding to the HES-1 binding site (-GTTACTGTGGGAAAGAAAG-) was used at a concentration 1.2× [CSL] in the cell. Protein concentrations were determined by both UV absorbance at 280 nm and BCA assay (Pierce). The data were analyzed using the ORIGIN software and fit to a one-site binding model.
Calculation of ΔCp Value for CTD-ANK Interface
To calculate a hypothetical ΔCp value corresponding to the interface between the CTD of CSL and the ANK domain of Notch, the CTD-ANK binary complex was extracted from the Protein Data Bank code 2F8X (human CSL-NICD-MAM-DNA complex crystal structure) (30). The amount of nonpolar and polar surface area buried at the CTD-ANK interface was determined by submitting the pseudo CTD-ANK coordinates to the GetArea server (31). ΔCp was calculated from the buried surface area using the equation given by Myers et al. (32). Comparable results were obtained using the CTD-ANK interface from the worm CSL-NICD-MAM-DNA complex crystal structure (33). A similar approach was successfully used by Johnson et al. (34) to calculate the ΔCp value for the BTD-RAM complex.
Circular Dichroism
CD measurements were taken in triplicate using an Aviv Circular Dichroism Spectrometer Model 215 at 25 °C in a 0.02-cm cuvette. Wavelength scans were performed between 190 and 290 nm using 1.0-nm increments. MINT, CSL, and NICD proteins were characterized in a buffer containing 50 mm sodium phosphate, pH 6.5, and 75 mm NaCl with protein concentrations ranging from 0.35 to 0.74 mg/ml (from 18 to 86 um). CD data were analyzed on Dichroweb using the CDSSTR analysis program and reference sets 7, 3, and 4 were used for MINT, CSL, and NICD data, respectively (35, 36).
Luciferase Reporter Assays
Mouse embryonic fibroblasts that were derived from CSL (rbp-j) knock-out embryos (MEFs; OT11) were kindly provided by Tasuku Honjo (37). The coding region for murine rbp-j, which corresponds to amino acids 1–526, and a C-terminal FLAG tag was PCR cloned into the MigR1 retroviral vector (38), using the BglII and EcoRI restriction sites. The CSL MigRI construct was cotransfected with pVSV-G into the retrovirus packaging cell line HEK GP2-293. Supernatants containing the retroviruses were collected 96 h post transfection and used to infect OT11 MEFs. Successful transduction was monitored by flow cytometry, which examined GFP expression from an internal ribosome entry site contained within MigR1, and immunoblots for the FLAG epitope. GFP-negative cells were removed from the population by fluorescence-activated cell sorting. CSL-transduced MEFs were maintained at 37 °C in 5% CO2 in Dulbecco's modified essential medium supplemented with 10% fetal bovine serum and penicillin-streptomycin.
For luciferase reporter experiments, MEFs were grown to ∼80% confluence in six-well plates and transiently transfected with a construct that expresses NICD1 (murine Notch1, residues 1744–2531) to activate signaling and the Notch responsive luciferase reporter 4xCBS, which contains four iterative CSL binding sites (39). The constructs were cotransfected with phRL, which expresses Renilla luciferase to normalize for transfection efficiency. MINT residues 2776–2833 were cloned into the plasmid pEGFP-C1 (Clontech) downstream of GFP, using the BamHI and XbaI restriction sites. To ensure nuclear localization of the GFP-MINT fusion protein, two nuclear localization sequences (DPKKRKV) were cloned into the XhoI and BamHI sites. The SatisFection (Stratagene) reagent was used for the transfections, following the manufacturers protocol, and the amount of transfected DNA was normalized using pBluescript (Stratagene). 48 h post-transfection, the cells were harvested and assayed for luciferase activity using the Dual-Luciferase kit (Promega). Firefly luciferase activity from the 4xCBS reporter was first normalized to Renilla luciferase expression and reported as relative activity by comparing cells transfected with and without GFP/GFP-MINT constructs. Average values, errors, and S.D. were determined from three independent experiments performed in duplicate.
RESULTS
Construct Rationale and Secondary Structure Analysis
Based on previous work by others (16, 17), we designed a construct of MINT, residues 2776–2833, which corresponds to a region that is highly conserved among all vertebrate MINT proteins and includes the residues identified by Oswald et al. (17) to be necessary for interacting with CSL (Fig. 2 and supplemental Fig. S1). The recombinant MINT (2776–2833) peptide, as well as variants of this construct, was isolated as a fusion protein from bacteria, cleaved from the fusion partner, purified to homogeneity by column chromatography, and verified by mass spectrometry. In some cases, certain isolated MINT peptides were insoluble in aqueous buffers, and therefore, were purified as fusion proteins for subsequent binding studies.
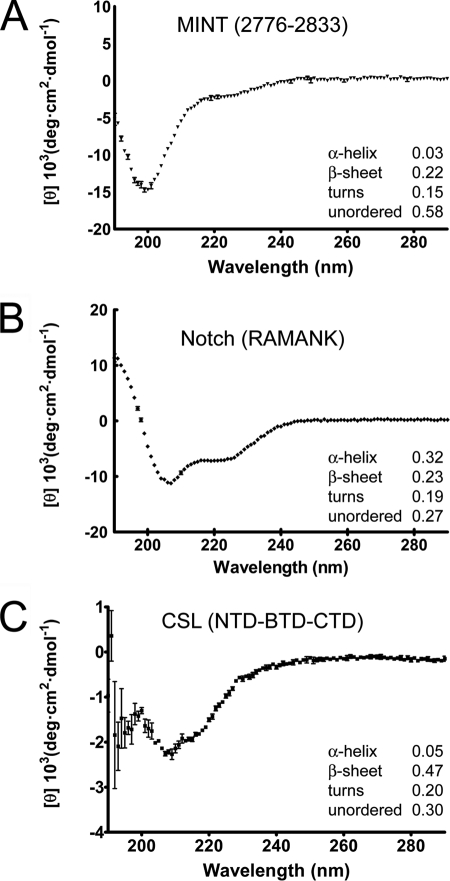
Secondary structure prediction programs suggest that the region of MINT corresponding to residues 2776–2833 is devoid of any secondary structure. To characterize the secondary structure content of MINT (2776–2833), as well as provide α-helical and β-strand protein samples for comparison, we used far-UV CD with purified samples of MINT, the RAMANK domains of Notch, and core CSL. Consistent with in silico analyses, the CD spectrum of MINT (2776–2833) has a distinct minima at ∼200 nm (Fig. 3A), which is characteristic of a random coil in solution. Consistent with previous reports (40–42), the CD spectra of CSL and RAMANK are consistent with these proteins being largely composed of β-sheet and α-helix, respectively (Fig. 3, B and C).
FIGURE 3.
Secondary structure analysis of CSL, NICD, and MINT. The figure shows far-UV circular dichroism spectroscopy for MINT (2776–2833), the RAMANK domains of Notch, and CSL (RBP-J) in A, B, and C, respectively. Relative amounts of secondary-structure were determined from CD data using Dichroweb and CDSSTR (35, 36). The normalized root mean square deviation parameter values for analysis of the MINT, RAMANK, and CSL CD data are 0.02, 0.04, and 0.11, respectively. A, the CD spectrum of Mint (2776–2833) has a distinct minimum at 199 nm, characteristic of random coil. B, consistent with its largely helical structure and previously published results (48), the CD spectrum of RAMANK shows characteristic minima for α-helix at 207 and 222 nm. C, the CD spectrum of CSL displays a minimum at 215 nm, consistent with it being largely composed of β-sheet (41, 51). Error bars, S.D.
Thermodynamics of CSL-MINT Interactions
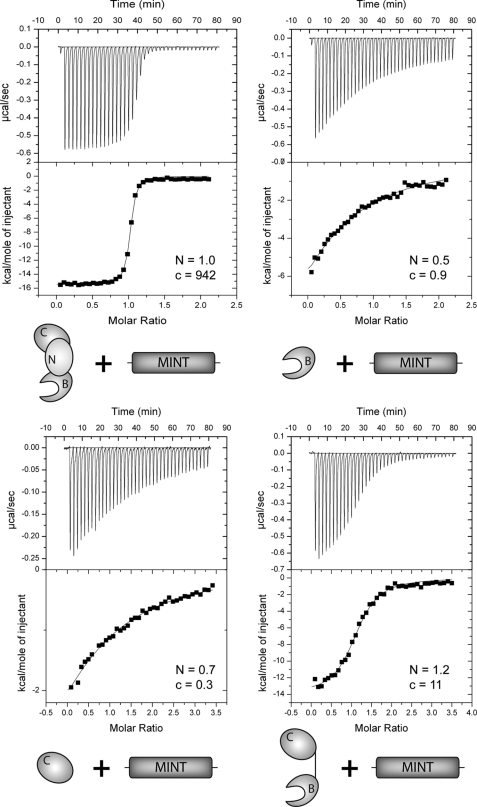
To characterize the equilibrium binding of complexes formed between MINT and CSL and define the thermodynamic parameters that underlie CSL-MINT interactions, we used ITC with our purified recombinant constructs of murine CSL and MINT. Unless otherwise noted, all experiments were carried out with CSL in the cell and MINT in the syringe (representative thermograms are displayed in Fig. 4). As shown in Table 1, MINT and CSL form a high affinity 1:1 complex that is enthalpically driven and characterized by a 11 nm Kd (dissociation constant) and a free energy of binding (ΔG°) of −10.9 kcal/mol. A control binding experiment was performed, in which CSL and MINT were placed in the syringe and cell, respectively, and as expected, similar overall affinities, thermodynamic parameters, and stoichiometries were obtained (supplemental Fig. S2). Titration of CSL prebound to a cognate DNA with MINT neither affected the overall free energy of binding nor the relative enthalpic or entropic contributions to complex formation (Table 1).
FIGURE 4.
CSL-MINT ITC binding assays. The figure shows representative thermograms, raw heat signal and nonlinear least squares fit to the integrated data, for CSL constructs (core, BTD, CTD, and BTD-CTD) interacting with MINT (2776–2833). Forty titrations were performed per experiment, consisting of 7-μl injections that were spaced 120 s apart. Average N (stoichiometry: ligand/macromolecule) and c (K[M]n) values for each subset of experiments are shown.
TABLE 1.
Calorimetric data for MINT binding to CSL
All experiments were performed at 25 °C. Values are the mean of at least three independent experiments, and errors represent the S.D. of multiple experiments.
| Macromolecule | MINT (ligand) | K | Kd | ΔG° | ΔH° | −TΔS° |
|---|---|---|---|---|---|---|
| m−1 | μm | kcal/mol | kcal/mol | kcal/mol | ||
| CSL | 2776–2833 | 9.4 ± 1.2 × 107 | 0.011 | −10.9 ± 0.1 | −14.6 ± 0.2 | 3.7 ± 0.2 |
| CSL + DNA | 2776–2833 | 7.3 ± 1.2 × 107 | 0.014 | −10.7 ± 0.1 | −14.8 ± 0.5 | 4.1 ± 0.6 |
| BTD | 2776–2833 | 2.5 ± 0.3 × 104 | 41 | −6.0 ± 0.1 | −14.9 ± 3.6 | 8.9 ± 3.5 |
| CTD | 2776–2833 | 1.7 ± 0.2 × 104 | 60 | −5.8 ± 0.1 | −7.7 ± 1.7 | 2.0 ± 1.9 |
| BTD-CTD | 2776–2833 | 1.0 ± 0.1 × 106 | 0.969 | −8.2 ± 0.1 | −14.2 ± 1.6 | 6.0 ± 1.7 |
Characterization of Interactions of BTD and CTD with MINT
Previous studies have implicated the BTD as the primary region of CSL that mediates interactions with transcriptional corepressors (21). We and others (27, 43) have also shown that the RAM domain of NICD forms a high affinity interaction with the BTD. To determine whether MINT also binds the BTD of CSL, we performed ITC binding studies using purified constructs of murine BTD with MINT (2776–2833). As shown in Table 1, MINT binds BTD; however, the interaction is at least a 1,000-fold weaker than what was observed for core CSL binding to MINT. Interestingly, the weaker binding is characterized by an approximate 5 kcal/mol entropic penalty, whereas the enthalpic contribution to complex formation is remarkably similar to what was observed for the core CSL-MINT (2776–2833) complex.
In light of these results, we sought to identify other domains of CSL that interact with MINT. Although constructs of NTD and NTD-BTD displayed poor solution properties under our binding conditions, precluding analysis by ITC, we were able to purify CTD and BTD-CTD constructs of CSL. As shown in Table 1, CTD also binds MINT (2776–2833), albeit weakly, with 60 μm affinity, similar to what was observed for BTD-MINT complexes. However, the enthalpic/entropic contributions to CTD-MINT binding differ from what was observed for BTD-MINT complexes (Table 1). Strikingly, the BTD-CTD construct of CSL bound MINT with ∼1 μm affinity, i.e. ∼50-fold tighter binding than what was observed for either BTD or CTD.
Delineating Regions of MINT That Bind BTD and CTD
We next sought to identify the regions of MINT (2776–2833) that interact with the BTD and CTD of CSL. Initially, we designed two additional constructs of MINT, (2776–2800) and (2801–2833), and pursued binding studies of these truncated peptides with CSL. The peptide that corresponds to MINT (2801–2833) was readily purified and, as shown in Table 2, binds the BTD with ∼10 μm affinity, comparable to the binding observed for MINT (2776–2833) to BTD. In contrast, no binding was detected with MINT (2801–2833) and CTD under any conditions/temperatures tested. In addition, MINT (2801–2833) binds core CSL with a similar overall affinity and free energy of binding, but with different enthalpic and entropic contributions to complex formation (Table 2). These data suggest that MINT residues 2801–2833 interact with the BTD of CSL.
TABLE 2.
Calorimetric data for MINT binding to the different domains of CSL
All experiments were performed at 25 °C. Values are the mean of at least three independent experiments, and errors represent the S.D. of multiple experiments. NBD, no binding detected.
| Macromolecule | MINT (ligand) | K | Kd | ΔG° | ΔH° | −TΔS° |
|---|---|---|---|---|---|---|
| m−1 | μm | kcal/mol | kcal/mol | kcal/mol | ||
| CSL | 2776–2800a | 2.5 ± 0.2 × 105 | 4.1 | −7.3 ± 0.1 | −4.9 ± 0.6 | −2.4 ± 0.6 |
| CSL | 2801–2833 | 1.1 ± 0.01 × 105 | 8.9 | −6.9 ± 0.01 | −11.3 ± 0.1 | 4.4 ± 0.2 |
| BTD | 2776–2800a | NBD | ||||
| BTD | 2801–2833 | 9.1 ± 3.2 × 104 | 12 | −6.7 ± 0.2 | −8.2 ± 1.6 | 1.4 ± 1.8 |
| CTD | 2776–2800a | 8.8 ± 2.5 × 103 | 121 | −5.4 ± 0.2 | −12.7 ± 3.2 | 7.4 ± 3.4 |
| CTD | 2801–2833 | NBD |
a Due to limited solubility in aqueous buffers, the peptide corresponding to MINT (2776–2800) was purified with the SMT3 fusion partner.
The purification of the peptide that corresponds to MINT (2776–2800) was problematic, as this peptide was neither soluble under our buffer conditions used to measure binding by ITC, nor was it soluble in other buffers that included DMSO. Alternatively, we purified the SMT3-MINT (2776–2800) fusion protein and determined its affinity for CTD, as well as for BTD and core CSL. SMT3-MINT (2776–2800) binds both CTD and core CSL weakly, with ∼100 μm and ∼4 μm affinity, respectively (Table 2). Complexes formed between CTD and SMT3-MINT (2776–2800) are characterized by a large and favorable enthalpic contribution to binding (∼13 kcal/mol), which is offset by a large and unfavorable entropic contribution to binding (∼7 kcal/mol). We were unable to detect any binding by ITC with the BTD of CSL and SMT3-MINT (2776–2800) (Table 2), suggesting that the residues that correspond to MINT (2776–2800) interact with the CTD.
Characterizing N- and C-terminal Truncations of MINT (2766–2833)
To further define the regions of MINT that are necessary and sufficient for interaction with CSL, we characterized the binding of several additional truncated MINT constructs with CSL. For these binding studies, we exclusively used SMT3-MINT fusion proteins for consistency and to avoid solubility issues with different peptide truncations. To control for the effects the SMT3 fusion partner may have on the binding reaction, we measured the binding of SMT3 and SMT3-MINT (2776–2833) with CSL. Although the isolated SMT3 protein does not bind CSL (data not shown), SMT3-MINT (2776–2833) binds core CSL with ∼8 nm affinity, which is comparable with the affinity obtained for the isolated MINT (2776–2833) peptide binding CSL (Tables 1 and 3). However, comparison of the enthalpic/entropic contributions with binding reveals differences as much as ∼4 kcal/mol with no clear trends with the different fusion proteins. In addition, for some of the peptide truncations, we noticed a moderate increase in the affinity of the SMT3 fusion protein for CSL when compared with the free peptide, e.g. MINT constructs (2776–2820) and (2801–2833).
TABLE 3.
Calorimetric data for SMT3-MINT fusion proteins binding to CSL
All experiments were performed at 25 °C. Values are the mean of at least three independent experiments, and errors represent the S.D. of multiple experiments.
| Macromolecule | SMT3-MINT (ligand) | K | Kd | ΔG° | ΔH° | −TΔS° |
|---|---|---|---|---|---|---|
| m−1 | μm | kcal/mol | kcal/mol | kcal/mol | ||
| CSL | 2776–2833 | 1.2 ± 0.02 × 108 | 0.008 | −11.0 ± 0.01 | −18.0 ± 0.6 | 7.0 ± 0.6 |
| CSL | 2776–2820 | 3.8 ± 1.1 × 108 | 0.003 | −11.7 ± 0.2 | −19.3 ± 0.5 | 7.6 ± 0.7 |
| CSL | 2776–2812 | 1.4 ± 0.5 × 108 | 0.008 | −11.1 ± 0.2 | −14.0 ± 1.1 | 2.9 ± 1.2 |
| CSL | 2791–2820 | 8.2 ± 1.8 × 104 | 13 | −6.7 ± 0.1 | −12.8 ± 1.9 | 6.1 ± 2.1 |
| CSL | 2791–2812 | 9.4 ± 1.3 × 104 | 11 | −6.8 ± 0.1 | −10.6 ± 1.3 | 3.8 ± 1.3 |
| CSL | 2752–2833a | 1.9 ± 0.1 × 108 | 0.005 | −11.3 ± 0.02 | −14.8 ± 1.0 | 3.5 ± 1.0 |
a The binding data reported for MINT (2752–2833) to CSL was done in the context of the free peptide and not the SMT3 fusion protein.
CD analysis of the corresponding MINT peptides and SMT3 fusion proteins did not reveal any significant differences in the secondary structure content of MINT when in the context of the SMT3 fusion protein (supplemental Fig. S3). However, we cannot formally exclude that a small change in secondary structure content, beyond the detection limits of our instrumentation, underlies the binding differences we observe for the free peptides and fusion proteins. A more complete comparison of the binding of MINT peptides and their corresponding SMT3-MINT fusion proteins with CSL are shown in supplemental Table S1. Taken together, this suggests that the SMT3-MINT fusion binding data is useful for qualitative comparisons; however, a detailed analysis of the enthalpic/entropic contributions to binding was not pursued due to the aforementioned caveats.
As shown in Table 3, the C-terminally truncated SMT3-MINT fusion constructs (2776–2820 and 2776–2812) retained comparable binding with CSL as the (2776–2833) MINT construct. Although the free MINT peptide (2776–2812) was insoluble, MINT (2776–2820) was soluble and bound CSL with similar affinity as (2776–2833) (supplemental Table S1). N-terminal truncations of our SMT3-MINT construct (2791–2820) and (2791–2812) resulted in a profound loss of affinity for CSL (Table 3). Analysis of a construct of MINT that was N-terminally extended by 24 residues (2752–2833) resulted in a free energy of binding that was indistinguishable from our original MINT (2776–2833) construct.
Heat Capacity Change Associated with CSL-MINT Complexation
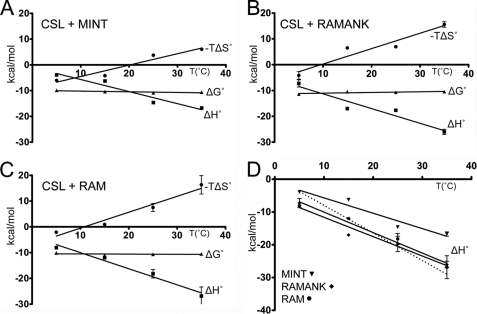
For the binding reactions of macromolecules, the change in heat capacity (ΔCp) largely correlates with the amount of nonpolar surface area buried upon complex formation, which can provide structural insights into the complex of interest (44–46). To determine the ΔCp associated with CSL-MINT interactions, we analyzed the enthalpy of binding as a function of temperature (5, 15, 25, and 35 °C; Table 4). In addition, because NICD competes with MINT for binding to CSL (16) and for comparative purposes, we also determined the ΔCp for the RAM and RAMANK domains of murine Notch1 binding to CSL. As shown in Fig. 5, the overall free energy of binding for CSL complexes with MINT, RAMANK, or RAM is independent of temperature. This is achieved by compensatory enthalpic and entropic contributions to binding at different temperatures. In all three cases, binding is enthalpically driven, which is consistent with previous calorimetric studies of CSL-NICD complexes (27, 43, 47). Moreover, there is excellent correspondence between our RAM and RAMANK thermodynamic data determined here, and our previously published results characterizing similar complexes (27). Interestingly, the thermodynamic profiles of RAM and RAMANK binding CSL are, within error, identical. The ΔCp values for MINT, RAMANK, and RAM binding to CSL are −0.47, −0.59, and −0.62 kcal/mol·K, respectively (Fig. 5D). In general, the ΔCp for these complexes is large and negative, which is indicative of the burial of significant amounts of nonpolar surface area upon complex formation and consistent with what is observed in the structures of CSL-RAM and CSL-NICD-MAM ternary complexes.
TABLE 4.
Temperature dependence of MINT, RAM, and RAMANK binding to CSL
Calorimetric data for the temperature dependence of MINT and RAM binding to CSL. For CSL-MINT and CSL-RAM binding experiments, CSL was placed in cells, and either MINT or RAM was placed in the syringe; however, for the CSL-RAMANK binding experiment, CSL and RAMANK were placed in the syringe and the cell, respectively. Values are the mean of at least three independent experiments, and the errors represent the S.D. of multiple experiments.
| Temperature | K | Kd | ΔG° | ΔH° | −TΔS° | |
|---|---|---|---|---|---|---|
| °C | m−1 | μm | kcal/mol | kcal/mol | kcal/mol | |
| CSL + MINT | 5 | 6.2 ± 1.4 × 107 | 0.017 | −9.9 ± 0.1 | −3.9 ± 0.4 | −6.0 ± 0.5 |
| 15 | 8.3 ± 2.3 × 107 | 0.013 | −10.4 ± 0.2 | −6.3 ± 0.3 | −4.2 ± 0.5 | |
| 25 | 9.4 ± 1.2 × 107 | 0.011 | −10.9 ± 0.1 | −14.6 ± 0.2 | 3.7 ± 0.2 | |
| 35 | 3.5 ± 0.6 × 107 | 0.029 | −10.6 ± 0.1 | −16.7 ± 0.7 | 6.1 ± 0.5 | |
| CSL + RAM | 5 | 1.1 ± 0.5 × 108 | 0.012 | −10.1 ± 0.3 | −8.1 ± 0.4 | −2.1 ± 0.4 |
| 15 | 2.5 ± 0.7 × 108 | 0.004 | −11.1 ± 0.2 | −12.0 ± 0.1 | 0.9 ± 0.2 | |
| 25 | 6.8 ± 0.8 × 107 | 0.015 | −10.7 ± 0.1 | −18.2 ± 1.7 | 7.5 ± 1.6 | |
| 35 | 2.9 ± 0.4 × 107 | 0.035 | −10.5 ± 0.1 | −26.8 ± 3.5 | 16.3 ± 3.6 | |
| CSL + RAMANK | 5 | 9.9 ± 4.4 × 108 | 0.001 | −11.4 ± 0.2 | −7.3 ± 1.4 | −4.2 ± 1.5 |
| 15 | 9.3 ± 1.6 × 107 | 0.011 | −10.5 ± 0.1 | −17.0 ± 0.1 | 6.5 ± 0.1 | |
| 25 | 7.2 ± 0.6 × 107 | 0.014 | −10.7 ± 0.1 | −17.7 ± 0.4 | 7.0 ± 0.3 | |
| 35 | 2.6 ± 0.2 × 107 | 0.039 | −10.4 ± 0.1 | −26.0 ± 1.0 | 15.6 ± 1.1 | |
FIGURE 5.
Thermodynamic profiles. The figure shows a summary of thermodynamic parameters (ΔG°, ΔH°, and −TΔS°) as a function of temperature (5, 15, 25, and 35 °C) for CSL interacting with the corepressor MINT (A), and the RAMANK (B), and RAM (C) domains of NICD. Over the temperature range examined, ΔG° is temperature-independent, highlighting the compensatory changes in enthalpy and entropy. D, enthalpies of binding for CSL complexes with MINT (triangles), RAMANK (diamonds), and RAM (circles). The ΔCp of binding for CSL-MINT, CSL-RAMANK, and CSL-RAM complexes is −0.47, −0.59, and −0.62 kcal/mol·K, respectively. The dashed line represents the expected additional contribution to ΔCp (−0.22 kcal/mol·K) for the CSL-RAMANK interaction based upon the buried surface area at the CTD-ANK interface.
MINT (2776–2833) Inhibits Notch Signaling in Cellular Transcription Assays
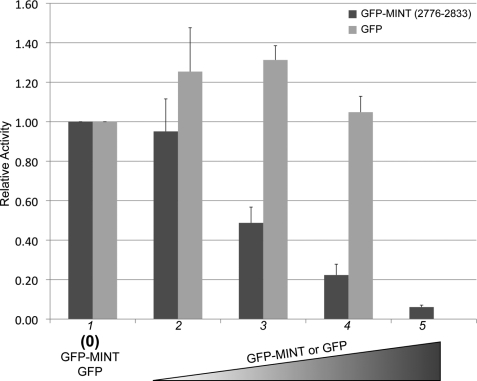
To determine whether MINT (2776–2833) can antagonize Notch signaling in cells, we performed luciferase reporter assays in MEFs that were derived from CSL-null embryos (OT11) (37). Retroviral transduction was used to express CSL in MEFs. MEFs were transiently transfected with the 4xCBS reporter, which contains four iterative CSL binding sites, and constructs that encode for an activated form of Notch1 (NICD1) and a GFP-MINT (2776–2833) fusion protein. A nuclear localization sequence was included in the GFP-MINT construct to ensure that the fusion protein was localized to the nucleus. As shown in Fig. 6 and reported similarly elsewhere (39), robust activation of the reporter is observed in cells transfected with NICD1. However, increasing amounts of transfected GFP-MINT DNA result in potent inhibition of activity from the reporter in a dose-dependent manner. Importantly, inhibition of the reporter was not observed in cells transfected with the control GFP vector. Additionally, similar results were obtained in wild-type MEFs (OT13) and HeLa cells (data not shown).
FIGURE 6.
MINT (2776–2833) competes with NICD for CSL binding in cells. Cultured MEFs were transiently transfected with an activated form of Notch1 (NICD1), the 4xCBS luciferase reporter, and increasing concentrations of GFP (light gray bars) or GFP-MINT (2776–2833) constructs (lanes 2–5). Transfection efficiency was normalized using a Renilla luciferase reporter construct. Robust activity is observed from the 4xCBS reporter in the absence of GFP or GFP-MINT (lane 1). However, increasing concentrations of GFP-MINT, but not the GFP control, result in strong inhibition from the reporter (lanes 2–5). Experiments were performed in duplicate in six-well plates with lanes 2, 3, 4, and 5 representing 25, 50, 100, and 250 ng of transfected GFP or GFP-MINT DNA, respectively. The y axis represents relative activity derived from normalizing the data to experiments performed in the absence of GFP or GFP-MINT (lane 1). Data are derived from three independent experiments (n = 3), and the error bars represent S.E.
DISCUSSION
The transcription factor CSL is the nuclear effector of the Notch pathway and is required for both the repression and activation of transcription from genes that are responsive to Notch signals (1). Previous structure-function studies have scrutinized the active transcription complex formed by CSL, NICD, and the transcriptional coactivator Mastermind (MAM), showing unequivocally that formation of this complex is the switch for activating transcription from Notch target genes (15). In light of these studies, however, much less is known regarding how CSL functions as a transcriptional repressor and how it is converted from a repressor to an activator. Although many corepressor proteins have been shown to interact with CSL and compete with NICD for CSL binding, the molecular details of these complexes are lacking. Given the widespread roles Notch signaling plays in biology and its therapeutic potential in treating many human diseases, most notably cancer, underscores the importance of a detailed understanding of these transcription complexes.
MINT is a large multidomain nuclear protein that has been shown to interact with CSL (16, 17), as well as other transcription factors and corepressor proteins, such as Msx2 (22), silencing mediator for retinoid and thyroid (48), and C-terminal binding protein (24). In vivo, MINT is an inhibitor of Notch signaling during lymphocyte development (49). Here, we used ITC to quantitatively define the thermodynamic parameters that underlie the complex formed between CSL and MINT. Our results define a highly conserved ∼44-residue region of MINT (2776–2820) that forms a high affinity 1:1 complex with CSL. Additional residues C-terminal to this region (2820–2833) contribute very little to binding (∼3-fold in Kd) and are conserved poorly; residues N-terminal to this region (2752–2776) do not contribute to binding and are not conserved. However, additional truncations of MINT (2776–2820) dramatically reduce complex formation. These data suggest that MINT residues 2776–2820 likely encompass the region that is necessary and sufficient for binding CSL. Interestingly, this region is only found in vertebrate MINT proteins, and therefore, it is unlikely that MINT orthologs in nematodes and flies function as repressors of the Notch pathway.
In addition, our CD analysis of MINT (2776–2833) suggests that in the absence of a binding partner MINT is largely unstructured in solution. Consistent with this result, and similar to other intrinsically disordered proteins (50), the binding of MINT to CSL is enthalpically driven and entropically unfavorable, which is likely due to the structural ordering of MINT upon complexation with CSL. Moreover, we demonstrate that MINT (2776–2833) is a strong inhibitor of Notch signaling in transcriptional reporter assays. These data suggest that in the absence of its other functional domains, MINT (2776–2833) can inhibit Notch signaling, potentially through a mechanism in which MINT and NICD directly compete for binding surfaces on CSL.
Our binding studies of individual domains of CSL with MINT suggest that the BTD and CTD interact with MINT. Although we were unable to characterize NTD constructs of CSL, a BTD-CTD construct recapitulates a substantial proportion of the CSL-MINT binding energy. A closer examination of the BTD-CTD/MINT thermodynamic parameters reveals a very similar enthalpy of binding, but an ∼2 kcal/mol additional entropic penalty, when compared with core CSL binding to MINT. This entropic penalty may be due to the additional degrees of freedom the BTD-CTD construct experiences in solution due to the absence of the NTD, which forms an extensive β-sheet with the strand between the BTD and CTD. This would suggest that the difference in affinity of BTD-CTD and core CSL for MINT is not due to additional contacts with the NTD but rather in vitro artifacts due to the protein constructs utilized. Moreover, given the number of MINT residues that are required for interacting with CSL (∼44), stereochemically, it is unlikely that these 44 residues could be interacting with BTD and CTD and simultaneously making contacts with the NTD; however, in the absence of a high resolution CSL-MINT structure, we cannot formally exclude this possibility.
In the context of previous work, what functional insights do our binding studies provide regarding the competitive binding of NICD and MINT for CSL and the interplay between these factors during transcriptional repression and activation? First, both MINT and NICD bind CSL with a remarkably similar overall affinity, and when present in excess, MINT can outcompete NICD for binding to CSL in cells. This suggests that the molecular mechanism by which NICD displaces/outcompetes MINT from CSL is not based solely on relative affinities, as it seems unlikely that nuclear concentrations of NICD would be significantly higher than MINT concentrations. Interestingly, the nuclear hormone receptor family of transcription factors also interacts with corepressors, such as silencing mediator for retinoid and thyroid, through short peptide-like regions (18). More recent models of transcriptional regulation by nuclear hormone receptors suggest that there is continuous cycling of corepressor and coactivator complexes on the promoter elements of active genes, which is required for transcriptional activation (18). Given the similar affinities of MINT and NICD for CSL, perhaps a similar cycling of corepressor/coactivator complexes occurs on Notch target genes. Nevertheless, it will be of future interest to complement our results obtained here with more quantitative studies that directly measure the competition of NICD and MINT for CSL, e.g. displacement ITC, and determine whether allostery or the presence of Mastermind makes NICD a more effective competitor for CSL.
Second, both MINT and NICD interact with the BTD and CTD of CSL. High resolution structures of CSL-NICD complexes reveal that the RAM and ANK domains of NICD interact with the BTD and CTD of CSL, respectively (27, 30, 33). In particular, a ∼15-residue segment of RAM binds the BTD of CSL with high affinity, primarily through a hydrophobic tetrapeptide motif (-LWFP- for Notch1); and ANK is a ∼31-kDa modular protein-protein interaction domain that in isolation interacts very weakly with CSL in some binding studies. The primary sequence of MINT bears no appreciable similarity to either RAM or ANK, and does not contain a hydrophobic tetrapeptide motif (supplemental Fig. S1). Nonetheless, our ΔCp analysis suggests that both RAM and MINT bury a significant amount of surface area upon complexation. This raises the question as to whether MINT and NICD utilize similar binding surfaces on CSL or does the CSL-MINT interface involve novel residues on BTD and CTD. Certainly, it will be of interest to determine whether mutations of CSL that abrogate NICD binding also affect interactions with MINT.
Third, it is interesting to note that although both NICD and MINT bind similar domains of CSL, the mode in which these proteins form high affinity complexes with CSL is strikingly different. The vast majority of the CSL-NICD binding energy is contributed by the BTD-RAM interaction, with CTD-ANK contributing very little binding energy to the overall CSL-NICD complex (27, 43, 52). In contrast, the individual complexes of MINT with BTD or CTD are of weak affinity, but when tethered together, the sum of these two weak interactions form a high affinity complex. As described originally by Jencks (53), this binding phenomena is known as the chelate effect. The chelate effect can be understood as the additional entropic penalty that is incurred for the two monovalent interactions, which is not experienced by the bivalent ligand, because the second binding event of the bivalent ligand does not incur the rotational and translational entropic cost associated with the first, thereby resulting in enhanced affinity. In the simple case, the overall free energies of binding for the two weak monovalent interactions would sum to give the free energy of binding for the bivalent ligand. However, this is not the case for CSL-MINT complexes (Table 1). The deviation from a “pure” chelate effect is likely due to unfavorable interactions that occur in the bivalent system that do not occur in the two monovalent binding events (53). Nonetheless, perhaps these two different modes of CSL-NICD and CSL-MINT binding are not coincidental, as the high affinity RAM-BTD interaction may be functionally important for NICD to ultimately dislodge MINT from CSL.
Fourth, it has been suggested that CSL-corepressor complexes may have lower affinity for DNA, which would account for the transient increase in CSL occupancy at target genes following Notch activation (54); however, our binding studies demonstrate that in the presence of a cognate DNA the affinity of CSL for MINT is unchanged. This suggests, due to the properties of linked equilibria (55), that MINT does not affect the affinity of CSL for DNA. Whether this is a common feature of all CSL-corepressor complexes remains to be determined.
Lastly, it should also be mentioned that our ΔCp analysis of RAM and RAMANK binding to CSL prompts a re-examination of the stepwise assembly of the transcriptionally active CSL-NICD-MAM ternary complex. The current model posits that RAM targets NICD to CSL via its high affinity interaction with BTD (15). Previously, we and others (27, 52) had measured a very small binding difference between RAM and RAMANK for CSL, with one group estimating a Kd of >20 μm for the CTD-ANK interaction; however, based on modeling RAM as a worm-like chain, it had been proposed that the local concentration of ANK, due to its tethering to CSL via RAM, would be in the low millimolar concentration range, which would offset the low affinity and promote CTD-ANK complexation (40). Thus, it was proposed that the RAMANK interaction with CSL would form a CTD-ANK interface very similar to what was observed in the structures of the CSL-NICD-MAM ternary complex, which was thought to be important for preforming the groove that MAM ultimately binds in the CSL-NICD binary complex.
Much to our surprise, comparison of the thermodynamic profiles for RAM and RAMANK binding CSL reveals that the two profiles are essentially identical. Importantly, the enthalpic (ΔH°) contribution to binding as a function of temperature is the same for RAM and RAMANK binding to CSL. This results in equivalent ΔCp values for CSL-RAM and CSL-RAMANK complexes, which is incompatible with the ∼880 Å2 of surface area that is buried between CTD-ANK, as observed in the human CSL-NICD-MAM ternary complex structure (30). Moreover, if we estimate the value of ΔCp for the CTD-ANK interface based solely on the amount of buried surface area, we obtain a value of −0.22 kcal/mol·K, which when combined with the −0.62 kcal/mol·K we measured for the CSL-RAM interaction, theoretically results in a ΔCp value of −0.84 kcal/mol·K for the CSL-RAMANK complex (Fig. 5D), a difference that is clearly larger than the error in the measurements. Although a more complicated explanation could be invoked, such as compensatory effects that coincidentally offset, resulting in equivalent ΔCp values, we suggest that the simplest explanation for the data is that the ANK domain of NICD does not interact with the CTD of CSL unless MAM is present.
One possibility to reconcile these results is that in the absence of MAM the CTD-ANK interactions only approximate the interface observed in the CSL-NICD-MAM structures, forming a subset of the interactions, which would be consistent with previous binding and mutational studies (27, 52). This step serves to additionally orient the CTD-ANK complex, which in conjunction with the increased local concentration of ANK due to the tethering of RAM to BTD, would be important for subsequent binding to MAM. Consistent with this idea is the cooperative binding of CSL-NICD-MAM dimers to paired binding sites, as typified by the HES1 promoter element (56, 57). Formation of the dimer on DNA is absolutely dependent on MAM inclusion in the complex, which likely points to its importance in consolidating the interface between CTD and ANK.
Supplementary Material
Acknowledgments
We thank the members of the Kovall laboratory for technical support and comments on the manuscript. We are grateful for the constructive criticism of binding data and manuscript provided by Andrew Herr.
This work was supported, in whole or in part, by National Institutes of Health Grant CA120199. This work was also supported by a Leukemia and Lymphoma Society award (to R. A. K.) and a predoctoral fellowship from the American Heart Association (to D. R. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1–S3.
- NICD
- Notch intracellular domain
- NTD
- N-terminal domain
- BTD
- β-trefoil domain
- CTD
- C-terminal domain
- RAM
- RBP-J-associated molecule
- ANK
- ankyrin repeats
- ITC
- isothermal titration calorimetry
- MAM
- Mastermind.
REFERENCES
- 1. Kopan R., Ilagan M. X. (2009) Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xue Y., Gao X., Lindsell C. E., Norton C. R., Chang B., Hicks C., Gendron-Maguire M., Rand E. B., Weinmaster G., Gridley T. (1999) Hum. Mol. Genet 8, 723–730 [DOI] [PubMed] [Google Scholar]
- 3. Jiang R., Lan Y., Chapman H. D., Shawber C., Norton C. R., Serreze D. V., Weinmaster G., Gridley T. (1998) Genes Dev. 12, 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oka C., Nakano T., Wakeham A., de la Pompa J. L., Mori C., Sakai T., Okazaki S., Kawaichi M., Shiota K., Mak T. W., Honjo T. (1995) Development 121, 3291–3301 [DOI] [PubMed] [Google Scholar]
- 5. Conlon R. A., Reaume A. G., Rossant J. (1995) Development 121, 1533–1545 [DOI] [PubMed] [Google Scholar]
- 6. Swiatek P. J., Lindsell C. E., del Amo F. F., Weinmaster G., Gridley T. (1994) Genes Dev. 8, 707–719 [DOI] [PubMed] [Google Scholar]
- 7. Bigas A., Robert-Moreno A., Espinosa L. (2010) Int. J. Dev. Biol. 54, 1175–1188 [DOI] [PubMed] [Google Scholar]
- 8. Chiba S. (2006) Stem Cells 24, 2437–2447 [DOI] [PubMed] [Google Scholar]
- 9. Gridley T. (2010) Curr. Top Dev. Biol. 92, 277–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koch U., Radtke F. (2010) Curr. Top Dev. Biol. 92, 411–455 [DOI] [PubMed] [Google Scholar]
- 11. MacGrogan D., Nus M., de la Pompa J. L. (2010) Curr. Top Dev. Biol. 92, 333–365 [DOI] [PubMed] [Google Scholar]
- 12. Gridley T. (2003) Hum. Mol. Genet 12, R9–13 [DOI] [PubMed] [Google Scholar]
- 13. Rizzo P., Osipo C., Foreman K., Golde T., Osborne B., Miele L. (2008) Oncogene 27, 5124–5131 [DOI] [PubMed] [Google Scholar]
- 14. Surendran K., Boyle S., Barak H., Kim M., Stomberski C., McCright B., Kopan R. (2010) Dev. Biol. 337, 386–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kovall R. A., Blacklow S. C. (2010) Curr. Top Dev. Biol. 92, 31–71 [DOI] [PubMed] [Google Scholar]
- 16. Kuroda K., Han H., Tani S., Tanigaki K., Tun T., Furukawa T., Taniguchi Y., Kurooka H., Hamada Y., Toyokuni S., Honjo T. (2003) Immunity 18, 301–312 [DOI] [PubMed] [Google Scholar]
- 17. Oswald F., Kostezka U., Astrahantseff K., Bourteele S., Dillinger K., Zechner U., Ludwig L., Wilda M., Hameister H., Knöchel W., Liptay S., Schmid R. M. (2002) EMBO J. 21, 5417–5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perissi V., Jepsen K., Glass C. K., Rosenfeld M. G. (2010) Nat. Rev. Genet 11, 109–123 [DOI] [PubMed] [Google Scholar]
- 19. Kao H. Y., Ordentlich P., Koyano-Nakagawa N., Tang Z., Downes M., Kintner C. R., Evans R. M., Kadesch T. (1998) Genes Dev. 12, 2269–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Waltzer L., Bourillot P. Y., Sergeant A., Manet E. (1995) Nucleic Acids Res. 23, 4939–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsieh J. J., Hayward S. D. (1995) Science 268, 560–563 [DOI] [PubMed] [Google Scholar]
- 22. Newberry E. P., Latifi T., Towler D. A. (1999) Biochemistry 38, 10678–10690 [DOI] [PubMed] [Google Scholar]
- 23. Tsuji M., Shinkura R., Kuroda K., Yabe D., Honjo T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oswald F., Winkler M., Cao Y., Astrahantseff K., Bourteele S., Knöchel W., Borggrefe T. (2005) Mol. Cell Biol. 25, 10379–10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Salat D., Liefke R., Wiedenmann J., Borggrefe T., Oswald F. (2008) Mol. Cell Biol. 28, 3502–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ariyoshi M., Schwabe J. W. (2003) Genes Dev. 17, 1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Friedmann D. R., Wilson J. J., Kovall R. A. (2008) J. Biol. Chem. 283, 14781–14791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mossessova E., Lima C. D. (2000) Mol. Cell 5, 865–876 [DOI] [PubMed] [Google Scholar]
- 29. Studier F. W. (2005) Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 30. Nam Y., Sliz P., Song L., Aster J. C., Blacklow S. C. (2006) Cell 124, 973–983 [DOI] [PubMed] [Google Scholar]
- 31. Fraczkiewicz R., Braun W. (1998) J. Comput. Chem. 19, 319–333 [Google Scholar]
- 32. Myers J. K., Pace C. N., Scholtz J. M. (1995) Protein Sci. 4, 2138–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilson J. J., Kovall R. A. (2006) Cell 124, 985–996 [DOI] [PubMed] [Google Scholar]
- 34. Johnson S. E., Ilagan M. X., Kopan R., Barrick D. (2010) J. Biol. Chem. 285, 6681–6692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sreerama N., Woody R. W. (2000) Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 36. Whitmore L., Wallace B. A. (2004) Nucleic Acids Res. 32, W668–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kato H., Taniguchi Y., Kurooka H., Minoguchi S., Sakai T., Nomura-Okazaki S., Tamura K., Honjo T. (1997) Development 124, 4133–4141 [DOI] [PubMed] [Google Scholar]
- 38. Pear W. S., Miller J. P., Xu L., Pui J. C., Soffer B., Quackenbush R. C., Pendergast A. M., Bronson R., Aster J. C., Scott M. L., Baltimore D. (1998) Blood 92, 3780–3792 [PubMed] [Google Scholar]
- 39. Ong C. T., Cheng H. T., Chang L. W., Ohtsuka T., Kageyama R., Stormo G. D., Kopan R. (2006) J. Biol. Chem. 281, 5106–5119 [DOI] [PubMed] [Google Scholar]
- 40. Bertagna A., Toptygin D., Brand L., Barrick D. (2008) Biochem. Soc. Trans. 36, 157–166 [DOI] [PubMed] [Google Scholar]
- 41. Nam Y., Weng A. P., Aster J. C., Blacklow S. C. (2003) J. Biol. Chem. 278, 21232–21239 [DOI] [PubMed] [Google Scholar]
- 42. Zweifel M. E., Barrick D. (2001) Biochemistry 40, 14357–14367 [DOI] [PubMed] [Google Scholar]
- 43. Lubman O. Y., Ilagan M. X., Kopan R., Barrick D. (2007) J. Mol. Biol. 365, 577–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spolar R. S., Livingstone J. R., Record M. T., Jr. (1992) Biochemistry 31, 3947–3955 [DOI] [PubMed] [Google Scholar]
- 45. Livingstone J. R., Spolar R. S., Record M. T., Jr. (1991) Biochemistry 30, 4237–4244 [DOI] [PubMed] [Google Scholar]
- 46. Sturtevant J. M. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 2236–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Johnson S. E., Ilagan M. X., Kopan R., Barrick D. (2010) J. Biol. Chem. 285, 6681–6692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zweifel M. E., Barrick D. (2001) Biochemistry 40, 14344–14356 [DOI] [PubMed] [Google Scholar]
- 49. Tanigaki K., Honjo T. (2007) Nat. Immunol. 8, 451–456 [DOI] [PubMed] [Google Scholar]
- 50. Dyson H. J., Wright P. E. (2005) Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 51. Kovall R. A., Hendrickson W. A. (2004) EMBO J. 23, 3441–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Del Bianco C., Aster J. C., Blacklow S. C. (2008) J. Mol. Biol. 376, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jencks W. P. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 4046–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krejcí A., Bray S. (2007) Genes Dev. 21, 1322–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wyman J., Gill S. J. (1990) Binding and Linkage: Functional Chemistry of Biological Macromolecules, University Science Books, Mill Valley, CA [Google Scholar]
- 56. Nam Y., Sliz P., Pear W. S., Aster J. C., Blacklow S. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2103–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Arnett K. L., Hass M., McArthur D. G., Ilagan M. X., Aster J. C., Kopan R., Blacklow S. C. (2010) Nat. Struct. Mol. Biol. 17, 1312–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.