Abstract
Aristaless related homeodomain protein (Arx) specifies the formation of the pancreatic islet α-cell during development. This cell type produces glucagon, a major counteracting hormone to insulin in regulating glucose homeostasis in adults. However, little is known about the factors that regulate Arx transcription in the pancreas. In this study, we showed that the number of Arx+ cells was significantly reduced in the pancreata of embryos deficient for the Islet-1 (Isl-1) transcription factor, which was also supported by the reduction in Arx mRNA levels. Chromatin immunoprecipitation analysis localized Isl-1 activator binding sites within two highly conserved noncoding regulatory regions (Re) in the Arx locus, termed Re1 (+5.6 to +6.1 kb) and Re2 (+23.6 to +24 kb). Using cell line-based transfection assays, we demonstrated that a Re1- and Re2-driven reporter was selectively activated in islet α-cells, a process mediated by Isl-1 in overexpression, knockdown, and site-directed mutation experiments. Moreover, Arx mRNA levels were up-regulated in islet α-cells upon Isl-1 overexpression in vivo. Isl-1 represents the first known activator of Arx transcription in α-cells, here established to be acting through the conserved Re1 and Re2 control domains.
Keywords: Development, Diabetes, Differentiation, Gene Regulation, Transcription Factors, Transcription Regulation, Glucagon
Introduction
Pancreatic islets play an important role in regulating carbohydrate metabolism through the production and secretion of hormones. The five cell types found in the islet are α-, β-, δ-, ϵ-, and pancreatic polypeptide cells that, respectively, produce the hormones glucagon, insulin, somatostatin, ghrelin, and pancreatic polypeptide (1). The hormonal products of the predominant α- and β-cells act in peripheral tissues in a counter-regulatory manner to control blood glucose homeostasis, with insulin promoting cellular glucose uptake and storage and glucagon promoting its release. Notably, although insulin resistance and β-cell dysfunction are the major causes of diabetes, the sustained, unregulated secretion of glucagon from α-cells also contributes to hyperglycemia and the associated complications (2–6). In fact, suppression of glucagon activity or levels has been shown to be a promising treatment for diabetics (3, 6–9). Unfortunately, and in contrast to islet β-cells, our understanding of the factors involved in controlling α-cell differentiation and function is quite limited (1, 10).
Glucagon+ cells first appear at embryonic day (E) 9.53 in mice, followed by insulin+ cells a day later (1). At around E13.5, a major expansion of a distinct population of hormone-producing cells begins to occur, with only these cells becoming mature islet α- and β-cells (11). A variety of distinct transcription factors are required during development for their production (1), including those that are necessary early in specifying the α- (e.g. Arx) and β- (e.g. Pax4 and Nkx6.1) lineages and others acting later in cell maintenance and maturation (e.g. MafB, Foxa2, Isl-1, and Pdx1) (12–20).
Arx plays an essential role in islet α-cell formation. Expression of this transcription factor is first detected in the pancreatic anlage at E9.5, then in differentiating pancreatic endocrine precursors, and subsequently in islet α-cells (15). Arx-null or pancreas-specific Arx-deficient mice display a complete loss of α-cells and a proportional increase in the number of β- and δ-cells (16, 21). Such changes are opposite to those found in Pax4-null mice, a transcription factor enriched in β-cell progenitors and islet β-cells (16, 20). Importantly, Pax4 and Arx function in an antagonistic manner and are both critical in the specification of these islet cell types during pancreatic development (15). For example, Arx misexpression in the pancreatic epithelium specifically converted β-cells into α- and pancreatic polypeptide cells, whereas Pax4 produced more β-cells at the expense of the α-cell population (22, 23). Significantly, as islet α-cells were also lost in humans with an ARX-null mutation (24), the collective evidence strongly suggests that this transcription factor serves as a master regulator of mammalian islet α-cell formation.
Thus far, the only established regulator of Arx transcription in the pancreas is Pax4, which inhibits expression through binding to a cis-acting element located ∼14.2 kb downstream of the Arx translation stop site (15). Recently, we have shown that removal of the Lin 1/Islet 1/Mec3-homeodomain Islet-1 (Isl-1) transcription factor in the pancreatic epithelium reduced both α- and β-cell numbers as well as key transcriptional regulators associated with their identity; specifically, MafA and Arx in β- and α-cells, respectively (17). These observations suggested that Isl-1 might be a good candidate for their regulation. The goal of this study was to investigate whether Arx is directly regulated by Isl-1 in α-cells. We identified two highly conserved regulatory regions (Re) within the Arx locus, termed Re1 and Re2, that are bound by Isl-1 and promote expression selectively in α-cells. Furthermore, we provide evidence that Isl-1 activates Arx transcription using a combination of in vitro and in vivo based loss-of-function and gain-of-function experiments. These results establish Isl-1 as a key activator of Arx transcription during α-cell formation and potentially also as a critical regulator for its regulation in adults.
EXPERIMENTAL PROCEDURES
Isl-1 Chromatin Immunoprecipitation Assays
Approximately 800 to 1000 islets were isolated from five 8-week-old CD1 male mice using the standard collagenase procedure as described previously (25). The islets were washed in PBS, cross-linked in 1.1% formaldehyde, and sonicated in buffer containing 50 mm Tris-HCl (pH8.1), 10 mm EDTA, 0.05% SDS, and complete protease inhibitor mixture (Roche). αTC6 (26) and βTC3 tissue-cultured cells were cross-linked under similar conditions, lysed in hypotonic buffer (50 mm Tris-HCl, 0.5% Nonidet P-40, 85 mm KCl with protease inhibitor mixture (Roche)), and sonicated. α-Isl-1 antibody (Developmental Studies Hybridoma Bank at the University of Iowa, 39.4D5) was added to the sonicated chromatin, and Isl-1 bound chromatin was isolated by incubation with protein A-agarose beads and eluted in buffer containing 1% SDS and 0.1 m Na2CO3. The immunoprecipitated DNA was purified using a PCR purification kit (Qiagen) and analyzed by either ChIP-Seq or real-time PCR analysis. ChIP PCR primer sequences are as follows: ArxRe1, 5′CCATTTGAAGGCAAAATGCT and 5′GTATGGGCTGCAAACACCTT; ArxRe2, 5′TGAAGTGGCTGAATGAGAGC and 5′AGTTGGAGCGCGTTTTGTAG; and PEPCK, 5′CAACAGGCAGGGTCAAAGTTTAG and 5′AGGCCTCAGGCCCCTCTAT.
Whole-genome ChIP-Seq Analysis
The purified immunoprecipitated DNA was prepared for next-generation high-throughput sequencing using the ChIP-Seq DNA sample prep kit (Illumina, San Diego, CA) following the manufacturer's instructions. The modified product was assayed for quantity and quality on an Agilent 2100 bioanalyzer (Agilent Tech). Sequencing and data processing were performed at the Diabetes Endocrinology Research Center. Functional Genomics Core at University of Pennsylvania. Isl-1-bound regions designated by peaks were identified using the Global Identifier of Target Regions algorithm (27).
Real-time PCR Analysis
RNA was isolated from islets or tissue-cultured cells using the RNAeasy kit (Qiagen), and cDNA was generated using Oligo(dT), Superscript II reverse transcriptase, plus the accompanying reagents (Invitrogen). Real-time PCR reactions were set up using the Brilliant SYBR Green PCR Master Mix (Stratagene). All reactions were performed in triplicate with reference dye for normalization. PCR primer sequences are as follows: Arx, 5′TCAAGCATAGCCGCGCTGAG and 5′ACACCTCCTTCCCCGTGCTG and Isl-1, 5′CGGAGAGACATGATGGTGGTT and 5′GGGCTGATCTATGTCGCTTTGC.
Luciferase Vector Construction and Reporter Assays
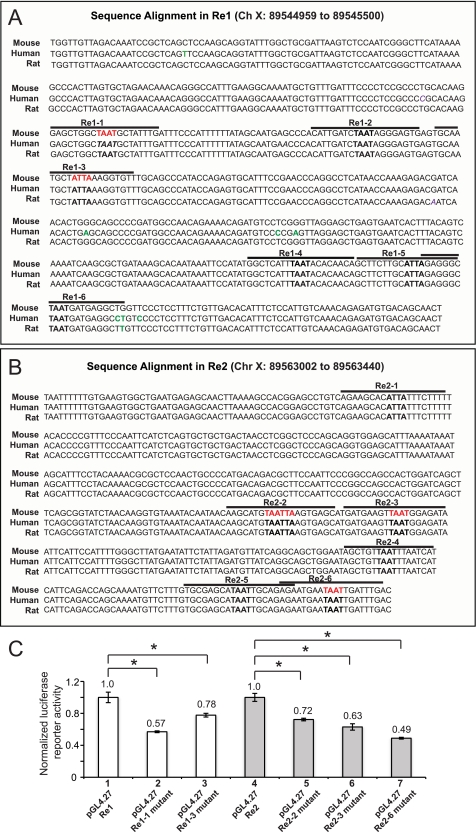
Arx Re1, and Re2 were cloned into the pGL4.27 luciferase vector (Promega). Site-directed mutants were generated with the QuikChange mutagenesis kit (Stratagene) to create Isl-1 binding site mutations (underlined). The corresponding wild-type nucleotide sequences for these sites are labeled in red in Fig. 5, A and B: Re1–1, 5′CCGCCCTGCACAAGGAGCTGGCGCCGGCTATTTGATTTCC; Re1–3, 5′GGGAGTGAGTGCAATGCTGCCGAAGGTGTTTGCAGCCC; Re2–2, 5′CAATAACAAGCATGGCCGTAAGTGAGCATGATGAAGTTAATGG; Re2–3, 5′GTGAGCATGATGAAGTGCCGGGAGATAATTCATTCC; and Re2–6, 5′GCATAATTGCAGAGAATGAAGCCGTGATTTGACAATTGTACCAG.
FIGURE 5.
The Isl-1 binding sites in Re1 and Re2 are necessary for αTC1–6 cell activity. Sequence identity within Arx Re1 (A) and Arx Re2 (B) between mouse, human, and rat. Core Isl-1 binding site sequences are shown in bold, and the lines above demarcate the EMSA probe. Nucleotide mutated for luciferase reporter (C) and EMSA (Fig. 6C) assays are labeled in red. Nonconserved nucleotides are labeled in green. C, the activity of wild-type and Isl-1 binding site mutants (red) of pGL4.27-Re1 and pGL4.27-Re2 in αTC1–6 cells. The pRL-SV40 normalized data were presented as the mean ± S.E. *, p <0.05.
αTC1–6 cells were cultured in 12-well plates (1 × 105 cells/well) and transfected with the Arx-driven expression vectors as well as the Renilla luciferase control expression plasmid (pRL-SV40) using Lipofectamine 2000 reagent (Invitrogen). Reporter assays were performed using the dual-luciferase reporter assay system (Promega) following the manufacturer's instructions. Arx-luciferase activity was normalized to that of Renilla luciferase.
Lentiviral Transduction
Lentiviral transduction particles (MISSION® shRNA, Sigma) targeting Isl-1 were applied to cultured αTC1–6 cells according to the manufacturer's instructions (multiplicity of infection = 5). Infected cells were selected for puromycin (5 μg/ml) resistance for 1 week before analysis. The Isl-1 shRNA sequence is as follows: 5′CCGGCGGCAATCAAATTCACGACCACTCGAGTGGTCGTGAATTTGATTGCCGTTTTTG.
Western Blotting
Whole cell lysate from control and Isl-1 shRNA transduced αTC1–6 cells as well as isolated islets (equivalent of ∼200 islets) were loaded and resolved on 10% Bis-Tris NuPAGE® Novex® mini gels (Invitrogen). The antibodies used for the Western blot analysis were Isl-1 (1:1000, 39.4D5, Developmental Studies Hybridoma Bank at the University of Iowa), tubulin (1:5000, Sigma), and Myc (1:1000, Cell Signaling Technology).
Immunofluorescence
Tissues were fixed in 4% paraformaldehyde overnight at 4 °C and then embedded in paraffin. Slides (8–10-μm sections) were subjected to microwave antigen retrieval in 10 mm citric acid buffer (pH 6.0) and treated with protein blocking reagent (Immunotech). Slides were incubated with primary antibodies overnight at 4 °C and secondary antibodies for 2 h at room temperature. Primary antibodies used were insulin (1:1000, Linco), glucagon (1:3000, Linco), Myc (1:100, Cell Signaling Technology), and Arx (1:2000, gift from Dr. Kitamura at Kyushu University, Japan).
EMSA
DNA binding reactions (20 μl total volume) included 10 μg of αTC1–6 nuclear extract (28) or 2 μl of in vitro translated Isl-1-Myc protein (17) plus 400 fmol of 32P end-labeled DNA oligo probe. Reactions were incubated at 4 °C in a buffer containing 10 mm HEPES (pH 7.9), 75 mm KCL, 2.5 mm MgCl2, 0.1 mm EDTA, 1 mm dithiothreitol, 3% (v/v) Ficoll, 1 mg/ml BSA, 500 ng poly(dI-dC), and 500 ng of poly(dA-dT). Competition experiments were performed using a 100-fold molar excess of the unlabeled oligonucleotide. α-Isl-1 (Developmental Studies Hybridoma Bank at the University of Iowa, 40.2D6) or α-Myc antibody (SC-40, Santa Cruz Biotechnology) was incubated with the nuclear extract for 10 min on ice prior to probe addition in the supershift assays. Reaction products were separated on 6% non-denaturing acrylamide gels in 0.5× TBE (89 mm Tris base, 89 mm boric acid, and 2 mm EDTA) buffer, and complexes were visualized by autoradiography. The double-stranded oligonucleotide sequences are as follows, with the putative homeodomain (HD) binding elements in bold lettering: Re1-1wild-type, 5′GAGCTGGCTAATGCTATTTG; Re1-2wild-type, 5′CATTGATCTAATAGGGAGTG; Re1-3wild-type, 5′GCAATGCTATTAAAGGTGTT; Re1-4wild-type, 5′GGCTCATTTAATACACAACA; Re1-5wild-type, 5′GCTTCTTGCATTAGAGGGC; Re1-6wild-type: 5′GAGGGCTAATGATGAGGCTG; Re2–1wild-type, 5′AGAAGCACATTATTTCTTTT; Re2–2wild-type, 5′AAGCATGTAATTAAGTGAGC; Re2–2mutant, 5′AAGCATGGCCGGCAGTGAGC (mutation is underlined); Re2–3wild-type, 5′GATGAAGTTAATGGAGATAC; Re2–3mutant, 5′GATGAAGTGCCGGGAGATAC (mutation is underlined); Re2–4wild-type, 5′AGCTGTTAATTTAATCATCAT; Re2–5wild-type, 5′TGCGAGCATAATTGCAGAGA; Re2–6wild-type, 5′AGAATGAATAATTGATTTGA; and Re2–6mutant, 5′AGAATGAAGCCGTGATTTGA (mutation is underlined).
Transgene Construction and Generation of Pdx1PB-Isl-1-Myc Mice
The Hnf6 cDNA (from Pdx1PB-Hnf6, gift from M. Gannon, 29) was replaced by rat Isl-1-Myc (gift from Sam Pfaff). DNA was SalI-digested, and the entire Pdx1PB-Isl-1-Myc transgene was injected into the pronuclei of C57BL6J embryos at the Children's Hospital of Philadelphia Transgenic Core Facility. Founder mice were identified by PCR analysis with the following genotyping primers: 5′CCCCTCTGCTAACCATGTTC and 5′TACCGCAACCAACACATAGG.
RESULTS
Arx+Cell Numbers Were Reduced in Isl-1-deficient Pancreata
Mice with pancreas-specific removal of Isl-1 were generated in our lab previously to investigate the requirement of Isl-1 during pancreas development (17). Arx mRNA levels were found previously to be decreased in the embryonic Pdx1-Cre;Isl-1LoxP/LoxP pancreata, conditions wherein removal of Isl-1 in pancreatic epithelium at E13.5 also resulted in reduced numbers of insulin+ and glucagon+ cells (17). In contrast, the steady-state levels of all other analyzed islet-enriched transcriptional regulators were unaffected, with the exception of MafA in insulin+ cells (17). Immunostaining for glucagon and Arx was performed with E15.5 control and Pdx1-Cre;Isl-1LoxP/LoxP pancreata to examine whether Arx+ cells were affected in the mutant (Fig. 1, A-D). Pax6 staining was performed on neighboring sections and used as an internal control for quantitative analysis because the mRNA expression level and number of Pax6+ cells were unaltered in the E15.5 Pdx1-Cre;Isl-1LoxP/LoxP pancreas (17).
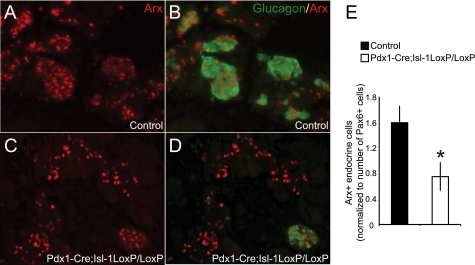
FIGURE 1.
The number of Arx+ cells is reduced in the Pdx1-Cre;Isl- 1LoxP/LoxP embryos. A–D, immunostaining with glucagon and Arx antibodies of E15.5 pancreatic sections from control and Pdx1-Cre;Isl-1LoxP/LoxP embryos. E, quantitative analysis of Arx+ cell populations as normalized to Pax6+ cells in the neighboring section. n = 6 for control and n = 5 for mutants. Data represents the mean ± S.E. *, p value <0.05.
From this analysis, we found that the total number of Arx+ cells was reduced by ∼50% in the Pdx1-Cre;Isl-1LoxP/LoxP pancreata (Fig. 1E), with the few remaining glucagon+ cells coexpressing Arx (Fig. 1D). Unlike the control pancreas, most of the Arx+ cells in the mutant pancreas did not produce glucagon, likely representing the significance of Isl-1 in activating glucagon production (17). Here we have focused on understanding how Isl-1 affects Arx mRNA expression in islet α-cells.
Transgenic Isl-1 Expression Increased Arx Expression in Mice
To further examine Isl-1 regulation in islet α-cell gene expression in vivo, a transgenic line was derived that expressed rat Isl-1-Myc from the Pst1 to Bst1 region of the mouse pdx-1 promoter, termed Pdx1PB-Isl-1-Myc (Fig. 2A). The transgenic activity of this pdx-1 control region is first observed at around E13.5 and is essential only in islet α, β, and δ cells throughout adulthood (29, 30). Isl-1 mRNA and protein levels were increased ∼2-fold in 8-week old Pdx1PB-Isl-1-Myc mice relative to age-matched controls (Fig. 2, B and C). Double immunostaining in islets for Myc and glucagon or insulin revealed that Isl-1-Myc was produced in 4% of glucagon+ and 32% insulin+ cells (Fig. 2, D–G). This pdx-1 fragment has been shown previously to drive stronger transgene expression in islet β-cells than α-cells (29).
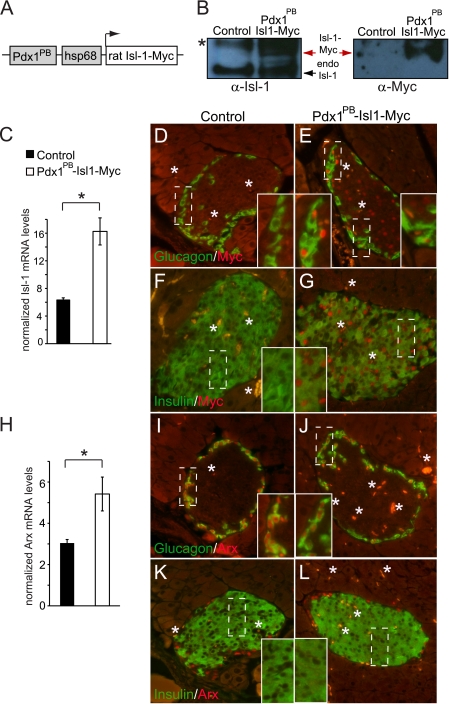
FIGURE 2.
Arx expression is elevated Pdx1PB-Isl-1-Myc mice. A, diagram representing the Pdx1PB-Isl-1-Myc transgenic construct, which contains the pdx1PB fragment of the pdx1 gene and the hsp68 minimal promoter. B, Western blot by α-Isl-1 and α-Myc analysis of total cell lysate from islets of 8-week-old control and Pdx1PB-Isl-1-Myc mice. The asterisk denotes a nonspecific band. C, Isl-1 mRNA levels in Pdx1PB-Isl-1-Myc and littermate control mice. Results are presented as mean ± S.E. *, p <0.05. D–G, immunohistochemical analysis for glucagon or insulin and Myc expression in 8-week-old Pdx1PB-Isl-1-Myc and control mice. H, Arx mRNA levels in Pdx1PB-Isl-1-Myc and control mice. Results are presented as mean ± S.E. *, p <0.05. I–L, costaining of Arx with glucagon or insulin in 8-week-old Pdx1PB-Isl-1-Myc and control mice. Insets show the magnified views of the outlined areas. The asterisk denotes autofluorescence from red blood cells.
Pdx1PB-Isl-1-Myc mice appeared indistinguishable from their control littermates at 8 weeks of age. In addition, we did not detect any gross abnormalities in the overall appearance of the pancreas. Immunohistochemical analysis of Pdx1PB-Isl-1-Myc pancreata showed no changes in islet morphology, size, or the ratio between glucagon- and insulin-expressing cells (Fig. 2, D–L). Furthermore, mRNA levels of glucagon and insulin were also comparable between Pdx1PB-Isl-1-Myc and control littermates (data not shown).
Interestingly, Isl-1-Myc overexpression elevated endogenous Arx mRNA levels by roughly 2-fold (Fig. 2H). Notably, Arx protein staining was only found in Pdx1PB-Isl-1-Myc α-cells and not the insulin+ β-cell population (Fig. 2, I and J). This in vivo result further suggested that Isl-1 stimulated Arx expression during α-cell development.
Isl-1 Binding Sequences Are Located in the Arx 3′-Noncoding Region
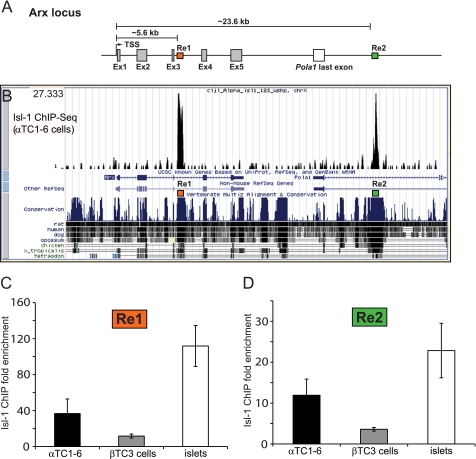
Isl-1 binding regions were identified in the Arx locus by whole-genome ChIP-Seq of α-Isl-1 antibody precipitates from nuclei of αTC1–6 cells, a transformed α-cell line (26). Binding was observed within two distinct conserved noncoding regions, one located ∼5.6 kb downstream from the transcription start site within intron 3 (termed Region 1: Re1, +5.6 to +6.1 kb) and the other ∼18.kb kb further away, within the last intron of Pola1 (Region 2: Re2, +23.6 to +24 kb) (Fig. 3A and B).
FIGURE 3.
Isl-1 binds to Re1 and Re2 of the mouse Arx gene. A, schematic diagram of the Arx locus illustrating the locations of Re1 (orange box, +5.6 to +6.1) and Re2 (green box, +23.6 to +24 kb). TSS, transcription start site; Ex, exon. B, the ChIP-Seq image depicts Isl-1 occupancy within Arx Re1 (chrX:89544959–89545500) and Re2 (chrX:89563002–89563440) in αTC1–6 cells. The conservation between mouse and other species within this region was shown using the University of California Santa Cruz genome browser. C and D, Re1 and Re2 binding to Isl-1 was readily detected in adult mouse islets and αTC1–6 cells by standard ChIP but to a lesser extent in βTC3 cells. Results were normalized to Isl-1 binding to the PEPCK promoter. Data are presented as the mean ± S.E. n = 3; p value <0.05.
Standard α-Isl-1 ChIP assays were performed over Re1 and Re2 in αTC1–6 cells, freshly isolated mouse islets, and βTC-3 (mouse insulinoma) cells to examine the specificity of binding (Fig. 3, C and D). These real-time PCR results were normalized to phosphoenolpyruvate carboxykinase promoter binding, an inactive gene in the pancreas. Isl-1 bound much more effectively to Re1 and Re2 in αTC1–6 than βTC-3 cells (Fig. 3, C and D). These data strongly suggested that the Isl-1 binding observed in islets is principally in α-cells and potentially important to Arx gene transcription.
Isl-1 Stimulated Re1- and Re2-directed Transcription in α-Cells
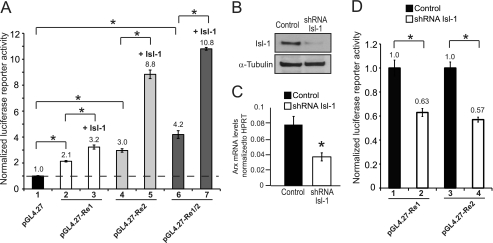
Re1, Re2, and Re1/Re2 combined were subcloned upstream of the minimal RNA polymerase II promoter in the pGL4.27 luciferase reporter vector, and their activation properties were analyzed in transfected αTC1–6 cells. All of these pGL4.27-Arx constructs were more active than pGL4.27 alone (Fig. 4A). Moreover, Isl-1 enhanced activation of each of the Arx-driven constructs in cotransfection assays, although pGL4.27-Re2 (2.9-fold) was more sensitive than either pGL4.27-Re1/2 (2.6-fold) or pGL4.27- Re1 (1.5-fold) (Fig. 4A).
FIGURE 4.
Re1 and Re2 mediate Arx transcription in αTC1–6 cells. A, pGL4.27-Re1, pGL4.27-Re2, and pGL4.27-Re1/2 were transfected into αTC1–6 cells in the presence or absence of a Isl-1-Myc expression plasmid. B, Western blot analysis using Isl-1 and control α-tubulin antibodies demonstrates a ∼70% reduction of Isl-1 protein levels in Isl-1 shRNA lentivirus-treated αTC1–6 cells. C, endogenous Arx mRNA expression was down-regulated relative to HPRT in Isl-1 shRNA treated αTC1–6 cells. D, Isl-1 shRNA knockdown reduced pGL4.27-Re1 and pGL4.27-Re2 reporter activity in αTC1–6 cells. Results are presented as the mean ± S.E. *, p <0.05. pGL4.27-Arx activity was normalized to that of the pRL-SV40 Renilla luciferase in A and D.
To further investigate the involvement of Isl-1 in Arx activity, αTC1–6 cells were infected with an Isl-1 shRNA expressing lentivirus. A nearly 70% reduction in cellular Isl-1 protein amount resulted in a 2-fold reduction in endogenous Arx mRNA levels (Fig. 4, B and C). Notably, Isl-1 knockdown also decreased pGL4.27-Re1 and pGL4.27-Re2 activity by ∼2-fold in αTC1–6 cells (Fig. 4D), strongly suggesting that Isl-1 control of Arx transcription is mediated through Re1 and Re2.
Isl-1 Binding Mutants Reduced Re1- and Re2-mediated Stimulation in α-Cells
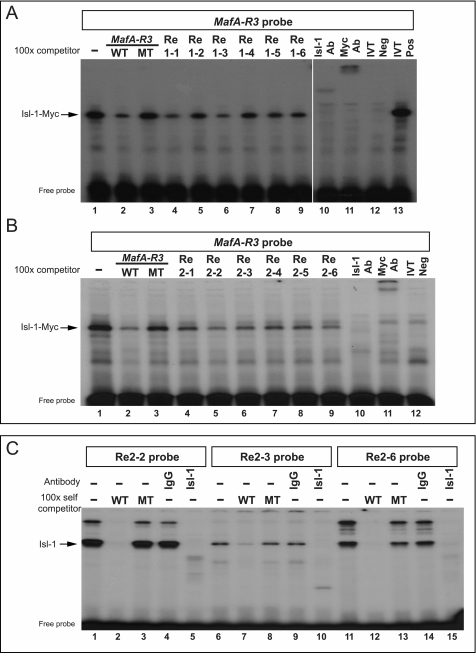
Several potential homeodomain-binding sites (TAAT/ATTA) were found within the highly conserved sequences of Re1 and Re2 (Fig. 5, A and B). EMSAs were performed to evaluate the binding ability of in vitro translated Myc-tagged Isl-1 protein to six Arx sites from each region (Fig. 6, A and B), with the well characterized binding element from MafA Region 3 (MafA-R3) serving as the positive control (17). Isl-1 and Myc antibodies were used to locate the Isl-1-Myc complex in these assays. Isl-1-Myc:MafA was competed to varying degrees by all of the wild-type Re1 and Re2 sequences (Fig. 6, A and B), and their binding differences were also evident upon analyzing endogenous Isl-1 binding to the Re2–2, Re2–3, and Re2–6 probes (Fig. 6C). These results suggested that Isl-1 was able to bind to multiple sites within Re1 and Re2.
FIGURE 6.
Isl-1 binding sites in Re1 and Re2. A and B, as a screen of putative Isl-1 binding elements, the radiolabeled MafA-R3 Isl-1 site probe (17) was used in reactions with in vitro translated Isl-1-Myc and MafA-R3, Re1, or Re2 competitor oligonucleotides. Isl-1 and Myc-epitope antibodies were used to localize the Isl-1-Myc:MafA complex. Ab, antibody; IVT, in vitro translation; Neg, without Isl-1-Myc; Pos, with Isl-1-Myc. C, Re2–2, Re2–3, and Re2–6 probes were incubated with αTC1–6 nuclear extract. The specificity of Isl-1:Arx-Re2 binding was determined by Isl-1 antibody addition and competition with excess of unlabeled wild-type (WT) and Isl-1 binding site mutant (MT).
Isl-1 binding mutations in Re1–1, Re1–3, Re2–2, Re2–3, and Re2–6 (marked in red, Fig. 5, A and B) were generated in the pGL4.27-Arx reporters to evaluate their effect on activation in αTC1–6 cells (Fig. 5C). These sites were chosen on the basis of the strength of binding to Isl-1 (Fig. 6). The relatively weak binding mutant Re2–3 reduced pGL4.27-Arx activity to the same extent as the strong Re1–1, Re1–3, Re2–2, and Re2–6 (Fig. 5C). These results indicated that Isl-1 activation of Arx transcription in α-cells is likely through many related cis-elements within the 3′-flanking Re1 and Re2 control domains.
DISCUSSION
The results of this study reveal for the first time that Isl-1, an islet transcription factor, is essential for Arx transcription in α-cells. Although the importance of Arx during α-cell formation has been demonstrated in mice and humans (15, 16, 22, 23, 24), the molecular mechanisms involved in Arx transcription in the islet α-cell have remained largely elusive. Here, we have demonstrated that Isl-1 is an activator for Arx transcription in α-cells through direct binding to multiple elements in the 3′-noncoding sequences. We found that Isl-1 overexpression in islet cells leads to enhanced Arx expression in α-cells. These findings not only shed new light on the transcriptional regulation of Arx in α-cell biology, but also identify Isl-1 as an essential islet transcriptional activator for α-cell formation.
Isl-1 proteins have been shown to directly regulate gene activation (e.g. glucagon, insulin, and MafA) in islet cells by binding to the 5′-control regions of these genes (17, 31, 32). It is intriguing that Isl-1 binds to the Arx locus in two distinct regions that are located in the intron and 3′-noncoding sequences that are ∼18 kb apart from each other. From our luciferase reporter assay, it is evident that these two regions have an additive effect when placed next to each other. Lin 1/Islet 1/Mec3-HD transcription factors, like Isl-1, have been shown to function in the context of coregulators that form a high-order transcriptional complex to exert their actions (33, 34). Whether coregulators like Lin 1/Islet 1/Mec3-domain binding proteins, Ldb1, or Ldb2, (33, 35) influence Isl-1 activation during pancreatic development remains to be investigated.
Interestingly, mice lacking ultraconserved elements (uc467) which encompass our Re2, have been generated by Ahituv et al. (36). Surprisingly, these mutant mice failed to reveal any overt abnormalities, although the specific impact on α-cell formation was not analyzed in their study (36). If no defect is observed, it is possible that Re1 might play a redundant role in mediating Arx transcription in developing islet α-cells. Outside of the pancreas, Arx expression is also detected in the developing ventral telencephalon of the brain, where its expression is directly regulated by the homeodomain factor Dlx2 (37, 38). Two Dlx2 binding sites (HD-1 and HD-2) have been identified in the highly conserved noncoding sequence of the Arx locus (referred to as mUAS3, 37), which encompasses the Re2 sequences. HD-1 and HD-2 are the equivalent of Re2–2 and Re2–4 in our study. Notably, Dlx2 is minimally expressed as compared with Isl-1 in the pancreas (data not shown), suggesting that Isl-1 is the major transcriptional activator of Arx in developing α-cells.
Isl-1 has widespread effects in many developing tissues including motor neurons, cardiac mesoderm, and the pancreas (17, 39–41). Our findings reveal a novel role for Isl-1 in controlling the islet α-cell phenotype by directly regulating Arx transcription. Although Arx is detected in both α-cell progenitor and fully differentiated glucagon-producing cell populations (16), Isl-1 is only found in the latter population, which is post-mitotic and fully differentiated (39, 42, 43). Therefore, Isl-1 is only necessary for the maintenance of Arx transcription in forming α-cells and not involved during the initial actions of Arx in early α-cell specification. Finally, although our current study also suggests that Isl-1 is likely to regulate Arx transcription in adult islet α-cells, the in vivo role Isl-1 plays in adult islet Arx expression and α-cell biology await to be investigated.
Acknowledgments
We thank Dr. John Le Lay and Jeff Raum for all scientific discussions and Dr. Nadav Ahituv for critical reading of the manuscript. We thank the members of the morphology Core in the Center for Molecular Studies in Digestive and Liver Diseases (P30-DK050306) for sample processing, and Dr. Jonathan Schug and the members of the Functional Genomics Core of the Penn Diabetes Center (DERC: P30-DK19525) for conducting ChIP-Seq experiments and data analysis. We also thank Dr. Sam Pfaff for generously providing the Isl-1 expression vector and Maureen Gannon for the Pdx1PB fragment.
This work was supported, in whole or in part, by National Institutes of Health Grants DK078606 (to C. L. M. and R. S.), F32DK083160 (to C. S. H.), T32GM07229 (to B. E.), and JDRF 2-2007-703 (to C. L. M.).
- E9.5
- embryonic day 9.5
- Isl-1
- Islet 1
- Re
- region(s)
- HD
- homeodomain.
REFERENCES
- 1. Oliver-Krasinski J. M., Stoffers D. A. (2008) Genes Dev. 22, 1998–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burcelin R., Katz E. B., Charron M. J. (1996) Diabetes Metab. 22, 373–396 [PubMed] [Google Scholar]
- 3. Dobbs R., Sakurai H., Sasaki H., Faloona G., Valverde I., Baetens D., Orci L., Unger R. (1975) Science 187, 544–547 [DOI] [PubMed] [Google Scholar]
- 4. Toft I., Gerich J. E., Jenssen T. (2002) Metabolism 51, 1128–1134 [DOI] [PubMed] [Google Scholar]
- 5. Unger R. H. (1971) Diabetes 20, 834–838 [DOI] [PubMed] [Google Scholar]
- 6. Yu X., Park B. H., Wang M. Y., Wang Z. V., Unger R. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14070–14075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerich J. E., Lorenzi M., Bier D. M., Schneider V., Tsalikian E., Karam J. H., Forsham P. H. (1975) N. Engl. J. Med. 292, 985–989 [DOI] [PubMed] [Google Scholar]
- 8. Liljenquist J. E., Bloomgarden Z. T., Cherrington A. D., Perry J. M., Rabin D. (1979) Diabetologia 17, 139–143 [DOI] [PubMed] [Google Scholar]
- 9. Raskin P., Unger R. H. (1978) N. Engl. J. Med. 299, 433–436 [DOI] [PubMed] [Google Scholar]
- 10. Gromada J., Franklin I., Wollheim C. B. (2007) Endocr. Rev. 28, 84–116 [DOI] [PubMed] [Google Scholar]
- 11. Pictet R. L., Clark W. R., Williams R. H., Rutter W. J. (1972) Dev. Biol. 29, 436–467 [DOI] [PubMed] [Google Scholar]
- 12. Ahlgren U., Jonsson J., Edlund H. (1996) Development 122, 1409–1416 [DOI] [PubMed] [Google Scholar]
- 13. Ahlgren U., Jonsson J., Jonsson L., Simu K., Edlund H. (1998) Genes Dev. 12, 1763–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Artner I., Le Lay J., Hang Y., Elghazi L., Schisler J. C., Henderson E., Sosa-Pineda B., Stein R. (2006) Diabetes 55, 297–304 [DOI] [PubMed] [Google Scholar]
- 15. Collombat P., Hecksher-Sørensen J., Broccoli V., Krull J., Ponte I., Mundiger T., Smith J., Gruss P., Serup P., Mansouri A. (2005) Development 132, 2969–2980 [DOI] [PubMed] [Google Scholar]
- 16. Collombat P., Mansouri A., Hecksher-Sorensen J., Serup P., Krull J., Gradwohl G., Gruss P. (2003) Genes Dev. 17, 2591–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Du A., Hunter C. S., Murray J., Noble D., Cai C. L., Evans S. M., Stein R., May C. L. (2009) Diabetes 58, 2059–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee C. S., Sund N. J., Behr R., Herrera P. L., Kaestner K. H. (2005) Dev. Biol. 278, 484–495 [DOI] [PubMed] [Google Scholar]
- 19. Sander M., Sussel L., Conners J., Scheel D., Kalamaras J., Dela Cruz F., Schwitzgebel V., Hayes-Jordan A., German M. (2000) Development 127, 5533–5540 [DOI] [PubMed] [Google Scholar]
- 20. Sosa-Pineda B., Chowdhury K., Torres M., Oliver G., Gruss P. (1997) Nature 386, 399–402 [DOI] [PubMed] [Google Scholar]
- 21. Hancock A. S., Du A., Liu J., Miller M., May C. L. (2010) Mol. Endocrinol. 24, 1605–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Collombat P., Hecksher-Sørensen J., Krull J., Berger J., Riedel D., Herrera P. L., Serup P., Mansouri A. (2007) J. Clin. Invest. 117, 961–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Collombat P., Xu X., Ravassard P., Sosa-Pineda B., Dussaud S., Billestrup N., Madsen O. D., Serup P., Heimberg H., Mansouri A. (2009) Cell 138, 449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Itoh M., Takizawa Y., Hanai S., Okazaki S., Miyata R., Inoue T., Akashi T., Hayashi M., Goto Y. (2010) Differentiation 80, 118–122 [DOI] [PubMed] [Google Scholar]
- 25. Gupta R. K., Gao N., Gorski R. K., White P., Hardy O. T., Rafiq K., Brestelli J. E., Chen G., Stoeckert C. J., Jr., Kaestner K. H. (2007) Genes Dev. 21, 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hohmeier H. E., Newgard C. B. (2004) Mol. Cell. Endocrinol. 228, 121–128 [DOI] [PubMed] [Google Scholar]
- 27. Tuteja G., White P., Schug J., Kaestner K. H. (2009) Nucleic Acids Res. 37, e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989) Nucleic Acids Res. 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gannon M., Ray M. K., Van Zee K., Rausa F., Costa R. H., Wright C. V. (2000) Development 127, 2883–2895 [DOI] [PubMed] [Google Scholar]
- 30. Wu K. L., Gannon M., Peshavaria M., Offield M. F., Henderson E., Ray M., Marks A., Gamer L. W., Wright C. V., Stein R. (1997) Mol. Cell. Biol. 17, 6002–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karlsson O., Thor S., Norberg T., Ohlsson H., Edlund T. (1990) Nature 344, 879–882 [DOI] [PubMed] [Google Scholar]
- 32. Wang M., Drucker D. J. (1995) J. Biol. Chem. 270, 12646–12652 [DOI] [PubMed] [Google Scholar]
- 33. Agulnick A. D., Taira M., Breen J. J., Tanaka T., Dawid I. B., Westphal H. (1996) Nature 384, 270–272 [DOI] [PubMed] [Google Scholar]
- 34. Matthews J. M., Visvader J. E. (2003) EMBO Rep. 4, 1132–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bach I., Carrière C., Ostendorff H. P., Andersen B., Rosenfeld M. G. (1997) Genes Dev. 11, 1370–1380 [DOI] [PubMed] [Google Scholar]
- 36. Ahituv N., Zhu Y., Visel A., Holt A., Afzal V., Pennacchio L. A., Rubin E. M. (2007) PLoS Biol. 5, e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Colasante G., Collombat P., Raimondi V., Bonanomi D., Ferrai C., Maira M., Yoshikawa K., Mansouri A., Valtorta F., Rubenstein J. L., Broccoli V. (2008) J. Neurosci. 28, 10674–10686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cobos I., Broccoli V., Rubenstein J. L. (2005) J. Comp. Neurol. 483, 292–303 [DOI] [PubMed] [Google Scholar]
- 39. Ahlgren U., Pfaff S. L., Jessell T. M., Edlund T., Edlund H. (1997) Nature 385, 257–260 [DOI] [PubMed] [Google Scholar]
- 40. Cai C. L., Liang X., Shi Y., Chu P. H., Pfaff S. L., Chen J., Evans S. (2003) Dev. Cell 5, 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pfaff S. L., Mendelsohn M., Stewart C. L., Edlund T., Jessell T. M. (1996) Cell 84, 309–320 [DOI] [PubMed] [Google Scholar]
- 42. Schwitzgebel V. M., Scheel D. W., Conners J. R., Kalamaras J., Lee J. E., Anderson D. J., Sussel L., Johnson J. D., German M. S. (2000) Development 127, 3533–3542 [DOI] [PubMed] [Google Scholar]
- 43. Thor S., Ericson J., Brännström T., Edlund T. (1991) Neuron 7, 881–889 [DOI] [PubMed] [Google Scholar]