Abstract
Dnmt1 (DNA methyltransferase 1) is the principal enzyme responsible for maintenance of cytosine methylation at CpG dinucleotides in the mammalian genome. The N-terminal replication focus targeting sequence (RFTS) domain of Dnmt1 has been implicated in subcellular localization, protein association, and catalytic function. However, progress in understanding its function has been limited by the lack of assays for and a structure of this domain. Here, we show that the naked DNA- and polynucleosome-binding activities of Dnmt1 are inhibited by the RFTS domain, which functions by virtue of binding the catalytic domain to the exclusion of DNA. Kinetic analysis with a fluorogenic DNA substrate established the RFTS domain as a 600-fold inhibitor of Dnmt1 enzymatic activity. The crystal structure of the RFTS domain reveals a novel fold and supports a mechanism in which an RFTS-targeted Dnmt1-binding protein, such as Uhrf1, may activate Dnmt1 for DNA binding.
Keywords: DNA Methyltransferase, DNA-Protein Interaction, Enzyme Inhibitors, Protein Domains, X-ray Crystallography
Introduction
DNA cytosine methylation, an epigenetic mark that occurs predominantly at CpG dinucleotides, is the principal eukaryotic DNA modification (1). DNA methylation is a key component of gene silencing, which makes significant contributions to cell phenotype. Establishment and maintenance of DNA methylation patterns are governed by three catalytically active DNA methyltransferases: Dnmt3a, Dnmt3b, and Dnmt1. The Dnmt3 isoforms are responsible for de novo methylation during germ cell and embryonic development and bind with high affinity to unmethylated DNA sequences (2). CpG methylation patterns are maintained in mammals by Dnmt1 with hemimethylated CpG dinucleotides serving as preferred substrates.
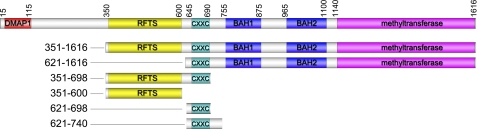
Human Dnmt1 (1616 amino acids) consists of a conserved C-terminal catalytic core (amino acids 1140–1616) and a large N-terminal region (amino acids 1–1139) harboring multiple globular conserved domains, including the DMAP1 (DNA methyltransferase-associated protein 1)-binding domain (3), the proliferating cell nuclear antigen-binding domain (4), the replication focus targeting sequence (RFTS)4 domain (residues 351–600) (5), the CXXC domain (6), and two bromo-adjacent homology (BAH) domains (see Fig. 1) (7).
FIGURE 1.
Dnmt1 protein domains. Full-length and truncated Dnmt1 constructs are shown with conserved domains colored as follows: DMAP1, red; RFTS domain, yellow; CXXC domain, teal; two BAH domains, blue; and methyltransferase domain, magenta.
The CXXC domain is understood to contribute to catalytic activity by interacting with unmethylated CpG DNA substrates (6, 8). This was observed in the recently solved crystal structures of Dnmt1, which encompass sequences from the CXXC domain to the C terminus (9). In the structures, the CXXC domain binds and holds unmethylated duplex CpG-containing DNA away from the active site, whereas the acidic linker between the CXXC and BAH1 domains is bound in the active site between the DNA segment and the S-adenosylhomocysteine product (9). This observation helps to explain the relationship between the CXXC and catalytic domains. However, it does not address the role of domains N-terminal to the CXXC domain, which are not present in the structures.
We set out to clarify the structure and function of the RFTS domain. The RFTS domain is conserved by sequence (supplemental Fig. S1) and contains the binding site for Uhrf1 (10), a Dnmt1-associated protein that recruits Dnmt1 to hemimethylated DNA (11–15). Despite the significance of the RFTS domain, progress in understanding its function has been limited by the availability of stable soluble protein fragments and robust DNA methyltransferase assays. Here, we generated soluble protein fragments of Dnmt1 and established activities for them. Strikingly, the binding of Dnmt1 to naked DNA oligonucleotides and native polynucleosomes was inhibited by the RFTS domain. Kinetic analysis established that Dnmt1 without the RFTS domain functioned with a Km of 1 nm for an internally quenched oligonucleotide substrate. This represents a >100-fold binding advantage with respect to recent assays with hemimethylated oligonucleotide substrates (9, 16). By comparison of kcat/Km terms between RFTS domain-containing and RFTS domain-lacking forms of Dnmt1, the RFTS domain is a 600-fold inhibitor of DNA methylation. Moreover, by titrating the RFTS domain into reactions with the RFTS domain-lacking Dnmt1 construct, we show that RFTS is a 100 nm inhibitor that is strictly competitive with DNA binding. Finally, the crystal structure of the RFTS domain reveals features that may allow it to occlude DNA substrate binding by the catalytic domain in a manner that could be relieved by a Dnmt1 activator, such as Uhrf1.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
Human Dnmt1 constructs schematized in Fig. 1 were cloned and purified with similar methods (17). Briefly, cDNA templates (MHS1768-98980929.pCR-XL-TOPO) from Open Biosystems were cloned into the pET28MHL or pNICCH vector using the In-Fusion CF Dry-Down PCR cloning kit (639605, Clontech), transformed into Escherichia coli BL21(DE3) cells, and grown in Terrific Broth in the presence of 50 μg/ml kanamycin at 37 °C. Selenomethionyl derivatives of the RFTS domain were expressed in M9 medium supplemented with glycerol using an M9 SeMet high yield growth media kit package (M2D045004–50L, Medicilon) according to the manufacturer's instructions. After lysis, cell supernatants were subjected to metal affinity chromatography using TALON columns (BD Biosciences). Protein was further purified by gel filtration (HiLoad 16/60 Superdex 200 column, GE Healthcare) equilibrated with buffer A (20 mm Tris-HCl (pH 8.0), 0.5 m NaCl, 5% glycerol, and 2 mm DTT) and by ion exchange chromatography on a 5-ml HiTrap Q column using a gradient of buffer A to 50% buffer B (20 mm Tris-HCl (pH 8.0), 1 m NaCl, 5% glycerol, and 2 mm DTT). Full-length Dnmt1 was obtained from New England Biolabs.
Nucleosome Purification
Polynucleosomes from chicken erythrocytes were obtained by micrococcal nuclease digestion as described (18). Briefly, cells were lysed and vortexed thoroughly in buffer containing 10 mm Tris (pH 8), 6 mm MgCl2, 0.1 mm PMSF, 0.05% Nonidet P-40, and 80 mg/ml sucrose. Nuclei were extracted and washed by a four-step sequential process and further digested by micrococcal nuclease (Sigma) for 10 min at 37 °C in 10 mm Tris (pH 8), 0.2 mm CaCl2, 0.1 mm PMSF, and 10 mg/ml sucrose. Purified native chromatin was extracted in 10 mm Tris (pH 8) and 0.25 mm EDTA and analyzed on agarose gels.
Electrophoretic Mobility Shift Assays
DNA binding was analyzed using duplex DNA oligonucleotides of varying lengths, sequences, and methylation states (supplemental Table 1). Duplex DNA (100 ng/lane) was incubated overnight at 4 °C with an appropriate concentration of protein (as indicated in the figure legends) in 10 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.5 mm DTT, and 0.1 mg/ml BSA and subjected to polyacrylamide gel electrophoresis (10% Tris borate/EDTA gels (Bio-Rad) run in 0.5× Tris borate/EDTA at 100 V for 1.5 h on ice). Gels were stained with 10,000-fold diluted SYBR Green I (Invitrogen) for 5 min, washed twice with 0.5× Tris borate/EDTA, and imaged using an excitation wavelength of 633 nm.
Dnmt1 Activity Assay
Dnmt1 activity was measured using an assay related to that developed by Wood et al. (16) in which a hemimethylated, internally quenched, fluorescent hairpin DNA substrate is incubated with Dnmt1 and AdoMet in the presence of GlaI, which cleaves the fully methylated product. Assays (0.1 ml) were performed in triplicate in 96-well half-area black plates at 37 °C. Triplicate reactions were prepared by making a solution of 10 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 1 mm DTT, 100 mm potassium glutamate, 0.105 mg/ml BSA, 1.05 mm AdoMet (HPLC-purified; Sigma), and 1.05× the desired concentration of oligonucleotide. For inhibition studies, 1.05× the desired concentration of the RFTS domain (amino acids 351–600) was added to the solution. AdoMet was added freshly to limit conversion to S-adenosylhomocysteine, which inhibits the Dnmt1 reaction. Ninety-five microliters of this solution were added per well and brought to 37 °C. A second solution was made containing Dnmt1 and GlaI (SibEnzyme, Novosibirsk, Russia) in 10 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 1 mm DTT, and 100 mm potassium glutamate such that 0.8 units of GlaI would be added to each well. Coupled reactions were initiated by adding 5 μl of enzyme solution, and the wells were covered with 5 μl of mineral oil. Data were collected in a Wallac VICTOR2 using excitation and emission wavelengths of 485 and 535 nm, respectively. A negative control containing GlaI without Dnmt1 was subtracted from each assay condition to correct for both substrate fluorescence and fluorescence from slow GlaI cleavage of the hemimethylated substrate. A standard curve was created using the fully cleaved oligonucleotide products to convert arbitrary fluorescence units to concentration. The oligonucleotide, custom-synthesized at Integrated DNA Technologies (Coralville, IA), was 5′-FAM-CCTATGCGmCATCAGTTTTCTGATGmCGmCATAGG-3′-Iowa Black, where mC denotes 5-methyldeoxycytidylate residues and underlined residues form the GlaI restriction site. Initial rate data were fit either to the Michaelis-Menten equation or to the quadratic velocity equation (Equation 1) for tight-binding substrates (19, 20) to determine Vmax and Km.
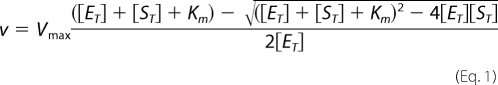 |
Data were fitted using either KaleidaGraph (Synergy, Inc.) or Prism (GraphPad Software, Inc).
Crystallization
Selenomethionyl derivatives of the RFTS domain (amino acids 351–600) were mixed with detergent Anapoe-80 solution (10%, v/v) in a 5:1 ratio (final protein and detergent concentrations of 16.5 mg/ml and 1.7%, respectively), and crystals were grown at 298 K using the hanging drop method by mixing 1 volume of this solution with 1 volume of well solution consisting of 23% polyethylene glycol 3350, 0.1 m bis-Tris (pH 6.0), 0.3 m sodium acetate, and 5 mm tris(2-carboxyethyl)phosphine. The crystals were cryoprotected by immersion in the well solution mixed 1:1 with 20% (w/v) sucrose, 4% (w/v) glucose, 18% (v/v) glycerol, and 18% (v/v) ethylene glycol and placed in liquid nitrogen.
Structure Determination
Diffraction data from a crystal of the selenomethionyl derivative of the Dnmt1 RFTS domain were collected at beamline 19-ID at the Advanced Photon Source. The data set, collected at the selenium peak wavelength, was integrated and scaled using the HKL2000 program suite (21). The structure was solved by single wavelength anomalous diffraction techniques using the program SOLVE/RESOLVE (22). A second data set collected at beamline 23-ID-B was used for refinement of the structure to 2.3-Å resolution. Iterative model building using the graphics program Coot (23) and maximum likelihood and translation/liberation/screw refinement with the program REFMAC5 (24) led to the final model. REFMAC5 parameters were generated using the TLS Motion Determination (TLSMD) web server. Statistics of data collection, processing, and refinement are provided in Table 1.
TABLE 1.
Crystallographic data and refinement statistics
PDB, Protein Data Bank; APS, Advanced Photon Source; r.m.s.d., root mean square deviation.
| Data | |
| Construct | Dnmt1 (residues 351–600) |
| PDB code | 3EPZ |
| Space group | P21 |
| Unit cell | a = 57.54, b = 59.77, c = 96.29 Å; β = 92.31° |
| Beamline | APS 23-ID-B |
| Wavelength (Å) | 1.0332 |
| Resolution (Å) | 2.3-50 |
| Unique reflections | 28,832 |
| Data redundancy | 3.700 (3.00) |
| Completeness (%) | 99.1 (91.6) |
| I/σI | 16.6849 (1.300) |
| Rsym | 0.08500 (0.97900) |
| Refinement | |
| Resolution (Å) | 2.31-48.11 |
| Reflections used | 27,330 |
| All atoms (solvent) | 3446 (99) |
| Rwork/Rfree | 0.216/0.264 |
| r.m.s.d. bond length (Å) | 0.009 |
| r.m.s.d. bond angle | 1.154° |
RESULTS AND DISCUSSION
The RFTS Domain Inhibits DNA Binding Independently of DNA Sequence, Length, Methylation Status, or CpG Content
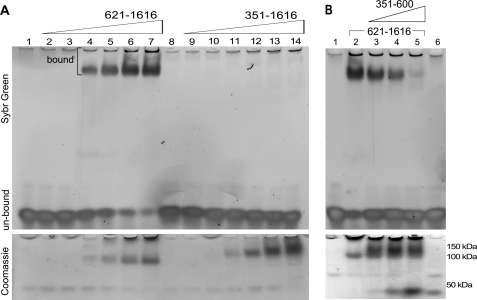
It has been proposed that the RFTS domain of Dnmt1 tunes the catalytic activity of the conserved DNA methyltransferase domain toward specific DNA substrates (6). To test this hypothesis, we titrated constructs containing the RFTS domain (amino acids 351–1616) or lacking the RFTS domain (amino acids 621–1616) against 12-bp hemimethylated DNA oligonucleotides and analyzed DNA binding using an EMSA (Fig. 2A). Surprisingly, at a range of Dnmt1 concentrations from 0.5 to 10 μm, the RFTS domain-lacking 621–1616 construct bound the DNA, but the RFTS domain-containing 351–1616 construct showed no detectable binding even at high protein concentrations (Fig. 2A). Moreover, DNA binding to the RFTS domain-lacking 621–1616 construct was inhibited by the RFTS domain (amino acids 351–600) added in trans (Fig. 2B). These data suggested that the RFTS domain might inhibit the ability of the Dnmt1 catalytic domain to bind particular naked DNA sequences. To determine whether the RFTS domain might be responsible for substrate selectivity and permit the catalytic domain to bind some but not all DNA sequences, we surveyed a series of different DNA substrates of various lengths, methylation states, CpG content, and sequences. As shown in supplemental Fig. S2, the 621–1616 construct had its electrophoretic mobility shifted by all DNA constructs, regardless of methylation status, length, or sequence, whereas the 351–1616 construct, containing the RFTS domain, showed impaired binding of all oligonucleotides investigated. Even non-CpG oligonucleotides were bound by the 621–1616 Dnmt1 construct in a manner that was inhibited in trans by the addition of the RFTS domain (supplemental Fig. S3).
FIGURE 2.
The RFTS domain is an inhibitor of naked DNA binding. A, RFTS domain-containing Dnmt1 (amino acids 351–1616) and RFTS domain-lacking Dnmt1 (amino acids 621–1616) (0.5–10 μm) were incubated with 100 ng of 12-bp hemimethylated oligonucleotide and analyzed for DNA binding via an EMSA. DNA was detected with SYBR Green (upper panel), and protein was detected with Coomassie Blue (lower panel). The RFTS domain-lacking protein exhibited superior DNA binding to the RFTS domain-containing protein as indicated by the shift in the electrophoretic mobility of the DNA observed in lanes 4–7. B, the RFTS domain (0.5–5.0 μm) was added in trans to assays containing 5 μm RFTS domain-lacking Dnmt1 and 100 ng of hemimethylated oligonucleotide. The RFTS domain inhibited DNA binding to the RFTS domain-lacking protein, which allowed the DNA to migrate rapidly. The sequences of all DNA oligonucleotides are presented in supplemental Table 1.
Dnmt1 Binding to Polynucleosomes Is Inhibited by the RFTS Domain
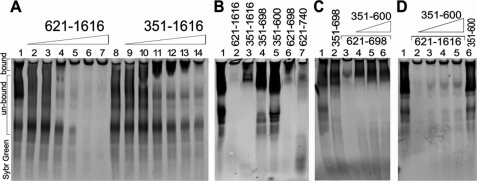
The CXXC and catalytic domains of Dnmt1 are both known to possess DNA-binding activities (8, 25, 26). To determine whether the RFTS domain interferes with the CXXC, catalytic, or both domain functions, we generated protein constructs containing only the CXXC domain (amino acids 621–698 and 621–740) or the RFTS domain with the CXXC domain (amino acids 351–698). Extensive attempts to express and purify fragments of Dnmt1, which begin C-terminal to the CXXC domain, were unsuccessful, suggesting that CXXC sequences may be required for folding and/or stability of the catalytic domain. Because naked DNA bound very poorly to CXXC domain constructs, we purified native polynucleosomes, which consist of epigenetically modified histone octamers and segments of DNA in various methylation states. EMSAs indicated that the 351–1616 construct was inhibited with respect to the polynucleosome-binding ability of the 621–1616 construct (Fig. 3A). In addition, the CXXC domain possesses a polynucleosome-binding capacity (Fig. 3B, lanes 6 and 7), which was inhibited by the RFTS domain both in cis (B, lane 4; and C, lane 2) and in trans (C, lanes 4–6). However, the binding to polynucleosomes by the 621–1616 construct, containing both the CXXC and catalytic domains, was not disrupted in trans by the addition of the RFTS-only domain (Fig. 3D). These data suggest that the polynucleosome-binding activity of the CXXC domain is additive with the DNA-binding activity of the catalytic domain and that simultaneous engagement of polynucleosomes by both domains renders the complex relatively resistant to exclusion of DNA by the RFTS domain.
FIGURE 3.
Dnmt1 binding to polynucleosomes is inhibited by the RFTS domain. A, RFTS domain-containing Dnmt1 (amino acids 351–1616) and RFTS domain-lacking Dnmt1 (amino acids 621–1616) (0.5–5.0 μm) were incubated with polynucleosomes (150 ng of DNA) and analyzed for binding via an EMSA. The SYBR Green-stained gel shows that polynucleosomal DNA ran as a typical smeary ladder (lane 1). The RFTS domain-lacking protein exhibited enhanced polynucleosome-binding ability compared with the RFTS domain-containing protein as indicated by the shift in mobility of the DNA bands, which were retained in the wells upon protein interaction. B, the polynucleosome-binding ability of all protein constructs was investigated via EMSA. Protein (5 μm) was incubated with 150 ng of polynucleosomal DNA. Constructs containing the RFTS domain (lanes 3–5) showed impaired polynucleosome binding. The CXXC domain-only constructs exhibited tight binding to polynucleosomes (lanes 6 and 7). C, the RFTS domain inhibited polynucleosome binding to the CXXC domain. The RFTS domain (0.5–5 μm) was added in trans to assays containing 5 μm CXXC domain (amino acids 621–698) and 150 ng of polynucleosomes. The RFTS domain inhibited DNA binding to the CXXC domain as indicated by increased mobility of the DNA. D, the RFTS domain (0.5–5.0 μm) was added in trans to assays containing 5 μm RFTS domain-lacking protein and 150 ng of polynucleosomes. The isolated RFTS domain did not dissociate polynucleosomes from the 621–1616 form of Dnmt1.
The RFTS Domain Is a Potent Inhibitor of Dnmt1 Enzymatic Activity
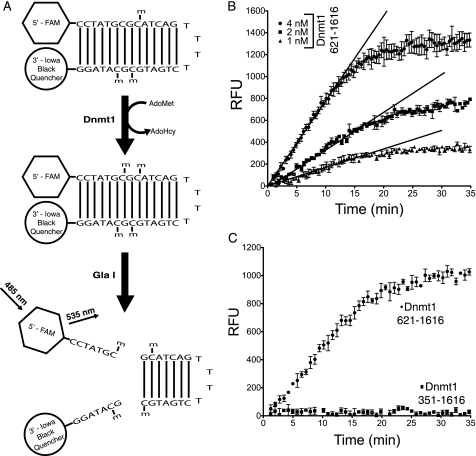
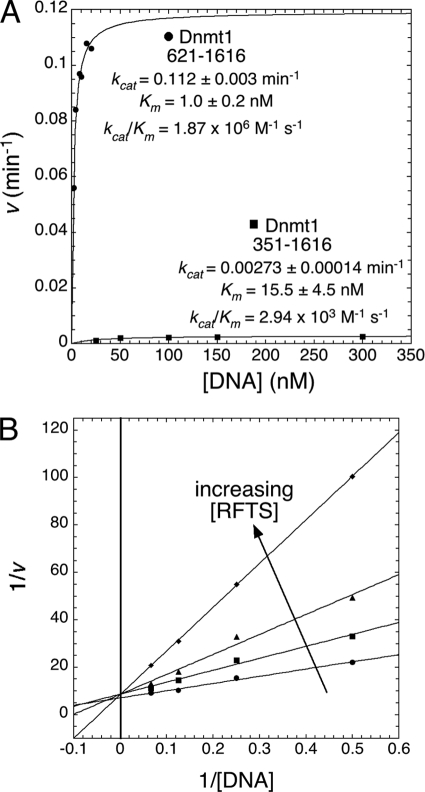
Data from EMSAs suggested that the RFTS domain is an intrinsic inhibitor of DNA binding by Dnmt1 sequences C-terminal to the RFTS domain. To test this hypothesis, we synthesized an internally quenched, fluorescent hairpin DNA substrate (16). In this restriction enzyme-coupled assay, methylation of a hemimethylated CpG site forms a GlaI site and generates fluorescence in real time (Fig. 4A). Analysis of the RFTS domain-lacking protein construct (amino acids 621–1616) with 20 nm DNA substrate indicated striking fluorescence-generating activity (Fig. 4B). Initial attempts to determine steady-state kinetic parameters with this enzyme and oligonucleotide indicated that the Km was in the low nanomolar range. Because assays were conducted with 2 nm Dnmt1, there was a need to test for the complication that the enzyme may titrate a significant proportion of substrate from the free form to an E-S complex at low substrate concentrations (19). To test for this behavior, substrate-dependent initial rates were measured at enzyme concentrations of 1–5 nm. As the enzyme concentration increased, data deviated significantly from the hyperbolic behavior of Michaelis-Menten enzyme kinetics. However, at all enzyme concentrations examined, the data fit well to the tight-binding equation (19, 20), giving essentially the same Km (1 nm) and kcat (∼0.1 min−1) values.
FIGURE 4.
The RFTS domain is an endogenous inhibitor of Dnmt1 activity. A, schematic of the GlaI-coupled Dnmt1 assay. B, time and Dnmt1 dependence of the GlaI-coupled assay with 20 nm DNA substrate. With the RFTS domain-lacking protein (amino acids 621–1616), robust increases in fluorescence that were dependent on the concentration of Dnmt1 were observed. C, autoinhibition of RFTS domain-containing Dnmt1. At 20 nm DNA substrate, 2 nm RFTS domain-lacking Dnmt1 construct (amino acids 621–1616) neared completion in 30 min, whereas no reaction progress was observed with 2 nm RFTS domain-containing Dnmt1 construct (amino acids 351–1616). RFU, relative fluorescence units.
Remarkably, Dnmt1 activity could not be detected with 2 nm RFTS domain-containing construct (amino acids 351–1616) and 20 nm DNA substrate (Fig. 4C). This led us to assay the RFTS domain-containing construct at higher enzyme and DNA substrate concentrations to test the hypothesis that the RFTS domain inhibits DNA binding and not some other step such as binding AdoMet. As shown in Fig. 5A, the second-order rate constant with respect to the DNA substrate was 1.87 × 106 m−1 s−1 for the RFTS domain-lacking construct. Inclusion of the RFTS domain increased the Km for DNA by ∼15-fold while depressing kcat by ∼40-fold (Fig. 5A and supplemental Fig. S4). Similar kinetic constants were determined for the full-length protein. Thus, the RFTS domain functions as an ∼600-fold inhibitor of binding and methyl group transfer to the DNA substrate. This finding is strikingly different from recently published results from a study that monitored transfer of a tritiated methyl group from AdoMet to DNA (9). In that study, the form of Dnmt1 with the first 645 amino acids deleted was only 2-fold more active than full-length Dnmt1 in kcat/Km (9). The greater activity in this study may be attributed to an improved assay and/or the greater activity of the Dnmt1 construct beginning at residue 621. Because full-length Dnmt1 has been assayed using an internally quenched fluorogenic substrate and found not to be saturated with the DNA substrate at 240 nm (16), we argue that the 621–1616 Dnmt1 construct is strongly derepressed for DNA binding and catalysis.
FIGURE 5.
The RFTS domain is a DNA-competitive inhibitor of methyltransferase activity. A, DNA substrate-dependent methyl transfer rates for the RFTS domain-lacking protein (amino acids 621–1616; ●) and RFTS domain-containing protein (amino acids 351–1616; ■) were measured at 1 mm AdoMet. The RFTS domain conferred an ∼40-fold reduction in kcat and an ∼15-fold increase in Km for the DNA substrate. B, the RFTS domain (amino acids 351–600) was used in trans as an inhibitor of the methyltransferase activity of the RFTS domain-lacking protein (amino acids 621–1616). The double reciprocal plot of the data with no RFTS domain (●), 100 nm RFTS domain (■), 250 nm RFTS domain (▲), and 500 nm RFTS domain (♦) clearly shows that the RFTS domain is competitive with respect to the DNA substrate. Using nonlinear regression to fit the alteration in Km(app), a Ki of 103 ± 11 nm was determined.
To further examine the inhibitory function of the RFTS domain, we titrated the RFTS polypeptide in trans to the RFTS domain-lacking Dnmt1 construct (amino acids 621–1616) and determined kinetic parameters. Plotting the data according to the Lineweaver-Burk method (Fig. 5B) clearly indicated that inhibition was competitive with the DNA substrate. As the concentration of the RFTS domain was increased, the Km(app) was increased without alteration of Vmax. By nonlinear regression, the RFTS domain-dependent effects on Km(app) indicated a Ki of 103 ± 11 nm. Thus, the RFTS domain is a previously unrecognized endogenous inhibitor of DNA binding within Dnmt1.
Crystal Structure of the Dnmt1 RFTS Domain and Model of Autoinhibition
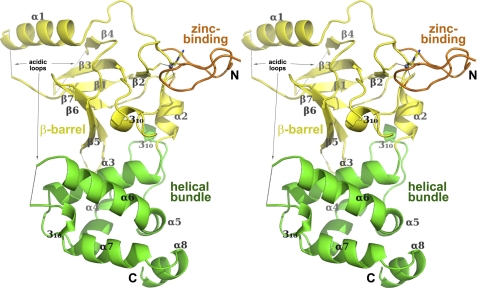
Having established discrete assays for the function of the RFTS domain, we aimed to determine whether the physical characteristics of the domain might account for inhibition of DNA binding. A selenomethionyl form of the RFTS domain crystallized in space group P21 and diffracted to 2.3-Å resolution. A refined model of the crystal structure is depicted in Fig. 6 (see Table 1 for refinement statistics). The RFTS domain consists of a zinc-binding motif, followed by a β-barrel and a helical bundle, which are closely associated. Using the secondary structure matching algorithm (27), the closest match to the β-barrel is the structure of the Src family kinase HCK Src homology 3 domain (Q score of 0.25; root mean square deviation of 2.36 Å for 46 equivalent Cα atom pairs). The six-stranded β-barrel lobe of the RFTS domain extends from residues 401 to 502, capped on one end by an α-helix (residues 496–499, α2) and on the other end by a helical bundle (residues 495–600). The nearest structural homolog of the helical bundle, as calculated by the DALI server (28), is the winged helix B lobe of the cullin protein Cdc53 (root mean square deviation of 1.7 Å across 68 backbone atoms). The role of the winged helix B lobe in these E3 ubiquitin ligases, which has recently been elucidated, involves regulating the enzyme via an autoinhibitory mechanism (29). The winged helix B lobe in the inhibited state binds to the RING (really interesting new gene) domain of the ligase, preventing the RING domain from reaching the substrate. Neddylation of the winged helix B relieves its interaction with the RING domain, allowing the RING domain to access substrate (29).
FIGURE 6.
Structure of the Dnmt1 RFTS domain. The RFTS domain is shown in a stereo ribbon representation with lobes color-coded: β-barrel, yellow; zinc-binding motif, orange; and α-helical bundle, green. The zinc atom is depicted as a gray sphere with interacting residues in stick format. Arrows point to disordered regions.
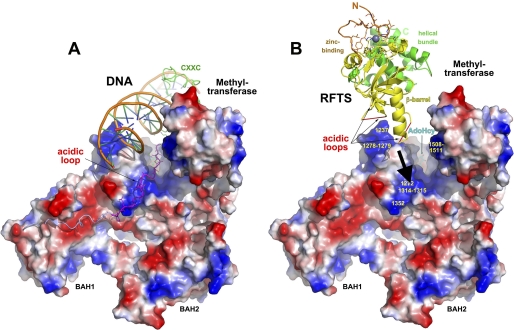
In recent crystal structures, the acidic linker between the CXXC and BAH1 domains associates with the DNA-binding site of the methyltransferase domain and prevents productive access of DNA to the active site, where S-adenosylhomocysteine is bound (Fig. 7A) (9). Deletion of the CXXC domain and linker region increased kcat/Km by 10-fold on unmethylated DNA substrates and increased activity on hemimethylated substrates by 20% (9). However, the 600-fold effect of the RFTS domain suggests that, were it present, the RFTS inhibitory activity would predominate over the mild effect of the CXXC-BAH1 linker in excluding DNA from the active site.
FIGURE 7.
Two models of autoinhibition in Dnmt1. Shown are electrostatic surface representations, generated with a gradient from −10 (red) to 10 (blue) kT/e, of Dnmt1 from BAH1 to the C terminus. In A, derived from Protein Data Bank code 3PT6, a structure not including the RFTS domain, the acidic linker (magenta) between BAH1 and the CXXC (green) domains blocks productive DNA access to the active site (9). In B, consistent with our observations (Figs. 2B and 5B) that the RFTS domain competes for DNA binding with CXXC domain-containing constructs such as the 621–1616 construct, Dnmt1 is displayed with the CXXC domain and acidic linker excluded as in Protein Data Bank code 3PTA. The RFTS domain is depicted in an orientation in which three acidic loops may descend on three basic patches in the methyltransferase domain. The DNA-competitive nature of RFTS domain inhibition suggests that the RFTS domain binds to the exclusion of DNA and may be released by an RFTS-targeted Dnmt1 activator, such as Uhrf1.
We sought to develop a model for Dnmt1 autoinhibition that would account for the 600-fold inhibition by the RFTS domain and account for how a positive regulator of Dnmt1 might relieve this inhibition. As shown in Fig. 7B, the RFTS domain has a size and electrostatic properties compatible with occlusion of the DNA substrate-binding site of Dnmt1. Three acidic loops, each disordered in our crystal structure of the RFTS domain, are positioned on the side of the RFTS domain we predict to associate with three basic patches of the methyltransferase domain of Dnmt1. We postulate that Uhrf1, a multidomain E3 ubiquitin ligase that is required for maintenance of DNA methylation and Dnmt1 localization in embryonic stem cells (11, 12), is the key mediator of Dnmt1 derepression. According to our model, Uhrf1 binding to the RFTS domain would release the inhibitory domain from the active site and allow DNA to bind productively. This model is attractive for two reasons. First, Uhrf1 has been reported to associate with Dnmt1 within the RFTS domain (10). Second, Uhrf1 is thought to function by binding hemimethylated DNA and recruiting Dnmt1 for methyl transfer to the non-modified cytosine of a hemimethylated site (13) or to nearby CpG dinucleotides (14, 15). If Uhrf1 binding to the RFTS domain is required to relieve inhibition of DNA binding, a mechanism is provided to activate Dnmt1 for processive methylation of hemimethylated regions of DNA identified by the essential activator Uhrf1.
Supplementary Material
Acknowledgments
We thank Yanjun Li and Peter Loppnau for plasmid constructions, Juan Pizzaro and Abdellah Allali-Hassani for technical assistance, Peter Lewis for assistance with chromatin preparation, and Bryce Plapp for conversations on enzyme inhibition.
The work performed at the University of Iowa was supported by National Institutes of Health Grant CA075954 and Contract HHSN261200433000C from NCI and a generous gift from the Roy J. Carver Foundation. Use of the Advanced Photon Source was supported by United States Department of Energy Office of Basic Energy Sciences Contract DE-AC02-06CH11357. The Structural Genomics Consortium is a registered charity (1097737) supported by the Canadian Institutes of Health Research, the Canadian Foundation for Innovation, and Genome Canada through the Ontario Genomics Institute.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table 1.
The atomic coordinates and structure factors (code 3EPZ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- RFTS
- replication focus targeting sequence
- BAH
- bromo-adjacent homology
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Suzuki M. M., Bird A. (2008) Nat. Rev. Genet. 9, 465–476 [DOI] [PubMed] [Google Scholar]
- 2. Yokochi T., Robertson K. D. (2002) J. Biol. Chem. 277, 11735–11745 [DOI] [PubMed] [Google Scholar]
- 3. Rountree M. R., Bachman K. E., Baylin S. B. (2000) Nat. Genet. 25, 269–277 [DOI] [PubMed] [Google Scholar]
- 4. Chuang L. S., Ian H. I., Koh T. W., Ng H. H., Xu G., Li B. F. (1997) Science 277, 1996–2000 [DOI] [PubMed] [Google Scholar]
- 5. Leonhardt H., Page A. W., Weier H. U., Bestor T. H. (1992) Cell 71, 865–873 [DOI] [PubMed] [Google Scholar]
- 6. Bestor T. H. (1992) EMBO J. 11, 2611–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Callebaut I., Courvalin J. C., Mornon J. P. (1999) FEBS Lett. 446, 189–193 [DOI] [PubMed] [Google Scholar]
- 8. Pradhan M., Estève P. O., Chin H. G., Samaranayke M., Kim G. D., Pradhan S. (2008) Biochemistry 47, 10000–10009 [DOI] [PubMed] [Google Scholar]
- 9. Song J., Rechkoblit O., Bestor T. H., Patel D. J. (2011) Science 331, 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Achour M., Jacq X., Rondé P., Alhosin M., Charlot C., Chataigneau T., Jeanblanc M., Macaluso M., Giordano A., Hughes A. D., Schini-Kerth V. B., Bronner C. (2008) Oncogene 27, 2187–2197 [DOI] [PubMed] [Google Scholar]
- 11. Bostick M., Kim J. K., Estève P. O., Clark A., Pradhan S., Jacobsen S. E. (2007) Science 317, 1760–1764 [DOI] [PubMed] [Google Scholar]
- 12. Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T. A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., Tajima S., Mitsuya K., Okano M., Koseki H. (2007) Nature 450, 908–912 [DOI] [PubMed] [Google Scholar]
- 13. Arita K., Ariyoshi M., Tochio H., Nakamura Y., Shirakawa M. (2008) Nature 455, 818–821 [DOI] [PubMed] [Google Scholar]
- 14. Avvakumov G. V., Walker J. R., Xue S., Li Y., Duan S., Bronner C., Arrowsmith C. H., Dhe-Paganon S. (2008) Nature 455, 822–825 [DOI] [PubMed] [Google Scholar]
- 15. Hashimoto H., Horton J. R., Zhang X., Bostick M., Jacobsen S. E., Cheng X. (2008) Nature 455, 826–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wood R. J., McKelvie J. C., Maynard-Smith M. D., Roach P. L. (2010) Nucleic Acids Res. 38, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bacik J. P., Walker J. R., Ali M., Schimmer A. D., Dhe-Paganon S. (2010) J. Biol. Chem. 285, 20273–20280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan S., Attisano L., Lewis P. N. (1988) J. Biol. Chem. 263, 15643–15651 [PubMed] [Google Scholar]
- 19. Cha S. (1970) J. Biol. Chem. 245, 4814–4818 [PubMed] [Google Scholar]
- 20. Goldstein A. (1944) J. Gen. Physiol. 27, 529–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 22. Terwilliger T. C. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 25. Svedruzić Z. M., Reich N. O. (2005) Biochemistry 44, 9472–9485 [DOI] [PubMed] [Google Scholar]
- 26. Jeltsch A. (2006) Epigenetics 1, 63–66 [DOI] [PubMed] [Google Scholar]
- 27. Krissinel E., Henrick K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 28. Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Duda D. M., Borg L. A., Scott D. C., Hunt H. W., Hammel M., Schulman B. A. (2008) Cell 134, 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.