Abstract
The phenylacetic acid (PAA) degradation pathway is the sole aerobic route for phenylacetic acid metabolism in bacteria and facilitates degradation of environmental pollutants such as styrene and ethylbenzene. The PAA pathway also is implicated in promoting Burkholderia cenocepacia infections in cystic fibrosis patients. Intriguingly, the first enzyme in the PAA pathway is present in two copies (paaK1 and paaK2), yet each subsequent enzyme is present in only a single copy. Furthermore, sequence divergence indicates that PaaK1 and PaaK2 form a unique subgroup within the adenylate-forming enzyme (AFE) superfamily. To establish a biochemical rationale for the existence of the PaaK paralogs in B. cenocepacia, we present high resolution x-ray crystal structures of a selenomethionine derivative of PaaK1 in complex with ATP and adenylated phenylacetate intermediate complexes of PaaK1 and PaaK2 in distinct conformations. Structural analysis reveals a novel N-terminal microdomain that may serve to recruit subsequent PAA enzymes, whereas a bifunctional role is proposed for the P-loop in stabilizing the C-terminal domain in conformation 2. The potential for different kinetic profiles was suggested by a structurally divergent extension of the aryl substrate pocket in PaaK1 relative to PaaK2. Functional characterization confirmed this prediction, with PaaK1 possessing a lower Km for phenylacetic acid and better able to accommodate 3′ and 4′ substitutions on the phenyl ring. Collectively, these results offer detailed insight into the reaction mechanism of a novel subgroup of the AFE superfamily and provide a clear biochemical rationale for the presence of paralogous copies of PaaK of B. cenocepacia.
Keywords: Enzyme Catalysis, Enzyme Kinetics, Enzyme Mechanisms, Enzyme Structure, X-ray Crystallography
Introduction
Aromatic compounds are ubiquitous in the environment and exist primarily in the form of recycled plant material. Specialized microbial degradative pathways have evolved to overcome the resonance stabilized aromatic nucleus, thereby serving an integral role in the global carbon cycle. Furthermore, characterization of the bioprocessing enzymes comprising such pathways offers opportunities to engineer novel bioremediation and biofuel production strategies. The ability to degrade aromatic compounds also is an influential factor in microbial pathogenesis. For example, the degradation of phenylacetic acid (PAA)3 is proposed to facilitate B. cenocepacia in establishing life-threatening infections in cystic fibrosis patients (1). Although the precise mechanism that governs this process is not yet understood, it is speculated that early PAA degradation pathway intermediates play a role in host cell damage (2, 3).
The PAA pathway is a highly conserved metabolic solution for the assimilation of PAA, with the corresponding genes identified in 16% of sequenced bacterial genomes (3). Moreover, many structurally related compounds, including aromatic pollutants styrene and ethylbenze, are converted to phenylacetate prior to metabolism via the PAA pathway (4, 5). Although this novel, hybrid pathway is the only known route for “aerobic” metabolism of PAA, it also exploits CoA activation, a feature typical of classical “anaerobic” aromatic degradation pathways. As such, the initial enzyme of the PAA pathway, phenylacetate-CoA ligase (PCL), plays a vital role in regulating entry of potential substrates into the pathway. An unusual feature of the PAA pathway in B. cenocepacia is that it encodes two copies of the pcl gene (paaK1 and paaK2), yet harbors only a single copy of each subsequent enzyme in the pathway (2). Although a rationale for the additional PCL in B. cenocepacia has not been established, studies of related bacterial PCLs suggest the potential for altered kinetics (6–8).
PaaK1 and PaaK2 are members of the adenylate-forming enzyme (AFE) family PFAM00501, which incorporates enzymes involved in metabolism of short-to-long chain fatty acids, aromatic compounds, biosynthesis of siderophores and peptide antibiotics, and luciferases (9). The family maintains a general architecture of two α/β domains with the active site formed at the interface of the N- and C-terminal domains (10–13). Sequences reveal noteworthy divergent features of PaaK1 and PaaK2 with respect to the superfamily, including a novel N-terminal sequence spanning ∼70 residues. The general mechanism of the AFE superfamily indicates that catalysis relies on a large conformational change (9, 14), yet it is rare that a single enzyme is captured in multiple conformations. Furthermore, there are very few ATP-bound structures (15) making it difficult to refine the general catalytic mechanism of these metabolically important enzymes.
The goal of our current study is to address the outstanding question of why B. cenocepacia encodes paralogous copies of PaaK1 and PaaK2. Through high resolution x-ray co-crystal structures with both ATP and the phenylacetyl adenylate intermediate, and kinetic assays, we propose a biochemical rationale for the presence of the two isozymes and include detailed analyses of the unique structural features such as the novel N-terminal microdomain. Collectively, these data offer rare insight into multifunctional roles for key active site residues and substructures and are discussed with respect to the general catalytic mechanism AFE superfamily enzymes.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
The paaK1 (YP_002229570) gene was amplified from B. cenocepacia J2315 genomic DNA provided by Dr. Silvia Cardona (University of Manitoba, Winnipeg, Canada). A synthetic paaK2 (YP_002234323) gene with reduced GC content was ordered from GenScript. Both genes were cloned into pET28a(+) (Novagen, Mississauga, ON, Canada) vector in frame with a hexahistidine tag and thrombin cleavage site. Escherichia coli BL21 Star (DE3) cells (Invitrogen) were used for protein production in Overnight Express Instant TB (EMD Chemicals) medium supplemented with 50 μg ml−1 kanamycin (Sigma) at 32 °C. Production of selenomethionine-derivatized PaaK1 was carried out in E. coli 834 (DE3) (a methionine auxotroph; Novagen) and grown in SelenoMet medium (AthenaES) supplemented with l-selenomethionine to a final concentration of 40 μg ml−1 (AthenaES). The cells were grown at 32 °C until reaching an A600 nm of 0.6 at which isopropyl-β,d-thiogalactopyranoside was added to a final concentration of 0.75 mm.
Following 22 h of growth, the cells were harvested and lysed, and the supernatants containing either PaaK1 (both native and selenomethionine-derivatized) or PaaK2 were applied to a HisTrapFF nickel affinity column (GE Healthcare) in buffer A (20 mm Hepes, pH 7.8, 500 mm NaCl, 30 mm imidazole) supplemented with 3 mm β-mercaptoethanol and 3% (PaaK1) or 10% (PaaK2) glycerol). An increasing imidazole gradient was used to elute the protein, and purity was assessed by SDS-PAGE. Fractions were then pooled, concentrated using Centricon (Millipore) spin concentrators, and the His tag was removed by thrombin digest. Digested samples were applied to a Superdex 200 Hi-load 16/60 size-exclusion column (GE Healthcare) in buffer B (20 mm Hepes, pH 7.8, 150 mm NaCl, 3 mm β-mercaptoethanol, and 3% (PaaK1), or 10% glycerol (PaaK2)). The final purification step incorporated a Source 30Q anion exchange column (GE Healthcare) equilibrated in buffer C (20 mm Hepes, pH 8.3, 10 mm NaCl, 3 mm β-mercaptoethanol, and 2% glycerol) and eluted with an increasing concentration of NaCl.
Crystallization and Data Collection
All crystallization trials were carried out using the sitting-drop, vapor diffusion method in 96-well plates. SeMet PaaK1 (at 12 mg ml−1) was initially incubated with 3 mm MgCl2 and ATP for 1 h prior to crystallization. Diffraction quality crystals were obtained in 20–25% PEG 3350, 200 mm potassium thiocyanate, and 5% glycerol. A single PaaK1 crystal was looped into cryo-protectant consisting of reservoir solution supplemented with 3 mm MgCl2, 3 mm ATP, and 25% glycerol for 30 s and flash cooled directly in the cryo-stream (100 K). Diffraction data were collected at the Stanford Synchrotron Radiation Lightsource on beamline 9-2 at a wavelength of 0.9792 Å.
To obtain the phenylacetyl adenylate co-structures, PaaK1 and PaaK2 were concentrated to 12 mg ml−1 protein and incubated for 1 h with 3 mm MgCl2, ATP, and 5 mm phenylacetic acid prior to crystallization trials. Crystals of PaaK1 were grown 10% (w/v) PEG 8000, 0.1 m Hepes, pH 7.5, and 8.0% (v/v) ethylene glycol, and crystals of PaaK2 were grown in 17% (w/v) PEG 6000, 0.1 m Hepes, pH 7.5, 0.1 m KCl, and 2.5% glycerol. A single PaaK1 crystal was flash cooled directly in the cryo-stream (100 K), and data were collected on a Rigaku R-axis IV++ area detector. For PaaK2, a single crystal was looped and stepped slowly into cryo-protectant solution consisting of reservoir solution supplemented with 3 mm MgCl2, 3 mm ATP, 5 mm phenylacetic acid, and 10 and 15% glycerol cooled directly on the cryo-stream (100 K). Diffraction data for PaaK2 was collected on beamline 9-2 at the Stanford Synchrotron Radiation Lightsource.
Data Processing, Structure Solution, and Refinement
All data sets were processed with iMosflm (16), scaled with Scala (17), and the models were refined with REFMAC using 5% of the reflections for calculation of Rfree (18). Phasing of PaaK1 (1.6 Å resolution) was carried out with ShelxC/D/E (19), and automated building was performed using Buccaneer (20), all within the CCP4 suite of programs (21). Manual building was completed in Coot (22). PaaK1 (1.92 Å resolution) and PaaK2 (1.90Å resolution) adenylated intermediate co-structures were solved by molecular replacement using MOLREP (23) with the ATP-bound PaaK1 as the search model. The adenylated phenylacetate ligand and accompanying library file were generated using the Dundee Prodrg2 Server (24). The majority of the PaaK2 model was built with Buccaneer (20), though the majority of the C-terminal domain required manual building in Coot (22). The high resolution of the data also permitted modeling of small molecules such as glycerol, thiocyanate, and β-mercaptoethanol. Data collection and refinement statistics are presented in Table 1.
TABLE 1.
Data collection and refinement statistics
Values in parentheses are for the highest resolution shell. r.m.s.d., root mean square deviation.
| PaaK1 ATP | PaaK1 intermediate | PaaK2 intermediate | |
|---|---|---|---|
| Data collection | |||
| Space group | P1 | P1 | P21 |
| a, b, c (Å) | 56.71, 62.47, 78.49 | 57.26, 62.38, 76.60 | 69.15, 81.97, 80.89 |
| α, β, γ | 90.97, 109.81, and 106.51° | 91.14, 108.35, and 106.28° | 90.00, 97.27, and 90.00° |
| Wavelength (Å) | 0.9792 | 1.5418 | 0.97884 |
| Resolution (Å) | 40.00-1.60 (1.69-1.60) | 27.01-1.92 (2.02-1.92) | 30.00-1.90 (2.0-1.90) |
| Measured reflections | 482,646 (69,360) | 262,037 (36,580) | 28,3830 (31,225) |
| Unique reflections | 121,839 (17,505) | 68,731 (9729) | 69,914 (9527) |
| Redundancy | 4.0 (4.0) | 3.8 (3.8) | 4.1 (3.1) |
| Completeness (%) | 95.8 (94.1) | 93.8 (90.8) | 96.0 (92.9) |
| I/σ(I) | 8.6 (2.2) | 15.2 (2.7) | 9.2 (3.1) |
| Rmergea | 0.084 (0.497) | 0.056 (0.444) | 0.094 (0.327) |
| Refinement statistics | |||
| Resolution range (Å) | 29.98–1.60 (1.64–1.60) | 27.01–1.92 (1.97–1.92) | 40.99–1.90 (1.95–1.90) |
| Rcrystb | 0.166 (0.302) | 0.1806 (0.327) | 0.168 (0.232) |
| Rfreec | 0.193 (0.331) | 0.2319 (0.433) | 0.214 (0.289) |
| No. of atoms | |||
| Protein (chain A, B) | 3344, 3366 | 3317, 3312 | 3339, 3327 |
| Solvent | 952 | 664 | 860 |
| ATP | 62 | N/A | N/A |
| Phenylacetyl Adenylate | N/A | 64 | 64 |
| Phenylacetate | N/A | 10 | N/A |
| Glycerol | 30 | N/A | N/A |
| Thiocyanate | 27 | N/A | N/A |
| β-mercaptoethanol | 20 | 12 | 4 |
| PEG | 12 | 17 | 32 |
| Mg | 6 | 4 | 2 |
| B-values (Å2) | |||
| Protein (chain A, B) | 21.33, 18.55 | 26.63, 32.98 | 19.74, 19.54 |
| Solvent | 34.34 | 38.45 | 31.12 |
| ATP | 14.14 | N/A | N/A |
| Phenylacetyl adenylate | N/A | 21.74 | 12.52 |
| Phenylacetate | N/A | 63.95 | N/A |
| Glycerol | 34.20 | N/A | N/A |
| Thiocyanate | 44.56 | N/A | N/A |
| β-mercaptoethanol | 55.46 | 47.31 | 44.40 |
| PEG | 45.17 | 51.53 | 48.34 |
| Mg | 19.78 | 38.46 | 22.12 |
| R.m.s.d. from ideality | |||
| Bond lengths (Å) | 0.009 | 0.005 | 0.007 |
| Bond angles | 1.199° | 0.798° | 0.962° |
| Ramachandran statistics (%) | |||
| Most favored | 92.7 | 91.4 | 92.1 |
| Allowed | 7.1 | 8.5 | 7.5 |
| Disallowed | 0.0 | 0.0 | 0.0 |
a Rmerge = ΣhklΣi|Ihkl,i − [Ihkl]|/ΣhklΣiIhkl,i, where [Ihkl] is the average of symmetry related observations of a unique reflection.
b Rcryst = Σ|Fobs − Fcalc/ΣFfobs, where Fobs and Fcalc are the observed and the calculated structure factors, respectively.
c Rfree is R using 5% of reflections randomly chosen and omitted from refinement.
Enzyme Assays and Kinetics
Enzyme activity measurements for PaaK1 and PaaK2 were carried out using the indirect spectrophotometric assay described by Ziegler et al. (25) and adapted to a 96-well plate (11). Briefly, the assay is based on linking production of AMP by PaaK1 and PaaK2 to the oxidation of NADH to NAD, which is measured spectrophotometrically at 365 nm. The assay utilized 3 μg of PaaK1 or PaaK2 per 200 μl of reaction volume. Concentrations ranging from 20 to 1000 μm phenylacetic acid were tested with saturating concentrations (2 mm) of co-substrates CoA and ATP. Similarly, for the substituted phenylacetate substrates, 0.1–3 mm concentrations were tested using 2 μg of PaaK1 or PaaK2. Activity is reported as none detected where change in absorbance with 3 mm substrate was not discernible from that of the negative control showing spontaneous decay of NADH.
RESULTS AND DISCUSSION
Dimerization Leads to Formation of an Intermolecular Extended β-Sheet
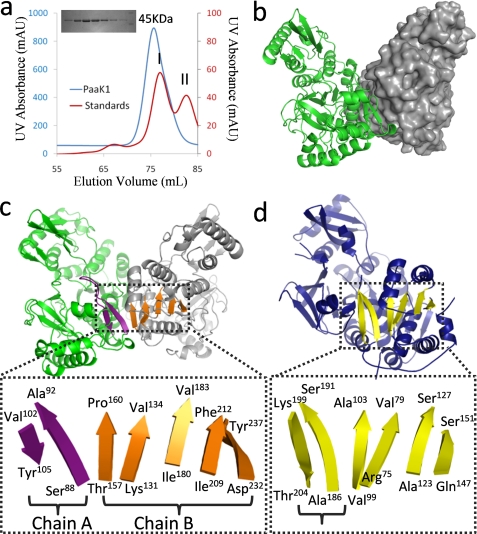
The size exclusion elution profile of PaaK1 indicated a dimeric organization (Fig. 1a) and accordingly, PaaK1 crystallized as an intimate dimer in space group P1 (Fig. 1b). The individual PaaK1 monomers are structurally equivalent with an root mean square deviation of 0.251 Å over 376 Cα atoms. In forming the dimer interface, ∼1383Å2 of surface area is buried, which is stabilized through extensive shape and chemical complementarity. Nonpolar residues comprise ∼55% of the interface and are accompanied by 17 hydrogen bonds and two salt bridges (Arg89 and Glu152). It is noteworthy that all of the residues participating in dimerization are derived from the N-terminal domain leaving the C-terminal domain free to undergo the conformational reorganization between the adenylation (phenylacetate + ATP → phenylacetyl adenylate + pyrophosphate) and thioesterification (phenylacetyl adenylate + CoA → phenylacetyl-CoA + AMP) reactions.
FIGURE 1.
Dimerization of PaaK1 reconstitutes β-sheet organization. a, size exclusion chromatogram with an inset SDS-PAGE gel. PaaK1 (molecular mass, 48 kDa) eluted as an apparent dimer (molecular mass, 82 kDa). Molecular mass (MW) standards conalbumin (I; 75 kDa) and ovalbumin (II; 43 kDa) are shown. b, secondary structure and surface representation of the symmetrical homodimer of PaaK1 with one monomer displayed in green and the second in gray. c, intimate dimerization interactions of PaaK1 create composite β-sheets spanning both monomers. The two-stranded sheet flanking the P-loop (purple) is abutted by the distorted, five-strand sheet (orange) of the second monomer. d, crystal structure of benzoate-CoA ligase (Protein Data Bank code 2V7B; blue) shows the typical familiar tertiary structure with the β-sheet highlighted yellow. The antiparallel strands flanking the P-loop (bracketed) form part of the sheet.
The assembly of the symmetrical PaaK1 dimer results in the formation of two extended, intermolecular seven-strand β-sheets each composed of a two-strand antiparallel β-sheet (Fig. 1c, purple) from one monomer and a five-strand distorted β-sheet from the second monomer (Fig. 1c, orange). Interestingly, the topology of the extended β-sheets observed in the PaaK1 dimer appear to mimic a similar arrangement defined within a single monomer of family members (Fig. 1d) (11, 26–28). The multimeric requirement of PaaK1 to recapitulate the extended β-sheet may indicate a branching point in the evolution of CoA ligases.
A Novel N-terminal Microdomain in PaaK1 and PaaK2
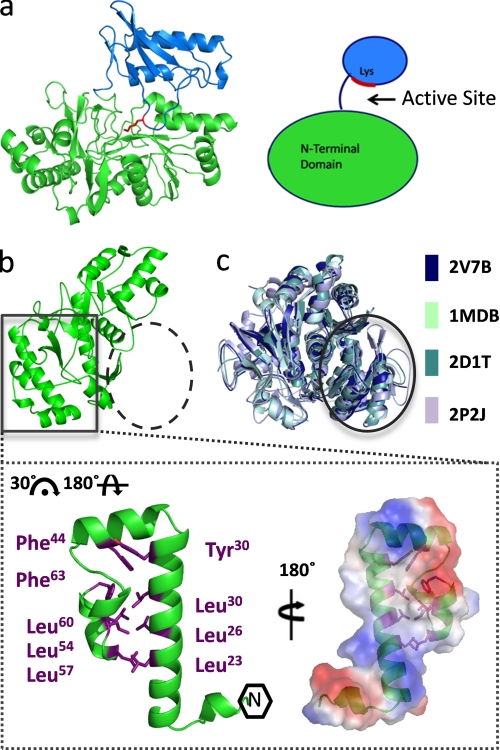
The PaaK1 monomer is composed of a larger N-terminal domain incorporating Pro4–Gly325 (Fig. 2a, green), which as described above, forms the dimer interface and a smaller C-terminal domain comprising residues Met331–Arg430 (Fig. 2a, blue). The domains are connected by a short, solvated linker (326RSDDM330), with the active site formed at the domain interface. The N-terminal domain of PaaK1 is comprised of three β-sheets sandwiched between nine α-helices, whereas the C-terminal domain is defined by two helices, a short two-strand antiparallel β-sheet, and a twisted four-stranded sheet. In the PaaK1 ATP co-structure, the C-terminal domain is properly oriented to position the invariant Lys422 (Fig. 2a, red) within the active site, consistent with superfamily members occupying conformation 1 (11, 14, 15, 26, 28).
FIGURE 2.
Novel N-terminal microdomain. a, monomer structure of PaaK1 with the larger N-terminal domain in green, the C-terminal domain in blue, and active site conserved lysine shown in red. b, PaaK1 contains a small helical bundle arrangement at the N terminus (boxed) but lacks the typical N-terminal arrangement exhibited by family members (dashed circle). Inset, helical bundle microdomain of PaaK1 is largely stabilized by hydrophobic interactions of Leu and Phe side chains. The electrostatic surface of the microdomain interface predicts a large hydrophobic patch surrounded by charged patches. c, overlays of four homologous family members (Protein Data Bank code 2V7B, benzoate-CoA ligase; Protein Data Bank code 1MDB, 2,4-dihydroxybenzoate AMP ligase; Protein Data Bank code 2D1T, firefly luciferase; Protein Data Bank code 2P2J, acetyl-CoA synthetase) demonstrate the typical α/β-sandwich arrangement at the N terminus (circle).
Although the general architectural features of PaaK1 conform to the homologous members in the AFE superfamily (14), extensive structural divergence is observed within the N-terminal ∼70 residues. Intriguingly, this region possesses negligible sequence or structural identity with any structurally characterized AFE family member, and thus, we propose that bacterial PCLs such as PaaK1 and PaaK2 constitute a separate subgroup within the superfamily. This region of PaaK1 adopts a continuous three helical bundle structure measuring ∼35 × 20 Å (Fig. 2b). Reminiscent of a leucine zipper, six leucine residues participate in hydrophobic interactions, with additional contributions from Tyr30, Phe44, and Phe63 (Fig. 2b). Collectively, these interactions stabilize what appears to be a compact microdomain. Despite the predicted stability of this substructure, it thoroughly integrated with the larger, conserved portion of the N-terminal domain (Thr67–Arg326) with a complexation significance score of 1.00 (29). The overall interface is formed through complementary basic and acid patches resulting in nine salt bridges and 26 hydrogen bonds. Hydrophobic interactions also have a significant presence with a hydrophobic segment defined by 295ALPII299 on the larger N-terminal domain inserting into the hydrophobic cavity formed at the center of the N-terminal helical bundle. Though no function is immediately apparent for this novel substructure, it may be utilized for protein-protein interactions, perhaps facilitating recruitment of subsequent PAA enzymes such as the multicomponent oxygenase, PaaABCDE. As a result of this remodeled N-terminal microdomain, PaaK1 is unable to adopt the typical familial arrangement of the initial ∼200 residues, normally consisting of four β-sheets sandwiched between five helices (Fig. 2c) (11, 12, 26, 28). Consequently, the P-loop flanking β-strands of PaaK1 are isolated in the monomer structure, necessitating dimerization for reconstitution of this β-sheet environment. However, apart from the N-terminal regions, the core N-terminal domain of PaaK is highly homologous with related family members such as benzoate-CoA ligase (Protein Data Bank code 2V7B) (11), displaying an root mean square deviation of 1.72 Å over 141 Cα atoms. The RCSB coordinate file and structure factor codes for the 1.60 Å crystal structure of PaaK1 in complex with ATP are 2Y27 and r2y27sf, respectively. The RCSB coordinate file and structure factor codes for the 1.92 Å crystal structure of PaaK1 in complex with the adenylated phenylacetate intermediate are 2Y4N and r2y4nsf, respectively. The RCSB coordinate file and structure factor codes for the 1.90 Å crystal structure of PaaK2 in complex with the adenylated phenylacetate intermediate are 2Y4O and r2y4osf, respectively.
Pre- and Postadenylation Complexes of PaaK1 Reveal Dynamic Enzyme-Substrate Interactions
The ability to discern dynamic information from crystal structures relies in part on being able to co-crystallize the enzyme at different stages of substrate turnover. In the case of an AFE, the first two stages can be defined as the pre- and postadenylation state. It is noteworthy that capturing these different states in the context of one enzyme is rare, with DltA from Bacillus cereus serving as the only reported example (15, 30). To explore the reorganization of key structural features in PaaK1 as the enzyme progresses through the adenylation reaction, we report the high resolution structures of PaaK1 in complex with ATP (Fig. 3a) and the adenylated phenylacetate intermediate (Fig. 3b).
FIGURE 3.
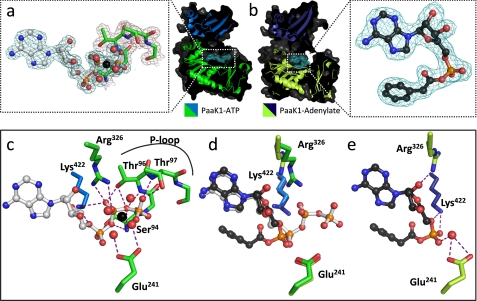
Comparison of conserved protein-substrate interactions pre- and post-adenylation reaction. a, secondary structure (green, N-terminal domain; marine blue, C-terminal domain) and surface (dark gray) of PaaK1 with ATP (teal electron density mesh) tightly bound within the active site. Inset, the well ordered P-loop of PaaK1 (green) envelopes the phosphates of the ATP (gray). Mg2+ coordinates the β- and γ-phosphates of ATP in addition to four water molecules. 2Fo − Fc electron density mesh is shown contoured to 1.0 σ. b, PaaK1 with phenylacetyl adenylate intermediate bound within the active site. (lime green, N-terminal domain; deep blue, C-terminal domain; dark gray, surface). Electron density mesh for the intermediate compound is shown (teal). Inset, the well ordered phenylacetyl-adenylate intermediate with 2Fo − Fc electron density mesh contoured to 1.0 σ. c, polar contacts (purple dashes) between conserved residues of PaaK1 (green) and ATP (gray). d, overlay comparison of conserved residues of PaaK1/ATP (green/marine blue) and PaaK1/adenylate (lime green/deep blue) structure. e, positions of conserved active site residues following the adenylation reaction with purple dashed lines representing polar interactions.
P-loop
The PaaK1 ATP co-structure displays a well ordered P-loop (93SSGTTGKPT101) that envelopes the β and γ phosphates of ATP (Fig. 3a, inset, green residues). Five hydrogen bonds between the backbone amides of the P-loop and the ATP phosphates stabilize the P-loop and are accompanied by three additional hydrogen bonds from Ser94, Thr96, and Thr97 side chains (Fig. 3c). Based on the stability imparted by this hydrogen bond network, it is not surprising that removal of the pyrophosphate in transitioning to the adenylate intermediate results in P-loop becoming disordered and therefore unmodeled (Fig. 3, d and e). These data are consistent with the classical expectations of the P-loop to bind the phosphates of ATP. Once the β and γ are removed, the P-loop does not contribute to binding the adenylate intermediate within the active site.
Lys/Arg Pair Orient Phosphates for Nucleophillic Attack
Initially, the catalytically essential lysine (Lys422) forms bifurcated hydrogen bonds with the α- and β-phosphates of ATP (Fig. 3c), where it is proposed to shield point charges and facilitate nucleophillic attack at the α-phosphate (15). Despite the flexibility of the lysine residue, only the ϵ-amino of Lys422 is reoriented in the PaaK1-phenylacetyl adenylate co-structure such that it forms two hydrogen bonds with the α phosphate (Fig. 3, d and e). These results are consistent with the lysine playing an essential catalytic role in the adenylation reaction, with the high density positive charge localized to the reactive center of the ATP, the α-phosphate.
In contrast, the active site arginine of the conserved interdomain linker, Rx(D/K)x6G, exhibits a much greater degree of movement between the two PaaK1 crystal structures (Fig. 3d). In the ATP/preadenylation complex, Arg326 of PaaK1 forms bidentate hydrogen bonds with the β-phosphate of ATP (Fig. 3a). In the adenylate intermediate co-structure, Arg326 is reoriented such that it hydrogen bonds with the 2′ ribose hydroxyl of the phenylacetyl adenylate intermediate (Fig. 3, d and e). Thus, our data is consistent with a proposed bifunctional role for Arg326 where initial stabilization of the ATP phosphates is replaced with a stabilizing role of the adenylated intermediate (15).
Coordinated Mg2+ Ion
The β- and γ-phosphates of ATP coordinate a single Mg2+ ion that is further coordinated by four water molecules in an overall octahedral geometry (Fig. 3, a and c). Abutting this arrangement is the conserved Glu241 that hydrogen bonds with two of the coordinated water molecules (Fig. 3c). Glu241 of the PaaK1-adenylate structure has essentially maintained its position from that of the ATP-bound structure, despite the departure of the β- and γ-phosphates and solvated Mg2+ from the active site (Fig. 3, d and e). Instead, a single bridging water molecule enables the Glu241 to interact indirectly with the α-phosphate in the PaaK1-adenylate structure (Fig. 3e).
It is clear that the β- and γ-phosphates require numerous interactions for correct orientation for in-line attack from the carboxylate oxygen of the phenylacetate substrate. The bifunctional role of the interdomain arginine and active site glutamate suggested by our structural data are consistent with DltA mutagenesis data where mutation of either the arginine or glutamate to glutamine reduced the kcat by ∼20- and 14-fold, respectively. The Km, however, was increased by 6- and 8-fold likely due to the critical role for these residues in anchoring the adenylate intermediate (15). Collectively, these data reveal bifunctional strategies for residues that initially contort the terminal phosphoryl groups of the ATP to adopt new catalytically important interactions as the reaction cycle progresses.
P-loop Functions to Stabilize C-terminal Domain in Conformation 2
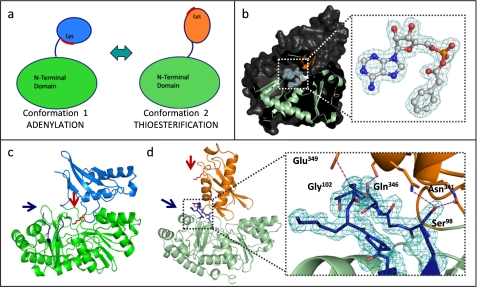
As members of the AFE superfamily, bacterial phenylacetate-CoA ligases such as PaaK1 and PaaK2 undergo large conformational changes to remodel the single active site for two distinct part reactions (Fig. 4a). Notably, PaaK2 was captured following the domain reorientation in conformation 2 (Fig. 4, b and d), thus presenting the opportunity for additional insight into structural reorganization during the course of catalysis. In contrast to both PaaK1 complexes, the invariant lysine (Lys429 in PaaK2) is now completely removed from the active site and shifted ∼29 Å from its previous position (Fig. 4, c and d). Unexpectedly, the P-loop is well ordered (Fig. 4d), which contrasts with the P-loop in the Paak1 adenylated intermediate capture in conformation 1. Stability of the reorganized P-loop in conformation 2 is mediated by hydrogen bonds between the backbone carbonyl of Thr100 and of Ser345 (Oγ), between Glu349 (Oϵ2) and the backbone amide nitrogen of Gly102, as well as a bifurcated hydrogen bond between Ser98 (Oγ), Gln346 (Nϵ2), and the backbone carbonyl of Asn341. These data strongly suggest that the P-loop plays a critical bifunctional role in the AFE superfamily where its role in coordinating ATP in the adenylation part reaction is replaced by a role in stabilizing the C-terminal domain in conformation 2 during the thioesterification part reaction. Indeed, an ordered P-loop has been observed previously for AFE enzymes captured in conformation 2 (9, 10, 31, 32), where polar interactions take place between the P-loop and C-terminal domain and often incorporate solvent molecules. Cycling of P-loop from ordered (PaaK1:ATP − conformation 1) to disordered (PaaK1:adenylate intermediate − conformation 1) back to ordered (PaaK2:adenylate intermediate − conformation 2) offers a rare and seamless view of the entire process of structural reorganization during substrate turnover.
FIGURE 4.
PaaK1 and PaaK2 phenylacetyl-adenylate co-structures captured in alternate conformations. a, schematic of the domain alternation hypothesis. Conformation 1 is utilized for the adenylation part reaction for which the invariant lysine is essential. The C-terminal domain then rotates relative to the N-terminal domain, clearing the lysine from the active site and presenting an alternate face for the thioesterification reaction. b, secondary structure (light green, N-terminal domain; orange, C-terminal domain) and surface (dark gray) representation of PaaK2 captured in conformation 2 with the adenylate intermediate bound within the active site. Inset, 2Fo − Fc electron density mesh (teal) is contoured to 1.0 σ. c, secondary structure representation of PaaK1 phenylacetyl adenylate co-structure in conformation 1 (green, N-terminal domain; light blue, C-terminal domain) with the invariant Lys (red arrow) positioned within the active site. The P-loop of this structure is disordered and not modeled in entirety (blue arrow). d, secondary structure representation of PaaK2 (N-terminal domain, light green; C-terminal domain, orange) in conformation 2. The P-loop is shown in blue, and Lys429 is shown in red. Inset, 2Fo − Fc electron density mesh contoured to 1.0 σ surrounding the P-loop of PaaK2. The P-loop forms four mutually stabilizing hydrogen bonding interactions (purple dashes) with the C-terminal domain.
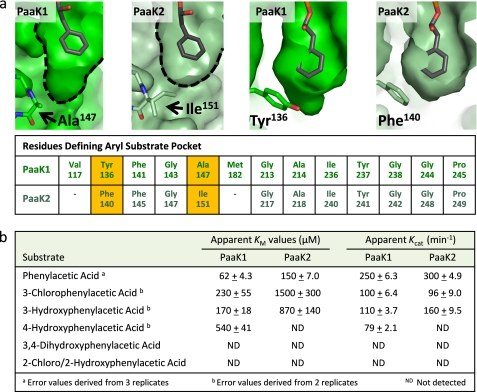
An Extended Aryl Binding Pocket in PaaK1 Contrasts with PaaK2
Both the PaaK1 and PaaK2 adenylate intermediate co-structures displayed well ordered electron density within the active site, permitting the phenylacetyl adenylate to be accurately modeled with low average B-factors of 26.69 and 12.12 Å2, respectively (Figs. 3b and 4c and Table 1). Detailed structural comparison of these forms revealed diversity in the aryl binding pockets (Fig. 5a). In PaaK1, the base of the pocket is defined by Ala147 resulting in a pocket ∼7 Å wide by 10 Å deep. In PaaK2, an isoleucine (Ile151) takes the place of Ala147 in PaaK1 resulting in the pocket being reduced in depth to ∼7 Å. Intriguingly, 4-chlorobenzoate-CoA ligase of Alcaligenes sp. AL3007, which accepts 4-chlorobenzoate as its native substrate, also incorporates an Ala residue (Ala213) at this position (33). Therefore, although the active site pockets in both PaaK paralogs accommodate the phenyl group of the phenylacetate intermediate, the extension in PaaK1 may endow the enzyme with the ability to bind a broader range of substrates with substitutions on phenyl ring. The only additional change in the active site pocket is the substitution of Tyr136 in PaaK1 for Phe140 in PaaK2 (Fig. 5a). These residues overlay nearly perfectly and do not contribute to the volume difference, however the hydroxyl group of Tyr136 would likely allow PaaK1 to better accommodate polar ring substitutions.
FIGURE 5.
Comparative analysis of the aryl substrate binding pockets of PaaK1 and PaaK2. a, top panels show surface representation of PaaK1 and PaaK2 pockets with bound phenylacetyl adenylate. Side chains of defining residues that differ between the paralogs are shown as sticks along with a tabular list of residues defining the aryl substrate binding pocket of the PaaK paralogs with substitutions highlighted in yellow. As a result of the Ala/Ile substitution between PaaK1 and PaaK2 and presence the bulkier Ile151 side chain of PaaK2, the equivalent residues of Val117 and Met182 in PaaK2 do not form part of the substrate binding pocket and are not included in the figure. b, shown are the kinetic parameters determined for PaaK1 and PaaK2.
PaaK1 Displays a Lower Km for Phenylacetic Acid and Broader Substrate Specificity Than PaaK2
Prior to this study, there was no rationale for the presence of two copies of PaaK in B. cenocepacia. One intriguing possibility was that the paralogs would exhibit different kinetic parameters and substrate specificities as a result of substrate pocket remodeling. This hypothesis was based on the previously observed wide range of Km values and substrate specificities for orthologous PCLs, coupled with the fact that PaaK1 and PaaK2 share only 69% identity and therefore may display substitutions within the substrate binding pocket. Indeed, two residue substitutions resulted in structural divergence between the paralogs. To determine how these structural differences translate to functional profiles, kinetic analyses were undertaken using phenylacetic acid as well as a variety of ring-substituted phenylacetic acids.
The most dramatic difference between the paralogs for the native phenylacetic acid substrate was observed for the Km, with PaaK2 (150 μm) more than double that of PaaK1 (62 μm) (Fig. 5b). The apparent Vmax and calculated kcat values were more similar, with PaaK2 displaying a 20% higher kcat at 300 min−1 (Fig. 5b). Collectively, these results show that despite the additional volume of the aryl substrate binding pocket, PaaK1 maintains a higher affinity for phenylacetic acid. Furthermore, PaaK1 was also able to better accommodate the substituted phenylacetic acid substrates, especially the 4-hydroxyphenylacetic acid, for which PaaK2 did not show detectable activity. Although both paralogs were able to accept 3′ substituted phenylacetic acid, PaaK1 exhibited a much lower Km (∼6.5-fold) than PaaK2, likely due to the extension of the aryl binding pocket. Not surprisingly, neither paralog was active toward 2′ substituted phenylacetic acid, which would position the substituent toward either the adenine ring or the helical backbone defining one side of the pocket. Similarly, 3,4-substituted phenylacetic acid was not accepted by either paralog, demonstrating that PaaK1 could accommodate either a 3′ or to a lesser extent a 4′ substituent, but not both, within the extended pocket. In the case of 4-chlorobenzoate-CoA ligase, it was possible to expand the substrate range to 3′ and 3′4′-substituted chlorobenzoate through mutation of the active site residue Ile303 to Ala and Gly (33). Mutation of the corresponding Ile of PaaK1 (Ile236) or PaaK2 (Ile240), which is positioned between the 3′ and 4′ positions of the phenyl ring, would likely also improve activity with of 3′- and 4′-substituted phenylacetate.
Conclusions
With PaaK1 and PaaK2 catalyzing the first and only committed step of the pathway (34), these enzymes are perfectly positioned to control the flow of phenylacetic acid into the PAA pathway thereby contributing to the nutritional requirements of B. cenocepacia in infected cystic fibrosis patients. High resolution crystal structures of PaaK1 and PaaK2 captured at different catalytic stages offer insight into the dynamic cycling of the P-loop, which likely serves to promote conformational change in the C-terminal domain, and the subtle, yet critical, repositioning of multipurpose active site residues such Arg326 and Glu241. Furthermore, our data provides a clear rationale for the existence of paralogous copies of PaaK1 and PaaK2 through divergent aryl binding pocket structure and corresponding kinetic profile. Incorporation of Ala at position 147 (Ile151 in PaaK2) and Tyr at position 136 (Phe140 in PaaK2) in the aryl binding pocket results in a lower Km for PaaK1 with the native substrate and a broader substrate specificity profile, relative to PaaK2.
It is conceivable that either the more (PaaK1) or less (PaaK2) active copy would be more beneficial depending on carbon source availability. Thus, although PaaK2 has been shown to be sufficient for growth on phenylacetic acid as a sole carbon source (2), a second more active copy (PaaK1) may be beneficial during infection. In fact, recent microarray studies of J2315 have shown that both paaK1 and paaK2 are up-regulated 10- and 8.9-fold respectively, during growth in synthetic cystic fibrosis media, whereas subsequent pathway genes such as paaF, paaG, paaJ, and paaI are only up-regulated only 2–5-fold (35). Intriguingly, high throughput RNA-seq studies using the closely related pathogenic strain HI2424 found increased transcript levels of the paaK1 ortholog during synthetic cystic fibrosis media growth (36). Because the PaaKs of J2315 are 98% identical to HI2424 with no substitutions in the aryl binding pocket, the HI2424 PaaKs likely display similar kinetic profiles further supporting a biological role for the structural and kinetic differences between PaaK1 and PaaK2 established by this study.
This work was supported by a research grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (to M. J. B.).
The atomic coordinates and structure factors (codes 2Y27, 2Y4N, and 2Y4O) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PAA
- phenylacetic acid
- PaaK1
- phenylacetate-CoA ligase 1
- PaaK2
- phenylacetate-CoA ligase 2
- AFE
- adenylate-forming enzyme
- PCL
- phenylacetate-CoA ligase.
REFERENCES
- 1. Hunt T. A., Kooi C., Sokol P. A., Valvano M. A. (2004) Infect. Immun. 72, 4010–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Law R. J., Hamlin J. N., Sivro A., McCorrister S. J., Cardama G. A., Cardona S. T. (2008) J. Bacteriol. 190, 7209–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Teufel R., Mascaraque V., Ismail W., Voss M., Perera J., Eisenreich W., Haehnel W., Fuchs G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14390–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Velasco A., Alonso S., García J. L., Perera J., Díaz E. (1998) J. Bacteriol. 180, 1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corkery D. M., O'Connor K. E., Buckley C. M., Dobson A. D. (1994) FEMS Microbiol. Lett. 124, 23–27 [DOI] [PubMed] [Google Scholar]
- 6. Erb T. J., Ismail W., Fuchs G. (2008) Curr. Microbiol. 57, 27–32 [DOI] [PubMed] [Google Scholar]
- 7. Martínez-Blanco H., Reglero A., Rodriguez-Aparicio L. B., Luengo J. M. (1990) J. Biol. Chem. 265, 7084–7090 [PubMed] [Google Scholar]
- 8. El-Said Mohamed M. (2000) J. Bacteriol. 182, 286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reger A. S., Wu R., Dunaway-Mariano D., Gulick A. M. (2008) Biochemistry 47, 8016–8025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gulick A. M., Starai V. J., Horswill A. R., Homick K. M., Escalante-Semerena J. C. (2003) Biochemistry 42, 2866–2873 [DOI] [PubMed] [Google Scholar]
- 11. Bains J., Boulanger M. J. (2007) J. Mol. Biol. 373, 965–977 [DOI] [PubMed] [Google Scholar]
- 12. Gulick A. M., Lu X., Dunaway-Mariano D. (2004) Biochemistry 43, 8670–8679 [DOI] [PubMed] [Google Scholar]
- 13. Conti E., Stachelhaus T., Marahiel M. A., Brick P. (1997) EMBO J. 16, 4174–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gulick A. M. (2009) ACS Chem. Biol. 4, 811–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Osman K. T., Du L., He Y., Luo Y. (2009) J. Mol. Biol. 388, 345–355 [DOI] [PubMed] [Google Scholar]
- 16. Leslie A. G. (1991) Recent Changes to the MOSFLM Package for Processing Film and Image Plate Data, SERC Laboratory, Daresbury, Warrington, United Kingdom [Google Scholar]
- 17. Evans P. (2006) Acta Crystallogr. D 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 18. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 19. Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 20. Cowtan K. (2006) Acta Crystallogr. D 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 21. Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D 50, 760–76315299374 [Google Scholar]
- 22. Emsley P., Cowtan K. (2004) Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 23. Vagin A., Teplyakov A. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 24. Schüttelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 25. Ziegler K., Buder R., Winter J., Fuchs G. (1989) Arch. Microbiol. 151, 171–176 [Google Scholar]
- 26. May J. J., Kessler N., Marahiel M. A., Stubbs M. T. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12120–12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reger A. S., Carney J. M., Gulick A. M. (2007) Biochemistry 46, 6536–6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakatsu T., Ichiyama S., Hiratake J., Saldanha A., Kobashi N., Sakata K., Kato H. (2006) Nature 440, 372–376 [DOI] [PubMed] [Google Scholar]
- 29. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 30. Du L., He Y., Luo Y. (2008) Biochemistry 47, 11473–11480 [DOI] [PubMed] [Google Scholar]
- 31. Hisanaga Y., Ago H., Nakagawa N., Hamada K., Ida K., Yamamoto M., Hori T., Arii Y., Sugahara M., Kuramitsu S., Yokoyama S., Miyano M. (2004) J. Biol. Chem. 279, 31717–31726 [DOI] [PubMed] [Google Scholar]
- 32. Yonus H., Neumann P., Zimmermann S., May J. J., Marahiel M. A., Stubbs M. T. (2008) J. Biol. Chem. 283, 32484–32491 [DOI] [PubMed] [Google Scholar]
- 33. Wu R., Reger A. S., Cao J., Gulick A. M., Dunaway-Mariano D. (2007) Biochemistry 46, 14487–14499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mohamed Mel-S., Ismail W., Heider J., Fuchs G. (2002) Arch. Microbiol. 178, 180–192 [DOI] [PubMed] [Google Scholar]
- 35. Yoder-Himes D. R., Konstantinidis K. T., Tiedje J. M. (2010) PLoS One 5, e8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoder-Himes D. R., Chain P. S., Zhu Y., Wurtzel O., Rubin E. M., Tiedje J. M., Sorek R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3976–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]