Abstract
Ataxia-telangiectasia (A-T) and Nijmegen breakage syndrome (NBS) are related genomic instability syndromes characterized by neurological deficits. The NBS1 protein that is defective in NBS is a component of the Mre11/RAD50/NBS1 (MRN) complex, which plays a major role in the early phase of the complex cellular response to double strand breaks (DSBs) in the DNA. Among others, Mre11/RAD50/NBS1 is required for timely activation of the protein kinase ATM (A-T, mutated), which is missing or inactivated in patients with A-T. Understanding the molecular pathology of A-T, primarily its cardinal symptom, cerebellar degeneration, requires investigation of the DSB response in cerebellar neurons, particularly Purkinje cells, which are the first to be lost in A-T patients. Cerebellar cultures derived from mice with different mutations in DNA damage response genes is a useful experimental system to study malfunctioning of the damage response in the nervous system. To clarify the interrelations between murine Nbs1 and Atm, we generated a mouse strain with specific disruption of the Nbs1 gene in the central nervous system on the background of general Atm deficiency (Nbs1-CNS-Δ//Atm−/−). This genotype exacerbated several features of both conditions and led to a markedly reduced life span, dramatic decline in the number of cerebellar granule neurons with considerable cerebellar disorganization, abolishment of the white matter, severe reduction in glial cell proliferation, and delayed DSB repair in cerebellar tissue. Combined loss of Nbs1 and Atm in the CNS significantly abrogated the DSB response compared with the single mutation genotypes. Importantly, the data indicate that Atm has cellular roles not regulated by Nbs1 in the murine cerebellum.
Keywords: Ataxia, Cell Division, DNA Damage, DNA Repair, Neuron, Atm, Nbs1, Nijmegen Breakage Syndrome, Ataxia-Telangiectasia
Introduction
Double strand breaks (DSBs)4 in the DNA are powerful activators of the DNA damage response (DDR) (1–4). Eukaryotic cells repair DSBs either by non-homologous end-joining, an error-prone ligation mechanism, or via a high fidelity process based on homologous recombination between sister chromatids (5, 6). However, the cellular response to this lesion is far greater than just the repair process (4). The DSB response is a multi-tiered process that is initiated by sensor/mediator proteins that are recruited to the damaged sites where they generate expanding nuclear foci (2, 3, 8, 9). These proteins are involved in the initial processing of the damage and activation of the transducers, nuclear protein kinases that convey the DNA damage alarm to numerous downstream effectors (2, 3, 11, 12).
An extensively documented DSB sensor/mediator is the Mre11/Rad50/Nbs1 (MRN) complex, composed of the Mre11 nuclease, the structural maintenance of chromosomes (SMC) protein Rad50, and the NBS1 protein. The complex is rapidly recruited to DSB sites where it tethers and processes the broken ends (13–16). ATM is a serine-threonine kinase with a wide range of substrates (11, 12) that is rapidly activated in response to DSBs (17, 18) and phosphorylates a plethora of key players in many damage response pathways (11, 12). ATM belongs to a conserved family of “PI3K-like protein kinases” (19, 20), which includes among others the catalytic subunit of the DNA-dependent protein kinase, which is involved in non-homologous end-joining of DSBs (21) and ATR (ATM- and Rad3-related) protein. ATR is activated primarily by stalled replication forks, UV damage, and single-stranded DNA stretches formed during DSB processing. ATR responds to DSBs later and with slower kinetics compared with ATM (22–25). Although ATM and ATR share substrates in the DSB response, they exhibit selective substrate specificities in response to different genotoxic stresses and different DSB inducers, and there is evidence of close cross-talk between them (25–30).
Loss or inactivation of ATM leads to a severe genomic instability syndrome, ataxia-telangiectasia (A-T). A-T is characterized by progressive cerebellar degeneration, immunodeficiency, genome instability, gonadal dysgenesis, extreme radiosensitivity, and a high incidence of lymphoreticular malignancies (31, 32). One of the most severe symptoms of A-T, cerebellar ataxia, develops progressively into general motor dysfunction; it is accompanied by a marked loss of Purkinje and granule neurons in the cerebellum (31, 33).
Functional relationships between ATM and the MRN complex were initially implied by a phenotypic similarity between the syndromes caused by their abrogation. A-T-like disease, which is a mild form of A-T with later age of onset and slower progression (34), is caused by hypomorphic mutations in the MRE11 gene (35–37). The phenotypic similarity between A-T and A-T-like disease reflects the requirement of the MRN complex for ATM activation (16, 18, 38–40). On the other hand, hypomorphic NBS1 mutations underlie the Nijmegen breakage syndrome (NBS), characterized by immunodeficiency, genomic instability, radiation sensitivity, and predisposition to lymphoid malignancies. Interestingly, the neurological manifestations of NBS include microcephaly and mental deficiency rather than cerebellar degeneration and ataxia (41). A recent study showed that DNA signaling in the nervous system differs between A-T-like disease and NBS and may explain their different neuropathologies (42).
A major tool in the investigation of human genetic disorders is the corresponding knock-out mice. Atm-deficient mice exhibit many of the characteristics of human A-T, such as retarded growth, immunodeficiency, cancer predisposition, radiosensitivity, infertility, and a cellular phenotype similar to that of A-T cells, but they barely show the most cardinal feature of A-T; that is, neuronal degeneration and associated neuromotor dysfunction (43–46). Careful analysis of these mice did show subtle neurological abnormalities in several instances (46–55) and a reduction in in vitro survival of Atm−/− Purkinje cells and their dendritic branching compared with wild type (WT) cells (56, 57). However, these defects were far from the severe human phenotype. A simple explanation for the intriguing lack of overt cerebellar degeneration in Atm-deficient mice might be that generation of the A-T cerebellar phenotype in the mouse requires suppression of the DNA damage response system beyond that achieved by eliminating ATM.
Support for this notion was obtained when the Nbs1-CNS-Δ mouse was generated by knocking out the Nbs1 gene in the murine nervous system that encodes the murine Nbs1 protein (58). This mouse exhibits a dramatic neurological phenotype that combines the microcephaly typical of human NBS patients with proliferation arrest of granule cell progenitors and apoptosis of post-mitotic neurons in the cerebellum that leads to severe ataxia. This growth arrest was p53-mediated, as p53 ablation rescued much of this phenotype. This phenotype can be explained by the requirement of the MRN complex for processes that are not under ATM control, such as ATR-dependent branches of the DDR (38, 40, 59–61). Thus, concomitant abrogation of both the ATM- and ATR-dependent axes of the DNA damage response might be necessary to evoke an effect in the murine CNS similar to that of ATM loss in humans. This valuable mouse model facilitates investigation of the role of the DDR in the nervous system, which is critical for understanding the molecular pathogenesis of A-T.
Here, we report that Atm loss in conjunction with conditional knock-out of the Nbs1 gene in the CNS exacerbates the effects of Nbs1 inactivation. Using cultured cerebellar slices, we investigate the functional relationships between Atm and Nbs1 in this tissue. Importantly, the results indicate that Atm has cellular functions that are not dependent on Nbs1.
EXPERIMENTAL PROCEDURES
Generation of Various Nbs1/Atm Genotypes
Atm+/− mice (43) were a generous gift from Dr. Anthony Wynshaw-Boris (University of California, San Diego, CA). Offspring of these mice were genotyped using PCR-based assays based on mouse-tail DNA prepared with the GenElute Mammalian Genomic DNA Miniprep kit (Sigma). Mice in this study have an SV129 background. Mice in which Nbs1 was deleted in the CNS (Nbs1-CNS-Δ mice) were generated as follows; NBS1 exon 6 was floxed by two loxP sites, resulting in Nbs1-floxed mice. Because the nestin protein is specifically expressed in the CNS (neurons and glia), the Nbs1 gene can be specifically deleted in the nervous system when crossed with Nestin-Cre transgenic mice (58). Nbs1-CNS-Δ//Atm−/− double mutant mice were generated by crossing Nbn-floxed mice (harboring two floxP sites in Nbn exon 6 (58)) with Atm+/−//nestin-Cre+ mice. All experiments with mice were compliant with minimal standards as defined by the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of Tel Aviv University.
Tissue Preparation
Mouse brains were dissected and fixed in 4% fixative (4% formaldehyde in PBS) for 24 h. The brains were then infiltrated for cryo-protection with 30% sucrose (Merck) for 24 h at 4 °C. Fixed brains were embedded in Tissue Freezing Medium (Leica Instruments GmbH, Nussloch, Germany) and quickly frozen in liquid nitrogen. Cross-sections (10 μm) were placed on subbed slides (0.5% gelatin, containing 0.05% chromium potassium sulfate) and stored at −20 °C.
Organotypic Slice Cultures from the Mouse Cerebellum
Unlike dissociated cell cultures, organotypic slice cultures remarkably preserve neuronal structure and cell-cell interaction. Cultures were prepared according to Noraberg et al. (63) and Inamura et al. (64) from 12-day-old WT and Atm−/−, Nbs1-CNS-Δ, and Nbs1-CNS-Δ//Atm−/− mice. Brain slices (400 μm thick) were mounted on Millicell-CM, PICM ORG50, 0.4 μm, 25 mm (Millipore, Bedford, MA) membranes.
Dissociated Primary Culture of Glial Cells
Dissociated cultures of total glial cells were prepared from the cortex of newborn mice (P1-P2) of the various Nbs1/Atm mutants. The meningeal tissues were removed, and cells were dissociated by trypsinization and plated in standard medium (Dulbecco's modified Eagle's medium, 10% heat inactivated fetal calf serum, 2 mm glutamine, 50 mg/ml gentamycin, and 250 ng/ml amphotericin B). The cultured glial cells were allowed to grow for 1 week, after which they were detached, collected, and re-plated in 12-well plates (6 × 104 per plate). At different time points the cells were detached with 250 μl of 0.25% trypsin, 0.05% EDTA and counted. The number of cells per well was then calculated.
Immunohistochemical Analysis of Cerebellar Sections
Sections were washed in PBS for 30 min and then incubated with blocking solution containing 1% BSA (Sigma) and 10% normal donkey serum (Jackson ImmunoResearch, West Grove PA) in PBS for 1 h at room temperature. The sections were incubated overnight with a primary antibody (see “Experimental Procedures” and “Results”) in 0.25% Triton X-100 at 4 °C. The slides were washed 3 times with PBS and incubated with the appropriate secondary antibody for 1 h at room temperature. After 1 wash with PBS and 2 washes in a buffer containing 10 mm Tris, 1 mm EDTA, the sections were incubated with the nucleic acid dye Sytox blue (Molecular Probes, Carlsbad, CA; Invitrogen) for 30 min. Slides were washed three times with the same buffer and mounted with aqueous mounting medium containing anti-fading agents (Biomeda, Burlingame, CA). Observations and photography were carried out with a Zeiss (Oberkochen, Germany) LSM 510 META confocal microscope.
Fluoro-Jade Staining
Fluoro-Jade staining was performed as described by Schmued et al. (65). Briefly, cryostat sections were incubated for 15 min in 0.06% potassium permanganate with gentle agitation, rinsed in distilled water, incubated for 30 min in Fluoro-Jade solution (0.001%; Histo-Chem, Jefferson, AR) in 0.1% acetic acid, rinsed in water, air-dried on a slide warmer, cleared in xylene for 10 min, and mounted in D.P.X. (Sigma). The staining was visualized by confocal microscopy.
Immunocytochemical Analysis of Organotypic Cultures
Cultures were fixed with 4% paraformaldehyde after permeabilization with 1% Triton X-100 or ice-cold methanol. Slides were then incubated with blocking solution (1% BSA, 10% serum, and 0.25% Triton X-100) and incubated overnight at 4 °C with primary antibodies as indicated below. The slides were washed 3 times with PBS and incubated with a secondary antibody for 1 h at room temperature. After 1 wash with PBS and 2 washes in a buffer containing 10 mm Tris, 1 mm EDTA, the organotypic cultures were incubated with the nucleic acid dye Sytox blue (Molecular Probes, Invitrogen) for 30 min, washed three times with the same buffer, and mounted with aqueous mounting medium containing anti-fading agents (Biomeda). Immunofluorescence was visualized using a Zeiss LSM-510 META confocal microscope.
Antibodies
The following primary antibodies were used for immunofluorescence analysis: mouse anti-calbindin D28K monoclonal antibody (Sigma), rabbit anti-calbindin D28K polyclonal antibody (Chemicon International, Inc., Temecula, CA), rabbit anti-γ-aminobutyric acid receptor α-6 subunit polyclonal antibody (Chemicon International), mouse anti-myelin 2′,3′-cyclic nucleotide 3′-phosphodiesterase monoclonal antibody (Covance Research Products, Berkeley, CA), mouse anti-GalC monoclonal antibody (a gift from Prof. Danielle Pham-Dinh, NMR S546, Paris), rabbit anti-phospho-Ser-1981 of ATM (Bethyl Laboratories, Inc., Montgomery, TX), mouse anti-γH2AX monoclonal antibody (Upstate Biotechnology, Inc., Waltham, MA), rabbit anti-53BP1 polyclonal antibody (Novus Biologicals, Inc., Littleton, CO). The following primary antibodies were used for Western blot analysis: mouse anti-tubulin monoclonal antibody (Sigma), mouse anti-phospho-Ser-1981 of ATM monoclonal antibody (Rockland Inc., Gilbertsville. PA), and mouse anti-ATM monoclonal antibody, MAT3 (produced in the laboratories of Y. Shiloh and N. Smorodinsky). The following secondary antibodies were used: donkey anti-rabbit rhodamine red X (RRx) conjugated; Donkey anti-rabbit Cy2 conjugated; donkey anti-mouse Cy2 conjugated; donkey anti-mouse rhodamine red X conjugated and donkey anti-goat Cy5 conjugated. All secondary antibodies were purchased from Jackson ImmunoResearch Laboratories.
Quantitative Analysis of γH2X and 53BP1 FXAoci and Measurements of Fluorescent Intensity
Nuclear H2AX or 53BP1 were counted using Image Pro software (Media Cybernetics, Bethesda, MD), and data are presented per Purkinje neuron.
Immunoblotting Analysis
Tissues were washed with ice-cold phosphate-buffered saline and homogenized in ice-cold homogenization buffer (150 mm NaCl, 10 mm Tris buffer, pH 7.6, 1% Triton X-100, 0.5% deoxycholic acid, 0.1% SDS, 1:50 phosphatase inhibitor mixture (I and II Sigma), and 1:100 protease inhibitor mixture). Protein concentration was determined according to Bradford (66) using BSA as standard. Blots were prepared as described by Harlow et al. (67) using 7.5% polyacrylamide gels. Each lane was loaded with 50 μg of protein extract, and after electrophoresis the proteins were transferred to an Immobilon polyvinylidene disulfide membrane for at least 12 h at 220 mA. Blots were stained with Ponceau to monitor protein amounts.
RESULTS
Markedly Reduced Life Span in Nbs1-CNS-Δ/Atm Double Mutant Mice
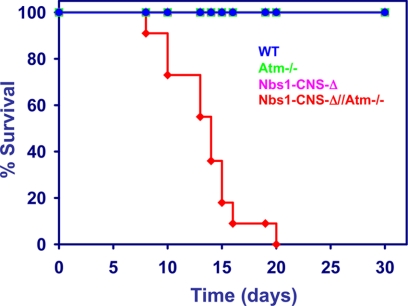
Specific Nbs1 inactivation in the CNS of Atm knock-out mice (Nbs1-CNS-Δ/Atm−/−) caused a markedly reduced life span compared with the single mutants. Fifty percent of the Nbs1/Atm double mutants died within 2 weeks after birth, and none survived beyond 3 weeks, whereas Atm deficiency or conditional disruption of Nbs1 in the CNS did not seem to affect survival for the first few months of life (Fig. 1).
FIGURE 1.
Life span of mice with different Nbs1 and Atm genotypes. Survival curves are shown for the different genotypes. n = 11 (84).
Conditional Nbs1 Deletion in the CNS on the Background of Atm Deficiency Exacerbated the Effect of Nbs1 Inactivation
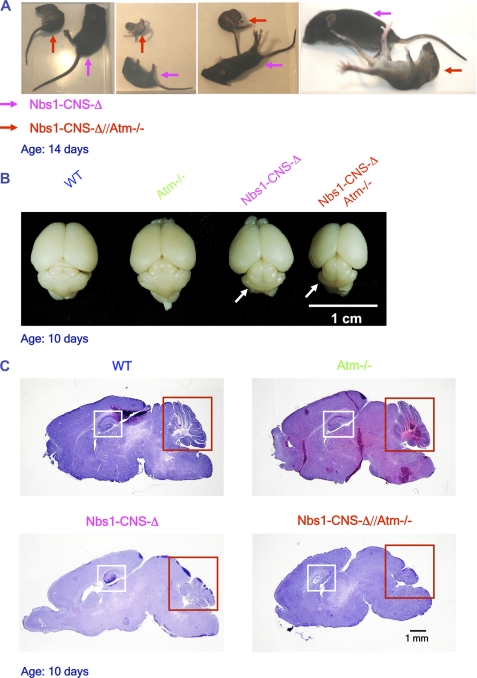
The recoveries of the Nbs1-CNS-Δ and the Nbs1-CNS-Δ//Atm−/− deviated slightly from the Mendelian distribution (from expected 6.25 to 5.49% for both genotypes). The Nbs1/Atm double mutant mice displayed marked growth retardation compared with the Nbs1-CNS-Δ (Fig. 2A and supplemental Fig. S1). At 14 days of age the weight of the Nbs1/Atm double mutant animals was around 40% that of the Nbs1-CNS-Δ mice (Fig. 2A and supplemental Fig. S1). Compared with the Nbs1-CNS-Δ animals, the postnatal development of the double mutants was retarded, and their neurological deficits including tremor and ataxia were more severe and appeared earlier. The Nbs1/Atm double mutants were unable to walk and displayed unsynchronized movements (Fig. 2A) and supplemental video. The internal organs of these mice showed no abnormalities or pathologies upon general inspection other than being smaller than those of WT animals. Collectively, the inactivation of Atm in Nbs1-CNS-Δ mice exacerbated the overall deficits caused by Nbs1 disruption in the CNS.
FIGURE 2.
Growth retardation, severe ataxia, microcephaly, and cerebellar developmental defects in Nbs1-CNS-Δ//Atm−/− mice. A, marked growth retardation was detected in14-day-old Nbs1-CNS-Δ//Atm−/− and Nbs1-CNS-Δ mice (red and pink arrows, respectively). The posture of the Nbs1-CNS-Δ//Atm−/− shows severe ataxia. B, reduced cerebellar size in Nbs1-CNS-Δ//Atm−/− compared with the rest of the Nbs1/Atm genotypes is shown. The brains were isolated from 10-day-old mice. C, histological analysis of P-10 mid-sagittal brain sections (20 μm) is shown. Hematoxylin and eosin staining. Bar = 1 mm.
Nbs1-CNS-Δ/Atm Double Mutant Mice Display Marked Cerebellar Disorganization, Reduced White Matter, and Decreased Glial Cell Proliferation
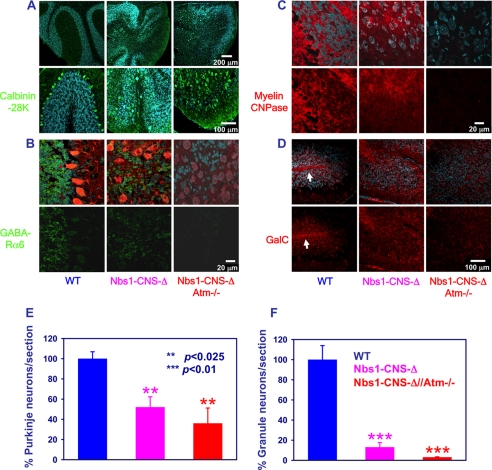
Atm-deficient brain sections and organotypic cultures were indistinguishable from those derived from the WT animals (not shown). However, dual inactivation of Nbs1 and Atm in the CNS severely exacerbated the Nbs1-CNS-Δ-deficient phenotype, especially in the cerebellum. The only conspicuous abnormality seen on gross anatomical inspection of the double mutant brain was atypical hippocampus and reduced cellular staining in the cortex (Fig. 2, B and C). Histological cerebellar analysis of P-14 Nbs1-CNS-Δ revealed markedly reduced foliation, although residual organization of the folia was still evident. In contrast, Nbs1-CNS-Δ//Atm−/− animals displayed a total lack of foliation (Figs. 2C and 3). Immunostaining using specific markers for Purkinje cells (calbindin 28K) and granule neurons (γ-aminobutyric acid receptor-α6) revealed ectopic presence of Purkinje cells throughout the folium, although some level of organization was maintained. Total disorganization of Purkinje cells was seen in the cerebella of the Nbs1-CNS-Δ//Atm−/− mice. Nbs1 deletion reduced the number of Purkinje neurons by 48% and granule neurons by 87%, whereas dual inactivation of Nbs1 and Atm reduced the Purkinje cell number by 64% and led to disappearance of 95% of the granule neurons (Fig. 3, A and B).
FIGURE 3.
The Nbs1-CNS-Δ/Atm−/− double mutant genotype exacerbates the effects of Nbs1 inactivation. A, the upper panel shows Purkinje cell organization in the various genotypes, and the lower panel shows a merged picture of Purkinje cells (calbindin K28, green), cell nuclei (Sytox blue) in higher magnification. Scale bars: 200 μm (upper) and 100 μm (lower). B, brain sections were prepared from 14-day-old Nbs1+/+//Atm+/+, Nbs1-CNS-Δ//Atm+/+, and Nbs1-CNS-Δ//Atm−/− mice after fixation with 4% paraformaldehyde and immunoreacted with anti-γ-aminobutyric acid receptor α-6 (GABA-Rα6; a marker of cerebellar granule neurons, green). Purkinje cells were labeled with calbindin K28 (red) and cell nuclei with Sytox blue. Scale bar = 20 μm. C, cerebellar sections were prepared from the three genotypes on postnatal day 14, stained with anti-myelin 2′,3′-cyclic nucleotide 3′-phosphodiesterase (a key enzyme in myelin formation, red) antibody, which labels the myelin, and co-stained with Sytox blue, which labels cell nuclei. Scale bar = 20 μm. D, cerebellar sections of the indicated genotypes were made on postnatal day 14, reacted with an anti-GalC antibody (red), which labels mature oligodendrocytes and co-stained with Sytox blue. The white arrows point to intrafoliar white matter. Scale bar = 100 μm. The number of Purkinje neurons (E) and granule cells (F) were counted for each genotype. The experiments were performed in cultures from at least three different mice for each genotype. Statistical analysis was performed using two-tailed Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.005 between WT and the rest of the genotypes).
We previously showed that conditional Nbs1 inactivation in the CNS led to reduced levels of myelin and white matter disorganization (68). No traces of white matter could be detected in the cerebella of Nbs1-CNS-Δ//Atm−/− mice, attesting to marked cerebellar disorganization. When we immunoreacted cerebellar sections with oligodendrocyte early stage marker (myelin 2′,3′-cyclic nucleotide 3′-phosphodiesterase) and mature stage marker (anti-GalC) antibody, we found typical distribution of oligodendrocytes in the folium of WT mice at the age of 2 weeks. Most of the oligodendrocytes were concentrated at the folium center and formed a white matter-rich region. Lack of Nbs1 led to increased disorganization of the folium. Combined Nbs1 and Atm inactivation further reduced the levels of oligodendrocytes and abolished organization completely (Fig. 3, C and D).
Division rate and apoptosis on P-5 and P-10 were measured to determine the effect of simultaneous inactivation of Nbs1 and Atm on postnatal cellular proliferation and death. Using the Ki67 protein as a marker of cellular proliferation (69), we found that cerebellar cell division was reduced by ∼60% on P-5 with no evidence of cell division on P-10 (Fig. 4, A and B). We next analyzed cell death at P-5 and P-10 and found in P-10 a 7.8% increase in active caspase-3 positive cells in Nbs1-CNS-Δ//Atm−/− cerebella. The active caspase-3-positive cells were mainly Purkinje neurons with no evidence of granule neurons death (Fig. 4, C and D). Neuronal degeneration usually involves concomitant changes in other cells, such as inflammatory response of astrocytes and microglial cells. Fluoro-Jade staining has been shown to co-localize with activated microglia and with astrocytes in APPSL/PS1 KI transgenic mice of Alzheimer disease (70). Using this method we found increased staining of Fluoro-Jade in Nbs1-CNS-Δ//Atm−/− cerebellar sections (Fig. 4E).
FIGURE 4.
The Nbs1-CNS-Δ/Atm−/− double mutant genotype reduces cell proliferation and increases cell death in the cerebellum. A, P5 cerebellar sections derived from the Nbs1-CNS-Δ//Atm−/− and WT were immunoreacted with Ki67 (marker for cell proliferation). EGL, external granular layer; IGL, internal granular layer. B, quantification of Ki67-positive cells is shown. C, P10 cerebellar sections derived from the Nbs1-CNS-Δ//Atm−/− and WT were immunoreacted with calbindin (marker of Purkinje neurons) active caspase 3 (marker for apoptosis). White arrows point at Purkinje neurons, which are active caspase 3-positive cells. D, quantification of active caspase 3-positive cells is shown. E, P10 cerebellar sections derived from the Nbs1-CNS-Δ//Atm−/− and WT were stained with Fluoro-Jade (marker of degenerative process). F, dissociated glial cell cultures were prepared from 1–2-day-old mice of various Nbs1/Atm genotypes. Bar = 25 μm. G, identical numbers of cells were taken from 1 week cultures, re-plated, and counted after 5, 10, and 15 days. The experiments were performed in cultures from at least three mice for each genotype. Statistical analysis was performed using two-tailed Student's t test. Error bars represent S.E. Green asterisks show the statistical significance between Atm−/− and Nbs1-CNS-Δ//Atm−/−; pink asterisks show the statistical significance between Nbs1-CNS-Δ and Nbs1-CNS-Δ//Atm−/−. *, p < 0.05; **, p < 0.02; ***, p < 0.01.
Because the total volume of the Nbs1-CNS-Δ//Atm−/− cerebellum is around 10% that of the WT, the total level of cell proliferation throughout the cerebellum is estimated to be reduced by ∼95% in the Nbs1/Atm double null cerebella. When the division rate of cultured glial cells isolated from the various Nbs1/Atm mutant cerebral cortex was measured, Nbs1 protein was significantly reduced (pointing at >95% knockdown), attesting to the efficiency of Cre expression in this tissue. Because Nbs1 was conditionally inactivated, efficiency of Nbs1 deletion was measured in astrocytes. One week after plating, WT glial cells were already confluent, whereas the proliferation rate of Atm−/− and Nbs1-CNS-Δ cultures was slower. The proliferation rate of Nbs1-CNS-Δ//Atm−/− glial cells was further reduced compared with the single mutants (Fig. 4, F and G). Taken together, these results indicate that dual inactivation of Nbs1 and Atm causes significant reduction in cell proliferation, particularly in granule neurons. A slight elevation in Purkinje neuronal cell death and degenerative process also plays a part, contributing to alterations in cerebellar size and morphology that lead to severe CNS (neuronal and glial) deficits that push the Nbs1/Atm double mutant mice beyond the threshold of survival.
Atm Autophosphorylation in Mouse Cerebellum Is Attenuated in the Absence of Nbs1
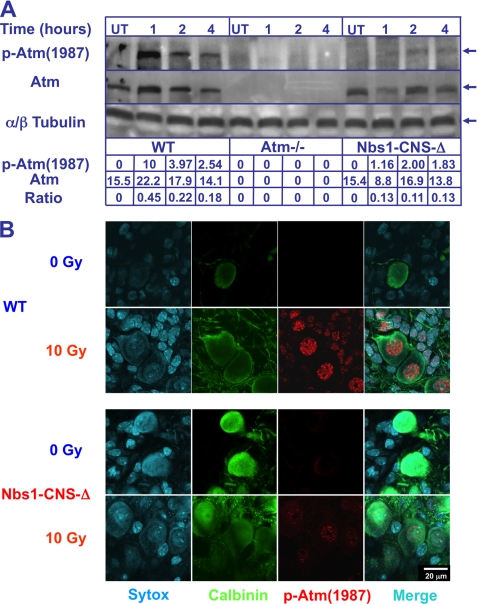
Evidence is accumulating that the cerebellar attrition in A-T results, like most other symptoms of this disease, from the defective response to DSBs (33, 71). However, because neurons (including Purkinje cells) differ in many respects from the cancer cell lines in which most DDR investigations are carried out, we asked whether functional relationships between DDR players identified in human cancer cell lines also exist in cerebellar cells, with emphasis on Purkinje cells. Damage-induced autophosphorylation of ATM is a hallmark of its activation (17, 72, 73). A commonly used marker of human ATM activation is the autophosphorylation on Ser-1981 (Bakkenist). The equivalent autophosphorylation on murine Atm occurs on Ser-1987. Atm activation in the murine cerebellum was examined after exposing Atm−/− and Nbs1-CNS-Δ mice to 10 gray (Gy; the international system unit of energy absorbed from ionizing radiation equal to the absorption of 1 J of radiation energy by 1 kg of matter; 1 Gy equals 100 rads)) of ionizing radiation by reacting cerebellar protein extracts with an anti-Ser(P)-1987 (Atm) antibody. Atm autophosphorylation in WT cerebella peaked at 1 h and subsequently declined, whereas Atm autophosphorylation in Nbs1-CNS-Δ cerebella was steady over 1–4 h after irradiation, with 70% reduction compared with WT tissue (Fig. 5A). Thus, in the mature cerebellum, Atm autophosphorylation is only partially dependent on Nbs1. When cerebellar organotypic cultures derived from WT and Nbs1-CNS-Δ were exposed to 10 Gy of ionizing radiation (Fig. 5B), Atm autophosphorylation was reduced, and focus formation by autophosphorylated Atm 2 h after damage induction was reduced but not completely abolished. No traces of Atm autophosphorylation were observed in cultures derived from Atm−/− animals (data not shown).
FIGURE 5.
Partial damage-induced Atm autophosphorylation in Nbs1-CNS-Δ cerebellar extract and Purkinje neurons. A, whole cerebella were isolated from untreated (UT) and irradiated (10 Gy) 1-month-old WT, Atm−/−, and Nbs1-CNS-Δ mice. Cerebellar proteins were extracted and immunoblotted with anti-Atm Ser(P)-1987. Total Atm and tubulin served as loading controls. Ratios represent the relative alterations of phosphorylated Atm compared with total Atm protein. B, cerebellar organotypic cultures derived from WT and Nbs1-CNS-Δ were exposed to 10 Gy, fixed, and 2 h later immunoreacted with an anti-Atm Ser(P)-1987 antibody (red). Purkinje neurons were labeled with calbindin D28K (green).
DNA Damage Responses in Cerebellar Tissue
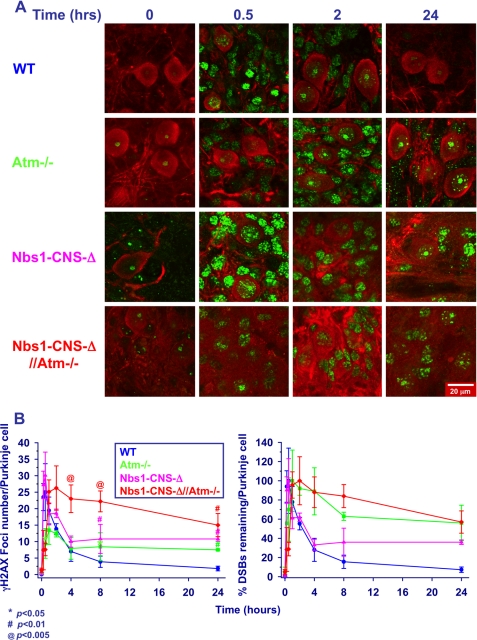
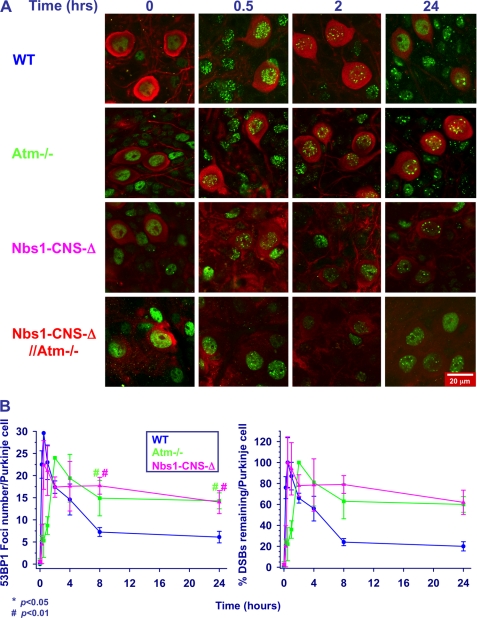
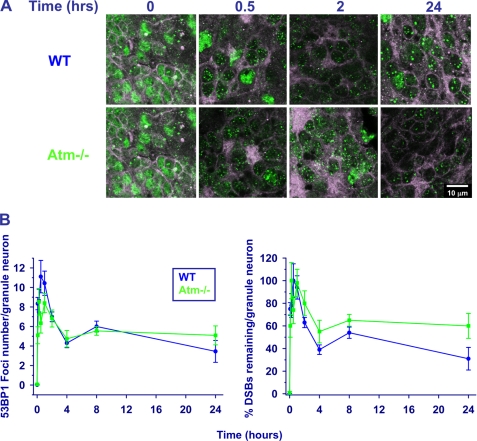
We used two markers of DSB formation and disappearance; that is, nuclear foci of the phosphorylated histone H2AX (γH2AX) (74) and of the damage sensor protein 53BP1(75). Organotypic cultures were irradiated after fixation at different time points and temporal analysis of the foci showed distinct time courses in the various Nbs1/Atm genotypes. Focus formation in Atm-deficient cultures was significantly delayed compared with WT and Nbs1-CNS-Δ cells (Fig. 6, A and B, and supplemental Fig. S2), and the number of foci was lower in Atm-deficient compared with WT and Nbs1-CNS-Δ Purkinje neurons (Fig. 6, A and B). Interestingly, the emergence and number of γH2AX foci in WT and Nbs1-CNS-Δ cerebella were similar (Fig. 6B). Atm inactivation on the background of Nbs1 CNS deletion significantly delayed the time course of γH2AX focus formation (Fig. 6, A and B). The main difference between WT cerebella and the rest of the Nbs1/Atm genotypes was the time course of the disappearance of γH2AX foci, which was significantly delayed in the various mutant mice (Fig. 6, A and B). Interestingly, the disappearance of γH2AX in irradiated Nbs1-CNS-Δ//Atm−/− Purkinje neurons was also significantly delayed compared with Atm−/− and Nbs1-CNS-Δ Purkinje neurons, suggesting additive and separate roles for Atm and Nbs1 in the process leading to their decay, presumably representing DSB repair. 53BP1 foci were rapidly formed in WT Purkinje neurons after ionizing radiation treatment, peaking 30 min thereafter. Formation of these foci was markedly delayed in Atm-deficient cultures, reaching its peak 2 h after irradiation, similar to γH2AX kinetics. 53BP1 focus disintegration in Nbs1-CNS-Δ and Atm−/− cultures was considerably delayed (Fig. 7 and supplemental Fig. S3). Combined inactivation of Atm and Nbs1 severely disrupted the ability of the cerebellar cells to form 53BP1 nuclear foci. Moreover, these foci were very diffuse, which restricted their counting. Because the persistence of γH2AX and 53BP1 foci is thought to denote unrepaired DSBs, the observed differences between the various Nbs1/Atm genotypes are likely to represent various delays in DSB sealing. Collectively, the data suggest that Atm and Nbs1 are required for generation of 53BP1 foci. Our impression is that the patterns of γH2AX and 53P1 focus formation and disappearance can serve as markers for the generation and repair of DSBs in neuronal cells, similar to their use in the commonly employed cell lines, including fibroblasts (76), murine embryonic fibroblasts (77), and cancer cell lines (78). The generation and repair of DSBs in granule neurons (the most abundant cells in the cerebellum) were studied by monitoring the kinetics of 53BP1 focus formation and disappearance, focusing on 53BP1 because the γH2AX foci were too fuzzy and could not be reliably counted. In contrast to Purkinje cells, WT and Atm−/− mice presented similar 53BP1 focus formation kinetics. The kinetics of DSB repair was slightly but not significantly slower in Atm−/− granule cells compared with WT cells (Fig. 8 and supplemental Fig. S4). Nbs1 inactivation in the cerebellum resulted in marked reduction in the number of granule neurons. After irradiation, the nuclei of these cells were atrophied, and the 53BP1 foci were highly diffused and could not be counted. Collectively, these experiments show that the kinetics of DSB repair in granule neurons is less dependent on Atm than in Purkinje neurons and that the kinetics of Atm-dependent DNA repair varies in different cell types.
FIGURE 6.
Dynamics of radiation-induced γH2AX foci in cerebellar organotypic cultures of various Nbs1/Atm mutants. A, after irradiation with 10 Gy of ionizing radiation, the cultures were fixed and reacted with an anti-γH2AX antibody (green) at the indicated time points. B, the number of γH2AX foci per nucleus of Purkinje neurons was measured using the Image Pro software in 10–20 nuclei for each treatment/genotype. The experiments were performed in cultures from at least three mice for each genotype. Statistical analysis was performed using two-tailed Student's t test (*, p < 0.05; #, p < 0.01; @, p < 0.005 between WT and the rest of the genotypes). Scale bar = 20 μm.
FIGURE 7.
Dynamics of radiation-induced 53BP1 foci in cerebellar organotypic cultures of various Nbs1/Atm mutants. A, after irradiation with 10 Gy of ionizing radiation, the cultures were fixed and reacted with an anti-53BP1 antibody (green) at the indicated time points. Purkinje neurons were labeled with calbindin D28K (red). B, the number of 53BP1 foci per nucleus was measured using the Image Pro software in 10–20 nuclei for each treatment/genotype at each culture. The experiments were performed in cultures from at least three mice per genotype. Statistical analysis was performed using two-tailed Student's t test (*, p < 0.05; #, p < 0.01 between WT and the rest of the genotypes). Scale bar = 20 μm.
FIGURE 8.
Dynamics of radiation-induced 53BP1 foci in granule neurons in organotypic cultures derived from various Nbs1/Atm mutants. A, after irradiation with 10 Gy of ionizing radiation, the cultures were fixed and reacted with an anti-53BP1 antibody (green) at the indicated time points. Purkinje neurons were labeled with calbindin D28K (red). B, the number of 53BP1 foci per nucleus was measured using the Image Pro software in 10–20 nuclei for each treatment/genotype at each culture. The experiments were performed in cultures from at least 3 mice per genotype. Statistical analysis was performed using two-tailed Student's t test (*, p < 0.05; #, p < 0.01 between WT and the rest of the genotypes). Scale bar = 20 μm.
Atm and DNA-PK Are Not the Only Kinases Phosphorylating Histone H2AX in Purkinje Neurons in Response to Ionizing Radiation
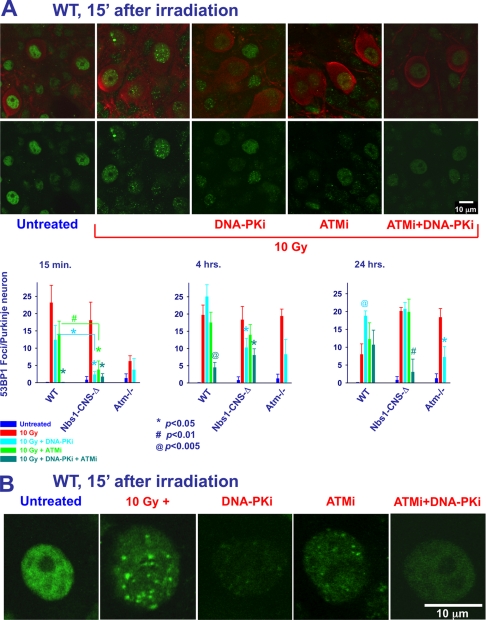
In dividing cells, ATM, ATR, and DNA-PK contribute jointly to H2AX phosphorylation (71, 79, 80). Atm and DNA-PK have overlapping and seemingly redundant functions in lymphocytes (81). We examined their cooperation in H2AX phosphorylation in non-dividing Purkinje cells by following γH2AX focus dynamics in response to 10 Gy of ionizing radiation in cerebellar cultures of various Nbs1/Atm genotypes using the ATM inhibitor KU55933 (ATMi) (82) and/or the DNA-PK inhibitor NU7026 (DNA-PKi) (83). Inhibition of DNA-PK at an early time point (15 min) had no measurable effect on the number of γH2AX foci formed after ionizing radiation (Fig. 9A and supplemental Fig. S5). Inhibition of Atm reduced the number of ionizing radiation-induced foci to 60% that of WT levels. Combined inhibition of Atm and DNA-PK resulted in foci reduction similar to that of Atm alone, although the difference compared with irradiated WT (without inhibitors) reached statistical significance (Fig. 9A and supplemental Fig. S5). This suggests that Atm is the major kinase in H2AX phosphorylation in cerebellar tissue soon after damage induction. Interestingly, separate or combined inhibition of Atm and DNA-PK significantly inhibited H2AX phosphorylation in Nbs1-deficient Purkinje neurons. DNA-PK inhibition in Atm−/− or Nbs1/Atm double null Purkinje neurons was more effective than their inhibition in WT cells at the 15-min time point. Atm or DNA-PK inhibitors led to increased amounts of γH2AX foci at 4 and 24 h after irradiation (Fig. 9A and supplemental Fig. S5). γH2AX foci inhibition was detected in the various Nbs1//Atm cultures with the exception of the Nbs1/Atm double null Purkinje neurons 4 h after irradiation. These results suggest that in Purkinje neurons, kinases other than Atm and DNA-PK are capable of phosphorylating H2AX in response to ionizing radiation.
FIGURE 9.
Proteins kinases other than Atm and DNA-PK are capable of phosphorylating H2AX in Purkinje neurons. A, organotypic cultures derived from various Nbs1 and Atm genotypes were incubated with 2 μg/ml Atm inhibitor KU55933 (ATMi) or 2 μg/ml DNA-PK inhibitor NU7026 (DNA-PKi). Cultures were then treated with 10 Gy of ionizing radiation, fixed at 15 min and 4 and 24 h after the irradiation and reacted with an anti-γH2AX antibody (green). Purkinje neurons were labeled with calbindin D28K (red). The number of γH2AX foci per nucleus of Purkinje neurons was measured using the Image Pro software in 10–20 nuclei for each treatment/genotype. The experiments were performed in cultures from at least 3 mice for each genotype (*, p < 0.05; #, p < 0.01; @, p < 0.005 between irradiated Purkinje neurons and Purkinje neurons that had been exposed to Atm and DNA-PK inhibitors). Scale bar = 10 μm. B, higher magnification of WT micrographs show the effect of Atm and DNA-PK inhibitors on the intensity and size of γH2AX nuclear foci.
Inhibition of Atm and DNA-PK Attenuates 53BP1 Recruitment to Damage Sites
The pattern of 53BP1 focus formation during inhibition of either Atm or DNA-PK reduced 53BP1 focus formation in Purkinje neurons 15 min after irradiation. Dual inhibition of these proteins abolished them entirely in WT organotypic cultures (Fig. 10A and supplemental Fig. S6). Four hours after irradiation, inhibition of either Atm or DNA-PK slightly affected 53BP1 focus formation in WT neurons. Their combined inhibition significantly reduced their ability to form 53BP1 foci in WT Purkinje neurons. Separate and combined inhibition of Atm and DNA-PK significantly reduced 53BP1 focus formation (85%) in Nbs1-CNS-Δ cultures. At a later time point (24 h), separate as well as combined inhibition of the proteins increased 53BP1 focus formation in WT cultures. Interestingly, combined inhibition in Nbs1-CNS-Δ cultures significantly attenuated 53BP1 recruitment at the 24-h point, whereas separate inhibition had almost no effect. Modest reduction in 53BP1 focus formation after DNA-PK inhibition was detected in Atm−/− Purkinje neurons. These results suggest that at least two processes affect the formation of 53BP1 foci; one at an early time point that is dependent on the activity of Atm and DNA-PK and one in a later process that is independent of these kinases.
FIGURE 10.
Involvement of Atm and DNA-PK in damage-induced 53BP1 foci in cerebellar neurons. A, organotypic cultures derived from various Nbs1 and Atm genotypes were incubated with 2 μg/ml Atm inhibitor KU55933 (ATMi) or 2 μg/ml DNA-PK inhibitor NU7026 (DNA-PKi). The cultures were then treated with 10 Gy of ionizing radiation, fixed at 15 min and 4 and 24 h after irradiation, and reacted with an anti-53BP1 antibody (green). Purkinje neurons were labeled with calbindin D28K (red). The number of 53BP1 foci per nucleus of Purkinje neurons was measured using the Image Pro software in 10–20 nuclei for each treatment/genotype. The experiments were performed in cultures from at least three mice for each genotype. Scale bar = 10 μm. B, higher magnification of WT micrographs show the effect of Atm and DNA-PK inhibitors on the intensity and size of 53BP1 nuclear foci.
Qualitatively, we observed that separate or combined inhibition of Atm and DNA-PK markedly reduced the intensity and the size of γH2AX and 53BP1 foci (Figs. 9B and 10B). Our results suggest that Atm and DNA-PK are major contributors to the process of H2AX phosphorylation and 53BP1 focus formation in cerebellar tissue.
DISCUSSION
The molecular interrelations between Nbs1 and Atm in neurons are important early steps in the DSB response in these cells and can enhance our understanding of the neuronal pathologies caused by DDR deficiencies. In this study we compared various aspects of the DDR in mice deficient for one or the other of these proteins and in double mutant animals. A commonly held notion is that Atm activation is dependent on the MRN complex, but our results indicate that Atm has biological functions in the CNS that are not dependent on the Nbs1 protein, whose presence is essential for MRN integrity and function (38, 84–86). Nbs1-CNS-Δ//Atm−/− double mutant mice exhibited a markedly shortened life span, and Atm loss on top of Nbs1-CNS-Δ exacerbated the effects of Nbs1 loss with regard to general cerebellar organization, loss of granule neurons, impairment of white matter development, and reduction of glial cell proliferation. Our finding that Atm-dependent phosphorylation of H2AX can go on in the absence of Nbs1 further suggests that the dependence of Atm activation on the MRN complex is not absolute.
Although the cerebellum is one of the first structures of the brain to differentiate, it achieves its mature configuration only many months after birth in humans and several weeks after birth in mice (87). This lengthy formation period makes the cerebellum particularly vulnerable to developmental irregularities in the form of structural and functional defects. We show here that co-inactivation of Nbs1 and Atm in the CNS severely exacerbated the neurological deficits in the Nbs1-CNS-Δ//Atm−/− double mutant mice, severely reducing cerebellar functionality compared with Nbs1-CNS-Δ mice. The marked reduction in the number of granule cells almost certainly reduced to a minimum the parallel fibers that normally form synapses on the dendritic tree of the Purkinje neurons. Disruption of the delicate interrelations between the climbing and the parallel fibers, which are the major excitatory input of the cerebellum, will severely affect the functionality of the Nbs1/Atm double null cerebellum. The granule neurons also affect the final maturation of the Purkinje neurons, thus, indirectly affecting the output of the cerebellum (88, 89). At birth, all the newborn mice (WT and mutants) are ataxic because the cerebellar maturation is in its initial stage. During the first weeks after birth, the WT animals quickly develop the motor skills critical for their survival; the Nbs1-CNS-Δ//Atm−/− animals do not. Our data indicate that lack of cerebellar development and functionality reduces motor skills in the Nbs1/Atm double mutant mice to a level that probably leads to their early death, compared with Nbs1-CNS-Δ mice.
Cerebellar size and morphology are dependent in part on cell proliferation and death. Our data indicate that combined Nbs1 and Atm inactivation leads mainly to significantly reduced postnatal cell proliferation and only slightly increased cell death. Reduced cell division, which reduces cell migration from the external to the internal granular layer is most likely the primary reason for the reduced number of granule neurons in the Nbs1-CNS-Δ//Atm−/− cerebella. Our data indicated that the neurons undergoing cell death were mainly Purkinje neurons with no evidence of granule neurons death. Collectively, our data suggest that reduced cell proliferation in the Nbs1/Atm double mutant mice and to some extent cell degeneration are the cause of reduced foliation. This scheme of reduced cell proliferation is also applicable to the dividing glial cells. The reduced proliferation of the inner folial cell is most likely the reason for the reduced number of astrocytes and oligodendrocytes in the Nbs1/Atm double mutant mice.
The MRN complex is a major, early DSB sensor/mediator whose recruitment to DSB sites is ATM-independent, but it is also necessary for proper ATM activation (16, 18). These mutual functional relationships lead to cyclic amplification of the DNA damage signal at DSB sites. Due to the physical association and functional cooperation between the three core components of the MRN complex, even partial loss of one of them leads to abrogation of the complex functions (90), making it difficult to assess the contribution of each component to the activities of the complex. Nbs1 loss is probably analogous to MRN inactivation.
Lee and Paull (91) have shown that in vitro Mre11 and Rad50 (“MR complex”) are sufficient to recruit ATM to the DNA, but only the whole MRN complex stimulates the kinase activity of ATM. We found that in the absence of Nbs1 (and presumably a functional MRN complex) murine Atm can undergo partial autophosphorylation in cerebellar cells in response to 10 Gy of ionizing radiation (Fig. 5), and phosphorylated Atm is recruited to damaged sites. Thus, in the absence of functional MRN, Atm is still capable of being activated and partially recruited to damage sites in post-mitotic neuronal cells.
Exposure to ionizing radiation can generate high levels of oxidative stress. In view of the recent finding of Guo et al. (92) that oxidative stress by itself can activate Atm, it is possible that in the absence of a functional MRN complex, reactive oxygen species generated by the ionizing radiation can directly activate Atm.
Nbs1 was shown to be involved in the recruitment and activation of Atm to damaged sites by interacting with Atm via a conserved carboxyl-terminal region (38). However, at least one other damage sensor, MDC1, assists in ATM recruitment and is specifically involved in ATM-mediated phosphorylation of H2AX (93). Therefore, it is not surprising that a lack of Nbs1 does not abolish ATM-mediated phosphorylation of H2AX. However, MDC1 was not reported to be involved in ATM activation. Additional unknown proteins may be involved in this process, and it will be important to compare the activation mechanism in proliferating cells and post-mitotic neurons.
Our findings strongly suggest that Purkinje neurons are highly dependent on Atm for DSB repair. On the other hand, the kinetics of DSB repair in Atm-deficient granule neurons was similar to that in WT cells. This unexpected result might provide partial explanation for why Atm deficiency does not lead to cerebellar attrition in mice. Whereas Atm inactivation does not seem to affect the number of granule neurons, cerebellar Nbs1 deficiency markedly reduces the amount of granule neurons. Interestingly, Atm inactivation on the background of Nbs1 deficiency led to massive loss of granule neurons. This demonstrates that under certain conditions, Atm deficiency can augment granule cell loss and exacerbate cerebellar attrition. These results support our notion that ATM has biological functions that are not dependent on NBS1.
Our findings also indicate that in murine cerebellar neurons Atm and DNA-PK are the major kinases that are involved in H2AX phosphorylation (Figs. 6 and 9). Atm-dependent phosphorylation of H2AX can occur in the absence of functional MRN; in Nbs1-CNS-Δ cultures the kinetics of H2AX phosphorylation were very similar to that of WT cells (Fig. 6).
In Purkinje neurons, inhibition of Atm and DNA-PK only partially inhibited γH2AX focus formation (Fig. 9A). This finding differs from that of Stiff et al. (79) in human fibroblasts where inactivation of both kinases completely ablated ionizing radiation-induced H2AX phosphorylation. In that study, DSB-induced H2AX phosphorylation in non-replicating cells was ATR-independent. In proliferating cells, ATR was indeed shown to be involved in H2AX phosphorylation (94). We assume that in Purkinje neurons as well, ATR may contribute to this phosphorylation. Recently, Shiotani and Zou (95) suggested that Atm is activated by DSBs with blunt ends or short single-strand overhangs, whereas ATR is activated after progressive end-resection of DSBs. It is plausible that DSBs are similarly resected in neurons.
Our results point to at least two phases of H2AX phosphorylation in cerebellar neurons; that is, an initial, Atm-dependent phase followed by an Atm-independent phase in which H2AX phosphorylation is stabilized (Fig. 6 and supplemental S2), with DNA-PK contributing to the maintenance of H2AX phosphorylation in the second phase. Interestingly, we also found that combined inhibition of Atm and DNA-PK ablated 53BP1 focus formation 15 min after ionizing radiation treatment, whereas it only partially inhibited H2AX phosphorylation (Figs. 9 and 10 and Figs. supplemental S5 and S6). This observation is consistent with that of Callén et al. (81) that DNA-PK inhibition abrogated 53BP1 in Atm−/− but not in WT B cells exposed to ionizing radiation. These results suggest that the early phase of 53BP1 focus formation is dependent on Atm and DNA-PK activities, whereas later (24 h after damage induction) it is independent of these protein kinases. It is possible that complete inhibition of Atm and DNA-PK in Purkinje neurons leads to the accumulation of unrepaired DSBs and that during this process redundant kinases are capable of phosphorylating H2AX, affecting the formation of 53BP1 foci. These results further suggest that H2AX phosphorylation and 53BP1 recruitment are necessary but insufficient for timely DSB repair.
Interestingly, the interplay between Atm and DNA-PK and other PI3K-like protein kinases seems to be dependent on Nbs1, as the efficacy of both Atm and DNA-PK inhibitors was increased in Nbs1-CNS-Δ cultures (Figs. 9 and 10 and supplemental Figs. S5 and S6). It is plausible that the MRN complex coordinates the activity of several PI3K-like protein kinases, and in its absence they may not be directed to the damage sites in a timely manner and possibly not retained at those sites as required. In its presence the PI3K-like protein kinases are capable of partially substituting each other. Our findings may explain in part why Atm deficiency in mice does not lead to the cerebellar degeneration seen in A-T patients. Atm inactivation or inhibition reduced the formation of γH2AX and 53BP1 foci to less than 40%, whereas the rest of the damage response may have been supplied by other PI3K-like protein kinases to an extent that is sufficient to slow down the degenerative process to become invisible during the life span of the animal. However, in the Nbs1-deficient cerebellum, the ability of other kinases to substitute Atm is markedly attenuated, leading to cerebellar mal-development and degeneration.
That Atm has biological functions independent of Nbs1 is suggested by the different phenotypes associated with their inactivation in humans and mice. Importantly, the human phenotypes corresponding to ATM loss (A-T) and partial loss of Mre11 (A-T-like disease) are much closer to each other than are A-T and NBS. This phenotypic similarity initially led to the assumption and subsequent observation that Mre11 is required for proper ATM activation, and Mre11 activity was specifically required for this process (40, 62). It was also recently suggested that oligonucleotides formed as a result of Mre11-mediated DNA resection as well as single-stranded stretches at the break sites (95) play a role in ATM activation. It appears, therefore, that different components of the MRN complex contribute in different ways to ATM activation and recruitment to damaged sites, with a critical contribution of Mre11 enzymatic activity. This notion is compatible with Shull et al. (42) who showed that Mre11 and Nbs1 activate different DDR signaling pathways in response to DNA damage.
We found that DSB repair in neurons is quite fast, with about 70% of DSBs repaired 4 h after 10 Gy of ionizing radiation (Figs. 6 and 7). Furthermore, we estimated that in Purkinje cells ∼60% of DSBs are dependent on Atm for their repair (Figs. 6 and 7). We attribute special importance to this result. It has been estimated that in human fibroblasts the majority of DSB repair is ATM-independent, and only about 10–15% of the breaks are not repaired in a timely manner in ATM-deficient cells, particularly in the vicinity of heterochromatin (7, 10, 76). The higher proportion of ATM-dependent DSB repair in Purkinje neurons may also explain the critical effect of ATM loss on their survival in humans.
Altogether, our study strongly supports the notion that the neurological deficits of A-T stem from malfunctioning DDR. The fact that the DDR machinery is very similar in neuronal and other types of cells argues in favor of this notion. This impression is further strengthened by the fact that the activity of the neural network is severely impaired as a function of DNA damage in Atm-deficient cultures.5
By concomitant ablation of various components of the DDR in the murine nervous system we were able to demonstrate their overlapping and distinct roles in maintaining the DDR and the DDR importance for tissue function and development. These results further demonstrate the value of animal models in the investigation of the molecular mechanisms underlying human disease.
Supplementary Material
This work was supported by research grants from the A-T Children's Project, the Israel Science Foundation, and the U. S.-Israel Binational Science Foundation (to A. B. and Y. S.) and by The German-Israeli Foundation (to A. B., Z.-Q. W., and Y. S.). Work in the laboratory of Y. S. was supported by research grants from the A-T Medical Research Foundation, The Israel Cancer Research Fund, and the A-T Ease Foundation.

The on-line version of this article (available at http://www.jbc.org) contains a supplemental video and Figs. S1–S6.
N. Levine-Small, Z. Yekutieli, J. Aljadeff, S. Boccaletti, E. Ben-Jacob, and A. Barzilai, submitted for publication.
- DSBs
- double strand break
- A-T
- ataxia telangiectasia
- ATM
- ataxia telangiectasia mutated
- ATR
- ATM and Rad3 Related protein
- DNA-PK
- DNA dependent protein kinase
- DDR
- DNA damage response
- Gy
- gray
- H2AX
- variant of histone 2
- γH2AX
- phospho-H2AX on serine 139
- MRN
- Mre11/Rad50/Nbs1
- MDC1
- mediator of DNA damage checkpoint protein 1
- NBS
- Nijmegen breakage syndrome
- 53BP1
- p53-binding protein 1.
REFERENCES
- 1. Bassing C. H., Alt F. W. (2004) DNA Repair 3, 781–796 [DOI] [PubMed] [Google Scholar]
- 2. Harrison J. C., Haber J. E. (2006) Annu. Rev. Genet. 40, 209–235 [DOI] [PubMed] [Google Scholar]
- 3. Bekker-Jensen S., Mailand N. (2010) DNA Repair 9, 1219–1228 [DOI] [PubMed] [Google Scholar]
- 4. Ciccia A., Elledge S. J. (2010) Mol. Cell 40, 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Gent D. C., van der Burg M. (2007) Oncogene 26, 7731–7740 [DOI] [PubMed] [Google Scholar]
- 6. Wyman C., Kanaar R. (2006) Annu. Rev. Genet. 40, 363–383 [DOI] [PubMed] [Google Scholar]
- 7. Deckbar D., Birraux J., Krempler A., Tchouandong L., Beucher A., Walker S., Stiff T., Jeggo P., Löbrich M. (2007) J. Cell Biol. 176, 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M. B., Bartek J., Lukas J. (2006) J. Cell Biol. 173, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Su T. T. (2006) Annu. Rev. Genet. 40, 187–208 [DOI] [PubMed] [Google Scholar]
- 10. Riballo E., Kühne M., Rief N., Doherty A., Smith G. C., Recio M. J., Reis C., Dahm K., Fricke A., Krempler A., Parker A. R., Jackson S. P., Gennery A., Jeggo P. A., Löbrich M. (2004) Mol. Cell 16, 715–724 [DOI] [PubMed] [Google Scholar]
- 11. Shiloh Y. (2006) Trends Biochem. Sci. 31, 402–410 [DOI] [PubMed] [Google Scholar]
- 12. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 13. Stracker T. H., Theunissen J. W., Morales M., Petrini J. H. (2004) DNA Repair 3, 845–854 [DOI] [PubMed] [Google Scholar]
- 14. Moreno-Herrero F., de Jager M., Dekker N. H., Kanaar R., Wyman C., Dekker C. (2005) Nature 437, 440–443 [DOI] [PubMed] [Google Scholar]
- 15. Lukas J., Bartek J. (2004) Cell 118, 666–668 [DOI] [PubMed] [Google Scholar]
- 16. Lavin M. F. (2007) Oncogene 26, 7749–7758 [DOI] [PubMed] [Google Scholar]
- 17. Bakkenist C. J., Kastan M. B. (2003) Nature 421, 499–506 [DOI] [PubMed] [Google Scholar]
- 18. Lee J. H., Paull T. T. (2007) Oncogene 26, 7741–7748 [DOI] [PubMed] [Google Scholar]
- 19. Abraham R. T. (2004) DNA Repair 3, 883–887 [DOI] [PubMed] [Google Scholar]
- 20. Lempiäinen H., Halazonetis T. D. (2009) EMBO J. 28, 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weterings E., Chen D. J. (2007) J. Cell Biol. 179, 183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cimprich K. A., Cortez D. (2008) Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. López-Contreras A. J., Fernandez-Capetillo O. (2010) DNA Repair 9, 1249–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith J., Tho L. M., Xu N., Gillespie D. A. (2010) Adv. Cancer Res. 108, 73–112 [DOI] [PubMed] [Google Scholar]
- 25. Jazayeri A., Falck J., Lukas C., Bartek J., Smith G. C., Lukas J., Jackson S. P. (2006) Nat. Cell Biol. 8, 37–45 [DOI] [PubMed] [Google Scholar]
- 26. Hurley P. J., Bunz F. (2007) Cell Cycle 6, 414–417 [DOI] [PubMed] [Google Scholar]
- 27. Helt C. E., Cliby W. A., Keng P. C., Bambara R. A., O'Reilly M. A. (2005) J. Biol. Chem. 280, 1186–1192 [DOI] [PubMed] [Google Scholar]
- 28. Paulsen R. D., Cimprich K. A. (2007) DNA Repair 6, 953–966 [DOI] [PubMed] [Google Scholar]
- 29. Kim J. A., Kruhlak M., Dotiwala F., Nussenzweig A., Haber J. E. (2007) J. Cell Biol. 178, 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Difilippantonio S., Celeste A., Fernandez-Capetillo O., Chen H. T., Reina San Martin B., Van Laethem F., Yang Y. P., Petukhova G. V., Eckhaus M., Feigenbaum L., Manova K., Kruhlak M., Camerini-Otero R. D., Sharan S., Nussenzweig M., Nussenzweig A. (2005) Nat. Cell Biol. 7, 675–685 [DOI] [PubMed] [Google Scholar]
- 31. Chun H. H., Gatti R. A. (2004) DNA Repair 3, 1187–1196 [DOI] [PubMed] [Google Scholar]
- 32. Lavin M. F. (2008) Nat. Rev. Mol. Cell Biol. 9, 759–769 [DOI] [PubMed] [Google Scholar]
- 33. Biton S., Barzilai A., Shiloh Y. (2008) DNA Repair 7, 1028–1038 [DOI] [PubMed] [Google Scholar]
- 34. Taylor A. M., Groom A., Byrd P. J. (2004) DNA Repair 3, 1219–1225 [DOI] [PubMed] [Google Scholar]
- 35. Stewart G. S., Maser R. S., Stankovic T., Bressan D. A., Kaplan M. I., Jaspers N. G., Raams A., Byrd P. J., Petrini J. H., Taylor A. M. (1999) Cell 99, 577–587 [DOI] [PubMed] [Google Scholar]
- 36. Delia D., Piane M., Buscemi G., Savio C., Palmeri S., Lulli P., Carlessi L., Fontanella E., Chessa L. (2004) Hum. Mol. Genet. 13, 2155–2163 [DOI] [PubMed] [Google Scholar]
- 37. Fernet M., Gribaa M., Salih M. A., Seidahmed M. Z., Hall J., Koenig M. (2005) Hum. Mol. Genet. 14, 307–318 [DOI] [PubMed] [Google Scholar]
- 38. Falck J., Coates J., Jackson S. P. (2005) Nature 434, 605–611 [DOI] [PubMed] [Google Scholar]
- 39. Carson C. T., Schwartz R. A., Stracker T. H., Lilley C. E., Lee D. V., Weitzman M. D. (2003) EMBO J. 22, 6610–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y. (2003) EMBO J. 22, 5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Digweed M., Sperling K. (2004) DNA Repair 3, 1207–1217 [DOI] [PubMed] [Google Scholar]
- 42. Shull E. R., Lee Y., Nakane H., Stracker T. H., Zhao J., Russell H. R., Petrini J. H., McKinnon P. J. (2009) Genes Dev. 23, 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J. N., Ried T., Tagle D., Wynshaw-Boris A. (1996) Cell 86, 159–171 [DOI] [PubMed] [Google Scholar]
- 44. Xu Y., Ashley T., Brainerd E. E., Bronson R. T., Meyn M. S., Baltimore D. (1996) Genes Dev. 10, 2411–2422 [DOI] [PubMed] [Google Scholar]
- 45. Elson A., Wang Y., Daugherty C. J., Morton C. C., Zhou F., Campos-Torres J., Leder P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13084–13089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Borghesani P. R., Alt F. W., Bottaro A., Davidson L., Aksoy S., Rathbun G. A., Roberts T. M., Swat W., Segal R. A., Gu Y. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3336–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chiesa N., Barlow C., Wynshaw-Boris A., Strata P., Tempia F. (2000) Neuroscience 96, 575–583 [DOI] [PubMed] [Google Scholar]
- 48. Eilam R., Peter Y., Elson A., Rotman G., Shiloh Y., Groner Y., Segal M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12653–12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kuljis R. O., Xu Y., Aguila M. C., Baltimore D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12688–12693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mount H. T., Martel J. C., Fluit P., Wu Y., Gallo-Hendrikx E., Cosi C., Marien M. R. (2004) J. Neurochem. 88, 1449–1454 [DOI] [PubMed] [Google Scholar]
- 51. Eilam R., Peter Y., Groner Y., Segal M. (2003) Neuroscience 121, 83–98 [DOI] [PubMed] [Google Scholar]
- 52. Watters D., Kedar P., Spring K., Bjorkman J., Chen P., Gatei M., Birrell G., Garrone B., Srinivasa P., Crane D. I., Lavin M. F. (1999) J. Biol. Chem. 274, 34277–34282 [DOI] [PubMed] [Google Scholar]
- 53. Watters D. J. (2003) Redox. Rep. 8, 23–29 [DOI] [PubMed] [Google Scholar]
- 54. Barlow C., Dennery P. A., Shigenaga M. K., Smith M. A., Morrow J. D., Roberts L. J., 2nd, Wynshaw-Boris A., Levine R. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9915–9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Allen D. M., van Praag H., Ray J., Weaver Z., Winrow C. J., Carter T. A., Braquet R., Harrington E., Ried T., Brown K. D., Gage F. H., Barlow C. (2001) Genes Dev. 15, 554–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen P., Peng C., Luff J., Spring K., Watters D., Bottle S., Furuya S., Lavin M. F. (2003) J. Neurosci. 23, 11453–11460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gueven N., Luff J., Peng C., Hosokawa K., Bottle S. E., Lavin M. F. (2006) Free Radic. Biol. Med. 41, 992–1000 [DOI] [PubMed] [Google Scholar]
- 58. Frappart P. O., Tong W. M., Demuth I., Radovanovic I., Herceg Z., Aguzzi A., Digweed M., Wang Z. Q. (2005) Nat. Med. 11, 538–544 [DOI] [PubMed] [Google Scholar]
- 59. Stiff T., Reis C., Alderton G. K., Woodbine L., O'Driscoll M., Jeggo P. A. (2005) EMBO J. 24, 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Myers J. S., Cortez D. (2006) J. Biol. Chem. 281, 9346–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Adams K. E., Medhurst A. L., Dart D. A., Lakin N. D. (2006) Oncogene 25, 3894–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dupré A., Boyer-Chatenet L., Sattler R. M., Modi A. P., Lee J. H., Nicolette M. L., Kopelovich L., Jasin M., Baer R., Paull T. T., Gautier J. (2008) Nat. Chem. Biol. 4, 119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Noraberg J., Kristensen B. W., Zimmer J. (1999) Brain Res. Brain Res. Protoc. 3, 278–290 [DOI] [PubMed] [Google Scholar]
- 64. Inamura N., Araki T., Enokido Y., Nishio C., Aizawa S., Hatanaka H. (2000) J. Neurosci. Res. 60, 450–457 [DOI] [PubMed] [Google Scholar]
- 65. Schmued L. C., Albertson C., Slikker W., Jr. (1997) Brain Res. 751, 37–46 [DOI] [PubMed] [Google Scholar]
- 66. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 67. Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual, pp. 471–510, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 68. Assaf Y., Galron R., Shapira I., Nitzan A., Blumenfeld-Katzir T., Solomon A. S., Holdengreber V., Wang Z. Q., Shiloh Y., Barzilai A. (2008) Exp. Neurol. 209, 181–191 [DOI] [PubMed] [Google Scholar]
- 69. Scholzen T., Gerdes J. (2000) J. Cell. Physiol. 182, 311–322 [DOI] [PubMed] [Google Scholar]
- 70. Damjanac M., Rioux Bilan A., Barrier L., Pontcharraud R., Anne C., Hugon J., Page G. (2007) Brain Res. 1128, 40–49 [DOI] [PubMed] [Google Scholar]
- 71. Barzilai A., Biton S., Shiloh Y. (2008) DNA Repair 7, 1010–1027 [DOI] [PubMed] [Google Scholar]
- 72. Bensimon A., Schmidt A., Ziv Y., Elkon R., Wang S. Y., Chen D. J., Aebersold R., Shiloh Y. (2010) Sci. Signal. 3, rs3. [DOI] [PubMed] [Google Scholar]
- 73. Lavin M. F., Kozlov S. (2007) Cell Cycle 6, 931–942 [DOI] [PubMed] [Google Scholar]
- 74. Fernandez-Capetillo O., Lee A., Nussenzweig M., Nussenzweig A. (2004) DNA Repair 3, 959–967 [DOI] [PubMed] [Google Scholar]
- 75. Mochan T. A., Venere M., DiTullio R. A., Jr., Halazonetis T. D. (2004) DNA Repair 3, 945–952 [DOI] [PubMed] [Google Scholar]
- 76. Goodarzi A. A., Noon A. T., Deckbar D., Ziv Y., Shiloh Y., Löbrich M., Jeggo P. A. (2008) Mol. Cell 31, 167–177 [DOI] [PubMed] [Google Scholar]
- 77. Kruhlak M. J., Celeste A., Dellaire G., Fernandez-Capetillo O., Müller W. G., McNally J. G., Bazett-Jones D. P., Nussenzweig A. (2006) J. Cell Biol. 172, 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mladenov E., Kalev P., Anachkova B. (2009) Radiat. Res. 171, 397–404 [DOI] [PubMed] [Google Scholar]
- 79. Stiff T., O'Driscoll M., Rief N., Iwabuchi K., Löbrich M., Jeggo P. A. (2004) Cancer Res. 64, 2390–2396 [DOI] [PubMed] [Google Scholar]
- 80. Burdak-Rothkamm S., Short S. C., Folkard M., Rothkamm K., Prise K. M. (2007) Oncogene 26, 993–1002 [DOI] [PubMed] [Google Scholar]
- 81. Callén E., Jankovic M., Wong N., Zha S., Chen H. T., Difilippantonio S., Di Virgilio M., Heidkamp G., Alt F. W., Nussenzweig A., Nussenzweig M. (2009) Mol. Cell 34, 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hickson I., Zhao Y., Richardson C. J., Green S. J., Martin N. M., Orr A. I., Reaper P. M., Jackson S. P., Curtin N. J., Smith G. C. (2004) Cancer Res. 64, 9152–9159 [DOI] [PubMed] [Google Scholar]
- 83. Nutley B. P., Smith N. F., Hayes A., Kelland L. R., Brunton L., Golding B. T., Smith G. C., Martin N. M., Workman P., Raynaud F. I. (2005) Br. J. Cancer 93, 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Paull T. T., Lee J. H. (2005) Cell Cycle 4, 737–740 [DOI] [PubMed] [Google Scholar]
- 85. Adelman C. A., De S., Petrini J. H. (2009) Mol. Cell. Biol. 29, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Galron R., Gruber R., Lifshitz V., Lu H., Kirshner M., Ziv N., Wang Z. Q., Shiloh Y., Barzilai A., Frenkel D. (2011) J. Mol. Neurosci., in press [DOI] [PubMed] [Google Scholar]
- 87. Wang V. Y., Zoghbi H. Y. (2001) Nat. Rev. Neurosci. 2, 484–491 [DOI] [PubMed] [Google Scholar]
- 88. Rakic P., Sidman R. L. (1973) J. Comp. Neurol. 152, 133–161 [DOI] [PubMed] [Google Scholar]
- 89. Baptista C. A., Hatten M. E., Blazeski R., Mason C. A. (1994) Neuron 12, 243–260 [DOI] [PubMed] [Google Scholar]
- 90. van den Bosch M., Bree R. T., Lowndes N. F. (2003) EMBO Rep. 4, 844–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lee J. H., Paull T. T. (2005) Science 308, 551–554 [DOI] [PubMed] [Google Scholar]
- 92. Guo Z., Kozlov S., Lavin M. F., Person M. D., Paull T. T. (2010) Science 330, 517–521 [DOI] [PubMed] [Google Scholar]
- 93. Lou Z., Minter-Dykhouse K., Franco S., Gostissa M., Rivera M. A., Celeste A., Manis J. P., van Deursen J., Nussenzweig A., Paull T. T., Alt F. W., Chen J. (2006) Mol. Cell 21, 187–200 [DOI] [PubMed] [Google Scholar]
- 94. Friesner J. D., Liu B., Culligan K., Britt A. B. (2005) Mol. Biol. Cell 16, 2566–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shiotani B., Zou L. (2009) Mol. Cell 33, 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.