Abstract
The endoplasmic reticulum (ER)-associated degradation (ERAD) pathway in the yeast Saccharomyces cerevisiae is mediated by two membrane-bound ubiquitin ligases, Doa10 and Hrd1. These enzymes are found in distinct multiprotein complexes that allow them to recognize and target a variety of substrates for proteasomal degradation. Although multiprotein complexes containing mammalian ERAD ubiquitin ligases likely exist, they have yet to be identified and characterized in detail. Here, we identify two ER membrane proteins, SPFH2 and TMUB1, as associated proteins of mammalian gp78, a membrane-bound ubiquitin ligase that bears significant sequence homology with mammalian Hrd1 and mediates sterol-accelerated ERAD of the cholesterol biosynthetic enzyme HMG-CoA reductase. Co-immunoprecipitation studies indicate that TMUB1 bridges SPFH2 to gp78 in ER membranes. The functional significance of these interactions is revealed by the observation that RNA interference (RNAi)-mediated knockdown of SPFH2 and TMUB1 blunts both the sterol-induced ubiquitination and degradation of endogenous reductase in HEK-293 cells. These studies mark the initial steps in the characterization of the mammalian gp78 ubiquitin ligase complex, the further elucidation of which may yield important insights into mechanisms underlying gp78-mediated ERAD.
Keywords: Cholesterol Metabolism, Cholesterol Regulation, Endoplasmic Reticulum (ER), Protein Degradation, Ubiquitination
Introduction
Ubiquitination is a key step in the endoplasmic reticulum (ER)2-associated degradation (ERAD) pathway, a highly conserved cellular process through which misfolded or unassembled proteins are selectively degraded from ER membranes by 26 S proteasomes (1–4). Most ERAD substrates are ubiquitinated through a reaction-mediated E3 ubiquitin ligase and E2 ubiquitin-conjugating enzymes, which transfer activated ubiquitin from the E1 ubiquitin-activating enzyme to specific lysine residues in the substrate or in a previously attached ubiquitin of a polyubiquitin chain. The polyubiquitination of substrates is an essential modification that ensures their efficient targeting to proteasomes for degradation. The specificity of substrate ubiquitination appears to be primarily determined by ubiquitin ligases. Thus, these enzymes have become a central focus of investigations into the molecular mechanisms underlying the selection of substrates for ubiquitination and subsequent ERAD (5, 6).
In the yeast Saccharomyces cerevisiae, two ERAD ubiquitin ligases have been described: Hrd1 and Doa10 (5, 7, 8). Both of these enzymes are anchored to the ER through a hydrophobic domain with multiple membrane-spanning segments, and they contain a cytosolic domain with a RING finger motif that directs ubiquitin ligase activity. Co-immunoprecipitation studies reveal that Hrd1 and Doa10 exist in distinct multiprotein complexes (7, 8). The Doa10 ubiquitin ligase complex consists of Doa10, the cytosolic ubiquitin-conjugating enzyme Ubc7 and its membrane anchor Cue1, and Ubx2, a ubiquitin regulatory X (Ubx)-domain containing protein that mediates recruitment of soluble cdc48 to membranes. Hrd1 associates with a large multiprotein complex that includes its cofactor Hrd3, Ubc7, and Cue1, the polytopic ER membrane protein Der1 and its recruitment factor Usa1, Ubx2/cdc48, and the Hsp70 chaperone Kar2 bound to the lectin Yos9. The presence of cdc48, which belongs to the family of ATPases associated with various cellular processes (AAA-ATPases), in Doa10 and Hrd1 complexes reflects the general function of the enzyme in the extraction of ubiquitinated ERAD substrates from membranes and their delivery to proteasomes (9, 10). Differences in the composition of Doa10 and Hrd1 complexes are thought to arise from the necessity of these enzymes to recognize distinct types of substrates. For example, Doa10-mediated ubiquitination appears to be restricted toward substrates with misfolded cytosolic domains, whereas Hrd1 recognizes substrates with both lumenal and intramembrane lesions (7, 8). Although mammalian ERAD ubiquitin ligase complexes have not been defined at the molecular level, they are likely to exist considering the degree of conservation of Hrd1 and Doa10 complex components across species and common features shared by the yeast and mammalian ERAD pathways.
The number of ubiquitin ligases that mediate ERAD in mammalian cells appears to far exceed that in yeast (5). One of these enzymes, termed gp78, mediates the sterol-accelerated ERAD of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase. The ER-resident reductase catalyzes the reduction of HMG-CoA to mevalonate, a rate-limiting reaction in the synthesis of cholesterol (11, 12). This sterol-accelerated ERAD contributes to a complex regulatory system that governs feedback regulation of reductase and prevents over-accumulation of cholesterol and other sterols (13). Under conditions of sterol overload, reductase binds to one of two ER membrane proteins called Insig-1 and Insig-2 (14). This binding bridges reductase to Insig-associated gp78 that, together with its cognate ubiquitin-conjugating enzyme Ubc7, mediates ubiquitination of reductase. This ubiquitination imparts the recognition of reductase by VCP/p97, the mammalian homolog of yeast cdc48, for extraction from membranes and delivery to proteasomes for degradation.
The cDNA for gp78 predicts a 643-amino acid protein that contains an N-terminal domain with 5–7 membrane-spanning segments (5). The C terminus of gp78 projects into the cytosol and contains a RING finger motif as well as binding sites for VCP/p97 and Ubc7. The membrane domain of gp78, which mediates its association with Insigs, is organized in membranes with a similar topology to that of human Hrd1. Although their membrane domains share significant amino acid homology, gp78, but not human Hrd1, binds to Insigs and mediates reductase degradation (15). Glycerol gradient centrifugation studies indicate that human Hrd1 exists in a large multiprotein complex (16); however, a complex containing gp78 has not been identified. In the current study we identify the ER membrane protein SPFH2 as an associated protein of gp78. Our results indicate that binding of SPFH2 to gp78 is mediated by a ubiquitin-like (UBL)-domain containing ER membrane protein called TMUB1. The relevance of this gp78-associated complex was indicated by the finding that RNAi-mediated knockdown of both SPFH2 and TMUB1 blunts sterol-induced ubiquitination and degradation of endogenous reductase. These studies indicate gp78 exists in a multiprotein complex containing an array of components that mediate various aspects of the ERAD pathway. The molecular characterization of this complex will likely have important implications for the ERAD of reductase and perhaps other gp78 substrates.
EXPERIMENTAL PROCEDURES
Materials
We obtained 25-hydroxycholesterol (25-HC) from Steraloids Inc. (Wilton, NH), MG-132 from Boston Biochem (Cambridge, MA) and Peptides International (Osaka, Japan), digitonin from Calbiochem, and horseradish peroxidase-conjugated donkey anti-mouse, anti-rabbit, and anti-goat from Jackson ImmunoResearch Laboratories (West Grove, PA). Lipoprotein-deficient serum (LPDS, d > 1.215 g/ml) was prepared from newborn calf serum by ultracentrifugation as described previously (17).
Expression Plasmids
The following plasmids have been described in the indicated references: pCMV-HMG-Red-T7 and pCMV-HMG-Red-Myc, which encode full-length hamster reductase followed by 3 tandem copies of a T7 epitope tag (MASMTGGQQMG) and 5 copies of a c-Myc epitope (EQKLISEEDL), respectively, under transcriptional control of the cytomegalovirus (CMV) promoter (14, 15); pCMV-Insig-1-Myc, which encodes human Insig-1 followed by 6 tandem copies of a c-Myc epitope (18); pCMV-gp78-Myc and pCMV-gp78-Myc (TM), which encode amino acids 1–643 and 1–308 of human gp78, respectively, followed by 5 copies of the c-Myc epitope (15); pCMV-UbxD2-Myc and pCMV-UbxD8-Myc, encoding full-length human UbxD2 and UbxD8, respectively, followed by 3 and 5 copies, respectively, of the c-Myc epitope (19). The following expression plasmids were generated in the pcDNA3.1 mammalian expression vector using standard PCR methods. pCMV-gp78-Myc (Cyto) was generated by fusing the membrane-spanning region of cytochrome P450 2C1 (amino acids 1–29) to the cytosolic domain of gp78 (amino acids 309–643). pCMV-gp78-TAP was generated by replacing the Myc epitope in pCMV-gp78-Myc with three copies of a T7 epitope followed by the tobacco etch virus (TEV) proteolytic cleavage site and Protein A. pCMV-SPFH1-T7 and pCMV-SPFH2-T7 were generated by fusing cDNAs encoding human SPFH1 and SPFH2 (kindly provided by Dr. Stephen Robbins) to a T7 epitope. pCMV-SPFH2-Myc was generated by fusing the human SPFH2 cDNA to one copy of the c-Myc epitope. pCMV-SPFH2-TAP was generated by replacing the T7 epitope in pCMV-SPFH2-T7 with a FLAG epitope followed by a TEV cleavage site and Protein A. pCMV-TMUB1-T7 was generated by fusing the cDNA for human TMUB1 (Open Biosystems) to three copies of the T7 epitope followed by into pcDNA3.1. The expression plasmids pCMV-TMUB1-T7 (ΔHD), (ΔTM1&2), and (ΔTM2) contain deletions of the N-terminal hydrophobic domain (amino acids 5–36), membrane-spanning regions 1 and 2 (amino acids 201–223 and 230–242), and region 2 (amino acids 230–242), respectively. pCMV-TMUB1-T7 (ΔUBL) contains a deletion of the UBL domain of TMUB1 (amino acids 103–171). pCMV-Hrd1-Myc was generated by removing the HA epitope from pCMV-HA-Hrd1 (15) and fusing the Hrd1 cDNA to three copies of the c-Myc epitope. pCMV-Trc8-Myc was generated by fusing the cDNA for human Trc8 (Invitrogen) to five copies of the c-Myc epitope.
Cell Culture
Stock cultures of Chinese hamster ovary 7 (CHO-7) cells, a line of CHO-K1 cells adapted for growth in LPDS, were maintained in monolayer in medium A (1:1 mixture of Ham's F-12 medium and Dulbecco's modified Eagle's medium containing 100 units/ml penicillin and 100 mg/ml streptomycin sulfate) supplemented with 5% (v/v) LPDS at 37 °C, 8–9% CO2. Stock cultures of SV-589 cells, a line of immortalized human fibroblasts expressing the SV40 large T antigen (20), human embryonic kidney (HEK)-293 cells, and HEK-293S cells (derivatives of HEK-293 cells adapted for growth in suspension culture) were grown in a monolayer at 37 °C, 5 and 8–9% CO2, respectively, in medium B (Dulbecco's modified Eagle's medium containing 1000 mg glucose/liter, 100 units/ml penicillin, and 100 mg/ml streptomycin sulfate) supplemented with 10% fetal calf serum (FCS).
Transient Transfection, Cell Fractionation, and Immunoblot Analysis
Cells were set up for experiments on day 0 at the density indicated in the legends to Figs. 1, 2, and 6. On day 1, transfections using the FuGENE 6 transfection reagent (Roche Applied Science) were performed as described (14). Conditions of the subsequent incubations are described in the legends to Figs. 1–6. After incubations, triplicate dishes of cells were harvested and washed with PBS. The resulting cell pellets were resuspended in buffer containing 10 mm HEPES-KOH, pH 7.4, 250 mm sucrose, 1.5 mm MgCl2, 10 mm KCl, 5 mm EDTA, 5 mm EGTA, 5 mm dithiothreitol, 0.1 mm leupeptin, and a protease inhibitor mixture consisting of 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 0.5 mm Pefabloc, 10 μg/ml leupeptin, 5 μg/ml pepstatin A, 25 μg/ml N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal, and 10 μg/ml aprotinin. The cell suspension was lysed by passing through a 22.5-gauge needle 30 times, and the resulting lysates were subjected to 1000 × g centrifugation for 7 min at 4 °C. The supernatant of this spin was subjected to centrifugation at 100,000 × g for 30 min at 4 °C to obtain the membrane pellet and cytosolic supernatant fractions. Aliquots of these fractions were fractionated by SDS-PAGE, after which the proteins were transferred to nitrocellulose membranes and subjected to immunoblot analysis. Primary antibodies used for immunoblotting were as follows: IgG-A9, a mouse monoclonal antibody against the catalytic domain of hamster reductase (21); IgG-740F, a rabbit polyclonal antibody against human gp78 (22); IgG-9E10, a mouse monoclonal antibody against c-Myc purified from the culture medium of hybridoma clone 9E10 (American Type Culture Collection); IgG-P4D1, a mouse monoclonal antibody against bovine ubiquitin (Santa Cruz Biotechnology); monoclonal anti-T7 Tag IgG (Novagen), goat polyclonal, anti-SPFH2 IgG (Novus Biologicals), rabbit polyclonal anti-apoptosis inducing factor (AIF) IgG and rabbit polyclonal anti-calnexin IgG (BD Biosciences), and monoclonal anti-VCP/p97 IgG (BD Transduction Laboratories). Rabbit polyclonal anti-SPFH1 and SPFH2 were generated by immunizing animals with keyhole limpet hemocyanin-conjugated peptides (Genemed Synthesis, Inc.) corresponding to amino acids 46–60 and 279–292 of human SPFH1 and SPFH2, respectively. IgG fractions were purified using Protein G-coupled Sepharose beads (GE Healthcare).
FIGURE 1.
Identification of SPFH2 as a gp78-associated protein. A, CHO/gp78-TAP cells were set up on day 0 at 4 × 105 cells/60-mm dish in medium A containing 5% LPDS. On day 1 the cells were switched to medium A supplemented with 5% LPDS, 10 μm sodium compactin, and 50 μm sodium mevalonate. After incubation for 16 h at 37 °C, the cells were re-fed the identical medium supplemented with 5% LPDS and 10 μm compactin in the absence or presence of 10 mm mevalonate plus 0.1–1.0 μg/ml 25-HC. After incubation for 5 h at 37 °C, the cells were harvested and subjected to subcellular fractionation as described under “Experimental Procedures.” Aliquots of the resulting membrane fractions were subjected to SDS-PAGE, the proteins were transferred to nitrocellulose membranes, and immunoblot analysis was carried out with monoclonal IgG-A9 (against reductase), monoclonal anti-T7 IgG (against gp78-TAP), and polyclonal anti-gp78 IgG. B, HEK-293 cells were set up on day 0 at 7 × 105 cells/100-mm dish in medium B supplemented with 10% FCS. On day 2 the cells were depleted of sterols through incubation in medium B containing 10% LPDS, 10 μm compactin, and 50 μm mevalonate. After incubation for 16 h at 37 °C, the cells were re-fed the identical medium in the absence or presence of 10 μm MG-132 for 2 h, after which they received 1 μg/ml 25-HC plus 10 mm mevalonate (Mev.) for additional 2 h as indicated. The cells were then harvested, lysed, and subjected to immunoprecipitation (Immunoppt.) with either control preimmune (Preim.) IgG or anti-gp78 IgG. Aliquots of the resulting pellet and supernatant fractions of the immunoprecipitation were subjected to SDS-PAGE followed by immunoblot analysis with polyclonal anti-gp78 IgG, monoclonal IgG-A9 (against reductase), monoclonal anti-VCP/p97, and polyclonal anti-SPFH2 IgG. Ab, antibody.
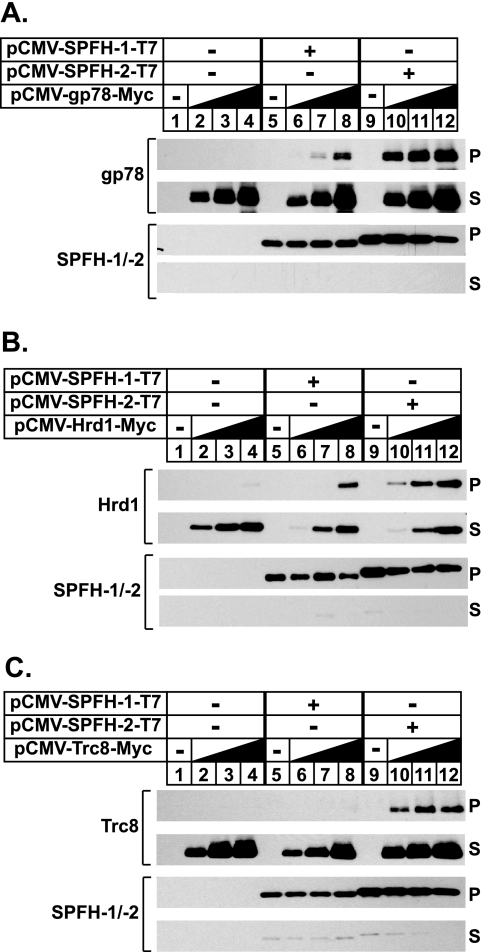
FIGURE 2.
Association of SPFH2 with gp78 and other membrane-bound ubiquitin ligases. A–C, CHO-7 cells were set up on day 0 at 5 × 105 cells/60-mm dish in medium A containing 5% LPDS. On day 1 the cells were transfected with 2 μg/dish of empty pcDNA3.1 vector (lanes 1–4), pCMV-SPFH1-T7 (lanes 5–8), or pCMV-SPFH2-T7 (lanes 9–12) together with increasing amounts (0.1, 0.3, and 1.0 μg) of pCMV-gp78-Myc (A), pCMV-Hrd1-Myc (B), or pCMV-Trc8-Myc (C) in medium A containing 5% LPDS as described under “Experimental Procedures.” The total amount of DNA/dish was adjusted to 2 μg by the addition of pcDNA3.1 mock vector. After incubation for 3–6 h at 37 °C, the cells were depleted of sterols by direct addition of medium A containing 5% LPDS, 10 μm compactin, and 50 μm mevalonate (final concentration). After 16 h at 37 °C, the cells were harvested, lysed, and immunoprecipitated with anti-T7 coupled beads. Aliquots of the pellet (P) and supernatant (S) fractions of the immunoprecipitation were subjected to SDS-PAGE, and immunoblot analysis was carried out with polyclonal anti-T7 IgG (against SPFH1 and SPFH2) and monoclonal IgG-9E10 (against gp78, Hrd1, and Trc8).
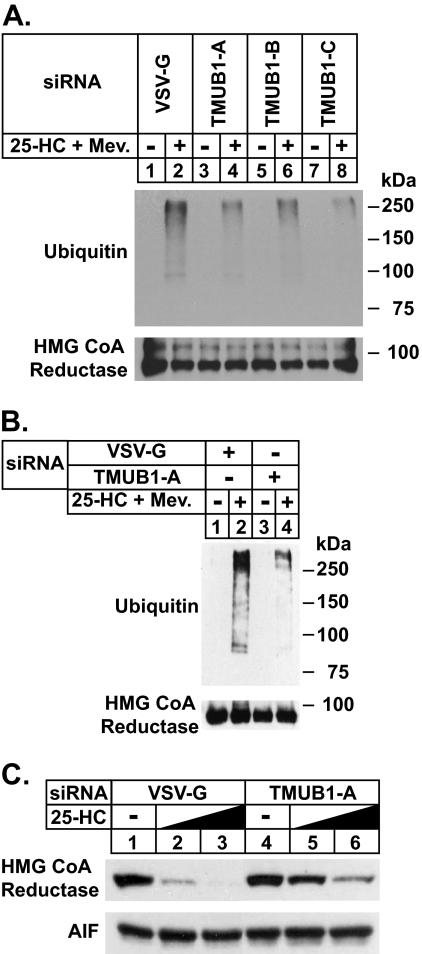
FIGURE 6.
RNAi-mediated knockdown of TMUB1 blunts degradation of HMG-CoA reductase and Insig-1. SV-589 cells were set up for experiments on day 0 at 2 × 105 cells/60-mm dish in medium B containing 10% FCS. On days 1 and 2 the cells were transfected with 600 pmol/dish of the indicated siRNA duplex in medium B containing 10% FCS as described under “Experimental Procedures.” A and B, after sterol depletion the cells were incubated for 30 min at 37 °C in medium B containing 10% LPDS, 10 μm compactin, and 10 μm MG-132 in the absence or presence of 1 μg/ml 25-HC plus 10 mm mevalonate (Mev.) as indicated. The cells were then harvested, lysed, and subjected to immunoprecipitation with polyclonal anti-reductase. Aliquots of the immunoprecipitates were subjected to SDS-PAGE followed by immunoblot analysis with monoclonal IgG-A9 (against reductase) and IgG-P4D1 (against ubiquitin). VSV-G, vesicular stomatitis virus glycoprotein. C, sterol-depleted cells were incubated for 2.5 h at 37 °C in medium B containing 10% LPDS and 10 μm compactin in the absence or presence of 25-HC (0.3 or 1.0 μg/ml). The cells were then harvested for subcellular fractionation. Aliquots of the membrane fractions were subjected to SDS-PAGE followed by immunoblot analysis with monoclonal IgG-A9 (against reductase) and polyclonal anti-apoptosis inducing factor (AIF) as a loading control.
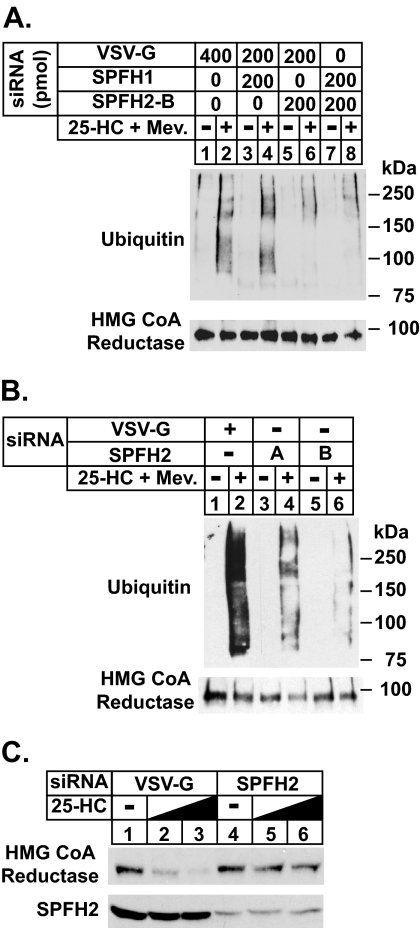
FIGURE 3.
RNAi-mediated knockdown of SPFH2 blunts ER-associated degradation of HMG-CoA reductase and Insig-1. HEK-293 cells were set up on day 0 in medium B containing 10% FCS at 2 × 105cells/60-mm dish. On days 1 and 3 the cells were transfected in medium B containing 10% FCS with the indicated amount of siRNA duplexes targeted SPFH1, SPFH2, or the control mRNA, vesicular stomatitis virus glycoprotein (VSV-G), as described under “Experimental Procedures.” A and B, after sterol depletion for 16 h at 37 °C, the cells were incubated for 1 h in medium B supplemented with 10% LPDS, 10 μm compactin, and 10 μm MG-132 in the absence or presence of 1 μg/ml 25-HC plus 10 mm mevalonate (Mev.). The cells were subsequently harvested and subjected to immunoprecipitation with polyclonal antibodies against reductase. Aliquots of the immunoprecipitates were subjected sequentially to SDS-PAGE and immunoblot analysis with monoclonal IgG-A9 (against reductase) and monoclonal IgG-P4D1 (against ubiquitin). C, after sterol depletion for 16 h at 37 °C, the cells were incubated for 2.5 h in medium B supplemented with 10% LPDS, 10 μm compactin, and 0, 0.3, or 1.0 μg/ml 25-HC as indicated. The cells were subsequently harvested for subcellular fractionation. Aliquots of the membrane fractions (normalized for equal protein loaded/lane) were subjected to SDS-PAGE followed by immunoblot analysis with IgG-A9 (against reductase) and anti-SPFH2 IgG.
FIGURE 4.
Organization of complex between HMG-CoA reductase, Insig-1, gp78, and SPFH2 as determined by co-immunoprecipitation. A, HEK-293 cells were set for experiments on day 0, depleted of sterols on day 2, and treated in the absence or presence of 10 μm MG-132 and 1 μg/ml 25-HC plus 10 mm mevalonate (Mev.) on day 3 as described in the legend to Fig. 1B. After treatments the cells were harvested, lysed, and subjected to immunoprecipitation (Immunoppt.) with polyclonal antibodies against reductase (Red.). Aliquots of the resulting pellet and supernatant fractions of the immunoprecipitation were subjected to SDS-PAGE, and immunoblot analysis was carried out with monoclonal IgG-A9 (against reductase), polyclonal anti-gp78, anti-SPFH2, and monoclonal anti-VCP/p97 IgGs. Ab, antibody; Preim., Preimmune. B, CHO-7 cells were set up for experiments on day 0, transfected with 1 μg of pCMV-HMG-Red-T7, 0.1 μg of pCMV-Insig-1-Myc, and 0.1 μg of pCMV-SPFH2-Myc, and depleted of sterols on day 1 as described in the legend to Fig. 2. The sterol-depleted cells were then incubated for 2 h at 37 °C in medium A containing 5% LPDS, 10 μm compactin, and 10 μm MG-132 in the absence or presence of 1 μg/ml 25-HC plus 10 mm mevalonate as indicated. The cells were subsequently harvested, lysed, and subjected to anti-T7 immunoprecipitation. Aliquots of the resulting supernatant and pellet fractions were subjected to SDS-PAGE followed by immunoblot analysis with polyclonal anti-T7 IgG (against reductase) and monoclonal IgG-9E10 (against Insig-1 and SPFH2). C, CHO-7 cells were set up on day 0, transfected with 0.1 μg of pCMV-Insig-1-T7, 0.01 μg of pCMV-gp78-Myc, and 0.1 μg of pCMV-SPFH2-Myc and depleted of sterols on day 1 as described in B. After sterol depletion, the cells were incubated for 2 h at 37 °C in medium A supplemented with 5% LPDS, 10 μm compactin, and 10 μm MG-132. The cells were subsequently harvested and lysed, and immunoprecipitation was carried out with anti-T7-coupled beads. Aliquots of the pellet and supernatant fractions of the immunoprecipitation were subjected to SDS-PAGE and immunoblot analysis with polyclonal anti-T7 IgG (against Insig-1) and monoclonal IgG-9E10 (against gp78 and SPFH2). D, CHO-7 cells were set up on day 0, transfected with 0.1 μg/dish pCMV-SPFH2-T7 together with 0.1 μg/dish pCMV-gp78-Myc (lanes 1 and 2, WT), (lanes 3 and 4, TM), or (lanes 5 and 6, Cyto.) as indicated and depleted of sterols on day 1 as described in A. After sterol depletion, the cells were subjected to incubation for 2 h in medium A containing 5% LPDS and 10 μm compactin with or without 1 μg/ml 25-HC plus 10 mm mevalonate as indicated. The cells were then harvested, lysed, and immunoprecipitated with anti-T7 beads. Aliquots of the pellet and supernatant fractions were subjected to SDS-PAGE followed by immunoblot analysis with polyclonal anti-T7 (against SPFH2) and monoclonal IgG-9E10 (against gp78).
FIGURE 5.
Identification of TMUB1 as an associated protein of SPFH2. A, shown is the domain structure of TMUB1. B–E, CHO-7 cells were set up for experiments on day 0, transfected with 0.1 μg of pCMV-SPFH2-T7, 1 μg of pCMV-TMUB1-Myc, 1 μg of pCMV-UbxD2-Myc, and 1 μg of pCMV-UbxD8-Myc (B), 0.1 μg of pCMV-SPFH2-Myc, 1 μg of pCMV-TMUB1 (WT), (ΔHD), (ΔTM1&2), and (ΔTM2)-Myc (C), 0.1 μg of pCMV-gp78-Myc, 0.5 μg of pCMV-TMUB1 (WT), 0.02 μg (ΔHD), 0.2 μg of (ΔTM1&2), 0.05 μg of (ΔTM2), and 1.2 μg of (ΔHD, TM1&2)-T7 (D), and 0.1 μg of pCMV-SPFH2-Myc, 1 μg of pCMV-TMUB1-T7, and 0.01 μg of pCMV-gp78-T7 (E) as indicated and depleted of sterols as described in Fig. 2. After sterol depletion, the cells were re-fed medium A containing 5% LPDS, 10 μm compactin, and 10 μm MG-132 in the absence or presence of 1 μg/ml 25-HC plus 10 mm mevalonate (Mev.) as indicated. The cells were subsequently harvested and subjected to immunoprecipitation with anti-Myc (B and E)- or anti-T7 (C and D)-coupled beads. Aliquots of the resulting pellet and supernatant fractions of the immunoprecipitations were subjected to SDS-PAGE followed by immunoblot analysis with polyclonal or monoclonal anti-T7 (against SPFH2 in B. TMUB1 in C, D, and E. gp78 in E) and monoclonal IgG-9E10 (against TMUB1, UbxD2, and UbxD8 in B, SPFH2 in C and E, and gp78 in D).
Generation of Stable Cell Lines for Tandem Affinity Purification (TAP)
CHO/gp78-TAP cells are derivatives of CHO-7 cells that stably overexpress gp78-TAP. These cells were generated as follows. CHO-7 cells were set up on day 0 in medium A supplemented with 5% LPDS at a density of 5 × 105 cells/100-mm dish. On day 1 the cells were transfected with 1 μg/dish pCMV-gp78-TAP using the FuGENE 6 transfection reagent as described above. On day 2 the cells were switched to medium A supplemented with 5% LPDS and 700 μg/ml G418. Fresh medium was added every 2–3 days until colonies formed after about 2 weeks. Individual colonies were isolated with cloning cylinders, and expression of gp78-TAP was determined by immunoblot analysis with monoclonal anti-T7 IgG or polyclonal anti-gp78 IgG. Cells from a single colony were cloned by limiting dilution and maintained in medium A supplemented with 5% LPDS and 700 μg/ml G418 at 37 °C, 8–9% CO2. A similar procedure was used to generate HEK-293S/SPFH2-TAP cells, derivatives of HEK-293S cells that stably overexpress SPFH2-TAP. These cells were maintained in medium B supplemented with 10% FCS and 700 μg/ml G418 at 37 °C, 8–9% CO2.
For gp78-TAP experiments, 100 dishes of CHO/gp78-TAP cells were set up on day 0 at 3 × 105 cells/100-mm dish in medium A containing 5% LPDS. On day 3 the cells were switched to medium A containing 5% LPDS, 50 μm compactin, and 10 μm mevalonate. After incubation for 16 h at 37 °C, the cells were pretreated with 10 μm MG-132 for 1 h, after which they received 1 μg/ml 25-HC, 10 μg/ml cholesterol, and 10 mm mevalonate and incubated for an additional 2 h at 37 °C. Cells were then harvested and lysed in Buffer A (PBS containing 1% digitonin, 5 mm EGTA, 5 mm EDTA, 0.1 mm leupeptin) supplemented with the protease inhibitor mixture. Clarified lysates were subjected to incubation with human IgG-conjugated Sepharose beads (GE Healthcare) for 16 h at 4 °C. After extensive washes with Buffer A, the beads were subjected to treatment with AcTEV protease (Invitrogen) for 16 h at 4 °C. To isolate gp78 and any associated proteins, the released material was subjected to immunoprecipitation with anti-T7-coupled agarose beads (Novagen) for 4 h at 4 °C. After several washes with Buffer A, bound proteins were eluted by incubating the beads in buffer containing 0.1 m citric acid, 1% digitonin, and 1% SDS.
For SPFH2-TAP experiments, HEK-293S/SPFH2-TAP cells were grown in suspension for 5 days in medium B supplemented with 10% FCS. On day 6, the cells received 10 μm MG-132, 1 μg/ml 25-HC, and 10 mm mevalonate and were incubated for an additional 2 h at 37 °C. The cells were subsequently harvested by centrifugation, lysed in Buffer A, and subjected to affinity chromatography with human IgG-conjugated Sepharose beads as described above. The resulting TEV protease-eluted material was then subjected to chromatography on anti-FLAG-coupled agarose beads (Sigma); bound proteins were eluted through incubation with the FLAG peptide (Sigma). Eluted material from the second affinity chromatographic steps (anti-T7 for gp78-TAP experiments and anti-FLAG for SPFH2-TAP experiments) were boiled and fractionated by SDS-PAGE, and the proteins were visualized by silver (ProteoSilverTM Silver Stain kit (Sigma) and Silver staining kit, (Invitrogen)) or colloidal blue (Invitrogen) staining. Segments of the gel that contained visible bands were excised, and the identities of the proteins were determined by tandem mass spectrometry in the Protein Chemistry Core Facility at the University of Texas Southwestern Medical Center.
Immunoprecipitation
Conditions of the incubations are described in legends to Figs. 1–6. At the end of the incubations, the cells were harvested and lysed in Buffer A supplemented with the protease inhibitor mixture. The resulting lysates were precleared for 1 h at 4 °C with irrelevant monoclonal or polyclonal IgG and Protein A/G-agarose beads. After centrifugation at 1000 × g for 5 min at 4 °C, the supernatant was transferred to a fresh tube and incubated overnight at 4 °C with anti-T7- or anti-Myc-coupled agarose beads. The beads were collected by centrifugation; the supernatant of the immunoprecipitation was transferred to a fresh tube. After several washes with Buffer A, the beads were mixed with 2× SDS loading buffer and boiled for 10 min. After centrifugation at 16,000 × g for 10 min at room temperature, the supernatant was transferred to a new tube and designated immunoprecipitation pellet. Aliquots of the supernatant and pellet fractions of the immunoprecipitations were subjected to SDS-PAGE followed by immunoblot analysis.
Ubiquitination of HMG-CoA Reductase
Conditions of the incubations are described in the legend to Figs. 3 and 6. At the end of the incubations, the cells were harvested, lysed in PBS containing 1% Nonidet P-40, 1% deoxycholic acid, 5 mm EDTA, 5 mm EGTA, 0.1 mm leupeptin, 10 mm N-ethylmaleimide, and the protease inhibitor mixture, and subjected to centrifugation at 16,000 × g for 15 min at 4 °C. Immunoprecipitation of the clarified lysates was carried out with polyclonal antibodies against the catalytic domain of human reductase as previously described (14). Aliquots of the immunoprecipitates were subjected to SDS-PAGE followed by immunoblot analysis with mouse monoclonal antibodies IgG-A9 (against reductase) and IgG-P4D1 (against ubiquitin).
RNA Interference
RNAi was carried out as described previously with minor modifications (23). Duplexes of small interfering RNAs (siRNAs) were designed and synthesized by Dharmacon/Thermo Fisher Scientific. SV-589 cells were set up on day 0 at 2 × 105 cells/60-mm dish in medium B containing 10% FCS. On day 1 the cells were washed with PBS and incubated with 200–600 pmol of siRNA duplexes (vesicular stomatitis virus glycoprotein (VSV-G), GGCUAUUCAAGCAGACGGU; SPFH1, GAUUAUGACAAGACCUUAA; SPFH2-A, GAATGTACCTTGTGGGACT; SPFH2-B, AGGTCTACATTGAGCTGTT; SPFH2-C, GAACTATACTGCTGACTAT; SPFH2-D, GCACAAGATAGAAGAGGGA; TMUB1-A, CCTCAATGATTCAGAGCAG; TMUB1-B, CTGAAATTCCTCAATGATT; TMUB1-C, GCAGCTACCGACAGCATGACCUCAAU) mixed with OligofectamineTM (Invitrogen) that was diluted in Opti-MEM I-reduced serum medium (Invitrogen) according to the manufacturer's procedure. After incubation for 6 h at 37 °C, FCS was added to a final concentration of 10%. On day 2, the RNAi procedure was repeated as described above, except that the cells were incubated for 16 h at 37 °C in medium B containing 10% LPDS, 50 μm compactin, and 50 μm mevalonate. The cells were subsequently treated and harvested for analysis as described in the legends to Figs. 3 and 6. RNAi experiments in HEK-293 cells were carried out similarly to those in SV-589 cells except that LipofectamineTM RNAiMAX (Invitrogen) diluted in Opti-MEM I was used to introduce siRNA into cells. Transfection of siRNA duplexes was performed on days 1 and 3 in medium B supplemented with 10% FCS and 10% LPDS plus 10 μm compactin and 50 μm mevalonate, respectively, as described above. After incubation for 16 h at 37 °C, the cells were treated and subsequently harvested for analysis as described in the legends to Figs. 3 and 6.
Real-time PCR
Total RNA was isolated using RNA STAT60 (Tel-Test) according to the manufacturer's procedures. Knockdown efficiency was verified by quantitative real time PCR using each specific primer for human SPFH1, SPFH2, TMUB1, and the control mRNA GAPDH.
RESULTS
To identify proteins that associate with gp78, we began by generating a line of CHO-7 cells that stably overexpress the full-length enzyme with a C-terminal TAP tag. The TAP tag is composed of three copies of a T7 epitope and Protein A separated by a cleavage site for the TEV protease. In previous studies (15), we found that appending epitope tags to the C terminus of gp78 impairs ubiquitin ligase activity. When overexpressed in cells, epitope-tagged gp78 exhibits dominant-negative activity toward reductase ERAD by competing with the endogenous enzyme for binding to Insig. In Fig. 1A, we compared the sterol-accelerated degradation of reductase in non-transfected cells with that in cells stably overexpressing gp78-TAP (designated CHO/gp78-TAP). The cells were first depleted of sterols through incubation for 16 h in medium containing lipoprotein-deficient serum and the reductase inhibitor compactin. The cells also received a low concentration of mevalonate (50 μm), which allows for the synthesis of essential nonsterol isoprenoids but not of cholesterol (24). The cells were then treated for 5 h with various concentrations of the regulatory oxysterol 25-HC plus 10 mm mevalonate, after which they were harvested and subjected to subcellular fractionation. Immunoblot analysis of the resulting membrane fractions with monoclonal anti-reductase antibody revealed that 25-HC stimulated degradation of reductase in a dose-dependent manner (Fig. 1A, top panel, lanes 1–4). In contrast, reductase resisted sterol-induced ERAD in CHO/gp78-TAP cells (lanes 5–8) even though the amount of transfected gp78 drastically exceeded that of the endogenous enzyme (bottom panel, lanes 5–8). This result implies that gp78-TAP is inactive but retains the ability to associate with Insigs or some other component required for sterol-accelerated ERAD of reductase.
We next conducted a large scale TAP experiment in which detergent lysates of CHO/gp78-TAP cells treated with the proteasome inhibitor MG-132, 25-HC, and mevalonate were subjected to affinity chromatography using IgG- and anti-T7-coupled agarose beads. The eluted proteins, which include gp78 and any associated proteins, were fractionated by SDS-PAGE and visualized by silver staining. Segments of the gel containing visible bands that were not present in mock-purified samples (supplemental Fig. 1A) were excised and digested with trypsin, and protein identities were determined by mass spectrometry. One of these proteins, SPFH2, was chosen for further study because of its recently appreciated role in the regulated ERAD of membrane-bound inositol trisphosphate receptors (25, 26).
SPFH2 (also known as Erlin-2) belongs to a family of proteins that contain a stretch of ∼340 amino acids of unknown function called the SPFH domain (named for the founding members Stomatin, Prohibitin, Flotillin, and HflC/HflK) (27). SPFH2 is a 45-kDa glycoprotein that localizes to lipid-rich regions of the ER and is anchored to membranes through a single N-terminal membrane-spanning segment (25, 27, 28). The remainder of the protein, including the SPFH domain, is located within the ER lumen. SPFH2 forms a high molecular weight complex with its close homolog SPFH1 (or Erlin-1); these proteins associate with activated inositol trisphosphate receptors before their ubiquitination and degradation. Both of these processes are significantly blunted when expression of SPFH1 and SPFH2 are reduced through RNAi (25). In addition to inositol trisphosphate receptors, SPFH2 associates with other ERAD substrates including the cystic fibrosis transmembrane conductance receptor ΔF508 mutant (26).
To confirm the association of gp78 with SPFH2, sterol-depleted cells were treated in the absence or presence of 25-HC plus mevalonate and MG-132 before lysis in detergent-containing buffer. The resulting lysates were then immunoprecipitated with polyclonal anti-gp78 IgG or control preimmune IgG. Immunoblot analysis of the resulting immunoprecipitates revealed specific pulldown of endogenous gp78 (Fig. 1B, top panel, compare lanes 1, 2, 5, and 6 with lanes 3, 4, 7, and 8). Endogenous SPFH2 co-immunoprecipitated with gp78 when cells were treated in both the absence and presence of 25-HC plus mevalonate (second panel, lanes 3 and 4); MG-132 had no effect on this interaction (lanes 7 and 8). We also probed the anti-gp78 precipitates for reductase and VCP/p97. Treatment of the cells with 25-HC plus mevalonate caused the disappearance of reductase from the supernatant fraction of the immunoprecipitate (third panel, lanes 9–12), indicating accelerated degradation of the enzyme. Regulated degradation of reductase was blocked by MG-132 (lanes 13–16), which also caused the appearance of enzyme in the pellet fraction of 25-HC and mevalonate-treated cells (compare lanes 7 and 8). In MG-132-treated cells, VCP/p97 also co-precipitated with gp78 in a sterol-independent manner (bottom panel, lanes 7 and 8). The constitutive, non-regulated interaction can be explained by our previous results that gp78 bridges the reductase-Insig complex to VCP/p97 (14).
In the next set of experiments we compared the association of SPFH2 and SPFH1 with gp78 and two other membrane-bound ubiquitin ligases, Trc8 and Hrd1. Cells were transfected with expression plasmids encoding T7-tagged SPFH1 or SPFH2 together with increasing amounts of plasmids encoding Myc-tagged gp78 (Fig. 2A), Hrd1 (Fig. 2B), or Trc8 (Fig. 2C). After transfection, the cells were harvested for preparation of detergent lysates, which were immunoprecipitated with anti-T7-coupled agarose beads to pull down transfected SPFH1 or SPFH2. The results show that SPFH1 co-immunoprecipitated with gp78 (Fig. 2A, top panel, lanes 5–8) and Hrd1 (Fig. 2B, top panel, lanes 5–8) but not with Trc8 (Fig. 2C, top panel, lanes 5–8). In contrast, SPFH2 formed a complex with all three ubiquitin ligases (Fig. 2, A–C, top panels, lanes 10–12). It should be noted that the affinity of SPFH2 for gp78 and Hrd1 exceeds that of SPFH1 as indicated by the appearance of the ubiquitin ligases in the SPFH2 immunoprecipitates when low amounts of the enzymes were transfected.
To appraise a role for SPFH2 in the sterol-induced ubiquitination of endogenous reductase, we transfected HEK-293 cells with various combinations of siRNAs targeting vesicular stomatitis virus glycoprotein (VSV-G), a control gene not expressed in the cells, SPFH1, and SPFH2 (Fig. 3A). After treatment with MG-132 in the absence or presence of 25-HC plus mevalonate, the cells were harvested, and detergent lysates were immunoprecipitated with polyclonal anti-reductase antibodies. Immunoblot analysis of the resulting precipitated material with anti-ubiquitin revealed that in control-transfected cells, reductase became ubiquitinated upon 25-HC plus mevalonate treatment (Fig. 3A, top panel, compare lanes 1 and 2). RNAi-mediated knockdown of SPFH1 had no effect on this reaction (lanes 3 and 4). In contrast, knockdown of SPFH2 significantly blunted the regulated ubiquitination of reductase (lanes 5 and 6); similar results were obtained when expression of both SPFH1 and SPFH2 were reduced by RNAi (lanes 7 and 8). To confirm this result, we conducted another RNAi experiment with an additional siRNA duplex targeting SPFH2 (Fig. 3B). The results show that the alternative siRNA blunted sterol-regulated ubiquitination of reductase, albeit the effect was reduced compared with the original duplex (Fig. 3B, top panel, compare lane 4 with lane 6). Quantitative real-time PCR revealed that SPFH1 and SPFH2 expression was reduced by 90 and 80%, respectively, by RNAi (data not shown). A similar experiment was conducted in the absence of MG-132 to examine whether SPFH2 knockdown blocks sterol-accelerated degradation of reductase. The results show that 25-HC stimulated reductase degradation in cells transfected with control vesicular stomatitis virus glycoprotein siRNA duplexes (Fig. 3B, top panel, lanes 1–3), but this degradation was significantly blunted in cells that received siRNA duplexes targeting SPFH2 (lanes 4–6).
Having established a role for SPFH2 degradation of reductase, experiments were next designed to define the interactions between gp78, SPFH2, and reductase. In the experiment of Fig. 4A, endogenous reductase was immunoprecipitated from lysates of cells treated in the absence or presence of 25-HC plus mevalonate and MG-132. In the absence of MG-132, 25-HC plus mevalonate caused reductase to become degraded, as indicated by reduced levels of the protein in the immunoprecipitate (Fig. 4A, first and fourth panels, compare lanes 1 and 2). This degradation was blocked by MG-132 (lanes 3 and 4), and immunoprecipitation of reductase pulled down endogenous SPFH2, gp78, and VCP/p97 in a manner that was enhanced by the presence of 25-HC plus mevalonate (second, third, and fifth panels, compare lanes 3 and 4). The low level of SPFH2-reductase binding in the absence of 25-HC is likely caused by residual sterols in the lipid-deprived cells.
The Insig requirement for SPFH2-reductase binding was examined by comparing co-immunoprecipitation of the two proteins with or without co-expression of pCMV-Insig-1-Myc encoding human Insig-1 followed by six copies of the Myc epitope. As shown in Fig. 4B, immunoprecipitation of overexpressed reductase brought down a small amount of SPFH2 from lysates of cells treated in both the absence and presence of 25-HC plus mevalonate (second panel, lanes 1 and 2). When Insig-1 was co-expressed, 25-HC plus mevalonate treatment caused a significant increase in the amount of SPFH2 that was brought down by reductase immunoprecipitation (compare lanes 5 and 6).
The experiment of Fig. 4C shows that the Insig-SPFH2 interaction is mediated in part by gp78. Immunoprecipitation of Insig-1-Myc brought down a small amount of SPFH2 (Fig. 4C, second panel, lane 3); this co-immunoprecipitation was markedly enhanced by the overexpression of gp78 (lane 4). Considered together, the results of Fig. 4, A–C, indicate the reductase-SPFH2 complex that forms in sterol-treated cells is mediated by interactions between gp78 and Insig-1.
To identify the region of gp78 that mediates its association with SPFH2, we conducted the co-immunoprecipitation experiment of Fig. 4D. In this experiment cells were transfected with SPFH2 together with full-length gp78, the truncated membrane domain of gp78, or the cytosolic domain of gp78 anchored to membranes through the membrane attachment region of cytochrome P450 2C1. As expected, precipitation of SPFH2 brought down full-length gp78 (Fig. 4D, top panel, lanes 1 and 2); a similar result was obtained for the cytosolic domain of gp78 (lanes 5 and 6). In contrast to this, the membrane domain of gp78 failed to co-precipitate with SPFH2 (lanes 3 and 4), indicating that its binding to gp78 is mediated by the cytosolic domain of the enzyme.
The observation that the cytosolic domain of gp78 mediates its association with SPFH2 is surprising considering the majority of SPFH2 resides in the ER lumen (25). This finding suggests the existence of a protein that bridges SPFH2 to gp78. To identify this protein, we employed HEK-293 cells stably overexpressing SPFH2 with a C-terminal TAP tag (designated SPFH2-TAP) and conducted a two-step affinity purification similar to that used to identify gp78-associated proteins (see Fig. 1). This procedure led to identification of several proteins that appeared in purifications from lysates of SPFH2-TAP-expressing cells (supplemental Fig. 1B). One of these proteins, TMUB1, was chosen for further study. The cDNA for TMUB1 predicts a 245-amino acid protein that contains one N-terminal and two C-terminal hydrophobic domains (Fig. 5A). Protease protection studies indicate that the first hydrophobic domain (HD) inserts into membranes in a hairpin fashion, whereas the two C-terminal hydrophobic domains traverse the membrane. Previous studies demonstrate that the TMUB1 UBL domain projects into the cytosol (29).
To confirm the interaction between SPFH2 and TMUB1, we transfected cells with an expression plasmid encoding T7-tagged SPFH2 together with plasmids encoding Myc-tagged TMUB1, UbxD2, and UbxD8. UbxD8 and UbxD2 are Ubx-domain containing proteins that are known to play key roles in ERAD (30). Overexpression of TMUB1 gave rise to two bands in immunoblots (Fig. 5B, top panel, lanes 1 and 2); the molecular weight of the slower migrating band corresponds to full-length TMUB1 as predicted by its cDNA. Mutagenesis studies show that the faster migrating band results from the use of an alternative site of translational initiation at methionine 56 (data not shown). Immunoprecipitation of transfected TMUB1 brought down SPFH2 (second panel, lanes 1 and 2) but not UbxD2 (lanes 3 and 4) or UbxD8 (lanes 5 and 6).
We next designed experiments to determine which region of TMUB1 mediates its binding to SPFH2. Wild type, full-length TMUB1 co-immunoprecipitated with SPFH2 as expected (Fig. 5C, second panel, lane 2). This binding was abolished by deletion of the first hydrophobic domain of TMUB1 (second panel, compare lanes 2 and 4). In contrast, mutant forms of TMUB1 lacking both TM1 and TM2 or only TM2 continued to bind SPFH2 (lanes 6 and 8, respectively). TMUB1 lacking the UBL domain also bound to SPFH2 (data not shown).
We next used co-immunoprecipitation to compare the binding of wild type and mutant forms of TMUB1 to gp78 (Fig. 5D). The results show that as expected, gp78 co-immunoprecipitated with wild type TMUB1 (second panel, lane 1). Deletion of the N-terminal HD completely abolished this interaction (lane 2), whereas deletion of TM1 and TM2 had an intermediate effect on gp78-TMUB1 binding (lane 3). In contrast, mutant TMUB1 lacking only TM2 continued to associate with gp78 (lane 4); deletion of all three hydrophobic domains in TMUB1 renders the protein cytosolic (data not shown) and abolished gp78 binding. Together, these results indicate that the N-terminal HD and TM1 of TMUB1 contribute significantly to the association of the protein with gp78.
The experiment of Fig. 5E was designed to establish that TMUB1 bridges SPFH2 to gp78. Immunoprecipitation of SPFH2 brought down a small amount of gp78 when the proteins were overexpressed together in cells (third and fourth panels, lanes 1 and 2). The co-immunoprecipitation between gp78 and SPFH2 was markedly enhanced when TMUB1 was co-expressed, indicating the protein bridges SPFH2 to gp78 (third and fourth panels, compare lanes 1 and 2 with 5 and 6). It should be noted that the larger form of TMUB1 corresponding to the full-length protein exhibited a significantly higher affinity for SPFH2 than the shorter form. Considering that the shorter form of TMUB1 is translationally initiated at methionine 56, this result is consistent with a role for the N-terminal hydrophobic domain of TMUB1 in mediating association with gp78 and SPFH2.
A role for TMUB1 in sterol-regulated ubiquitination and degradation of reductase was addressed in Fig. 6. In cells transfected with control siRNA duplexes, reductase became ubiquitinated (Fig. 6, A and B, top panels, lane 2) and degraded (Fig. 6B, top panel, lanes 1–3) in the presence of 25-HC and mevalonate. The RNAi-mediated knockdown of TMUB1 significantly blunted both the sterol-induced ubiquitination (Fig. 6, A, top panel, lanes 4, 6, and 8, and B, top panel, lane 4) and sterol-accelerated degradation (Fig. 6B, top panel, lanes 4–6) of endogenous reductase.
DISCUSSION
In the current studies techniques of tandem affinity purification and mass spectrometry were used to identify SPFH2 as an associated protein of the ERAD ubiquitin ligase gp78 (Figs. 1 and 2). Two lines of evidence indicate that binding of SPFH2 to gp78 is a physiologically relevant event in the sterol-accelerated ERAD of HMG-CoA reductase in HEK-293 cells used in this study. First, 25-HC plus mevalonate stimulated the binding of endogenous SPFH2 to reductase (Fig. 4A), and the interaction was found to be enhanced by the overexpression of Insig-1 (Fig. 4B). The binding of Insig-1 to SPFH2 was found to be mediated by the association of SPFH2 with gp78 (Fig. 4C). Second, RNAi-mediated knockdown of SPFH2, but not the related SPFH1, blunted the sterol-induced ubiquitination and degradation of endogenous reductase (Fig. 3). These results are consistent with a previous report from Wojcikiewicz and co-workers (25) who found that knockdown of SPFH2 stabilized reductase in cycloheximide chase studies. However, it should be noted that in a subsequent study, this group found that SPFH2 knockdown had no effect on regulated ubiquitination and degradation of reductase (31). The discrepancy between these results may be explained by varying reductase/Insig ratios among the different cell lines used in these studies. The regulatory actions of Insigs on reductase are known to be critically dependent on the ratio of the two proteins (18, 32). High levels of Insigs render cells more sensitive to sterol-accelerated reductase ERAD and presumably more refractory to RNAi-mediated knockdown of factors involved in the process. Thus, comparison of Insig levels in cells used in this and previous work will be required to begin the complete resolution of this issue.
SPFH2 is an ER-localized glycoprotein with a single N-terminal membrane-spanning domain followed by a lumenal C-terminal domain. Co-immunoprecipitation experiments reveal that SPFH2 binds to gp78 through the enzyme cytosolic domain (Fig. 4D), a surprising result considering that the majority of SPFH2 resides in the ER lumen. This result suggested the existence of an intermediary protein that bridges SPFH2 to gp78. Indeed, TAP experiments reveal that the UBL-containing protein TMUB1 associates with SPFH2 (Fig. 5, B and C) and facilitates the binding of SPFH2 to gp78 (Fig. 5, D and E). RNAi-mediated knockdown of TMUB1 blunts sterol-regulated ubiquitination and degradation of reductase (Fig. 6, A and B), which is consistent with a role for the protein in the action of SPFH2. The identification of SPFH2 and TMUB1 as gp78-associated proteins marks the beginning of the characterization of this membrane-bound ubiquitin ligase complex. The molecular definition of the gp78 ubiquitin ligase complex will likely yield a set of factors that play important roles in the degradation of not only reductase but of other targets of gp78-mediated ubiquitination.
A major question for future studies is the mechanism through which SPFH2 and TMUB1 facilitate the activity of gp78. The binding of SPFH2 and TMUB1 is not restricted to gp78; these proteins bind to two other membrane-bound ubiquitin ligases, Hrd1 and Trc8 (Fig. 2 and supplemental Fig. 2). This indicates that SPFH2 and TMUB1 may play a general role in the ERAD pathway. A clue to the function of SPFH2 may be provided by the previous finding that the protein localizes to a lipid-enriched region of the ER (28). Thus, the possibility exists that SPFH2 directs gp78 and other ERAD ubiquitin ligases to specific subdomains of the ER where substrate ubiquitination is initiated. This would be consistent with our recent finding that sterols partition reductase to a subdomain of ER membranes that is closely associated with organelles called lipid droplets from which the ubiquitinated enzyme becomes extracted and delivered to proteasomes for degradation (33). Importantly, a fraction of gp78 localizes to lipid-droplet associated subdomains of the ER. Thus, an obvious avenue for future investigation will be to determine the role of SPFH2 and TMUB1 in the localization of gp78 (and perhaps other ERAD ubiquitin ligases) to these lipid droplet-associated ER membranes. Such studies may yield important insights into mechanisms for the degradation of reductase and other ERAD substrates.
Acknowledgments
We thank Drs. Michael S. Brown and Joseph L. Goldstein for continued encouragement and insightful advice. We also thank Dr. Jin Ye for critical reading of the manuscript, Tammy Dinh for excellent technical assistance, and Lisa Beatty, Angela Caroll, Muleya Kapaale, Shomanike Head, and Ijeoma Onwuneme for help with tissue culture.
This work was supported, in whole or in part, by National Institutes of Health Grants HL20948 and GM090216. This work was also supported by the Perot Family Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- 25-HC
- 25-hydroxycholesterol
- LPDS
- lipoprotein-deficient serum
- TAP
- tandem affinity purification
- UBL
- ubiquitin-like
- TEV
- tobacco etch virus
- SPFH
- Stomatin, Prohibitin, Flotillin, and HflC/HflK
- HD
- hydrophobic domain
- Ubx
- ubiquitin regulatory X
- TM
- transmembrane domain.
REFERENCES
- 1. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hampton R. Y., Garza R. M. (2009) Chem. Rev. 109, 1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 4. Jarosch E., Lenk U., Sommer T. (2003) Int. Rev. Cytol. 223, 39–81 [DOI] [PubMed] [Google Scholar]
- 5. Kostova Z., Tsai Y. C., Weissman A. M. (2007) Semin. Cell Dev. Biol. 18, 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deshaies R. J., Joazeiro C. A. P. (2009) Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 7. Carvalho P., Goder V., Rapoport T. A. (2006) Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 8. Denic V., Quan E. M., Weissman J. S. (2006) Cell 126, 349–359 [DOI] [PubMed] [Google Scholar]
- 9. Vij N. (2008) J. Cell. Mol. Med. 12, 2511–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ye Y., Meyer H. H., Rapoport T. A. (2001) Nature 414, 652–656 [DOI] [PubMed] [Google Scholar]
- 11. DeBose-Boyd R. A. (2008) Cell Res. 18, 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goldstein J. L., DeBose-Boyd R. A., Brown M. S. (2006) Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 13. Brown M. S., Goldstein J. L. (1980) J. Lipid Res. 21, 505–517 [PubMed] [Google Scholar]
- 14. Sever N., Yang T., Brown M. S., Goldstein J. L., DeBose-Boyd R. A. (2003) Mol. Cell 11, 25–33 [DOI] [PubMed] [Google Scholar]
- 15. Song B. L., Sever N., DeBose-Boyd R. A. (2005) Mol. Cell 19, 829–840 [DOI] [PubMed] [Google Scholar]
- 16. Schulze A., Standera S., Buerger E., Kikkert M., van Voorden S., Wiertz E., Koning F., Kloetzel P. M., Seeger M. (2005) J. Mol. Biol. 354, 1021–1027 [DOI] [PubMed] [Google Scholar]
- 17. Goldstein J. L., Basu S. K., Brown M. S. (1983) Methods Enzymol. 98, 241–260 [DOI] [PubMed] [Google Scholar]
- 18. Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. (2002) Cell 110, 489–500 [DOI] [PubMed] [Google Scholar]
- 19. Lee J. N., Zhang X., Feramisco J. D., Gong Y., Ye J. (2008) J. Biol. Chem. 283, 33772–33783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamamoto T., Davis C. G., Brown M. S., Schneider W. J., Casey M. L., Goldstein J. L., Russell D. W. (1984) Cell 39, 27–38 [DOI] [PubMed] [Google Scholar]
- 21. Liscum L., Luskey K. L., Chin D. J., Ho Y. K., Goldstein J. L., Brown M. S. (1983) J. Biol. Chem. 258, 8450–8455 [PubMed] [Google Scholar]
- 22. Lee J. N., Song B., DeBose-Boyd R. A., Ye J. (2006) J. Biol. Chem. 281, 39308–39315 [DOI] [PubMed] [Google Scholar]
- 23. Sever N., Song B. L., Yabe D., Goldstein J. L., Brown M. S., DeBose-Boyd R. A. (2003) J. Biol. Chem. 278, 52479–52490 [DOI] [PubMed] [Google Scholar]
- 24. Nakanishi M., Goldstein J. L., Brown M. S. (1988) J. Biol. Chem. 263, 8929–8937 [PubMed] [Google Scholar]
- 25. Pearce M. M., Wang Y., Kelley G. G., Wojcikiewicz R. J. (2007) J. Biol. Chem. 282, 20104–20115 [DOI] [PubMed] [Google Scholar]
- 26. Brodsky J. L., Wojcikiewicz R. J. H. (2009) Curr. Opin. Cell Biol. 21, 516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Browman D. T., Hoegg M. B., Robbins S. M. (2007) Trends Cell Biol. 17, 394–402 [DOI] [PubMed] [Google Scholar]
- 28. Browman D. T., Resek M. E., Zajchowski L. D., Robbins S. M. (2006) J. Cell Sci. 119, 3149–3160 [DOI] [PubMed] [Google Scholar]
- 29. Yang H., Takagi H., Konishi Y., Ageta H., Ikegami K., Yao I., Sato S., Hatanaka K., Inokuchi K., Seog D. H., Setou M. (2008) PLoS One 3, e2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schuberth C., Buchberger A. (2008) Cell. Mol. Life Sci. 65, 2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Y., Pearce M. M., Sliter D. A., Olzmann J. A., Christianson J. C., Kopito R. R., Boeckmann S., Gagen C., Leichner G. S., Roitelman J., Wojcikiewicz R. J. (2009) Biochim. Biophys. Acta 1793, 1710–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee P. C., Liu P., Li W. P., Debose-Boyd R. A. (2007) J. Lipid Res. 48, 1944–1954 [DOI] [PubMed] [Google Scholar]
- 33. Hartman I. Z., Liu P., Zehmer J. K., Luby-Phelps K., Jo Y., Anderson R. G., DeBose-Boyd R. A. (2010) J. Biol. Chem. 285, 19288–19298 [DOI] [PMC free article] [PubMed] [Google Scholar]