Abstract
Cell surface proteoglycans on T cells contribute to retroviral infection, binding of chemokines and other proteins, and are necessary for some T cell responses to the matricellular glycoprotein thrombospondin-1. The major cell surface proteoglycans expressed by primary T cells and Jurkat T cells have an apparent Mr > 200,000 and are modified with chondroitin sulfate and heparan sulfate chains. Thrombospondin-1 bound in a heparin-inhibitable manner to this proteoglycan and to a soluble form released into the medium. Based on mass spectrometry, knockdown, and immunochemical analyses, the proteoglycan contains two major core proteins as follows: amyloid precursor-like protein-2 (APLP2, apparent Mr 230,000) and CD47 (apparent Mr > 250,000). CD47 is a known thrombospondin-1 receptor but was not previously reported to be a proteoglycan. This proteoglycan isoform of CD47 is widely expressed on vascular cells. Mutagenesis identified glycosaminoglycan modification of CD47 at Ser64 and Ser79. Inhibition of T cell receptor signaling by thrombospondin-1 was lost in CD47-deficient T cells that express the proteoglycan isoform of APLP2, indicating that binding to APLP2 is not sufficient. Inhibition of CD69 induction was restored in CD47-deficient cells by re-expressing CD47 or an S79A mutant but not by the S64A mutant. Therefore, inhibition of T cell receptor signaling by thrombospondin-1 is mediated by CD47 and requires its modification at Ser64.
Keywords: Heparan Sulfate, Proteoglycan Structure, Receptor Structure-Function, T Cell Receptor, Thrombospondin
Introduction
Cell surface proteoglycans play critical roles in cell-matrix interactions. They act as co-receptors for some pathogens and growth and motility factors and regulate the functions of other cell surface signaling receptors (reviewed in Refs. 1–4). Chondroitin sulfate or heparan sulfate chains are attached through a conserved core oligosaccharide sequence to specific Ser residues on the core proteins (5). The most prevalent cell surface heparan sulfate proteoglycans (HSPG)5 are the syndecans and glypicans, but a number of additional cell surface proteins can be specifically modified with heparan sulfate glycosaminoglycans (GAG) in certain cells and tissues (6).
CD4+ T cells express highly sulfated heparan sulfate chains because of their high expression of the N-deacetylase/N-sulfotransferase NDST2 and the 3-O-sulfotransferases 3-OST3 (7). T cell HSPGs have been identified as receptors for histones (8) and cyclophilin B (9, 10) and serve as co-receptors for heparin-binding chemokines such as regulated on activation normal T cell expressed and secreted (RANTES) (11).
T cell HSPGs are also co-receptors for retroviral infection. An unidentified cell surface HSPG expressed by the H9 T cell line interacts with the V3 region of the HIV1 envelope gp120-gp41, and heparitinase treatment inhibited binding and entry of the virus into H9 cells (12). The fusion peptide domain of gp41 also interacts specifically with an HSPG on T cells (13). A subsequent study concluded that syndecan functions in trans to promote HIV infection of T cells (14). Similar HSPG binding was demonstrated for the human T cell leukemia virus type 1 (HTLV-1) surface envelope glycoprotein gp46 (15). HSPG functions together with neuropilins to mediate HTLV1 internalization (16).
These GAGs may be present on several core proteins. mRNAs encoding syndecan-1, -2, and -4, CD44v3 isoform, and betaglycan were detected at low levels in peripheral blood CD4+/CD45RO+ T cells (17). Heparan sulfate modification of CD44 requires alternative splicing to include exon V3 (18), but V3 splice isoforms are poorly expressed in tumor-infiltrating lymphocytes (19). Syndecan-4 expressed on T cells serves as a receptor for the heparan sulfate proteoglycan-dependent integrin ligand (DC-HIL) on antigen-presenting cells (20). Syndecan-1 on CD4+/CD45RO+ T cells serves as a receptor for adhesion and chemotaxis responses to cyclophilin B (17).
The proteoglycan agrin is important for signaling through the immunological synapse formed between T cells and antigen-presenting cells (21). Agrin has specific modification sites for chondroitin sulfate and heparan sulfate chains (22), but the occurrence of such GAG modifications on T cell agrin has not been demonstrated, and T cell agrin is primarily expressed as low molecular weight forms (23).
The matricellular glycoprotein thrombospondin-1 (TSP1) binds to heparin and HSPG primarily via its N-terminal domain (24). In Jurkat T cells, TSP1 induces phosphorylation of ERK and AP-1-dependent transcription (25). These responses were inhibited by heparin or growth in the presence of chlorate to inhibit HSPG sulfation. TSP1 also inhibits T cell receptor signaling by binding to an unidentified HSPG (26, 27).
In addition to HSPG, TSP1 interacts with the α4β1 integrin and CD47 on T cells (28, 29). Engaging each of these receptors elicits specific signals in T cells (25, 27). Somatic mutants of the Jurkat T cell line lacking β1 integrins or CD47 have been useful to define signaling pathways mediated by these thrombospondin receptors (27, 30). However, further defining how TSP1 and TSP2 regulate T cell function is limited by not knowing the identity of the HSPG receptor. We have now purified and identified two major cell surface proteoglycans expressed by T cells. We report here that T cells express high molecular weight proteoglycan isoforms of the transmembrane proteins amyloid precursor-like protein-2 (APLP2) and CD47 and examine their roles in mediating T cell responses to TSP1.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Jurkat T cells and the CD47-deficient Jurkat somatic mutant JinB8 (31) were routinely cultured in RPMI 1640 medium supplemented with 10% FBS (Biofluids, Rockville, MD, or Gemini BioProducts), penicillin/streptomycin, and glutamine (Invitrogen). For metabolic labeling studies, Jurkat cells were grown in serum-free medium containing 90% Ham's F-12, 10% RPMI 1640 medium, 5 mm HEPES, 2 mm glutamine, 0.1% BSA, 5 μg/ml insulin, 5 ng/ml sodium selenite, 5 μg/ml transferrin, 200 nm hydrocortisone, and 100 μCi/ml [35S]sulfate as described previously (32). Human umbilical vein endothelial cells (HUVEC) at passages 2–10 (Lonza, Walkersville, MD) and bovine aortic endothelial cells (BAEC) (33) were cultured at 37 °C with 5% CO2 using EGM2 (endothelial growth medium, Lonza). Vascular smooth muscle cells were also from Lonza. Parental and GAG-deficient CHO K1 cell lines (34) were cultured with Ham's F-12 medium (Invitrogen) supplemented with 10% FBS and antibiotics. TSP1 was purified from human platelets as described previously (35). Heparitinase and chondroitinase ABC were from Seikagaku, Associates of Cape Cod, Inc., East Falmouth, MA.
The following antibodies were used: anti-human CD47 (B6H12, Abcam, Cambridge, MA); rabbit anti-human/murine CD47 (H-100, Santa Cruz Biotechnology); anti-cleaved CD47 (Dr. Laura Maile); mouse CD47-miap301 (Pharmingen); anti-FLAG antibody clone M2 (Sigma); anti-DDK monoclonal antibody (OriGene, Rockville, MD); anti-APLP2 (clone D2-II, EMD Calbiochem); anti-Δ-heparan sulfate (clone 3G10, Seikagaku); anti-chondroitin ΔDi-4S (2B6, Seikagaku); anti-chondroitin ΔDi-6S (3B3, Seikagaku); anti-agrin (K-17, Santa Cruz Biotechnology); anti-syndecan-1 (1D4, Sanquin Reagents, Netherlands); syndecan-2 (Santa Cruz Biotechnology); syndecan-4 (5G9, Santa Cruz Biotechnology); anti-carbonic anhydrase 1× (S-20, Santa Cruz Biotechnology); anti-inter-α-trypsin inhibitor (1:2000; Dako A/S, Denmark); and anti-GFP (Santa Cruz Biotechnology). EZ-Link Sulfo-NHS-LC-biotin was purchased from Thermo Scientific.
Proteoglycan Purification
35S-Labeled proteoglycan fractions were isolated from Jurkat cells, primary T cells, and conditioned medium essentially as described previously (32). Briefly, conditioned medium was prepared for ion exchange chromatography by addition of 8 m urea and 0.15 m NaCl, and the pH was adjusted to 4.5 using acetic acid. Cellular proteoglycans were extracted with 8 m urea, 2% Triton X-100, 0.05 m sodium acetate, pH 6.0, and protease inhibitors. After clarification by centrifugation, the extracts were applied to DEAE-Sephacel or Q-Sepharose fast flow columns equilibrated in 8 m urea, 0.05 m sodium acetate ± 0.5% Triton X-100, pH 6.0, and eluted with linear NaCl gradients from 0.15 to 1.3 m. Fractions were analyzed by scintillation counting and pooled for further use. For some experiments, the proteoglycan fractions were concentrated by diluting 8-fold in 8 m urea and applying to 0.2-ml columns of Q-Sepharose fast flow. After washing with column buffer, the proteoglycan was eluted by a step gradient to 1.3 m NaCl in column buffer ± 0.5% Triton X-100 for cell or medium proteoglycans, respectively.
For LC/MS, Jurkat cell proteoglycans from large scale purifications were subjected to a second round of ion exchange chromatography and further purified by SDS-gel electrophoresis. The excised protein bands were digested in gel as described previously (36), and the released peptides were identified using MALDI-TOF mass spectrometry and Mascot searches by the NCI-Frederick Advanced Technology Program.
Purified 35S-labeled proteoglycan fractions were digested with 0.7 milliunits/ml chondroitinase ABC in 0.050 m Tris, 0.050 m sodium acetate, pH 8.0, 0.1 mg/ml BSA, 0.5 mm PMSF, 0.1 mm N-ethylmaleimide for 2–3 h at 37 °C. Digestions with 12.5 milliunits/ml heparitinase or heparitinase plus chondroitinase were performed in 0.005 m Tris, 0.050 m sodium acetate, pH 7.0, 0.1 mg/ml BSA, 0.5 mm PMSF, and 5 mm CaCl2 for 2–3 h at 37 °C. Proteoglycan purification and digestion was monitored by SDS-gel electrophoresis using Tris-glycine SDS gels (Bio-Rad) or BisTris NuPAGE LDS gels (Invitrogen) as indicated and detected by fluorography after treating the gels with Enlightning (PerkinElmer Life Sciences) and drying under vacuum.
Real Time PCR
Jurkat T cells were activated by culturing for 2 h with immobilized anti-CD3 antibody (5 μg/ml, Pharmingen) in the presence or absence of immobilized TSP1 (12.5 μg/ml). Total RNA was isolated using TRIzol following instructions provided by the manufacturer (Invitrogen). For each analysis, 2 μg of RNA was reverse-transcribed using a Fermentas Maxima RT kit (Glen Burnie, MD) and oligo(dT) primers. PCR primers for human CD69 (NR_026671) (5′-CTG GTC ACC CAT GGA AGT GG-3′/5′-ACT TTC CAT GCT GCT GAC CT-3′) and human HPRT1 (NM_000194) (5′-ATT GTA ATG ACC AGT CAA CAG GG-3′/5′-GCA TTG TTT TGC CAG TGT CAA-3′) were identified using FastPCR on-line software (37). Real time PCR was performed using SYBR Green (Thermo Scientific, MD) on an MJ Research Opticon I instrument (Bio-Rad) with the following amplification program: 95 °C for 15 min, followed by 40 cycles of 95 °C for 15 s, 58 °C for 20 s, 72 °C for 25 s, and 72 °C for 1 min. Melting curves were performed for each product from 30 to 95 °C, reading every 0.5 °C with a 6-s dwell time. Fold change in mRNA expression was calculated normalized to HPRT1 mRNA levels.
Glycosaminoglycan Analysis
Jurkat cells were cultured with [34S]sulfate in DMEM containing 10% FBS. After homogenizing cells with a Polytron, proteins in the homogenized cells were digested with Streptomyces griseus protease (Sigma P5147), and the released glycosaminoglycans were isolated by chromatography on DEAE-Sephacel. Glycosaminoglycans were freed of core peptide by β-elimination followed by DEAE-Sephacel purification and then quantified by an HPLC method as described previously (38). Disaccharide compositions were measured by digestion of glycosaminoglycans with a combination of heparin lyase I–III or chondroitinase ABC lyase (EC 4.2.2.4) followed by LC/MS analysis as described previously (39).
Expression Vector Construction
The CD47-GFP expression plasmid pN3-CD47-GFP, encoding an in-frame fusion of EGFP at the C terminus of full-length human CD47 isoform 2, was obtained from Dr. Dennis E. Discher (University of Pennsylvania, Philadelphia) (40) and used as a template. An internal FLAG-like sequence was inserted into the CD47 coding sequence 10 amino acids upstream of the first transmembrane domain between residues Arg132 and Val133 (ELKYR-132DYKDDDDK133-VVSWFS) by PCR amplification of primers containing the FLAG-like coding sequence. A CD47 N-terminal PCR construct was derived using primers 5′-ATTCTCGAGATGTGGCCCCTGGTAGCGG-3′ (XhoI underlined) and 5′-CTTATCGTCGTCATCCTTGTAATCACGATATTTTAGCTCG-3′ (Hpy99I underlined). This procedure resulted in an XhoI site just upstream of the CD47 ATG start site with a FLAG-like coding sequence at the 3′ end that contained a distal Hyp99I site. A C-terminal product was also derived that contained a proximal Hpy99I site (underlined) in the FLAG-like coding region at the 5′ end (5′-GATGACGACGATAAGGTTGTTTCATGGTTTTGTCC-3′) and the remainder of the CD47 coding region (5′-ACTAGTCTATCAGTTATTCCTAGGAGG-3′). These two PCR constructs were cloned in the pCR4-TOPO vector (Invitrogen) and sequenced to verify the absence of mutations. The cloned PCR fragments were digested with XhoI/Hyp99I and PstI (vector-derived)/Hyp99I, respectively, and a three-way ligation for 16 h at 14 °C was performed with pBluescript SK+ that had been digested with XhoI/PstI. The resulting plasmid pSK-CD47iFLAG was used as a template to mutate predicted GAG attachment sites using the QuickChange system following instructions provided by the manufacturer (Stratagene La Jolla, CA). Primer reverse complements are not shown. S64A was constructed using the primer CCACTGTCCCCACTGACTTTGCTAGTGCAAAAATTGAAGTC and S79A using GGAGATGCCGCCTTGAAGATGGATAAGAGTG. For expression cloning, either wild type or mutated CD47 FLAG sequences were released from the pSK-CD47iFLAG vector by digestion with XhoI/NotI, and the purified fragments were ligated into pN3-CD47-EGFP, which had been digested with XhoI/NotI to release the CD47-EGFP fusion sequence.
Transfection and Immunoprecipitation
For transient expression in CHO proteoglycan-biosynthesis mutants, cells were plated overnight in 6-well plates. 2 μg of DNA (CD47-GFP) and 8 μl of TurboFect (Fermentas, MD) were used for transfection. Antisense CD47-morpholino (Gene Tools) or CD47-FLAG was transfected into Jurkat cells using an Amaxa nucleofection kit (Lonza). Transfection was performed for 24 h before cells were lysed or total RNA was extracted.
For immunoprecipitation, cell were lysed using IP buffer (150 mm NaCl, 1% Nonidet P-40, and 50 mm Tris, pH 8.0) or RIPA buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mm EGTA, 1 mm NaF) and ProteoBlock proteinase inhibitor mixture (Fermentas, Glen Burnie, MD). Cell lysates and conditioned media were incubated with 50-μl suspensions of Dynabeads (Invitrogen), washed, and activated according to the manufacturer's protocol. Immunoprecipitation was performed using B6H12 antibody (1:500) and incubated overnight at 4 °C on a shaker. CD47-IP beads were washed with lysis buffer three times. The CD47-IP beads were heated with 50 μl of 4× NuPAGE LDS Sample Buffer (Invitrogen) without reducing agent at 95 °C for 5 min. A 20-μl volume of the IP samples was applied to each lane, and Western blotting was performed using CD47 antibody B6H12 (1:1000) and secondary protein G-horseradish peroxidase conjugate (1:10,000) on 4–12% BisTris gels. Chemiluminescent substrate was used for detection.
For transient expression of CD47 FLAG mutants, JINB8 cells were transfected with CD47 FLAG WT and the respective serine mutants. Immunoprecipitation was performed using Dynabeads with M2-FLAG antibody overnight at 4 °C on a shaker. Beads were washed with lysis buffer three times. The lysates were heated in loading buffer at 95 °C for 5 min and applied to 4–12% BisTris gels. CD47 FLAG was detected using anti-DDK antibody followed by protein G-horseradish peroxidase conjugate (Millipore, 1:5000).
Western Blots
Cells were plated in 6-well plates 1 day before transfection, and both conditioned media and cell lysates were collected for experiments as indicated. Where indicated, the media were concentrated using Centricon filters (Amicon Bioseparations). Cell lysates were prepared using RIPA buffer or IP buffer along with 1× Complete mini-protease inhibitor mixture (Roche Applied Science). The cells were then centrifuged, and the supernatant was collected. Total protein was measured using a bicinchoninic acid assay (Thermo Scientific, Rockford, IL). Equal amounts of total protein (30–40 μg) were loaded for each lane. The cell lysates were mixed with 2× SDS protein gel loading solution (Quality Biological, Inc., Gaithersburg, MD) and heated at 95 °C for 5 min. The proteins were separated on BisTris 4–12% gels (Invitrogen). Western blots were performed using primary antibodies specific for CD47 (B6H12), green fluorescent protein (GFP), or FLAG M2 antibody-horseradish peroxidase (1:1000) in 5% BSA or 5% milk solution and incubated overnight at 4 °C. Secondary antibodies against mouse or rabbit IgG conjugated to HRP were used at 1:5000. SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, Rockford, IL) was used to detect bound antibodies.
To detect heparan sulfate and chondroitin sulfate cleavage epitopes on purified proteoglycans, samples were treated with 12.5 milliunits/ml of heparitinase and/or 0.7 milliunits/ml chondroitinase ABC for 2–3 h at 37 °C. Enzyme treatments were terminated by adding SDS-PAGE sample buffer (with reducing agent) and heating to 100 °C. Samples were resolved by SDS-PAGE and transferred to PVDF. Membranes were blocked with 3% BSA/DPBS/Tween 20 for at least 1 h at room temperature or overnight at 4 °C. They were probed with three antibodies recognizing neo-epitopes created by GAG lyases as follows: anti-Δ-heparan sulfate (clone 3G10, 1:2000), monoclonal anti-proteoglycan ΔDi-4S (2B6, 1:3000), or monoclonal anti-proteoglycan ΔDi-6S (3B3, 1:3000). After incubating for 1–4 h at room temperature with shaking and thorough washing, membranes were incubated with secondary antibody in 3% BSA for 1 h at room temperature. After extensive washing, chemiluminescent substrate was used as to visualize the Western blots.
Heparitinase and Chondroitinase Digestion of Conditioned Media
HUVEC, BAEC, and T cells were plated overnight in 10-cm tissue culture dishes. Approximately 10 ml of media from HUVEC, BAEC, and T cells was harvested and concentrated using Centricon YM-10 filters. To 100 μl of concentrated medium was added 12.5 mIU/ml heparitinase in 100 μl of heparitinase buffer as above or 100 μl of 0.7 mIU/ml of chondroitinase ABC in the corresponding buffer, pH 8.0, as above. Heparitinase or chondroitinase ABC was used either separately for single digestion or together for double digestion. The samples were incubated at 37 °C for 3 h. Immunoprecipitation with CD47 antibody (B6H12) was performed using Dynabeads overnight at 4 °C on a shaker. The samples were heated, and immunoprecipitation and Western blotting were performed using 4–12% BisTris gels.
Isolation of Primary T Cells
Human CD4+ and CD8+ T cells were purified using an autoMACS magnetic isolation system (Miltenyi Biotec) from buffy coat or leukopaks obtained under an approved protocol from the National Institutes of Health Blood Bank. Spleens were removed from anesthetized mice and dispersed through a nylon mesh to generate single cell suspensions. The splenocytes were washed followed by removing erythrocytes using ACK lysis buffer (Lonza). The T cell-enriched splenocytes were isolated using a pan-T cell isolation kit (MACS Miltenyi Biotech) according to the manufacturer's instructions. The cells were lysed using IP buffer and centrifuged at 15,000 rpm for 10 min. Supernatant was collected, and Western blotting was performed using 4–12% BisTris gels with anti-mouse CD47 antibody (1:1000).
TSP1 Binding to T Cell Proteoglycans
Immulon-2 HB microtiter strips were incubated with 10–20 μg/ml TSP1 in 50 μl/well DPBS without Ca2+, Mg2+ for 18 h at 4 °C. Nonspecific binding was blocked by incubating with 3% BSA in DPBS, 200–300 μl/well for 30 min at room temperature. 35S-Labeled proteoglycans diluted 1:10 in DPBS/SA buffer, 50 μl/well, were added and incubated for 3 h at 4 °C. The wells were washed four times with cold DPBS. The bound proteoglycans were solubilized with SDS sample buffer, analyzed by SDS-PAGE, and visualized by fluorography, or 35S-labeled proteoglycans were added in the presence 1–100 μg/ml of heparin (Lilly).
In a reverse assay, pooled proteoglycan fractions exchanged into buffer containing CHAPS in place of Triton X-100 were adsorbed onto plastic, and 125I-TSP1 binding was determined as described previously (32).
Flow Cytometry
Expression of syndecans and agrin was analyzed by flow cytometry. Briefly, cultured Jurkat cells were washed twice with PBS and stained with FITC-conjugated antibodies in staining buffer (PBS, 0.5% BSA, 0.02% sodium azide) at 4 °C for 30 min. After two washes with PBS and fixing with 500 μl of 1% paraformaldehyde (Electron Microscopy Sciences, Ft. Washington, PA), cells were acquired by a FACSCalibur flow cytometer (BD Biosciences). Flow cytometry data were then analyzed by FlowJo software (TreeStar, San Jose, CA). Jurkat cells were gated based on cell optical characteristics (FSC versus SSC). The expression of syndecans and agrin was gated based on antibody staining. All assays were performed in at least three independent experiments and yielded similar results.
RESULTS
T Cells Express a High Molecular Weight HSPG
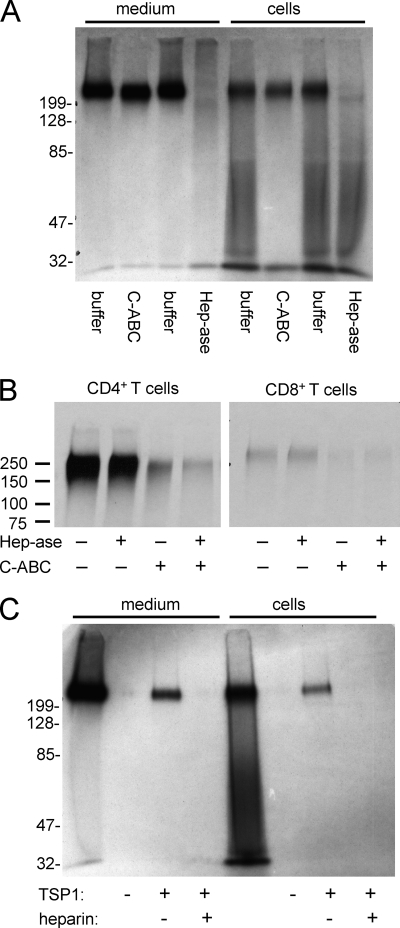
Heparan sulfate and chondroitin sulfate GAGs are known to be released into the medium of Jurkat T cells, but their identities were not determined (41). Metabolic labeling of Jurkat T cells using [35S]sulfate revealed a prominent proteoglycan that migrated with an apparent Mr >200,000 in the cell lysates and was released into conditioned medium (Fig. 1A). Heterogeneous proteoglycans of lower molecular weight were also seen in the cellular fraction but were not released into conditioned medium. Digestion with heparitinase eliminated most 35S labeling of the high molecular weight proteoglycan in both the cell and medium fractions. Migration of the high molecular weight proteoglycan in the cellular fraction was not visibly altered by chondroitinase ABC digestion alone, but a slight decrease in size was reproducibly detected when the medium fraction was treated with this enzyme, suggesting that the same core protein may bear both chondroitin sulfate and heparan sulfate chains. The heterogeneous lower molecular weight species in the cellular fraction appears to be largely chondroitin/dermatan sulfate based on its sensitivity to chondroitinase ABC digestion.
FIGURE 1.
A high molecular weight proteoglycan is expressed by primary human and Jurkat T cells and binds to thrombospondin-1. A, total 35S-labeled proteoglycan fractions extracted from Jurkat T cells and conditioned medium and purified by ion exchange were digested in solution as indicated, separated on 10% acrylamide Tris-glycine SDS gels, and visualized by fluorography. Proteoglycans were incubated for 3 h at 37 °C without (1st and 5th lanes) or with (2nd and 6th lanes) 0.1 IU/ml chondroitinase ABC (C-ABC) without (3rd and 7th lanes) or with (4th and 8th lanes) 12.5 mIU/ml heparitinase (Hep-ase). B, human peripheral blood CD4+ or CD8+ T cells purified by negative selection with magnetic beads were metabolically labeled with 35S at 2 × 106 cells/ml for 24 h. Proteoglycan fractions purified by ion exchange were analyzed on 7.5% gels after incubating for 2.5 h at 37 °C in buffer containing chondroitinase ABC, heparitinase, or buffer alone. C, 35S-proteoglycan from Jurkat cells and medium were bound to immobilized TSP1 in the presence or absence of heparin. After washing, the bound material was eluted with SDS loading buffer and analyzed on 10% acrylamide Tris-glycine SDS gels and detected by fluorography. The 1st and 5th lanes are the respective unfractionated medium and cellular proteoglycan fractions.
Based on HPLC quantification, the Jurkat T cell proteoglycans have 76% heparan sulfate and 24% chondroitin sulfate. The composition of the released 34S-labeled GAG was determined by LC/MS analysis (Table 1). The heparan sulfate contains a high percentage of the trisulfated disaccharide ΔUA2SGlcNS6S. The chondroitinase ABC digestion used for digestion cannot distinguish dermatan sulfate from chondroitin sulfate, but the presence of a significant amount (7%) of the disulfated disaccharide UA2SGalNac4S suggests that Jurkat cells make some dermatan sulfate-like structures. Chondroitin sulfates, including chondroitin sulfate A and chondroitin sulfate C, always contain small amounts of iduronic acid residues, which indicate the general presence of dermatan sulfate in chondroitin sulfate (42).
TABLE 1.
Heparan sulfate (HS) and chondroitin sulfate (CS) disaccharide compositions of Jurkat cells
Results were calculated by LC/MS analyses and represent three independent determinations (mean ± σ).
| HS | ||
| D0A0 | (ΔUAGlcNAc) | 11 ± 2% |
| D0S0 | (ΔUAGlcNS) | 2 ± 0.3% |
| D0A6 | (ΔUAGlcNAc6S) | 5 ± 0.5% |
| D0S6 | (ΔUAGlcNS6S) | 18 ± 1% |
| D2S0 | (ΔUA2SGlcNS) | 6 ± 0.4% |
| D2A6 | (ΔUA2SGlcNAc6S) | 1 ± 0.3% |
| D2S6 | (ΔUA2S GlcNS6S) | 57 ± 3% |
| CS | ||
| D0a0 | (ΔUAGalNAc) | 10 ± 3% |
| D0a4 | (ΔUAGalNAc4S) | 78 ± 5% |
| D0a6 | (ΔUAGalNAc6S) | 5 ± 1% |
| D2a4 | (ΔUA2SGalNAc4S) | 7 ± 0.7% |
Metabolic labeling of CD4+ T cells from human peripheral blood revealed the same prominent high molecular weight proteoglycan (Fig. 1B). Compared with the Jurkat cell proteoglycan, it was more sensitive to chondroitinase digestion, but it also showed partial sensitivity to heparitinase. The same proteoglycan was present in CD8+ T cells but at lower abundance than in CD4+ T cells (Fig. 1B, right panel). It was primarily sensitive to chondroitinase ABC digestion.
Thrombospondin affinity purification demonstrated that the high molecular weight proteoglycan from the cell fraction and that released into the medium bind to TSP1 (Fig. 1C). No binding of lower molecular weight proteoglycans in the cellular fraction to TSP1 was detected. Binding of both the cellular and released HSPG to TSP1 was inhibited by heparin (Fig. 1C). Conversely, soluble radiolabeled TSP1 bound saturably to immobilized proteoglycan, which was inhibited in the presence of heparin (results not shown). Thus, binding is probably mediated by the GAG rather than TSP1 interaction with the core protein, which is denatured during isolation of the proteoglycan.
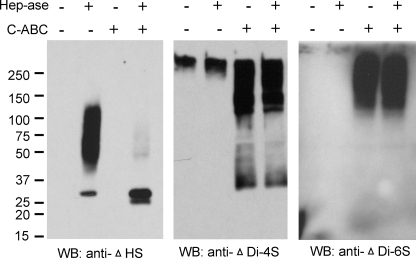
Using antibodies that recognize specific GAGs and neoepitopes formed upon enzymatic cleavage of the GAG chains (43, 44), the secreted high molecular weight proteoglycan fraction was shown to contain heparan sulfate, chondroitin 4-sulfate, and chondroitin 6-sulfate chains (Fig. 2). Digestion with heparitinase alone yielded a Mr 50,000–120,000 species reactive with the Δ-heparan sulfate antibody (3G10), which is specific for nonreducing terminal desaturated uronates (43). Combined digestion with heparitinase plus chondroitinase ABC reduced this to a Mr ∼30,000 species that was recognized by both the Δ-heparan sulfate (3G10) and ΔDi-4S (2B6) antibodies, indicating that heparan sulfate and chondroitin sulfate modifications occur on the same core protein. Recognition of the undigested high molecular weight proteoglycan by 2B6 is consistent with previous reports that this antibody recognizes undigested chondroitin 4-sulfate in some tissues (45). Thus, the major high molecular weight proteoglycan is modified with both heparan sulfate and chondroitin 4-sulfate chains. The chondroitin 6-sulfate may be on a distinct core protein because combined digestion with heparitinase and chondroitinase ABC did not result in any further decrease is size relative to chondroitinase ABC digestion alone, and the Mr ∼30,000 core protein fragment revealed by the Δ-heparan sulfate (3G10) and ΔDi-4S (2B6) antibodies was not detected using the ΔDi-6S (3B3) antibody.
FIGURE 2.
Detection of GAG-modified core protein fragments released from Jurkat T cells. A pooled proteoglycan fraction purified by ion exchange chromatography from conditioned medium was analyzed by Western blotting (WB) on 10% acrylamide gels without digestion or after digestion with the indicated combinations of heparitinase (Hep-ase) and chondroitinase ABC (C-ABC). Blotting with anti-ΔHS (clone 3G10) detects enzymatically cleaved heparan sulfate chains. Further reduction in size following simultaneous digestion with chondroitinase ABC demonstrates core protein fragments modified with both GAG chains. The antibody reacts with a heparan sulfate neo-epitope, generated by digesting heparan sulfate with heparitinase. Blotting with anti-ΔDi-4S (clone 2B6) detects the undigested chondroitin 4-sulfate chains as described elsewhere (45) and detects cleaved chondroitin 4-sulfate chains, some of which are reduced in size by simultaneous heparitinase digestion (compare 3rd with 4th lane). Blotting with anti-ΔDi-6S (clone 3B3) detects cleaved chondroitin 6-sulfate chains only after chondroitinase ABC digestion.
Identification of APLP2 as a Proteoglycan Core Protein
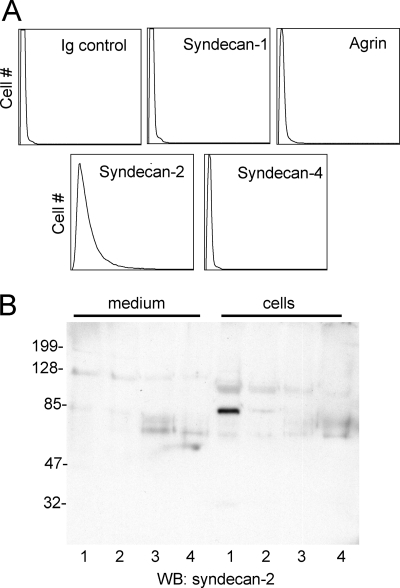
To identify the core protein or proteins associated with this high molecular weight HSPG, we first examined known leukocyte cell surface proteoglycans. We confirmed expression of agrin in peripheral blood T cells by flow cytometry (data not shown), but Jurkat cells lacked detectable agrin expression (Fig. 3A). Because syndecan-1 is a known TSP1 receptor on B cells and endothelial cells (46, 47), we examined syndecan expression in Jurkat cells. Jurkat cells express low levels of syndecan-2 but not detectable levels of syndecan-1 or -4 (Fig. 3A). However, syndecan-2 expressed by Jurkat cells had an apparent Mr of 80,000, which did not coincide with the major TSP-binding HSPG detected by metabolic labeling (Fig. 3B). Also, syndecan-2 was not detected in the conditioned medium. Glypicans were not examined, because their known sizes were not consistent with the major HSPG we observed.
FIGURE 3.
Syndecan and agrin expression by Jurkat T cells. A, cells were analyzed by flow cytometry using antibodies recognizing syndecan-1, syndecan-2, syndecan-4, agrin, or a control IgG. B, proteoglycan fractions extracted from Jurkat cells or conditioned medium were applied to DEAE-Sephacel columns and eluted with increasing NaCl concentrations (0.6–1 m, lanes 1–4). The fractions were separated on a 10% acrylamide Tris-glycine SDS gel and analyzed by Western blotting (WB) using a syndecan-2 antibody.
Mass spectrometry of the cell-associated proteoglycans purified from Jurkat cells by two sequential ion exchange chromatography gradients and excised from SDS gels coincident with the major metabolically labeled HSPG (Mr ∼ 230,000) identified several core protein candidates for the TSP1 receptor (Table 2). Among these, APLP2 was the most abundant.
TABLE 2.
Tryptic peptides identified by LC-MS in a high molecular weight proteoglycan fraction purified from Jurkat T cells
Arrows indicate trypsin cleavage sites.
| Identity | Accession no. | Raw cross-correlation score | δ correlation score | Sequence |
|---|---|---|---|---|
| Carbonic anhydrase IX precursor | Q16790 | |||
| 4.65 | 0.587 | R↓VIEASFPAGVDSSPR↓A | ||
| 3.041 | 0.406 | K↓GGVSYRPAEVAETGA↓ | ||
| Amyloid-like protein 2 precursor | Q06481 | |||
| 4.109 | 0.41 | K↓EAASEKQQLVETHLAR↓V | ||
| 2.222 | 0.317 | R↓WYFDLSK↓G | ||
| 3.312 | 0.425 | R↓VKKEWEEAELQAK↓N | ||
| 3.074 | 0.179 | K↓AKEQLEIR↓H | ||
| 5.199 | 0.575 | K↓ALEKEAASEKQQLVETHLAR↓V | ||
| Neuroglycan C | O95196 | |||
| 3.128 | 0.547 | R↓EAGSAVEAEELVK↓G | ||
| PI3K-related protein kinase | Q96Q15 | |||
| 2.445 | 0.226 | K↓AVEHNIQIGKFSQLVMNR↓A | ||
| Plexin-B1/SEP receptor precursor | Q9UJ93 | |||
| 5.056 | 0.473 | R↓YGLIQAAAVATSR↓E | ||
| Similar to hypothetical protein FLJ20508 | Q8WVD1 | |||
| 2.285 | 0.141 | R↓SMSNTQTQLALMLSCSLQQ↓ |
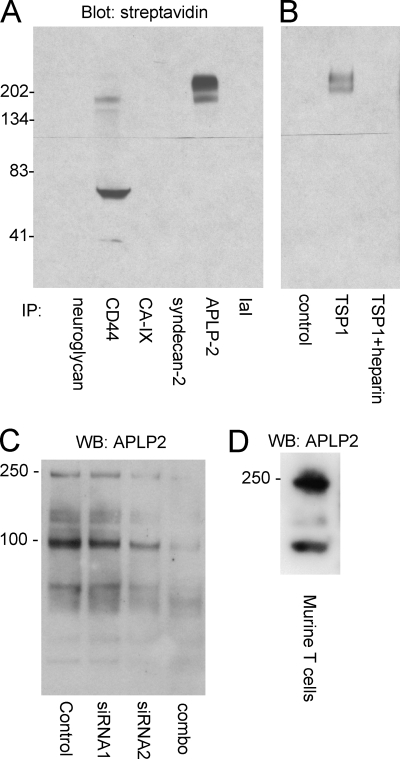
Immunoprecipitation followed by streptavidin blotting of a total proteoglycan fraction purified by ion exchange confirmed the presence of APLP2 with a molecular weight consistent with the metabolic labeling results (Fig. 4A). CD44 was also detected by immunoprecipitation of the proteoglycan fraction, but its apparent molecular weight was lower than that of the major HSPG. Several other candidate core proteins were not detected using the respective antibodies, although this might reflect the relative affinities of these antibodies rather than the total absence of other core proteins. The biotinylated high molecular weight proteoglycan was also pulled down by immobilized TSP1 and migrated as a doublet as detected by blotting with streptavidin (Fig. 4B). The lower band in this doublet coincided with the major band of APLP2 immunoreactivity.
FIGURE 4.
APLP2 is expressed as a high molecular weight proteoglycan by Jurkat and primary mouse T cells. A, immunoprecipitation (IP) of biotin-labeled proteoglycan fraction purified on Q-Sepharose with the indicated antibodies and detected using HRP-streptavidin as follows: neuroglycan, CD44, CA-IX (carbonic anhydrase-IX), syndecan-2, APLP2, IaI (inter-α-trypsin inhibitor). B, same biotinylated proteoglycan fraction was incubated in wells coated with or without immobilized TSP1 ± heparin, and the bound fraction was analyzed by blotting on 10% acrylamide Tris-glycine gels with HRP-streptavidin. C, Jurkat cells were treated with a control siRNA or with individual or combined siRNAs specific for APLP2 for 36 h. Whole cell lysates were resolved on a 4–12% gradient acrylamide BisTris gel and analyzed by Western blotting (WB) using APLP2 antibody. D, murine splenic T cell extract was separated on a 4–12% gradient gel and blotted with APLP2 antibody.
Blotting of total Jurkat cell lysates using an APLP2 antibody was consistent with expression of a GAG-modified isoform at Mr ∼ 230,000 and unmodified forms of APLP2 at Mr ∼ 90,000 (Fig. 4C). The lower band is consistent with the behavior of unmodified APLP2 on SDS gels reported previously, and the former is somewhat larger than previously reported for a chondroitin sulfate-modified isoform of APLP2 (48). The high molecular weight doublet is typically not resolved in this type of gel.
A combination of two siRNAs complementary to APLP2 mRNA was found to suppress expression of both bands relative to a scrambled siRNA control (Fig. 4C). Both isoforms of APLP2 were also detected in primary T cells purified from mouse spleen (Fig. 4D). Therefore, the proteoglycan isoform of APLP2 is not limited to the transformed Jurkat T cell line.
A High Molecular Weight Isoform of CD47
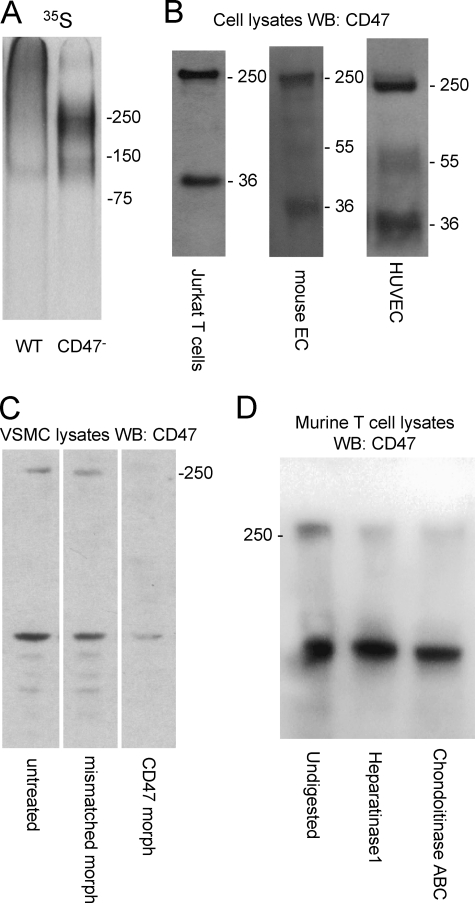
We performed parallel 35S-labeling studies using a somatic mutant Jurkat cell line JinB8 that lacks the known TSP1 receptor CD47 (31), and we noticed that a higher molecular weight proteoglycan was diminished relative to that seen in WT Jurkat cells (Fig. 5A). This suggested that CD47 might also be expressed as a proteoglycan in Jurkat cells. Western blotting of CD47 in Jurkat cells using antibody B6H12 revealed very little CD47 at its expected Mr of 55,000 (Fig. 5B). Rather, most immunoreactivity migrated with an apparent Mr > 250,000. The band at ∼ 36,000 is probably the previously described proteolytic cleavage product of CD47 (49). This assignment was confirmed using the cleavage-specific CD47 antibody R569 mA (data not shown) (49).
FIGURE 5.
CD47 is widely expressed as a high molecular weight sulfated isoform. A, 32S-labeled proteoglycan fractions from WT Jurkat cells (1st lane) and JinB8 CD47-deficient cells (2nd lane) were purified on Q-Sepharose and analyzed by electrophoresis on a 4–12% acrylamide gel and fluorography. B, samples extracted from Jurkat cells (1st lane), mouse lung endothelial cells (2nd lane), and HUVEC (3rd lane) were immunoprecipitated using CD47 anti-human or mouse CD47 antibodies and analyzed by Western blotting (WB) on 4–12% gradient acrylamide BisTris gels. The Mr 36,000 bands are a C-terminal membrane-bound proteolytic fragment of CD47 (49). C, vascular smooth muscle cells were transfected with CD47 morpholino and control morpholino. After transfection, the CD47 Western blot was performed. Untransfected cells (1st lane), mis-matched morpholino (2nd lane), and CD47-morpholino (3rd lane) are shown. D, enzymatic digestion of CD47 proteoglycans from primary murine T cells. Undigested (1st lane), 12.5 mIU/ml heparitinase (2nd lane), and 0.1 IU/ml chondroitinase ABC (3rd lane) were detected by blotting with miab301.
Similar patterns of staining were obtained using a different CD47 antibody H100 with lysates from mouse lung endothelial cells and using B6H12 with lysates from HUVEC (Fig. 5B) and vascular smooth muscle cells (Fig. 5C, left lane). Note that HUVEC do show the typical Mr 55,000 isoform of CD47 but at relatively low abundance. Therefore, the high molecular weight isoform of CD47 may generally be the most abundant form of the protein. The high molecular weight isoform was confirmed to be CD47 by knockdown using a previously validated translation-blocking antisense morpholino oligonucleotide (50). The CD47 morpholino decreased expression of both isoforms, but a 4-base mismatched control morpholino at the same concentration was inactive (Fig. 5C).
The high molecular weight isoform of CD47 was also detected by Western blotting of lysates from purified murine splenic T cells (Fig. 5D, left lane). This band was sensitive to heparitinase and chondroitinase digestion, confirming it to be a proteoglycan and expressed in primary T cells.
Proteoglycan Isoform of CD47 Is Released from Cells
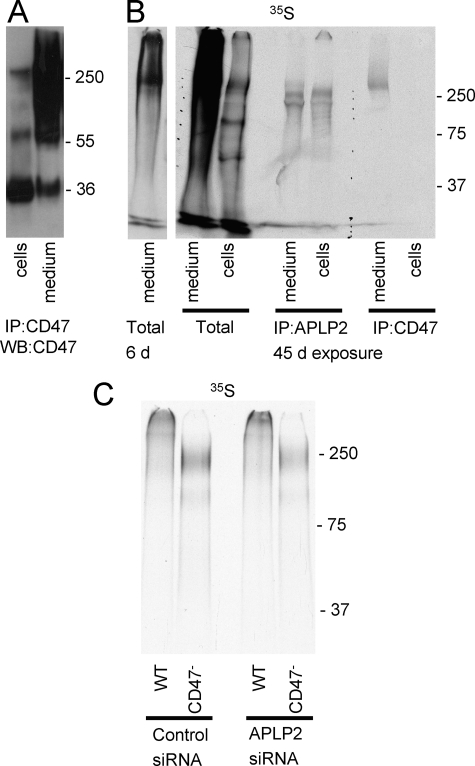
In numerous experiments comparing [35S]sulfate metabolic labeling of conditioned media and cell lysates, we found that the large isoform of CD47 is variably released from Jurkat T cells (results not shown). Similar results were seen by Western blotting. Equivalent amounts of conditioned medium and cell lysate were immunoprecipitated with B6H12 and analyzed by Western blotting (Fig. 6A). In most experiments CD47 immunoreactivity at Mr ∼ 250,000 was much greater in conditioned medium than in cell lysates. When the same experiment was repeated following metabolic labeling with [35S]sulfate and analyzed by immunoprecipitation, sulfated forms of APLP2 were detected at similar levels in the cell lysate and conditioned medium (Fig. 6B). In contrast, 35S incorporation into CD47 was readily detectable in the conditioned medium but only with prolonged exposure in the cell lysate. Consistent with the data in Fig. 5A, the proteoglycan isoform of CD47 migrated slower than that of APLP2. Thus, proteoglycan forms of both APLP2 and CD47 are released from the cell surface. What controls this rate of CD47 release in Jurkat cells is unclear, but it is consistent with the variable proteolytic cleavage of CD47 recently reported in vascular smooth muscle cells (49).
FIGURE 6.
Jurkat T cells express proteoglycan isoforms of both CD47 and APLP2. A, CD47-HSPG expression in cell lysates and conditioned media of Jurkat cells was detected by immunoprecipitation (IP) and Western blotting (WB) after separation on 4–12% gradient BisTris gels. B, Jurkat T cells were metabolically labeled with [32S]sulfate, and incorporation into APLP2 and CD47 in the cell pellet and conditioned medium was detected by fluorography after immunoprecipitation using the respective antibodies and separation on a 7.5% acrylamide Tris-glycine gel. The 1st lane is a 6-day (6 d) exposure, and the 2nd lane is a 45-day (45 d) exposure of the total extract from medium. C, Jurkat and JinB8 cells were transfected with APLP2 siRNA or scrambled control siRNA. [35S]Sulfate was added to the medium 24 h post-transfection, and proteoglycans in the conditioned media harvested after 24 h were purified by Q-Sepharose chromatography. After desalting, pooled samples corresponding to equal cell numbers were applied to 4–12% gradient NuPAGE gels as follows: control siRNA, 1st lane (WT), and 2nd lane, (CD47-deficient cells), APLP2-siRNA, 3rd lane (WT), and 4th lane (CD47-deficient cells).
The absence of the Mr >250,000 proteoglycan in the CD47− mutant indicates that CD47 is the major core protein for this species. To confirm that APLP2 represents the major core protein for the Mr 230,000 species, siRNA knockdown was combined with metabolic labeling in WT and CD47− Jurkat cells (Fig. 6C). Proteoglycan purified by Q-Sepharose chromatography from equal cell numbers was loaded for each lane. In repeated experiments, 35S incorporation into the major 230,000 band in CD47− cells decreased 32–37% in the APLP2 knockdown relative to the scrambled siRNA control. In WT cells, 35S incorporation at the same molecular weight decreased 20–31% with the specific siRNA, whereas 35S incorporation into the >250,000 species presumed to be CD47 was unchanged (98–102% relative to scrambled control). Overall, 35S incorporation was similar for the WT and CD47− cell lines. GAG modification of APLP2 was higher in the cells lacking CD47, but no causal relationship can be assigned from this observation. These results indicate that CD47 and APLP2 account for a majority of the high molecular weight proteoglycan expressed by these cells.
CD47 Is Modified with Heparan and Chondroitin Sulfate Chains
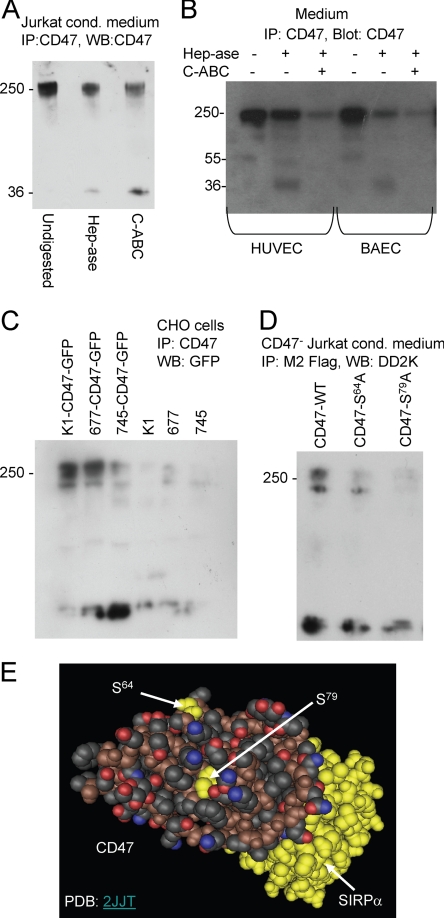
To further confirm that the high molecular weight CD47 isoform detected on Western blots is a proteoglycan, Jurkat cell conditioned medium was analyzed by immunoprecipitation with CD47 antibody followed by blotting with an HRP-conjugated CD47 antibody before and after enzymatic digestion with chondroitinase ABC or heparitinase (Fig. 7A). The intensity of the high molecular weight isoform was decreased by both enzymes, and a new species at Mr 36,000 appeared. This is consistent with the Mr ∼ 30,000 core species detected using anti-cleaved GAG antibodies in Fig. 2.
FIGURE 7.
CD47 is modified by heparan and chondroitin sulfate chains attached to Ser residues in the IgV domain. A, proteoglycans from Jurkat conditioned (cond) medium were digested in solution without enzymes (1st lane), with 12.5 mIU/ml heparitinase (Hep-ase) (2nd lane), or with 0.1 IU/ml chondroitinase ABC (C-ABC) (3rd lane) and separated on a 4–12% NuPAGE gel. B, concentrated conditioned media from the indicated cells were analyzed without digestion (1st and 4th lanes), after digestion with 0.1 IU/ml chondroitinase ABC (3rd and 6th lanes), or 12.5 mIU/ml heparitinase with (2nd and 5th lanes) by electrophoresis on 4–12% gradient acrylamide BisTris gels. After blotting, CD47 was detected using anti-CD47-HRP conjugate and chemiluminescence. C, transient expression of CD47-GFP in Chinese hamster ovary (CHO) cells was analyzed by Western blotting (WB) with GFP antibody. Wild type CHO K1 cells show high molecular weight immunoreactivity characteristic of GAG modification, which is absent in CHO mutants lacking the N-acetylglucosaminyltransferase and glucuronosyltransferase activities (677) (51) or with diminished xylosyltransferase activity (745) (34). D, transient expression of FLAG-tagged CD47 in CD47-deficient JinB8 Jurkat T cells. Cells were transfected with WT CD47-FLAG (lane 2) or CD47 constructs with Ala mutations at Ser64 or and Ser79. Conditioned media were analyzed by immunoprecipitation (IP) with M2 FLAG antibody and blotted with HRP-conjugated DD2K FLAG antibody. E, crystal structure of the IgV domain of CD47 (foreground) complexed with the extracellular domain of its counter-receptor SIRPα (yellow) showing the locations of two Ser residues in CD47 identified as potential GAG modification sites. Structure is from data set Protein Data Bank (PDB) code 2JJT rendered using Cn3D software version 4.1.
The high molecular weight isoform of CD47 immunoprecipitated from conditioned media of HUVEC and BAEC was also partially sensitive to digestion by heparitinase and further decreased or abolished with double digestion (Fig. 7B). Therefore, CD47 was modified with both heparan sulfate and chondroitin sulfate GAGs, although the relative extent of modification with heparan sulfate was higher in the bovine endothelial cells. The residual core fragment of CD47 was poorly reactive with B6H12 after removal of the GAG chains but again is consistent with the core protein size implied by Fig. 2.
To further confirm that CD47 is a proteoglycan, a CD47-GFP construct was transiently expressed in CHO K1 cells or CHO mutant cell lines lacking the N-acetylglucosaminyltransferase and glucuronosyltransferase activities required for heparan sulfate biosynthesis (pgsD-677) (51) or a mutant with diminished xylosyltransferase required for GAG biosynthesis (S745) (Fig. 7C) (34). High molecular weight CD47/GFP immunoreactivity was expressed by CHO K1 cells. This was slightly diminished in the 677 mutant, which lacks heparan sulfate but still can synthesize chondroitin sulfate proteoglycans. However, the abundance of this species was lower in the 745 mutant, which expresses diminished levels of the xylosyltransferase required for priming all GAG synthesis. Therefore, CD47 also exhibits heparan sulfate and chondroitin sulfate modification when transiently expressed in CHO cells.
Identification of GAG Modification Sites on CD47
Based on the previously defined amino acid sequence parameters for GAG modification (52) and a crystal structure for the recombinant extracellular domain of CD47 (53), we identified Ser64 and Ser79 as surface-exposed residues that could be potential acceptors for GAG addition (Fig. 7E). Both are located away from the surface of CD47 known to interact with SIRPα (Fig. 7E, yellow). To investigate whether these residues are involved in CD47 modification, we mutated each Ser residue to Ala in an expression plasmid encoding an internal FLAG-tagged CD47. We transiently expressed these constructs in JinB8 cells and found that mutation of Ser64 or Ser79 greatly diminished expression of the high molecular weight isoform of CD47 (Fig. 7D). Similar results were obtained when the constructs were transiently expressed in BAEC, except some high molecular weight immunoreactivity was retained in the S79A mutant (results not shown). Given our evidence that CD47 is subject to both chondroitin and heparan sulfate modification, it is possible that one molecule of CD47 could bear both GAG chains.
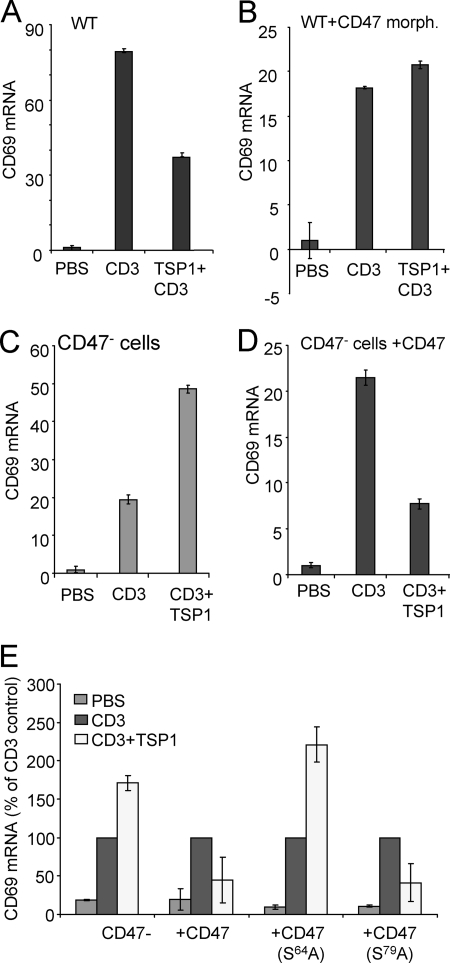
Ser64 of CD47 Is Essential for Inhibiting T Cell Activation
Previous studies have shown that TSP1 inhibits T cell activation induced by antibody engagement of the T cell receptor component CD3 (26, 27). Activation of WT Jurkat cells using CD3 antibody increased expression of mRNA for the early activation marker CD69 by 80-fold at 2 h (Fig. 8A). As expected, this was inhibited more than 50% in the presence of TSP1. We then suppressed CD47 expression using a previously validated CD47 antisense translation-blocking morpholino (50, 54). Induction of CD69 mRNA by anti-CD3 was reduced to 20-fold by this treatment, but this induction was no longer inhibited by the addition of TSP1 (Fig. 8B). Conversely, TSP1 did not inhibit CD69 expression in the CD47-deficient Jurkat cell mutant JinB8 (Fig. 8C). The net induction of CD69 by TSP1 in this mutant is probably mediated by its binding to α4β1 integrin, other ligands of which have been shown to co-stimulate CD69 expression (55, 56). Transient re-expression of CD47 in this cell line restored sensitivity to TSP1 inhibition (Fig. 8D). Because we have shown that APLP2 is expressed as a proteoglycan in the JinB8 cells, this establishes that APLP2 binding is not sufficient for TSP1 to inhibit T cell activation in the absence of CD47. Thus, T cells express two major HSPGs that can bind to TSP1, but only CD47 is essential for TSP1 to inhibit T cell receptor signaling.
FIGURE 8.
CD47 expression is necessary for TSP1 inhibition of T cell activation. A and B, control-treated (A) and CD47 translation-blocking morpholino (morph.)-treated WT Jurkat cells (B) were stimulated by plating on immobilized anti-CD3 for 2 h in the presence or absence of immobilized TSP1, and CD69 mRNA expression was analyzed by real time PCR. C and D, CD47-deficient JinB8 Jurkat cells (C) or the same cells transiently transfected with CD47-FLAG vector (D) were plated on immobilized anti-CD3 with or without co-immobilized TSP1, and mRNA was isolated at 2 h to analyze CD69 expression. Results are ± S.D. and normalized to hypoxanthine-guanine phosphoribosyltransferase mRNA levels as an internal standard. E, plasmids encoding WT, S64A, or S79A mutants of CD47-FLAG were transfected into CD47-deficient cells. After 24 h, the transfected cells were plated on immobilized CD3 antibody in the presence or absence of TSP1 for 2 h. CD69 mRNA expression was analyzed by real time PCR and normalized to HPRT1 mRNA levels. Results are presented as a percentage of the CD69 expression in CD3 activated cells for each set of samples (±S.D.).
To define a specific role for GAG modification of CD47 in this inhibitory activity, CD47-deficient Jurkat cells were transiently transfected with the S64A or S79A mutants and then plated on immobilized CD3 ± TSP1. Expression of the S79A mutant restored sensitivity to inhibition of CD69 induction by TSP1, but cells expressing the S64A mutant were resistant (Fig. 8E). Therefore, Ser64 modification of CD47 is necessary for TSP1-mediated inhibition of T cell activation.
DISCUSSION
We have identified CD47 and APLP2 as the two major cell surface HSPGs expressed by Jurkat T cells. CD47 is expressed as a large Mr > 250,000 proteoglycan, and APLP2 is expressed as a Mr ∼ 230,000 proteoglycan. These proteins bear both heparan and chondroitin sulfate GAG chains, and specific modification sites were identified in the extracellular IgV domain of CD47 that are distal from its SIRPα-binding surface. GAG-modified forms of both APLP2 and CD47 were also found in conditioned medium, implying that ectodomains of CD47 and APLP2 HSPG are shed from the cell surface, as has been frequently observed for syndecans (57, 58). Modification of APLP2 with chondroitin sulfate has been reported previously (59, 60), but its modification with heparan sulfate is novel. To confirm that CD47 is a proteoglycan, we showed that this modification is decreased when CD47-GFP is expressed in CHO cell mutants lacking enzymes required for heparan sulfate biosynthesis. Based on site-directed mutagenesis of several serine residues in the IgV domain of CD47, Ser64 and Ser79 are important for GAG chain addition. Although proteoglycan isoforms of both APLP2 and CD47 probably bind to TSP1, only CD47 is necessary for inhibition of T cell receptor signaling by TSP1. In CD47, Ser64 is required for inhibition of T cell activation by TSP1.
CD47 lacks a conventional Ser-Gly consensus for GAG modification (52), but Ser64 and Ser79 have other features that suggested this function. Both are flanked by Ala residues, which is common for acceptor sites. Furthermore, both Ser residues in human CD47 are flanked on each side by acidic residues characteristic of GAG acceptor sites (Asp62/Glu69 and Asp77/Asp83, respectively) (52). Acidic amino acid side chains are also proximal to both of these exposed Ser residues in the published structure for the extracellular domain of human CD47 (Fig. 7E).
An alternately spliced isoform of APLP2, APLP2–751, was identified as a proteoglycan core protein when expressed in CHO or COS-1 cells (61). A subsequent survey showed that chondroitin sulfate-modified APLP2 is expressed by many cell lines (48), and our studies add primary and transformed T cells to this list. To our knowledge, however, heparan sulfate modification of APLP2 has not been previously reported. APLP2 contains a consensus GAG modification site, and modification of a single site by chondroitin and heparan sulfate chains is possible (52, 62). Chondroitin sulfate modification of APLP2 was found to alter chemotaxis and adhesion responses (60), suggesting that GAG modification of APLP2 could also alter signaling in T cells. However, we found that expression of this form of APLP2 in JinB8 cells is not sufficient to mediate inhibitory signaling by TSP1.
TSP1 does not inhibit T cell receptor signaling in CD47 null T cells. This confirms our previous findings that TSP1 and other CD47 ligands inhibit T cell receptor signaling (26, 27). CD47 is a well known signaling receptor (63, 64), but its ability to interact with ligands via GAG recognition was not previously appreciated. Further studies are needed to determine whether this modified CD47 contributes to T cell responses to heparin-binding chemokines and growth factors and in the pathogenesis of retroviral infections.
This study was conducted primarily using transformed T cells, but our data show that CD47 also exists as a high molecular weight proteoglycan in primary T cells, endothelial cells CHO cells, and vascular smooth muscle cells. Therefore, CD47 is probably widely expressed as a proteoglycan. Notably, in some early CD47 publications a similar high molecular weight CD47 band can be seen on Western blots that were not cropped (65, 66). Given that CD47 plays important roles to regulate NO/cGMP signaling and in physiological processes, including hemostasis, cardiovascular dynamics, and survival of ischemic and radiation injury responses (64, 67, 68), its GAG modification could potentially play a major role in CD47 function. Shedding of this modified CD47 also has the potential to regulate these responses, and identifying conditions that modulate the shedding of CD47 merits further study.
The mechanism by which TSP1 binds to CD47 has been controversial. CD47 was first identified as a TSP1 receptor based on its affinity purification on an immobilized peptide derived from the C-terminal lectin-like domain of TSP1 (69). Further studies have confirmed this binding and binding of recombinant C-terminal regions of TSP1 to cell surface CD47 (70). However, a recombinant extracellular domain of CD47 expressed in bacteria failed to bind to TSP1 (71). This may be explained by incorrect folding of the recombinant CD47 domain or involvement of a long range disulfide bond not present in that construct (72, 73), but our results suggest that post-translational modification with heparan sulfate may also be required for high affinity binding of TSP1 to CD47. Such glycosylation would not occur when recombinant CD47 is expressed in bacteria.
Based on displacement of another CD47 ligand and the dose response for inhibiting cGMP signaling, binding of TSP1 to cell surface CD47 appears to have a picomolar dissociation constant (70), whereas TSP1 binding to highly sulfated heparin is generally of nanomolar affinity and is mediated primarily by the N-terminal rather than the C-terminal domain of TSP1 (74, 75). Notably, a recombinant trimeric fragment of TSP1 containing the N-terminal heparin-binding sites does not inhibit T cell activation (27) or CD47-dependent inhibition of cGMP signaling in vascular cells (76–78). Therefore, TSP1 binding to heparan sulfate via its N-terminal heparin binding domain is not sufficient to elicit an inhibitory signal through CD47. GAG-mediated interactions of cells with the C-terminal region of TSP1 were suggested from cell adhesion studies using myoblasts (79), but the mechanism and receptors involved were not identified. The GAG chains on CD47 may be specifically modified to generate a higher affinity binding site for the C-terminal domain of TSP1, or TSP1 may bind with high affinity by simultaneously engaging carbohydrate and protein determinants on the HSPG isoform of CD47. Such cooperative binding has been documented for some cell surface lectins (80). Further studies are needed to determine the relative contributions of protein-protein versus protein-GAG interactions in the binding of TSP1 to cell surface CD47 and the resulting signaling through this receptor in both immune and vascular cells.
Acknowledgments
We thank Drs. Laura Maile, Jeff Esko, Eric Brown, and Dennis Discher for providing reagents and Tim Veenstra and Tom Conrads for the mass spectrometric analysis.
This work was supported, in whole or in part, by National Institutes of Health Intramural Research Program of the NCI.
- HSPG
- heparan sulfate proteoglycan
- BAEC
- bovine aortic endothelial cells
- GAG
- glycosaminoglycan
- HUVEC
- human umbilical vein endothelial cells
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- DPBS
- Dulbecco's PBS.
REFERENCES
- 1. Park P. W., Reizes O., Bernfield M. (2000) J. Biol. Chem. 275, 29923–29926 [DOI] [PubMed] [Google Scholar]
- 2. Couchman J. R. (2003) Nat. Rev. Mol. Cell Biol. 4, 926–937 [DOI] [PubMed] [Google Scholar]
- 3. Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 4. Schaefer L., Schaefer R. M. (2010) Cell Tissue Res. 339, 237–246 [DOI] [PubMed] [Google Scholar]
- 5. Zhang L., David G., Esko J. D. (1995) J. Biol. Chem. 270, 27127–27135 [DOI] [PubMed] [Google Scholar]
- 6. Kirn-Safran C., Farach-Carson M. C., Carson D. D. (2009) Cell. Mol. Life Sci. 66, 3421–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deligny A., Denys A., Marcant A., Melchior A., Mazurier J., van Kuppevelt T. H., Allain F. (2010) J. Biol. Chem. 285, 1701–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Watson K., Gooderham N. J., Davies D. S., Edwards R. J. (1999) J. Biol. Chem. 274, 21707–21713 [DOI] [PubMed] [Google Scholar]
- 9. Allain F., Vanpouille C., Carpentier M., Slomianny M. C., Durieux S., Spik G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2714–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vanpouille C., Deligny A., Delehedde M., Denys A., Melchior A., Liénard X., Lyon M., Mazurier J., Fernig D. G., Allain F. (2007) J. Biol. Chem. 282, 24416–24429 [DOI] [PubMed] [Google Scholar]
- 11. Oravecz T., Pall M., Wang J., Roderiquez G., Ditto M., Norcross M. A. (1997) J. Immunol. 159, 4587–4592 [PubMed] [Google Scholar]
- 12. Roderiquez G., Oravecz T., Yanagishita M., Bou-Habib D. C., Mostowski H., Norcross M. A. (1995) J. Virol. 69, 2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cladera J., Martin I., O'Shea P. (2001) EMBO J. 20, 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bobardt M. D., Saphire A. C., Hung H. C., Yu X., Van der Schueren B., Zhang Z., David G., Gallay P. A. (2003) Immunity 18, 27–39 [DOI] [PubMed] [Google Scholar]
- 15. Piñon J. D., Klasse P. J., Jassal S. R., Welson S., Weber J., Brighty D. W., Sattentau Q. J. (2003) J. Virol. 77, 9922–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lambert S., Bouttier M., Vassy R., Seigneuret M., Petrow-Sadowski C., Janvier S., Heveker N., Ruscetti F. W., Perret G., Jones K. S., Pique C. (2009) Blood 113, 5176–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pakula R., Melchior A., Denys A., Vanpouille C., Mazurier J., Allain F. (2007) Glycobiology 17, 492–503 [DOI] [PubMed] [Google Scholar]
- 18. Greenfield B., Wang W. C., Marquardt H., Piepkorn M., Wolff E. A., Aruffo A., Bennett K. L. (1999) J. Biol. Chem. 274, 2511–2517 [DOI] [PubMed] [Google Scholar]
- 19. Weimann T. K., Wagner C., Goos M., Wagner S. N. (2003) Exp. Dermatol. 12, 204–212 [DOI] [PubMed] [Google Scholar]
- 20. Chung J. S., Dougherty I., Cruz P. D., Jr., Ariizumi K. (2007) J. Immunol. 179, 5778–5784 [DOI] [PubMed] [Google Scholar]
- 21. Khan A. A., Bose C., Yam L. S., Soloski M. J., Rupp F. (2001) Science 292, 1681–1686 [DOI] [PubMed] [Google Scholar]
- 22. Winzen U., Cole G. J., Halfter W. (2003) J. Biol. Chem. 278, 30106–30114 [DOI] [PubMed] [Google Scholar]
- 23. Jury E. C., Eldridge J., Isenberg D. A., Kabouridis P. S. (2007) J. Immunol. 179, 7975–7983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carlson C. B., Lawler J., Mosher D. F. (2008) Cell. Mol. Life Sci. 65, 672–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilson K. E., Li Z., Kara M., Gardner K. L., Roberts D. D. (1999) J. Immunol. 163, 3621–3628 [PubMed] [Google Scholar]
- 26. Li Z., He L., Wilson K., Roberts D. (2001) J. Immunol. 166, 2427–2436 [DOI] [PubMed] [Google Scholar]
- 27. Li Z., Calzada M. J., Sipes J. M., Cashel J. A., Krutzsch H. C., Annis D. S., Mosher D. F., Roberts D. D. (2002) J. Cell Biol. 157, 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yabkowitz R., Dixit V. M., Guo N., Roberts D. D., Shimizu Y. (1993) J. Immunol. 151, 149–158 [PubMed] [Google Scholar]
- 29. Brown E. J., Frazier W. A. (2001) Trends Cell Biol. 11, 130–135 [DOI] [PubMed] [Google Scholar]
- 30. Barazi H. O., Li Z., Cashel J. A., Krutzsch H. C., Annis D. S., Mosher D. F., Roberts D. D. (2002) J. Biol. Chem. 277, 42859–42866 [DOI] [PubMed] [Google Scholar]
- 31. Reinhold M. I., Green J. M., Lindberg F. P., Ticchioni M., Brown E. J. (1999) Int. Immunol. 11, 707–718 [DOI] [PubMed] [Google Scholar]
- 32. Roberts D. D. (1988) Cancer Res. 48, 6785–6793 [PubMed] [Google Scholar]
- 33. Chandrasekaran L., He C. Z., Al-Barazi H., Krutzsch H. C., Iruela-Arispe M. L., Roberts D. D. (2000) Mol. Biol. Cell 11, 2885–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Esko J. D., Stewart T. E., Taylor W. H. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 3197–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roberts D. D., Cashel J., Guo N. (1994) J. Tissue Cult. Methods 16, 217–222 [Google Scholar]
- 36. Barazi H. O., Zhou L., Templeton N. S., Krutzsch H. C., Roberts D. D. (2002) Cancer Res. 62, 1541–1548 [PubMed] [Google Scholar]
- 37. Kalendar R., Lee D., Schulman A. H. (2009) Genes Genomes and Genomics 3, 1–14 [Google Scholar]
- 38. Studelska D. R., Giljum K., McDowell L. M., Zhang L. (2006) Glycobiology 16, 65–72 [DOI] [PubMed] [Google Scholar]
- 39. Lu H., McDowell L. M., Studelska D. R., Zhang L. (2010) Glycobiol. Insights 2010, 13–28 [PMC free article] [PubMed] [Google Scholar]
- 40. Subramanian S., Boder E. T., Discher D. E. (2007) J. Biol. Chem. 282, 1805–1818 [DOI] [PubMed] [Google Scholar]
- 41. Makatsori E., Karamanos N. K., Papadogiannakis N., Hjerpe A., Anastassiou E. D., Tsegenidis T. (2001) Biomed. Chromatogr. 15, 413–417 [DOI] [PubMed] [Google Scholar]
- 42. Pan J., Qian Y., Zhou X., Pazandak A., Frazier S. B., Weiser P., Lu H., Zhang L. (2010) Nat. Biotechnol. 28, 203–207 [DOI] [PubMed] [Google Scholar]
- 43. David G., Bai X. M., Van der Schueren B., Cassiman J. J., Van den Berghe H. (1992) J. Cell Biol. 119, 961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Caterson B., Christner J. E., Baker J. R., Couchman J. R. (1985) Fed. Proc. 44, 386–393 [PubMed] [Google Scholar]
- 45. Asari A., Akizaki S., Itoh T., Kominami E., Uchiyama Y. (1996) Osteoarthritis Cartilage 4, 149–152 [DOI] [PubMed] [Google Scholar]
- 46. Lebakken C. S., Rapraeger A. C. (1996) J. Cell Biol. 132, 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adams J. C., Kureishy N., Taylor A. L. (2001) J. Cell Biol. 152, 1169–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pangalos M. N., Shioi J., Robakis N. K. (1995) J. Neurochem. 65, 762–769 [DOI] [PubMed] [Google Scholar]
- 49. Maile L. A., Capps B. E., Miller E. C., Allen L. B., Veluvolu U., Aday A. W., Clemmons D. R. (2008) Mol. Endocrinol. 22, 1226–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Isenberg J. S., Romeo M. J., Abu-Asab M., Tsokos M., Oldenborg A., Pappan L., Wink D. A., Frazier W. A., Roberts D. D. (2007) Circ. Res. 100, 712–720 [DOI] [PubMed] [Google Scholar]
- 51. Lidholt K., Weinke J. L., Kiser C. S., Lugemwa F. N., Bame K. J., Cheifetz S., Massagué J., Lindahl U., Esko J. D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 2267–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Esko J. D., Zhang L. (1996) Curr. Opin. Struct. Biol. 6, 663–670 [DOI] [PubMed] [Google Scholar]
- 53. Hatherley D., Graham S. C., Turner J., Harlos K., Stuart D. I., Barclay A. N. (2008) Mol. Cell 31, 266–277 [DOI] [PubMed] [Google Scholar]
- 54. Maxhimer J. B., Soto-Pantoja D. R., Ridnour L. A., Shih H. B., Degraff W. G., Tsokos M., Wink D. A., Isenberg J. S., Roberts D. D. (2009) Sci. Transl. Med. 1, 3ra7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Billard M. J., McIntyre B. W. (2008) Immunol. Cell Biol. 86, 381–384 [DOI] [PubMed] [Google Scholar]
- 56. Neto E. H., Coelho A. L., Sampaio A. L., Henriques M. G., Marcinkiewicz C., De Freitas M. S., Barja-Fidalgo C. (2007) Biochim. Biophys. Acta 1773, 176–184 [DOI] [PubMed] [Google Scholar]
- 57. Alexopoulou A. N., Multhaupt H. A., Couchman J. R. (2007) Int. J. Biochem. Cell Biol. 39, 505–528 [DOI] [PubMed] [Google Scholar]
- 58. Bass M. D., Morgan M. R., Humphries M. J. (2009) Sci. Signal. 2, pe18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guo J., Thinakaran G., Guo Y., Sisodia S. S., Yu F. X. (1998) Invest. Ophthalmol. Vis. Sci. 39, 292–300 [PubMed] [Google Scholar]
- 60. Li X. F., Thinakaran G., Sisodia S. S., Yu F. S. (1999) J. Biol. Chem. 274, 27249–27256 [DOI] [PubMed] [Google Scholar]
- 61. Thinakaran G., Sisodia S. S. (1994) J. Biol. Chem. 269, 22099–22104 [PubMed] [Google Scholar]
- 62. Shintani Y., Takashima S., Asano Y., Kato H., Liao Y., Yamazaki S., Tsukamoto O., Seguchi O., Yamamoto H., Fukushima T., Sugahara K., Kitakaze M., Hori M. (2006) EMBO J. 25, 3045–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Frazier W. A., Isenberg J. S., Kaur S., Roberts D. D. (Feb. 16, 2010) Nature Signaling Gateway 10.1038/mp.a002870.01 [DOI] [Google Scholar]
- 64. Isenberg J. S., Frazier W. A., Roberts D. D. (2008) Cell. Mol. Life Sci. 65, 728–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brown E., Hooper L., Ho T., Gresham H. (1990) J. Cell Biol. 111, 2785–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mawby W. J., Holmes C. H., Anstee D. J., Spring F. A., Tanner M. J. (1994) Biochem. J. 304, 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Isenberg J. S., Martin-Manso G., Maxhimer J. B., Roberts D. D. (2009) Nat. Rev. Cancer 9, 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Isenberg J. S., Qin Y., Maxhimer J. B., Sipes J. M., Despres D., Schnermann J., Frazier W. A., Roberts D. D. (2009) Matrix Biol. 28, 110–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gao A. G., Lindberg F. P., Finn M. B., Blystone S. D., Brown E. J., Frazier W. A. (1996) J. Biol. Chem. 271, 21–24 [DOI] [PubMed] [Google Scholar]
- 70. Isenberg J. S., Annis D. S., Pendrak M. L., Ptaszynska M., Frazier W. A., Mosher D. F., Roberts D. D. (2009) J. Biol. Chem. 284, 1116–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Adams J. C., Bentley A. A., Kvansakul M., Hatherley D., Hohenester E. (2008) J. Cell Sci. 121, 784–795 [DOI] [PubMed] [Google Scholar]
- 72. Floquet N., Dedieu S., Martiny L., Dauchez M., Perahia D. (2008) Arch. Biochem. Biophys. 478, 103–109 [DOI] [PubMed] [Google Scholar]
- 73. Rebres R. A., Vaz L. E., Green J. M., Brown E. J. (2001) J. Biol. Chem. 276, 34607–34616 [DOI] [PubMed] [Google Scholar]
- 74. Sun X., Kaesberg P. R., Choay J., Harenberg J., Ershler W. B., Mosher D. F. (1992) Semin. Thromb. Hemost. 18, 243–251 [DOI] [PubMed] [Google Scholar]
- 75. Yu H., Tyrrell D., Cashel J., Guo N. H., Vogel T., Sipes J. M., Lam L., Fillit H. M., Hartman J., Mendelovitz S., Panel A., Roberts D. D. (2000) Arch. Biochem. Biophys. 374, 13–23 [DOI] [PubMed] [Google Scholar]
- 76. Isenberg J. S., Ridnour L. A., Perruccio E. M., Espey M. G., Wink D. A., Roberts D. D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13141–13146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Isenberg J. S., Wink D. A., Roberts D. D. (2006) Cardiovasc. Res. 71, 785–793 [DOI] [PubMed] [Google Scholar]
- 78. Isenberg J. S., Romeo M. J., Yu C., Yu C. K., Nghiem K., Monsale J., Rick M. E., Wink D. A., Frazier W. A., Roberts D. D. (2008) Blood 111, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Adams J. C., Lawler J. (1994) Mol. Biol. Cell 5, 423–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Rosen S. D. (2004) Annu. Rev. Immunol. 22, 129–156 [DOI] [PubMed] [Google Scholar]