Abstract
Human vitamin K 2,3-epoxide reductase complex subunit 1-like 1 (VKORC1L1), expressed in HEK 293T cells and localized exclusively to membranes of the endoplasmic reticulum, was found to support both vitamin K 2,3-epoxide reductase (VKOR) and vitamin K reductase enzymatic activities. Michaelis-Menten kinetic parameters for dithiothreitol-driven VKOR activity were: Km (μm) = 4.15 (vitamin K1 epoxide) and 11.24 (vitamin K2 epoxide); Vmax (nmol·mg−1·hr−1) = 2.57 (vitamin K1 epoxide) and 13.46 (vitamin K2 epoxide). Oxidative stress induced by H2O2 applied to cultured cells up-regulated VKORC1L1 expression and VKOR activity. Cell viability under conditions of no induced oxidative stress was increased by the presence of vitamins K1 and K2 but not ubinquinone-10 and was specifically dependent on VKORC1L1 expression. Intracellular reactive oxygen species levels in cells treated with 2,3-dimethoxy-1,4-naphthoquinone were mitigated in a VKORC1L1 expression-dependent manner. Intracellular oxidative damage to membrane intrinsic proteins was inversely dependent on VKORC1L1 expression and the presence of vitamin K1. Taken together, our results suggest that VKORC1L1 is responsible for driving vitamin K-mediated intracellular antioxidation pathways critical to cell survival.
Keywords: Antioxidant, Endoplasmic Reticulum (ER), Enzyme Kinetics, Membrane Proteins, Reactive Oxygen Species (ROS), Vitamin K, VKOR, VKORC1, VKORC1L1, VKR
Introduction
Aging and age-related chronic diseases including Alzheimer disease, amyotrophic lateral sclerosis, atherosclerosis, cancer, diabetes, Parkinson disease, and rheumatoid arthritis among others, are widely recognized as resulting from cellular damage inflicted by reactive oxygen species (ROS)3 (1), yet cells produce ROS as a result of normal aerobic metabolism including oxidative phosphorylation in mitochondria and oxidative protein folding in the endoplasmic reticulum (ER). Intracellular antioxidant defense systems, evolved to combat ROS-induced damage, include direct enzymatic scavengers (e.g. superoxide dismutases and peroxidases) and non-enzymatic small-molecule antioxidants (e.g. vitamins C, E, and glutathione) that are maintained in their active, reduced forms by antioxidant-regenerating enzymes (2). Recently, vitamin K hydroquinone (KH2) was recognized as a potent biological antioxidant (3, 4), but regenerative antioxidative enzymatic mechanisms have not been identified for this required trace nutrient.
In vertebrates, K vitamins and their 2,3-epoxides are recognized substrates of vitamin K 2,3-epoxide reductase complex subunit 1 (VKORC1), an enzyme required for blood clotting and bone homeostasis (5). Recently, the first high-resolution structure of a prokaryotic vitamin K epoxide reductase (VKOR) enzyme from the fully sequenced thermophilic cyanobacterial Synechococcus species JA-2-3′a(2–13) (for which the VKOR enzyme will be subsequently identified as “proVKOR” in this article) (6, 7) was solved to 3.6 Å resolution; it is responsible for passing reducing equivalents from de novo oxidative protein folding in the periplasmic space to lipidic quinones in the cell outer membrane (8). In contrast to genomes of archaea, eubacteria, plants, protists, and lower animals that include a single VKOR protein ortholog, vertebrate genomes include two paralogous enzymes, VKORC1 and VKORC1-like 1 (VKORC1L1), likely resulting from a gene duplication of an early common VKOR ancestor (9–12). Although the function and subcellular location of VKORC1 were well characterized as early as two decades before the gene was identified (13), there has been no informative study of function or location for VKORC1L1.
Here we show that VKORC1L1 is responsible for vitamin K-mediated increased survival of oxidatively stressed cells and for limiting the amount of intracellular ROS, and that it plays a key role in mitigating oxidative damage to membrane proteins. Thus, VKORC1L1 apparently plays a ubiquitous, fundamental role in intracellular antioxidation.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK 293T human embryonic kidney cells (ATCC cell line CRL-11268) were cultured in minimum Eagle's medium, 10% FBS, in a humidified atmosphere under 5% CO2 at 37 °C. Cells (80–90% confluent) were transfected with pCEP4 (Invitrogen) mammalian expression vector constructs with integrated cDNA open reading frames encoding human VKORC1L1, VKORC1, or γ-glutamyl carboxylase (GGCX) proteins using FuGENE HD transfection reagent (Roche Applied Science) according to the manufacturer's instructions. For VKORC1L1 transcription knockdown, 100 μm VKORC1L1 siRNA in culture medium was applied for transfection (Qiagen predesigned siRNA, catalog number S104138407).
Antioxidant Supplementation
Concentrations of vitamins K1 and K2 were measured for neat fetal bovine serum (FBS) by extraction into isopropanol:hexane (3:2), and a standard workup was done for HPLC-based determination as for the VKOR enzymatic assay (see following VKOR assay method). The mean nominal Q10 concentration in FBS was taken to be 50 nm from a previous report (14). Nominal concentrations of antioxidants in culture media supplemented to 10% FBS were: K1, 24 pm; K2, undetectable; Q10, 5 nm. For experiments requiring elevated antioxidant concentrations, antioxidants were additionally supplemented with 1 μm in culture media.
Relative Quantification of mRNA by Real-time RT-PCR
Total cDNA of control and hydrogen peroxide-treated (75 μm) HEK 293T cells was synthesized by reverse transcription using random primers and hexanucleotides (Omniscript RT kit, Qiagen) after isolating mRNA from cells using the RNeasy Mini kit (Qiagen). Transcription rates of VKORC1, VKORC1L1, and the porphobilinogen deaminase (PBGD) gene, an endogenous, constitutively transcribed control, were analyzed by hydrolysis probe-based real-time PCR (ABSOLUTETM QPCR, Abgene; see supplemental “Methods” for primers, probes, and temperature cycling details). Real-time detection of amplification was performed in triplicate on an ABI Prism 7500 sequence detection system (Applied Biosystems). VKORC1 and VKORC1L1 transcription rates were calculated by the comparative Ct (ΔΔCt) method and normalized to PBGD expression levels relative to the zero time (t = 0) control samples (15).
VKOR and Vitamin K Quinone Reductase (VKR) Activity Assays for HEK 293T Cell Crude Membranes
VKOR activity of crude cell membranes was measured by the standard dithiothreitol (DTT)-driven method (16). Epoxidation of vitamins K1 and K2 (Sigma-Aldrich) was performed according to Tishler et al. (17) to yield the respective vitamin K 2,3-epoxides (K>O). VKR activity was determined by the same method as for VKOR activity but with vitamin K quinone replacing K>O as substrate. Enzymatic activities to reduce K vitamin epoxides to the respective quinones, or to reduce K1 quinone to K1 hydroquinone, were driven by the addition of 5 mm DTT for 1 h at 30 °C as described previously (16). After organic phase extraction with isopropanol:hexane (3:2) and dry-down in vacuo, products were dissolved in methanol, resolved by C18 RP-HPLC chromatography (Supelcosil LC-18-S column, 150 × 4.6 mm, 5 μm), subjected to isocratic methanol elution, and quantitated by diode array detection at 254 nm (LaChrom Elite HPLC system, VWR-Hitachi, Darmstadt, Germany). K vitamins and epoxides were resolved by RP-HPLC (LaChrom Elite®, VWR-Hitachi) and quantified by flow-cell spectrophotometry (L-2455 diode array detector) with integration of peak area A248 nm (telution ∼10 min). Vitamin K hydroquinone was detected by flow-cell fluorometry (L-2485 fluorescence detector; λex 240 nm, λem 430 nm, telution ∼2.4 min).
Protein concentrations were determined by standard Lowry protein assay to normalize activities with respect to total protein concentration (18). VKOR kinetics for VKORC1L1 were determined for various concentrations of K vitamin epoxides (0–16 μm); pseudo first-order reaction conditions (i.e. quinone product concentration negligible compared with epoxide substrate concentration) at maximal velocities were verified by Eisenthal and Cornish-Bowden direct linear plots. The apparent kinetic constants Km and Vmax were calculated from the intersection loci of linear regression line fits to the raw data (velocity versus −[substrate]): x intercept, mean measured velocity (nmol/mg enzyme/h); negative y intercept, mean substrate concentration (μm). Error ranges were ±S.E. for the range of data at each intersection locus. VKR activity of VKORC1L1 was semiquantitatively confirmed by comparison of reaction products separated by C18 RP-HPLC monitored by flow-cell fluorometry detection of the K1 hydroquinone product (LaChrom Elite L-2485 fluorometer, λex = 246 nm, λem = 430 nm; VWR-Hitachi). Fluorescence peaks at 2.4 min elution time are characteristic of fully reduced vitamin K hydroquinone as determined for a chemically reduced external standard.
Cell Viability Analysis
Cell viability was measured using the CellTiter 96® AQueous cell proliferation assay (Promega, Mannheim, Germany) according to the manufacturer's instructions. HEK 293T cells overexpressing VKORC1L1, anti-VKORC1L1 siRNA (HP GenomeWide siRNA, Qiagen, Hilden, Germany), or random sequence siRNA (negative control) were cultured for 24 h. Subsequently, cells were plated into 96-well plates (2 × 104 cells/well), cultured for 4 h in the presence or absence of antioxidants (K1, K2, or Q10 dissolved in dimethyl sulfoxide; neat solvent without antioxidants used as control) at 37 °C in minimum Eagle's medium, and subsequently exposed to 0 or 25 μm hydrogen peroxide for 18 h. Viability determination began with the addition of 15 μl of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxy-methoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt (MTS) reagent and incubation at 37 °C for 2 h followed by microplate reader absorbance measurement (490 nm) (MRX, Dynatech Laboratories, Denkendorf, Germany) to detect the colored formazan product produced by endogenous dehydrogenases in metabolically active cells.
Reactive Oxygen Species Quantitation
Intracellular ROS was quantitated by fluorometric detection of intracellular H2DCF-DA dye (Invitrogen) (19, 20). Accordingly, 2 × 104 cells/well were preincubated for 3 h in minimum Eagle's medium supplemented with 1 μm K1, K2 (MK-4), or Q10 (Sigma-Aldrich) or with neat solvent ([dimethyl sulfoxide]final < 0.1% v/v). Cells were loaded for 30 min at 37 °C with H2DCF-DA, 8 μm final concentration, in Hanks' balanced salt solution and washed three times with Hanks' balanced salt solution before the addition of the aqueous-soluble free radical generator 2,3-dimethoxy-1,4-naphthoquinone (DMNQ; Merck, Nottingham, UK) in culture medium without FBS. ROS formation was detected by conversion of 2′,7′-dichlorodihydrofluorescein to fluorescent dichlorofluorescein (λex = 480 nm, λem = 510 nm) (19) at 37 °C using a microplate fluorescence reader (Fluoroskan Ascent FL, ThermoFisher Scientific) (20, 21).
Intracellular Protein Carbonylation Quantitation
Protein carbonyl moieties were measured in crude membranes of HEK 293T cells, with or without VKORC1L1 overexpression or knockdown and/or 1 μm supplemented K1, using the BioCell protein carbonyl assay kit (Cell Biolabs, Inc., San Diego) according to the manufacturer's instructions for low protein level and fluorescence plate reader (Fluoroscan Acent FL). Dinitrophenylhydrazine (DNP)-derivatized carbonyl groups were detected by a fluorescent DNP-specific antibody-based ELISA (λex 485 nm, λem 538 nm) (Gentaur, Brussels, Belgium). Forty-eight hours post-transfection with VKORC1L1, anti-VKORC1L1 siRNA, or empty plasmid, cells were washed twice with 0 °C PBS, harvested, and mechanically lysed in 25 mm imidazole (pH 7.5), 0.5% CHAPS.
Prediction of VKORC1L1 Secondary Structure and Topology
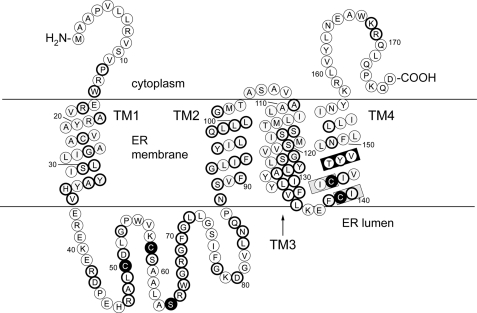
VKORC1L1 was recently reported to be exclusively localized to ER membranes in HeLa cells (22). We confirmed similar results for subcellular localization of VKORC1L1 in PtK2 cells (supplemental Fig. S1). Because of high sequence homology to VKORC1, for which we reported topology and secondary structure predictions in a recent study (23), we were able to predict secondary structural elements and a topological model for VKORC1L1 (Fig. 1) by alignment of VKORC1 and VKORC1L1 primary sequences to the proVKOR sequence (supplemental Fig. S2) for which a high-resolution structural model was published (8).
FIGURE 1.
A topological model for human VKORC1L1 based on sequence alignment (see supplemental Fig. S2) to a prokaryotic VKOR homolog protein structure determined to 3.6 Å resolution. Circles, represent amino acid residues; bold circles, indicate sequence identity shared by VKORC1L1 and paralog VKORC1; TM1–TM4, first through fourth transmembrane α-helices; black-filled circles, residues completely conserved among all VKOR family proteins; gray-boxed regions, the catalytic CIIC motif; black-boxed region, putative warfarin binding site.
Bioinformatics and Data Analysis
Human and mouse tissue- and cell-specific mRNA expression profiles for VKORC1L1 and VKORC1 were obtained from the BioGPS web server whole-genome gene expression array results (44,775 human and 36,182 mouse transcripts; 79 human and 69 mouse tissues/cell types) of the Genomics Institute of the Novartis Research Foundation (24). Multiple sequence alignment was performed using Jalview 2.6.1 (25). Statistical analyses were performed using KaleidaGraph 4.04 (Synergy Software, Reading, PA). S.E. was calculated for all replicate data and is indicated by error bars for all figures except where specifically noted in the figure legends. Unpaired Student's t test (for time course data) and analysis of variance with post hoc Fisher's least significant difference test (for matched data groups) were used to assess statistical significance between data mean values as indicated by calculated p values.
RESULTS
VKORC1L1 Catalyzes Both De-epoxidation of Vitamin K-Epoxide and Reduction of Vitamin K Quinone
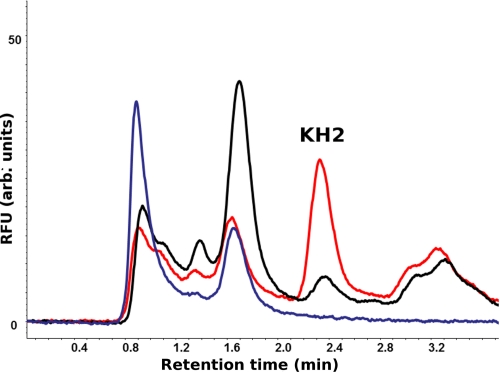
To directly assess VKORC1L1 function, we began by exploring whether VKORC1L1 could support VKOR and vitamin K quinone reductase (VKR) enzymatic activities similar to those catalyzed by VKORC1 and known to be critical for hemostasis (11, 26). Table 1 summarizes our experimentally determined effective Michaelis-Menten enzyme kinetics for DTT-driven VKOR activity under pseudo first-order conditions and maximal initial velocity for VKORC1L1 overexpressed in HEK 293T cells but where concentration of the K quinone product does not appreciably compete for enzyme as a result of the alternative VKR reaction catalyzed by the enzyme. Under these conditions we found VKORC1L1 to have 2.2- and 7.3-fold lower affinity for the K1>O and K2>O substrates, respectively, than VKORC1, suggesting that VKORC1 has evolved greater efficiency for VKOR enzymatic activity than VKORC1L1. We also measured warfarin inhibition (5 μm inhibitor in culture medium) for VKORC1L1, which was 1.8-fold lower than that for the equivalent amount of VKORC1 overexpressed in the same HEK 293T cell line (Table 1). Additionally, we semiquantitatively confirmed VKR activity for VKORC1L1 (Fig. 2).
TABLE 1.
VKOR in vitro reaction enzymatic parameters for human enzymes overexpressed in HEK 293T cells
| Enzyme | Substratea | Inhibitor | Kmb | Vmaxb | % Inhibitionc |
|---|---|---|---|---|---|
| μm | nmol·mg−1·h−1 | ||||
| VKORC1L1 | K1>O | 4.15 ± 0.10 | 2.57 ± 0.05 | 0 | |
| VKORC1L1 | K1>O, 50 μm | Warfarin, 5 μm | 29.2 ± 7.9 | ||
| VKORC1L1 | K2>O | 11.24 ± 0.23 | 13.46 ± 0.22 | 0 | |
| VKORC1 | K1>O | 1.88 ± 0.13 | 1.13 ± 0.03 | 0 | |
| VKORC1 | K1>O, 50 μm | Warfarin, 5 μm | 52.9 ± 6.6 | ||
| VKORC1 | K2>O | 1.55 ± 0.55 | 1.72 ± 0.10 | 0 |
a Range of K > O substrate concentrations: 1–16 μm; DTT-driven (5 μm) VKOR enzymatic reaction run under pseudo first-order conditions at maximum velocity for each substrate concentration.
b Apparent Km and Vmax mean values ± graphically determined error range of data scatter from Eisenthal & Cornish-Bowden direct linear plots.
c Values for % Inhibition are means ± S.E.
FIGURE 2.
VKORC1L1-mediated reduction of K1 quinone to K1 hydroquinone in HEK 293T whole cell membranes. C18 RP-HPLC chromatograms (overlaid) show the fluorescence peak of K1H2 at ∼2.4 min for wild-type HEK 293T cell membranes incubated with DTT in the absence of K1 substrate (blue trace) and with DTT and K1 (black trace) and for HEK 293T cells overexpressing VKORC1L1 incubated with DTT and K1 (red trace).
However, we had recently reported that VKORC1L1 expression in vivo cannot support sufficient physiological VKOR activity in VKORC1−/− knock-out mice to rescue a dyscoagulation phenotype that results in early post-partum death (27). Thus, VKOR enzymatic activity does not appear to be the primary physiological function of VKORC1L1.
Oxidative Stress Induces VKORC1L1 Expression
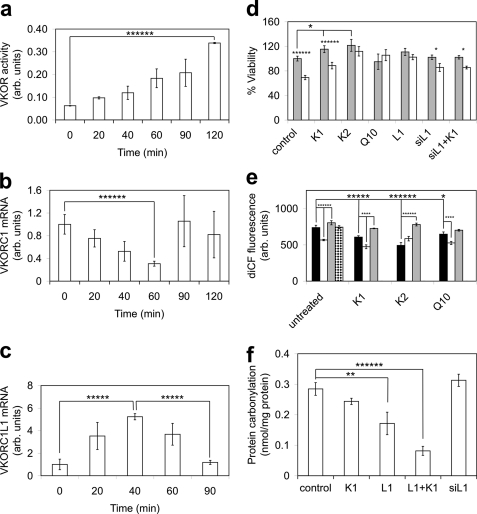
We measured in vitro VKOR activity to confirm functional expression of VKORC1L1 in HEK 293T cells treated with 75 μm H2O2, which induces intracellular oxidative stress, and found VKOR activity increased nearly 5.5-fold (p < 0.0001) by 120 min (Fig. 3a). To determine whether VKORC1L1 or VKORC1 contributed to the elevated VKOR activity, we measured VKORC1 transcript levels upon H2O2 treatment by quantitative PCR of split samples used for the VKOR activity measurements and found that VKORC1 expression was actually down-regulated by 70%4 (p < 0.0001) by 60 min before returning to basal levels from 90 to 120 min (Fig. 3b), suggesting that increased VKOR activity was due to increased VKORC1L1 expression. Direct measurement of VKORC1L1 transcript levels confirmed a 5.2-fold (p < 0.0005) increase by 40 min after inducing oxidative stress with H2O2 (Fig. 3c). Taken together, these data confirmed that up-regulated VKORC1L1 expression was responsible for the rise in intracellular VKOR activity induced by H2O2 treatment.
FIGURE 3.
Experimentally measured time-dependent VKOR activity, VKORC1L1 and VKORC1 mRNA levels, cell viability, intracellular ROS levels, and membrane protein carbonylation from lysates of HEK 293T cells overexpressing VKORC1L1 with or without antioxidant supplementation. a, DTT-driven VKOR activity (n, number of replicate measurements; n = 2 for each time point) for crude membranes of VKORC1L1-expressing cells prepared at 0–120 min post-treatment with 75 μm H2O2. b, quantitative PCR data (n = 3 for each time point) for VKORC1 mRNA expression from aliquots of same cells assayed for VKOR activity in panel a at 0–120 min post-treatment with 75 μm H2O2. c, quantitative PCR data (n = 3 for each time point) for VKORC1L1 mRNA expression from aliquots of same cells assayed for VKOR activity in panel a at 0–90 min post-treatment with 75 μm H2O2. d, cell viability assay data (n = 21 for control, K1, L1, siL1, siL1+K1; n = 5 for K2, Q10) for HEK 293T cells exposed to biological quinones or with exogenously overexpressed or silenced VKORC1L1 in the presence or absence of 25 μm H2O2. Control, untreated cells; K1, cells with 1 μm phylloquinone in the medium; K2, cells with 1 μm menaquinone-4 in the medium; Q10, cells with 1 μm ubiquinone-10 (coenzyme Q10) in the medium; L1, cells overexpressing VKORC1L1; siRNA L1, cells expressing silencing RNA for VKORC1L1; siRNA L1+K1, cells expressing silencing RNA for VKORC1L1 with 1 μm phylloquinone in the medium; gray-filled bars, culture medium without H2O2; open bars, culture medium with 25 μm H2O2. e, influence of VKORC1L1 expression and applied antioxidants on ROS generation induced by 100 μm DMNQ in HEK 293T cells measured as dichlorohydrofluorescein fluorescence (λex 488, λem 525). Bars indicate mean values (n = 8 for all samples except siL1+Q10 for which n = 7) of intracellular ROS measured by H2DCF-DA fluorescence for cells with the following attributes: black bars, wild-type cells; open bars, cells overexpressing VKORC1L1; gray bars, cells expressing anti- VKORC1L1 siRNA; bar with horizontal hatching, cells overexpressing GGCX. Groups of bars represent the following antioxidants added to culture media: control, without added antioxidant; K1, with added 1 μm phylloquinone; K2, with added 1 μm menaquinone-4; Q10, with added 1 μm ubiquinone-10 (coenzyme Q10). f, influence of VKORC1L1 overexpression and antioxidants on protein carbonylation. Bar heights represent mean DNP-derivatized carbonyl groups measured by anti-DNP ELISA assay (n = 4) for HEK 293T cells with the following culture conditions: control, wild-type cells with no added antioxidants; control+K1, wild-type cells with added 1 μm phylloquinone; L1, cells overexpressing VKORC1L1; L1+K1, cells overexpressing VKORC1L1 with added 1 μm phylloquinone; siL1, cells expressing siRNA against VKORC1L1; siL1+K1, cells expressing siRNA against VKORC1L1 with added 1 μm phylloquinone. All measurements reported as arbitrary units were normalized to the initial mean control values. Error bars, ±S.E. Statistical significance: *, p < 0.05; **, p < 0.01; ****, p < 0.001; *****, p < 0.0005; ******, p < 0.0001.
VKORC1L1-mediated Intracellular Antioxidative Effects Are Vitamin K-dependent
We next established the contributions of lipidic quinone antioxidants and VKORC1L1 expression level, alone and in concert, to cell viability in the absence or presence of H2O2-induced ROS stress. In the absence of applied H2O2, HEK 293T cell viability was affected by the supplemented antioxidants according to the increasing rank order: control ≈ Q10 < K1 (115% of control, p < 0.05) < K2 (122% of control, p < 0.05) (cf. Fig. 3, four leftmost gray bars), suggesting that K1 and K2, but not Q10, could alleviate experimentally elevated levels of intracellular oxidative stress, leading to increased viability. Upon H2O2 treatment, cell viability was affected in increasing rank order: control (69% of the untreated value, p < 0.0005) < K1 (77% of the untreated K1-supplemented value, p < 0.0001) < K2 (∼100%, i.e. not significantly different from the untreated K2-supplemented value) ≈ Q10 (∼100%, i.e. not significantly different from the untreated Q10-supplemented value) (cf. Fig. 3, four leftmost gray/white pairs of bars). Taken together, the results show that lipidic quinones supplemented at a supraphysiological concentration of 1 μm in cell culture medium affect cell viability in the presence of physiological or H2O2-induced, elevated intracellular ROS levels in identical rank order: K2 > K1 > Q10. Thus, the positive effect of quinone antioxidants on cell viability is, at least in part, VKORC1L1-dependent.
Although the viability of unstressed cells that overexpress VKORC1L1 was not statistically different than that for untreated control cells (Fig. 3d, cf. control and L1 gray bars), H2O2 treatment did not reduce viability for cells that overexpress VKORC1L1 (Fig. 3d, cf. gray and open L1 bars), suggesting that VKORC1L1 mediates vitamin K-dependent cellular homeostasis mechanisms during oxidative stress and that trace amounts of bioavailable K1 in FBS-supplemented culture medium (24 pm measured; see “Experimental Procedures”) are not limiting to the VKORC1L1-mediated antioxidant effect. In contrast, although the viability of VKORC1L1 knockdown cells in both the presence and absence of 1 μm supplemented K1 was not significantly different from the value for untreated control cells (Fig. 3d, cf. control, siL1, and siL1+K1 gray bars), there was a small but significant measurable difference in the viability for H2O2-stressed VKORC1L1 knockdown cells (Fig. 3d, 84% of untreated siL1 viability, p < 0.05; cf. gray/white pair of bars for siL1) and for H2O2-stressed VKORC1L1 knockdown cells supplemented with K1 (Fig. 3d, 84% of untreated siL1+K1 viability, p < 0.05; cf. gray/white pair of bars for siL1+K1) compared with the respective unstressed controls. Taken together, these results suggest that VKORC1L1 knockdown does not have a negative impact on cell viability during unstressed growth, although VKORC1L1 is apparently required for the vitamin K-mediated effect on cell viability because 1 μm supplemented K1 had no effect on the siL1+K1 cells.
Also, upon H2O2 treatment, 1 μm supplemented K1 did not significantly increase viability relative to VKORC1L1 knockdown cells without supplemented K1 (Fig. 3d, cf. two rightmost white bars). This suggests that VKORC1L1 is required for vitamin K-dependent increased cell proliferation.
Increased VKORC1L1 Expression Level Correlates with Reduced Intracellular ROS Level
We further investigated the effects of the various lipidic small-molecule antioxidants and VKORC1L1 expression level on increased intracellular ROS level induced by cell uptake and redox cycling of DMNQ. The intracellular ROS level was assessed by fluorescence report of oxidation of intracellular 2′,7′-dichlorodihydrofluorescein to 2′,7′-dichlorofluorescein by free radicals. In the presence of nominal antioxidant concentrations in the culture medium, and relative to wild-type control cells, ROS levels were reduced by 23% (p < 0.0001) or elevated by 9% (p < 0.0001) in cells overexpressing VKORC1L1 or transfected with anti-VKORC1L1 siRNA, respectively (Fig. 3e, untreated (leftmost group), white and gray bars). We found that protein overexpression, as such, was not responsible for elevated ROS levels when we overexpressed GGCX, a vitamin K cycle enzyme that oxidizes vitamin K hydroquinone, as a control (Fig. 3e, untreated, stippled bar). Taken together, these results indicate that the ROS-scavenging antioxidative effect of endogenous K1 and/or Q10 (both nominally supplied from culture medium, with Q10 also endogenously synthesized in cells) is potentiated in a dose-dependent manner by VKORC1L1. Supplementation of 1 μm K1, K2, and Q10 to DMNQ-treated cells resulted in ROS levels lowered by 18% (p < 0.0005), 33% (p < 0.0001), and 12% (p < 0.05), respectively, compared with the untreated wild-type control (Fig. 3e, black bars). Thus, for HEK 293T cells expressing constitutive levels of protein, the rank order for effectiveness of lipidic antioxidants in lowering DMNQ-reported free radicals is K2 > K1 > Q10.
Additionally, the combined effects of VKORC1L1 protein overexpression or knockdown together with supplementation of individual lipidic antioxidants (Fig. 3e, compare triplets of black, open, and gray bars for each antioxidant) revealed altered ROS levels of −16% (L1, p < 0.001) and +20% (siL1, p < 0.001) for K1-treated cells, +58% (siL1, p < 0.0001) for K2-treated cells (L1 data did not reach statistical significance), and −19% (L1, p < 0.001) for Q10-treated cells (siL1 data did not reach statistical significance). These results strongly suggest that the antioxidant effects are dependent on VKORC1L1 expression level. Thus, where differences in the measured levels of DMNQ-reported ROS were found to be statistically significant between constitutively expressing cells and VKORC1L1 overexpressing or knockdown cells, the same trends in VKORC1L1 overexpression resulting in lowered ROS levels, as well as in VKORC1L1 knockdown resulting in increased ROS levels, were observed for each of the three supplemented lipidic antioxidants. These results, taken together with the co-localization of VKORC1L1 and lipidic quinones in ER membranes, strongly suggest that VKORC1L1 plays a fundamental role in mediating antioxidative effects in cellular membranes.
Interestingly, although Q10 supplementation alone had no significant effect on cell viability relative to untreated controls (see Fig. 3d), VKORC1L1 overexpression together with applied 1 μm Q10 resulted in a considerably lower ROS level (−19%, p < 0.001) relative to the Q10-treated wild-type control (Fig. 3e, cf. rightmost black and open bars). However, Q10 has been reported to be a substrate for a homologous prokaryotic VKOR enzyme (8). Thus, the antioxidant effect of Q10 revealed in this study might be synergistic with VKORC1L1-mediated reduction of vitamin K, or Q10 might be a physiological substrate for VKORC1L1 in addition to K1 and K2. Our study does not discriminate between these two possibilities.
Taken together, the results presented in Fig. 3e indicate that the VKORC1L1-dependent antioxidative effect, reflected in ROS level as a biomarker, scales with VKORC1L1 expression level and is mediated by K vitamins, and possibly also by Q10, over the experimentally tested ranges for both factors.
VKORC1L1 Expression Level Inversely Correlates with Oxidative Protein Damage
To directly assess the role of VKORC1L1 in alleviating intracellular ROS-mediated damage, we directly measured membrane protein carbonylation (Fig. 3f), the result of free radical attack on unsaturated bonds in proteins. Overexpression of VKORC1L1 alone resulted in a 40% (p < 0.01) reduction in carbonylation level, whereas overexpression of VKORC1L1 together with 1 μm K1 resulted in a 71% (p < 0.0001) reduced carbonylation, compared with untreated control cells in the presence of endogenous K1 in normal culture medium. Knockdown of VKORC1L1 resulted in a marginal (10%) increase of mean measured protein carbonylation relative to untreated controls, but the level of statistical significance for the data does not provide confidence on this point.
DISCUSSION
Previous work by Li et al. (4) demonstrated that cell death caused by oxidative stress can be prevented by low nanomolar concentrations of vitamins K1 and K2 in cultured neurons and oligodendrocytes. Furthermore, seminal studies by Mukai et al. (28, 29) reported that KH2 has a greater capacity than ubihydroquinone-10 (Q10H2) to regenerate α-tocopherol (αT) from the α-tocopheryl radical (αT•) resulting from the major physiological free radical scavenging pathway. Therefore, we hypothesized that the specific function of VKORC1L1 is to keep the intracellular intramembranous pool of K vitamins in the reduced, antioxidant-active hydroquinone form.
Here we studied cells exposed to sustained, supraphysiological ROS (H2O2 and DMNQ) levels to reveal a direct role for VKORC1L1 in mediating intracellular antioxidative effects. Contrastingly, there have been numerous reports confirming the roles of transient, low ROS levels in cell signaling pathways involving complex regulation of transcription factor networks (30, 31). Most recently, studies have shown ROS signaling to be important to cell survival and proliferation (32, 33). We recognize that, in addition to the VKORC1L1-mediated antioxidative function we revealed, various signaling pathways sensitive to the lipidic quinone antioxidants used in our study may also play a role in the effects on cell viability. For example, the steroid and xenobiotic receptor (SXR), upon binding vitamin K2, was found to up-regulate CYP3A4 expression and to regulate transcription factor expression involved in bone homeostasis (34–36). However, the data we have presented here on the actual intracellular ROS level and oxidative protein damage, complemented by cell viability data, are all consistent with a direct role for VKORC1L1 in antioxidation.
Evidence That VKORC1L1 Functions to Maintain Vitamin K in the Reduced Active Antioxidant State
For wild-type cells we found that oxidative stress induced by H2O2 dramatically up-regulated VKORC1L1 expression (Fig. 3c) accompanied by increased VKOR enzymatic activity (Fig. 3a). We also found that VKORC1L1 supports VKR activity (Fig. 2), implying that VKORC1L1-dependent VKR activity also increases upon H2O2 exposure. Taken together, these results suggest that oxidative stress increases VKORC1L1 expression, which can increase intracellular levels of reduced vitamin K cofactors that, in turn, directly or indirectly lower intracellular ROS levels (Fig. 3e) and ROS-induced protein damage (Fig. 3f). Additionally, we determined that increased VKORC1L1 expression mediates increased cell viability, which is vitamin K-dependent under conditions of experimentally induced oxidative stress (Fig. 3d); this strongly suggests a regulatory role for VKORC1L1 in intracellular redox homeostasis.
In support of a fundamental homeostatic role for VKORC1L1, the results from a previous high-throughput human genome-wide expression study indicate that VKORC1L1 is expressed at uniform levels in nearly all tissues and cell types, whereas VKORC1 exhibits greater tissue-specific variation in expression level (supplemental Fig. S3 and Ref. 24). Ubiquitous, constitutive expression of VKORC1L1 in nearly all investigated human tissues likely points to an important metabolic housekeeping function (supplemental Fig. S3a). In contrast, there were significantly high VKORC1L1 expression levels (supplemental Fig. S3a, light green bar >3-fold above the mean) in highly redox-active brown fat adipocytes and significantly elevated levels in monocytic CD34+ cell lines and B lymphoblasts (supplemental Fig. S3a, blue bars). Surprisingly, these results correlate well with recent reports of intense and protracted ROS bursts, which are characteristic of these cell types during normal physiological function (37–40). Thus, independent results from these studies further point to specific roles for VKORC1L1 in intracellular redox homeostasis as well as in response to acute ROS-induced oxidative stress.
We found further in vivo evidence for VKORC1L1 playing a central role in intracellular antioxidation in previous study data of transcription levels in mouse mast cells. Mast cells have recently been shown to generate prolonged, high concentrations of ROS upon activation by binding IgE on the cell surface (39, 40). Several years earlier, the results of a genome-wide high density oligonucleotide array study of mouse mRNA expression included data for both VKORC1L1 and VKORC1 in mast cells at rest or activated by IgE (supplemental Fig. S4 and Ref. 24). These data showing up-regulation of VKORC1L1, but not VKORC1, expression during IgE binding-induced ROS stress in mouse mast cells are in agreement with our finding that VKORC1L1 expression is up-regulated during oxidative stress, further supporting our conclusion that VKORC1L1 plays a fundamental role in intracellular antioxidation. Therefore, VKORC1L1 up-regulation during oxidative stress could effectively increase the fraction of KH2 in the intracellular vitamin K pool, which would serve to protect membrane lipids and proteins from oxidative damage either directly or indirectly through regenerative cycling of lipid membrane-resident ubihydroquinone-10 (Q10H2), E vitamins, and ascorbate antioxidant species (supplemental Fig. S5). In fact, K vitamins recently have been shown to directly regenerate reduced ubihydroquinone (Q10H2) and αT in both polar and nonpolar environments, the latter at a significantly faster rate (8 × 106 m−1s−1) than the well characterized αT/ascorbate regenerative system (3 × 10−6 m−1s−1; see supplemental Fig. S5 comparing rates of free radical scavengers) (28, 29). This suggests that overexpressed VKORC1L1 might indirectly mediate reduction of Q10 by directly reducing endogenous K1, which in turn could keep the intracellular Q10 pool reduced.
Various Physiological Partner Oxidoreductases Provide Reducing Equivalents to VKOR Enzymes
All prokaryotic and eukaryotic VKOR primary protein sequences share highly similar predicted structural elements including their VKOR core domains, which comprise a four-transmembrane α-helical bundle, two conserved cysteines and a conserved serine/threonine in the large 40–50-residue loop connecting the first two transmembrane helices, and two additional conserved cysteines in a CXXC reaction center located in the fourth transmembrane helix (cf. Fig. 1 and supplemental Fig. S2). However, distinct functional differences have already been reported between proVKOR and VKORC1.
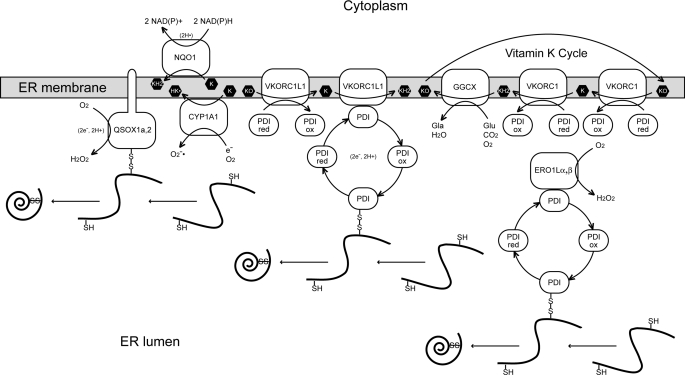
For example, VKORC1 has been shown to obtain reducing equivalents from the ER lumen from protein disulfide isomerase (PDI), the major ER-resident oxidoreductase responsible for oxidative protein folding in rat hepatocytes (41), and to transfer these to K > O or to K in the ER membrane (42) (Fig. 4). Very recently, membrane-intrinsic thioredoxin-like TMX, TMX4, and ERp18 oxidoreductases were shown to be the preferred physiological redox partners for human VKORC1 heterologously expressed in monkey-derived COS-7 cells (43). In fact, VKORC1 and VKORC1L1 might functionally interact with as many as 19 PDI family proteins in human cells (44). In contrast, in vivo proVKOR obtains reducing equivalents from its C-terminal thioredoxin-like domain or from separate, soluble, periplasmic oxidoreductases such as DsbA to reduce ubiquinone to ubihydroquinone in the bacterial periplasmic membrane (45). However, as K>O is not found in any known prokaryotic lipidome (46) and functional enzymes responsible for γ-glutamyl carboxylation of proteins are absent from prokaryotic genomes (47), proVKOR function must be distinctly different from that of VKORC1.
FIGURE 4.
Enzymatic pathways involving oxidative protein folding and K vitamins in the ER. The ER membrane bilayer is represented as a gray bar with embedded proteins (open white shapes) and K vitamin species (black hexagons). Soluble ROS molecules (O2˙̄ and H2O2) and proteins are indicated in the ER lumen below the ER membrane. PDI is protein disulfide isomerase; proteins that can be oxidatively folded incorporating disulfide bonds are indicated as black curves (reduced forms) and spirals (folded, oxidized forms). (Conflicting reports suggest that PDIs in microsomes from rat liver cells (41) or human TMX, TMX4, and ERp18 heterologously expressed in monkey kidney-derived COS-7 cells (43) are the physiological redox partner proteins for rat VKORC1 and human VKORC1, respectively.) Arrows indicate the directional flow of reducing equivalents between enzymes and K vitamin species and transitions between reduced and oxidized states for PDI (ox, oxidized; red, reduced). The exchanged reducing equivalent species are indicated in parentheses. Enzymes (left to right): QSOX1a,2, quinone:sulfide oxidoreductase paralogs 1a and 2 (both bitopic membrane proteins anchored in the membrane by a single transmembrane α-helix; NQO1, NAD(P)H:quinone oxidoreductase 1 (formerly DT-diaphorase); CYP1A1, cytochrome P450 oxidase isoform 1A1; PDI, protein disulfide isomerase 1A (equivalent to proline 4-hydroxylase β subunit); ERO1Lα,β, endoplasmic oxidoreductin-1 paralogs α and β. Gla, γ-carboxyglutamyl residues of vitamin K-dependent post-translationally modified proteins; Glu, unmodified target glutamic acid residues of vitamin K-dependent post-translationally modified proteins. The PDI redox cycle is explicitly depicted only for EroL1α,β and VKORC1L1; VKORC1 interacts similarly with PDI where only the oxidized and reduced forms are shown for economy of space. The classical vitamin K cycle heretofore described in the literature involving diffusion of K vitamins between VKORC1 and GGCX is shown at the right.
Substrate Specificity Varies among VKOR Enzymes
Heterologously expressed proVKOR from Escherichia coli is reported to pass reducing equivalents in vitro to vitamins K1 and K2 in addition to Q10 but not to K > O (8). Therefore, proVKOR apparently does not possess VKOR (de-epoxidase) activity and should technically not be called proVKOR, but rather proVKR, as it catalyzes only quinone to hydroquinone reduction. Ultimately, both VKORC1 and proVKOR enzymatic activities result in the transfer of reducing equivalents from cysteines incorporated into protein disulfides to quinone cofactors in the ER membrane. Thus, quinone cofactor usage in VKORC1 appears to have been neofunctionalized or subfunctionalized after its divergence from the last universal common ancestor (LUCA) common to itself and proVKOR.
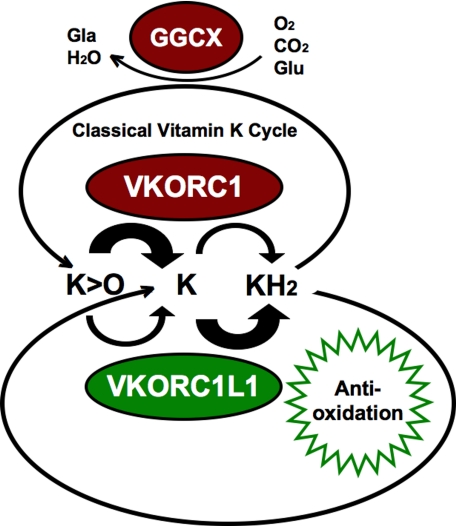
A Proposed Grand Vitamin K Cycle in Vertebrates
In the presence of intracellular dissolved O2, GGCX incorporates dissolved CO2 into γ-glutamyl carboxylate groups of post-translationally modified vitamin K-dependent proteins, consuming KH2 and O2 to produce K > O and water as byproducts. KH2 is also likely consumed in the ER membrane through its antioxidative action, ultimately resulting in conversion to oxidized K, likely via collisional dismutation or possibly nonenzymatic reduction of intermediate K• radicals. VKOR paralog enzymes are coexpressed in all tissues but at relatively uniform levels for VKORC1L1, whereas at widely varied tissue-specific levels for VKORC1 (cf. supplemental Fig. S3, a and b). Both enzymes possess both VKOR and VKR activities, although VKORC1 appears to be specialized to perform the VKOR reaction at high turnover rates relative to VKR activity (26) (Fig. 5). We propose that VKORC1L1 is specialized to carry out the VKR reaction at high turnover rates relative to VKORC1L1-specific VKOR activity measured in the present study. Consistent with this view, we measured higher apparent Km and Vmax values for the VKOR activity of VKORC1L1 compared with values measured for VKORC1 (Table 1). Additionally supporting this scheme are the turnover rates we calculated from the previous results of Chu et al. (26) for measured VKOR (46.4·10−3 molK1>O·gVKORC1−1·h−1) and VKR (85.7·10−6 molK1·gVKORC1−1·h−1) enzymatic activities of purified, reconstituted VKORC1, indicating that VKR activity represents a turnover rate that is ∼2.5 orders of magnitude (541-fold) slower than that for VKOR activity. Surprisingly, we measured a relatively low VKOR activity for VKORC1L1 with a turnover of 2.57·10−6 molK1>O·gVKORC1−1·h−1, equivalent to >4 orders of magnitude (18,054-fold) slower turnover than for VKORC1. To explain this low VKOR activity, and as VKORC1L1 was reported unable to support adequate VKOR activity to substitute for the function of VKORC1 in VKORC1−/− mice (27), we hypothesize that VKR enzymatic activity is the primary physiological function of VKORC1L1. This hypothesis is consistent with our findings that VKORC1L1 supports vitamin K-mediated intracellular antioxidative mechanisms. In summary, Fig. 5 illustrates our current working hypothesis of how both VKOR enzyme paralogs catalyze the same pair of reactions, yet possess complementary functions to maintain the intracellular vitamin K pool in the fully reduced hydroquinone form. Accordingly, VKORC1 is more efficient at de-epoxidation (i.e. VKOR activity), whereas VKORC1L1 is more efficient at quinone to hydroquinone reduction (i.e. VKR activity).
FIGURE 5.
A proposed grand vitamin K cycle. Shown are proposed functional relationships among vertebrate VKORC1L1 and VKORC1 paralog enzymes, GGCX responsible for post-translational protein γ-glutamyl carboxylation, and reducing equivalent exchange functions of K vitamins in the ER membrane. Glu, glutamyl protein residue; Gla, γ-carboxylated glutamyl protein residue; K, vitamin K quinone. Enzymes depicted as ovals: GGCX, γ-glutamyl carboxylase; VKORC1, red ovals represent enzymes of the “classical” vitamin K cycle; VKORC1L1, green oval. Thick arrows indicate relative turnover rates of naphthoquinone substrates K > O and vitamin K quinone by VKORC1L1 (lower pair of arched arrows) and by VKORC1 (upper pair of arched arrows). Antioxidative function of KH2 is depicted as a green sunburst.
In conclusion, VKORC1L1 appears to be responsible for the transfer of reducing equivalents generated as byproducts of oxidative protein folding to vitamin K-mediated intracellular antioxidation pathways (Fig. 4). The involvement of VKORC1L1 and K vitamins in antioxidation pathways warrants closer investigations to answer open questions about relative amounts and dynamics of small-molecule antioxidants in various subcellular compartments. Similarly, the localization, expression levels, and regulatory control of other enzymes involved in maintaining lipidic quinones in the respective reduced, active, antioxidant forms deserve closer scrutiny. Moreover, the specific impact of VKORC1L1 and the physiological levels of K vitamins on cellular damage inflicted by reactive oxygen species, as well as their general impact on aging and age-related chronic diseases including cancer, cardiovascular disease, and neurodegeneration, require future detailed assessment. Finally, the discovery of the antioxidant-regenerative role of VKORC1L1 will require a major reappraisal of the fundamental concepts of intracellular redox homeostasis.
Supplementary Material
This work was supported by Grant BMBF/DLR-01GS0424/NHK-S12T21 from the National Genome Research Net Cardiovascular Diseases, Germany.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5, “Methods,” “Discussion” relating to Fig. 4, and references.
All numerical percentage (%) values appearing under “Results” refer to comparisons between mean data values for the respective groups where statistical significance is reported as a calculated p value in each case.
- ROS
- reactive oxygen species
- αT
- α-tocopherol
- DMNQ
- 2,3-dimethoxy-1,4-naphthoquinone
- DNP
- dinitrophenylhydrazine
- ER
- endoplasmic reticulum
- FBS
- fetal bovine serum
- GGCX
- γ-glutamyl carboxylase
- H2DCF-DA
- dichlorodihydrofluorescein diacetate
- K1
- vitamin K1 (phylloquinone)
- K2
- vitamin K2 (menaquinone)
- KH2
- vitamin K hydroquinone
- K > O
- vitamin K 2,3-epoxide
- PDI
- protein disulfide isomerase
- proVKOR
- prokaryotic VKOR enzyme
- Q10
- ubiquinone-10
- Q10H2
- ubihydroquinone-10
- VKOR
- vitamin K 2,3-epoxide reductase
- VKORC1
- vitamin K 2,3-epoxide reductase complex subunit 1
- VKORC1L1
- VKORC1-like1
- VKR
- vitamin K quinone reductase.
REFERENCES
- 1. Balaban R. S., Nemoto S., Finkel T. (2005) Cell 120, 483–495 [DOI] [PubMed] [Google Scholar]
- 2. Gutteridge J. M., Halliwell B. (2010) Biochem. Biophys. Res. Commun. 393, 561–564 [DOI] [PubMed] [Google Scholar]
- 3. Vervoort L. M., Ronden J. E., Thijssen H. H. (1997) Biochem. Pharmacol. 54, 871–876 [DOI] [PubMed] [Google Scholar]
- 4. Li J., Lin J. C., Wang H., Peterson J. W., Furie B. C., Furie B., Booth S. L., Volpe J. J., Rosenberg P. A. (2003) J. Neurosci. 23, 5816–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oldenburg J., Bevans C. G., Müller C. R., Watzka M. (2006) Antioxid. Redox Signal. 8, 347–353 [DOI] [PubMed] [Google Scholar]
- 6. Allewalt J. P., Bateson M. M., Revsbech N. P., Slack K., Ward D. M. (2006) Appl. Environ. Microbiol. 72, 544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhaya D., Grossman A. R., Steunou A. S., Khuri N., Cohan F. M., Hamamura N., Melendrez M. C., Bateson M. M., Ward D. M., Heidelberg J. F. (2007) ISME J. 1, 703–713 [DOI] [PubMed] [Google Scholar]
- 8. Li W., Schulman S., Dutton R. J., Boyd D., Beckwith J., Rapoport T. A. (2010) Nature 463, 507–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oldenburg J., von Brederlow B., Fregin A., Rost S., Wolz W., Eberl W., Eber S., Lenz E., Schwaab R., Brackmann H. H., Effenberger W., Harbrecht U., Schurgers L. J., Vermeer C., Müller C. R. (2000) Thromb. Haemost. 84, 937–941 [PubMed] [Google Scholar]
- 10. Fregin A., Rost S., Wolz W., Krebsova A., Muller C. R., Oldenburg J. (2002) Blood 100, 3229–3232 [DOI] [PubMed] [Google Scholar]
- 11. Rost S., Fregin A., Ivaskevicius V., Conzelmann E., Hörtnagel K., Pelz H. J., Lappegard K., Seifried E., Scharrer I., Tuddenham E. G., Müller C. R., Strom T. M., Oldenburg J. (2004) Nature 427, 537–541 [DOI] [PubMed] [Google Scholar]
- 12. Goodstadt L., Ponting C. P. (2004) Trends Biochem. Sci. 29, 289–292 [DOI] [PubMed] [Google Scholar]
- 13. Hildebrandt E. F., Preusch P. C., Patterson J. L., Suttie J. W. (1984) Arch. Biochem. Biophys. 228, 480–492 [DOI] [PubMed] [Google Scholar]
- 14. Kishi T., Okamoto T., Takahashi T., Goshima K., Yamagami T. (1993) Clin. Investig. 71, S71–75 [DOI] [PubMed] [Google Scholar]
- 15. Schefe J. H., Lehmann K. E., Buschmann I. R., Unger T., Funke-Kaiser H. (2006) J. Mol. Med. 84, 901–910 [DOI] [PubMed] [Google Scholar]
- 16. Rost S., Fregin A., Hünerberg M., Bevans C. G., Müller C. R., Oldenburg J. (2005) Thromb. Haemost. 94, 780–786 [DOI] [PubMed] [Google Scholar]
- 17. Tishler M., Fieser L. F., Wendler N. L. (1940) J. Am. Chem. Soc. 60, 2866–2871 [Google Scholar]
- 18. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 19. Hempel S. L., Buettner G. R., O'Malley Y. Q., Wessels D. A., Flaherty D. M. (1999) Free Radic. Biol. Med. 27, 146–159 [DOI] [PubMed] [Google Scholar]
- 20. Halliwell B., Whiteman M. (2004) Br. J. Pharmacol. 142, 231–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horke S., Witte I., Wilgenbus P., Krüger M., Strand D., Förstermann U. (2007) Circulation 115, 2055–2064 [DOI] [PubMed] [Google Scholar]
- 22. Schaafhausen A., Rost S., Oldenburg J., Muller C. R. (2011) Thromb. Haemost. 105, 285–294 [DOI] [PubMed] [Google Scholar]
- 23. Watzka M., Geisen C., Bevans C. G., Sittinger K., Spohn G., Rost S., Seifried E., Müller C. R., Oldenburg J. (2011) J. Thromb. Haemost. 9, 109–118 [DOI] [PubMed] [Google Scholar]
- 24. Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M. P., Walker J. R., Hogenesch J. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chu P. H., Huang T. Y., Williams J., Stafford D. W. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19308–19313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spohn G., Kleinridders A., Wunderlich F. T., Watzka M., Zaucke F., Blumbach K., Geisen C., Seifried E., Müller C., Paulsson M., Brüning J. C., Oldenburg J. (2009) Thromb. Haemost. 101, 1044–1050 [PubMed] [Google Scholar]
- 28. Mukai K., Itoh S., Morimoto H. (1992) J. Biol. Chem. 267, 22277–22281 [PubMed] [Google Scholar]
- 29. Mukai K., Morimoto H., Kikuchi S., Nagaoka S. (1993) Biochim. Biophys. Acta 1157, 313–317 [DOI] [PubMed] [Google Scholar]
- 30. Landriscina M., Maddalena F., Laudiero G., Esposito F. (2009) Antioxid. Redox Signal. 11, 2701–2716 [DOI] [PubMed] [Google Scholar]
- 31. Ma Q. (2010) Pharmacol. Ther. 125, 376–393 [DOI] [PubMed] [Google Scholar]
- 32. Morgan M. J., Liu Z. G. (2011) Cell Res. 21, 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dickinson B. C., Peltier J., Stone D., Schaffer D. V., Chang C. J. (2011) Nat. Chem. Biol. 7, 106–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tabb M. M., Sun A., Zhou C., Grün F., Errandi J., Romero K., Pham H., Inoue S., Mallick S., Lin M., Forman B. M., Blumberg B. (2003) J. Biol. Chem. 278, 43919–43927 [DOI] [PubMed] [Google Scholar]
- 35. Ichikawa T., Horie-Inoue K., Ikeda K., Blumberg B., Inoue S. (2006) J. Biol. Chem. 281, 16927–16934 [DOI] [PubMed] [Google Scholar]
- 36. Horie-Inoue K., Inoue S. (2008) J. Bone Miner. Metab. 26, 9–12 [DOI] [PubMed] [Google Scholar]
- 37. Tanaka T., Halicka H. D., Traganos F., Darzynkiewicz Z. (2006) Cell Cycle 5, 2671–2675 [DOI] [PubMed] [Google Scholar]
- 38. Gregor M. F., Hotamisligil G. S. (2007) J. Lipid Res. 48, 1905–1914 [DOI] [PubMed] [Google Scholar]
- 39. Sly L. M., Kalesnikoff J., Lam V., Wong D., Song C., Omeis S., Chan K., Lee C. W., Siraganian R. P., Rivera J., Krystal G. (2008) J. Immunol. 181, 3850–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suzuki Y., Yoshimaru T., Inoue T., Ra C. (2009) Mol. Immunol. 46, 2200–2209 [DOI] [PubMed] [Google Scholar]
- 41. Wajih N., Hutson S. M., Wallin R. (2007) J. Biol. Chem. 282, 2626–2635 [DOI] [PubMed] [Google Scholar]
- 42. Jin D. Y., Tie J. K., Stafford D. W. (2007) Biochemistry 46, 7279–7283 [DOI] [PubMed] [Google Scholar]
- 43. Schulman S., Wang B., Li W., Rapoport T. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 15027–15032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Appenzeller-Herzog C., Ellgaard L. (2008) Biochim. Biophys. Acta 1783, 535–548 [DOI] [PubMed] [Google Scholar]
- 45. Dutton R. J., Boyd D., Berkmen M., Beckwith J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11933–11938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Collins M. D., Jones D. (1981) Microbiol. Rev. 45, 316–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Berkner K. L. (2008) Vitam. Horm. 78, 131–156 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.