Abstract
Trametes cervina lignin peroxidase (LiP) is a unique enzyme lacking the catalytic tryptophan strictly conserved in all other LiPs and versatile peroxidases (more than 30 sequences available). Recombinant T. cervina LiP and site-directed variants were investigated by crystallographic, kinetic, and spectroscopic techniques. The crystal structure shows three substrate oxidation site candidates involving His-170, Asp-146, and Tyr-181. Steady-state kinetics for oxidation of veratryl alcohol (the typical LiP substrate) by variants at the above three residues reveals a crucial role of Tyr-181 in LiP activity. Moreover, assays with ferrocytochrome c show that its ability to oxidize large molecules (a requisite property for oxidation of the lignin polymer) originates in Tyr-181. This residue is also involved in the oxidation of 1,4-dimethoxybenzene, a reaction initiated by the one-electron abstraction with formation of substrate cation radical, as described for the well known Phanerochaete chrysosporium LiP. Detailed spectroscopic and kinetic investigations, including low temperature EPR, show that the porphyrin radical in the two-electron activated T. cervina LiP is unstable and rapidly receives one electron from Tyr-181, forming a catalytic protein radical, which is identified as an H-bonded neutral tyrosyl radical. The crystal structure reveals a partially exposed location of Tyr-181, compatible with its catalytic role, and several neighbor residues probably contributing to catalysis: (i) by enabling substrate recognition by aromatic interactions; (ii) by acting as proton acceptor/donor from Tyr-181 or H-bonding the radical form; and (iii) by providing the acidic environment that would facilitate oxidation. This is the first structure-function study of the only ligninolytic peroxidase described to date that has a catalytic tyrosine.
Keywords: Crystal Structure, Electron Paramagnetic Resonance (EPR), Electron Transfer, Enzyme Kinetics, Enzyme Mechanisms, Site-directed Mutagenesis, Trametes cervina, Compounds I and II, Lignin Peroxidase, Tyrosyl Radical
Introduction
Lignin is one of the most abundant and recalcitrant natural polymers, in which various nonphenolic phenylpropanoid units form a complex three-dimensional network (1). Therefore, lignin degradation is a key step for carbon recycling in forest ecosystems and a central issue in lignocellulose-based industries (e.g. in paper pulp manufacture and production of bioethanol and other renewable chemicals) (2). In nature, extracellular heme peroxidases from fungi of the basidiomycetes group are responsible for the one-electron oxidation of lignin units that initiates the biodegradation process, being of interest as industrial biocatalysts (3, 4).
Ligninolytic peroxidases are included into class II of the plant, fungal, and bacterial peroxidase superfamily (5). They comprise lignin peroxidases (LiP;4 EC 1.11.1.14); manganese peroxidases (MnP; EC 1.11.1.13), which were first described in Phanerochaete chrysosporium; and versatile peroxidases (VP; EC 1.11.1.16), described later in Pleurotus and Bjerkandera species (6, 7). LiP exhibits the high redox potential and ability to oxidize large molecules required for direct oxidation of the lignin polymer and other nonphenolic aromatic substrates. The latter includes veratryl (3,4-dimethoxybenzyl) alcohol (VA), the LiP physiological substrate with a mediating role in lignin biodegradation (8). MnP specifically oxidizes Mn2+ to Mn3+, which acts as a diffusible mediator for oxidation of phenolic substrates (9) and as a starter of lipid peroxidation reactions generating strong oxidizers (10). Finally, VP is a hybrid enzyme with the catalytic properties of LiP, MnP, and phenol-oxidizing fungal peroxidases, such as Coprinopsis cinerea peroxidase (11).
The overall catalytic cycle of ligninolytic peroxidases is similar to that of classical peroxidases, such as horseradish peroxidase (12). Resting enzyme (with ferric heme) is two-electron-oxidized by H2O2 and converted into Compound I (with oxo-ferryl, Fe4+=O, heme containing a porphyrin π-cation radical). The activated enzyme sequentially performs one-electron oxidations of two substrate molecules, coming back to the resting state via Compound II (the one-electron-oxidized intermediate with Fe4+=O heme).
Mn2+ oxidation by MnP and VP is produced by direct electron transfer to the peroxide-activated heme (13, 14), as also found for oxidation of phenols and dyes in classical peroxidases (15). However, oxidation of high redox potential substrates (including lignin models) by LiP and VP occurs at a catalytic tryptophan exposed on the protein surface (e.g. Trp-171 of P. chrysosporium LiP and Trp-164 of Pleurotus eryngii VP) via long-range electron transfer to heme (16–18). Spin-trapping and more recently electron paramagnetic resonance (EPR) studies have demonstrated that the peroxide-activated forms (Compounds I and II) of the above peroxidases contain a radical at the catalytic tryptophan (19–21). Moreover, different LiP and VP cDNA sequences (up to more than 30 sequences available) have shown that the catalytic tryptophan is conserved in all of them (5–7). Therefore, this residue is considered to be essential for the LiP-type activities exhibited by both LiP and VP enzymes.
A new type of LiP was recently found in the white rot basidiomycete Trametes cervina (22). This LiP exhibits oxidative activities toward high redox potential substrates such as VA and 1,4-dimethoxybenzene (1,4-DMB) (1.3–1.4 V), and large molecules such as ferrocytochrome c (Cc2+), as reported previously for P. chrysosporium LiP (23, 24). However, its first molecular characterization showed that T. cervina LiP lacks the conserved catalytic tryptophan and contains one tyrosine residue (Tyr-181) at a position where tyrosine residues were never found in other LiP and VP sequences (25). Homology molecular modeling showed that this tyrosine is located near the heme and accessible to the solvent (25), suggesting that it could represent an alternative substrate oxidation site, in agreement with previous chemical modification results (26). Mechanistic studies on T. cervina LiP catalysis, including Tyr-181 role, are of high interest, but they could not be produced so far because of the low enzyme levels obtained from T. cervina cultures.
Recently, Escherichia coli production of recombinant T. cervina LiP (LiP*), followed by in vitro activation, was optimized (27), enabling site-directed mutagenesis and other studies. In the present work, the crystal structure of native (wild-type) LiP* was determined at high resolution, and several site-directed variants at three substrate oxidation site candidates exposed to the solvent were investigated by spectroscopic and kinetic techniques. Low temperature EPR of the peroxide-activated T. cervina LiP* successfully detected a protein radical at Tyr-181. These results provide the first structure-function information on the only ligninolytic peroxidase described to date that has a catalytic tyrosine.
EXPERIMENTAL PROCEDURES
Production of LiP* and Site-directed Variants
The previously constructed pET-mlip expression vector (27), which contains the cDNA of mature T. cervina LiP (GenBankTM entry AB191466) in pET23 (Merck), was used as template for site-directed mutagenesis with the QuikChange kit (Stratagene). Mutations were introduced by polymerase chain reaction (PCR) using the following primers: 5′-CCTCGTTCCCGGCCCGACTGACT-3′ for D146G; 5′-CTCCCACTGAGGTCGTTTCGCTGCTCGCTTCCCACAG-3′ for H170S; and 5′-CACAGTATTGCTGCTCAAAACGAAGTGGACACGGACGTC-3′ for Y181N (only direct primers are shown, with the mutated codons in bold and the mutated bases underlined). The PCR reactions (20 μl) contained 10 ng of pET-mlip, 50 ng of each (forward and reverse) primer, 50 μm of each dNTP, and 1 unit of PfuTurbo DNA polymerase (Stratagene) in 1× PCR reaction buffer. The reactions were as follows: 95 °C for 1 min, 55 °C for 1 min, and 68 °C for 10 min in 16 cycles, with a final 10-min extension at 68 °C.
The expression vectors were transformed into E. coli BL21(DE3)pLysS (Merck). Recombinant proteins were produced by isopropyl-β-d-thiogalactopyranoside induction, recovered from inclusion bodies, in vitro-refolded, and purified in a single anion-exchange chromatography step, as described previously (27). The concentrations of native LiP* and its mutated variants were calculated spectrophotometrically (ϵ407 147 mm−1 · cm−1).
Steady-state Reactions
Absorbance changes during oxidation of VA and 2,2′-azino-bis(3-ethylbenzo-thiazoline-6-sulfonate) (ABTS) were recorded with a Shimadzu UV-1600 spectrophotometer. VA oxidation rates were estimated from maximal veratraldehyde (ϵ310 9.3 mm−1 · cm−1) formation during 5 s in 20 mm sodium succinate buffer (pH 3.5) containing 50 nm enzyme and 0.1 mm H2O2. ABTS oxidation rates were estimated from maximal ABTS cation radical (ϵ420 36 mm−1 · cm−1) formation during 5 s in 20 mm succinate buffer (pH 3.5) containing 5 nm enzyme and 0.1 mm H2O2.
The apparent affinity constant (Michaelis-Menten constant, Km) and maximal enzyme turnover (kcat) for oxidation of VA and ABTS were obtained by nonlinear least-squares fitting of the experimental values (reaction rates v and substrate concentrations [S]) to the Michaelis-Menten equation.
Spectral changes during oxidation of 1,4-DMB and Cc2+ were recorded with an Agilent 8453 diode array spectrophotometer, showing 1,4-benzoquinone and ferricytochrome c (Cc3+) formation, respectively (27). 1,4-Benzoquinone concentration was followed at 245 nm in 20 mm sodium succinate buffer (pH 3.5) containing 0.2 mm 1,4-DMB, 100 nm enzyme, and 0.1 mm H2O2. Cc2+ concentration was followed at 550 nm in 20 mm sodium succinate buffer (pH 4.0) containing 15 μm Cc2+ (prepared by reducing Cc3+ with sodium dithionite and excess dithionite removal with Sephadex G-25 prior to use), 0.1 μm enzyme, and 0.1 mm H2O2. All reactions were carried out at 25 °C and initiated by the addition of H2O2.
Compound I Formation and Spontaneous Decay
Spectroscopic and kinetic studies of Compound I formation and decay were performed with an SFM 300 stopped-flow device (Bio-Logic) equipped with a MOS-DA rapid-scanning diode array detector (J&M). Five equivalents of H2O2 were used to ensure pseudo first-order kinetics. Typically, one syringe contained 2 μm enzyme in 10 mm sodium succinate buffer (pH 4.5), the other contained 10 μm H2O2 in 50 mm sodium succinate buffer (pH 3.5), and equal volumes were mixed. Apparent second-order rate constants of Compound I formation (k1app) were determined from the slope of the plots of H2O2 concentration versus kobs1, calculated from traces at 399 nm (Compound I and II isosbestic point). First-order rate constants of Compound I decay (kobs-1) were calculated from the absorbance changes at 416 nm (Compound II and resting state isosbestic point).
EPR Measurements
Solutions of native T. cervina LiP* and its Y181N variant (0.2 mm) were prepared in 20 mm sodium succinate buffer (pH 4.5) containing 1 mm CaCl2 and 0.1 n NaCl. Eight or nineteen equivalents of H2O2 were added to the EPR tubes containing the native LiP* and Y181N solutions, respectively, that were rapidly frozen in liquid nitrogen. CW-X-band (9.4 GHz) EPR measurements were carried out with a Bruker E500 Elexsys series using the Bruker ER 4122 SHQE cavity and an Oxford helium continuous flow cryostat (ESR900). Spin quantitation was performed by double integration of the experimental EPR radical signal compared with the iron signal. EPR simulations were performed using the X-Sophe package (Bruker).
Crystallographic Study
Crystals of native T. cervina LiP* were obtained with the sitting-drop vapor diffusion method. The protein solution (10 mg/ml, in 20 mm sodium succinate buffer, pH 4.5, containing 1 mm CaCl2) was mixed (1:1) with a solution containing 30% 2-methyl-2,4-pentanediol (MPD) and 10% polyethylene glycol 4000 in 0.1 m imidazole-HCl buffer (pH 8.0). Crystals appeared after 7 days at 22 °C and were frozen in the crystallization medium containing 15% polyethylene glycol 400. A complete data set was collected at the beamline ID29 of the European Synchrotron Radiation Facility (ESRF, Grenoble, France). Data were processed using XDS (28) and merged and scaled with SCALA, from the CCP4 package (29). The structure was solved by molecular replacement with MOLREP (30) using the P. eryngii VP structure (Protein Data Bank (PDB) entry 3FM4) as a search probe. Consecutive cycles of refinement and manual rebuilding were done using REFMAC (31) and COOT (32). The statistics of data collection, processing, and refinement are shown in Table 1. The structure was validated using MolProbity (33).
TABLE 1.
Statistics (data collection and refinement) of native LiP* crystal structure (PDB entry 3Q3U)
| Data collection | |
| Space group | P 21 21 21 |
| Unit cell dimensions | |
| a, b, c (Å) | 43.90, 72.93, 95.23 |
| Resolution (last shell) (Å) | 43.90-1.85 (1.95-1.85) |
| Observations (unique) | 128,680 (26055) |
| Rmeasa (last shell) | 0.084 (0.256) |
| Completeness (last shell) (%) | 96.6 (77.2) |
| Multiplicity (last shell) | 4.9 (3.2) |
| 〈I/σ(I)〉b (last shell) | 7.1 (2.4) |
| Refinement | |
| Non H-atoms (solvent mol.) | 2889 (343) |
| Heteroatoms | |
| Heme | 1 |
| MPD | 1 |
| Calcium | 2 |
| Sodium | 1 |
| Chlorine | 1 |
| Rworkc (Rfreed) (%) | 0.149 (0.195) |
| r.m.s.d. bond lengthe (Å)/angles (°) | 0.025/1.839 |
| B-factor (Å) | |
| Main/side chain | 10.3/11.5 |
| Solvent | 19.5 |
| Ramachandran regions (%) | |
| Favored/outliers | 99.4/0.6 |
a Rmeas = {〈hkl 〈N/(N − 1)〉1/2 〈i |Ii(hkl) − <I(hkl)>|}/〈hkl 〈i Ii(hkl), where Ii(hkl) are the observed intensities, <I(hkl)> are the average intensities, and N is the multiplicity of reflection hkl.
b 〈I/σ(I)〉 is the relation between the intensity of the diffraction and the background.
c Rfactor = 〈hkl {〈Fobs(hkl)〉 − 〈Fcalc(hkl)〉}/〈hkl 〈Fobs(hkl)〉, where Fobs(hkl) and Fcalc(hkl) are the structure factors observed and calculated respectively.
d Rfree corresponds to Rfactor calculated using 5% of the total reflections selected randomly and excluded during refinement.
e r.m.s.d. is the root mean square deviation.
RESULTS
Crystal Structure of T. cervina LiP*
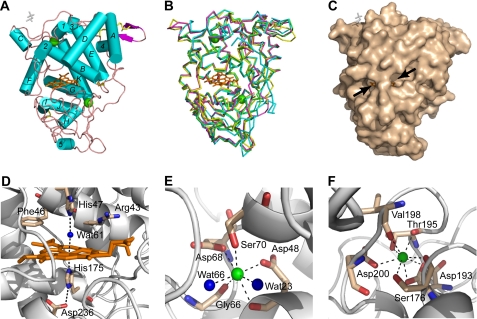
Native T. cervina LiP* was successfully crystallized, the crystals containing one molecule per asymmetric unit. The structure was determined by molecular replacement and refined against data to 1.85 Å. The LiP* refined structure consists of 337 residues (from residue 1 to 337, of a total of 338) and contains one heme molecule and two Ca2+ ions (as well as one molecule of MPD and two more ions from the crystallization medium, all of them located at the protein surface) (Fig. 1A). The C-terminal amino acid (Gly-338) and the aromatic ring of Phe-89 were not solved because they are highly disordered in the crystal. The overall structure is mostly helical (Fig. 1A), including 11 α-helices and one short antiparallel β-sheet. Four disulfide bridges (Cys-3/Cys-15, Cys-14/Cys-283, Cys-34/Cys-120, and Cys-247/Cys-314) were also identified.
FIGURE 1.
Crystal structure of T. cervina LiP*. A, scheme for the overall structure of the enzyme showing 11 (A–K) α-helices and five (1–5) short helices (as cyan cylinders), one small antiparallel β-sheet (magenta arrow), the heme cofactor (orange sticks), two structural calcium ions (green balls), four disulfide bonds (yellow sticks), and one MPD molecule (white sticks) on the molecular surface (PDB entry 3Q3U). B, schematic representation of the T. cervina LiP* backbone folding (yellow), showing also the heme (orange sticks), two calcium ions (green balls), and disulfide bonds (yellow sticks), superimposed with the backbones of P. chrysosporium LiP (magenta), P. eryngii VP (green), and P. chrysosporium MnP (cyan) (from PDB entries 1LLP, 3FKG, and 1MNP, respectively). C, protein surface with indication of two channels (arrows) enabling access to the heme cofactor (orange sticks) (an MPD molecule is also shown as white sticks). D, consensus structure of the heme cavity, including proximal histidine and conserved aspartate on the lower side and distal histidine, together with conserved arginine and phenylalanine and a water molecule (blue ball), on the upper side, with bonds as dashed lines. E and F, structure of the distal and proximal, respectively, calcium (green balls) binding sites, with bonds as dashed lines.
The overall folding of T. cervina LiP* is largely superimposable with those of other ligninolytic peroxidases such as P. chrysosporium LiP and MnP and P. eryngii VP (Fig. 1B). The heme cofactor is entirely buried in the protein but accessible to the solvent via two channels (Fig. 1C). Its iron ion is coordinated at the lower side of the heme by Nϵ2 of one of two axial histidines (proximal histidine) with a bond length of 2.11 Å (Fig. 1D). The sixth coordination site of the iron (at the upper side of the heme) is occupied by a water molecule (Wat-61) with a bond length of 2.55 Å, which is hydrogen-bonded (2.65 Å) to Nϵ2 of the second axial histidine (distal histidine). In addition to the proximal (His-175) and distal (His-47) histidines, other residues conserved in the T. cervina LiP* heme pocket are Arg-43 and Phe-46 at the upper (distal) side and Asp-236, hydrogen-bonded (2.84 Å) to Nδ1 of the proximal histidine, at the lower (proximal) side (Fig. 1D). The distal and proximal side Ca2+ ions in the T. cervina LiP* structure are, respectively, hepta-coordinated with Asp-48, Gly-66, Asp-68, Ser-70, and two water molecules (Fig. 1E) and octa-coordinated with Ser-176, Asp-193, Thr-195, Val-198, and Asp-200 (Fig. 1F).
Possible Substrate Oxidation Sites and Design of Site-directed Variants
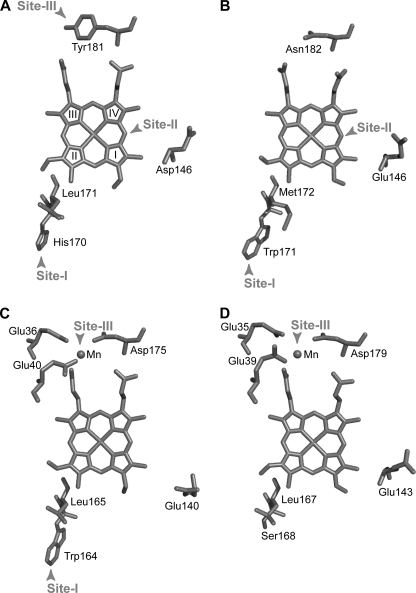
According to the substrate oxidation sites and electron transfer pathways described in other ligninolytic peroxidases, including P. chrysosporium LiP and MnP and P. eryngii VP (13–17), three putative substrate oxidation sites (all of them accessible from the solvent) were identified in the T. cervina LiP* crystal structure (Fig. 2A). Site-I would involve electron transfer from His-170, homologous to P. chrysosporium LiP Trp-171 and VP Trp-164 (Fig. 2, B and C), to methyl-3 of heme ring II via Leu-171. This corresponds to the long-range electron transfer pathway for high redox potential substrate oxidation at the conserved catalytic tryptophan of LiP and VP (16, 17). Site-II would be at the δ-carbon of the heme (i.e. that between rings I and IV) involving Asp-146, homologous to Glu-146 contributing to a second substrate oxidation site in P. chrysosporium LiP (16) (Fig. 2B), being also similar to the site for oxidation of phenols and dyes by classical peroxidases (15). Finally, site-III would be located at Tyr-181 and involves electron transfer from this exposed residue to the porphyrin macrocycle via the internal heme propionate (bond to carbon-6 of heme ring III), its final part being similar to the propionate electron transfer pathway for Mn2+ oxidation by MnP and VP (13, 14) (Fig. 2, C and D).
FIGURE 2.
Possible substrate oxidation sites in T. cervina LiP (A) as compared with those described in P. chrysosporium LiP (B), P. eryngii VP (C), and P. chrysosporium MnP (D) (from PDB entries 3Q3U, 1LLP, 3FKG, and 1MNP, respectively). Shown is an upper view of the heme cofactors with indications of the following. Site-I, long-range electron transfer pathway from the exposed catalytic tryptophan described in P. chrysosporium LiP (Trp-171) and VP (Trp-164) and corresponding residues in T. cervina LiP (His-170) and MnP (Ser-168). Site-II, oxidation site at the heme edge described in classical peroxidases and P. chrysosporium LiP involving Glu-146 and equivalent acidic residues in T. cervina LiP (Asp-146), VP (Glu-140), and MnP (Glu-143). Site-III, Mn2+ oxidation site described in VP and MnP formed by two glutamates and one aspartate, where electron transfer from the substrate (Mn2+) to the porphyrin macrocycle and iron center is produced via the internal propionate, and corresponding residues in LiP of T. cervina (Tyr-181) and P. chrysosporium (Asn-182). Heme ring numbering is shown in A.
To evaluate these possible substrate oxidation sites, site-directed mutagenesis of T. cervina LiP* was performed. Mutations were introduced to block the three sites as detailed below. His-170 was replaced by serine because MnP, lacking site-I, includes Ser-168 at this position (Fig. 2D). Asp-146 was substituted by glycine because it has been demonstrated that the E146G mutation in P. chrysosporium LiP affects VA oxidation activity at site-II (Fig. 2B) (16). Finally, Tyr-181 was replaced by asparagine because LiP without site-III has Asn-182 at this position (Fig. 2B).
The same E. coli expression and in vitro refolding conditions previously optimized for native LiP* were used for the three mutated variants (D146G, H170S, and Y181N). These were obtained as inactive proteins, which accumulated in inclusion bodies with high purities. Their in vitro refolding yields, up to 5% activation of the total recombinant protein, were similar to that obtained for the native LiP*. The purified variants appeared electrophoretically homogeneous, and their electronic absorption spectra were almost identical to the native LiP* spectrum, indicating no significant changes of the heme environment. The changes in T. cervina LiP catalytic properties caused by the three point mutations are described below.
Catalytic Properties of Native LiP* and Mutated Variants
Oxidation of VA and ABTS by native LiP* and its mutated variants was investigated. The kinetic constants obtained are shown in Table 2. The Y181N mutation completely removes VA oxidation activity and also decreases (7-fold) the catalytic efficiency for ABTS oxidation. The H170S variant shows the same kinetic constants for VA oxidation of native LiP*, although it slightly decreases (32%) the catalytic efficiency on ABTS. Finally, the D146G variant shows slightly lower VA oxidation efficiency (30% decrease) than native LiP*, whereas its kinetic parameters for ABTS oxidation are almost identical to those of native LiP*.
TABLE 2.
Steady-state kinetic parameters for oxidation of VA and ABTS by native T. cervina LiP* and its site-directed variants D146G, H170S, and Y181N
| VA oxidation |
ABTS oxidation |
|||||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| mm | s−1 | s−1 · mm−1 | μm | s−1 | s−1 · mm−1 | |
| LiP* | 6.0 ± 0.5 | 23.5 ± 0.6 | 3.92 ± 0.10 | 868 ± 790 | 210 ± 7 | 242 ± 8 |
| D146G | 5.6 ± 0.7 | 15.3 ± 0.8 | 2.73 ± 0.14 | 814 ± 170 | 231 ± 2 | 284 ± 2 |
| H170S | 6.0 ± 0.7 | 22.1 ± 1.0 | 3.68 ± 0.16 | 1125 ± 100 | 184 ± 7 | 164 ± 6 |
| Y181N | NDa | NDa | NDa | 1773 ± 112 | 60 ± 1 | 34 ± 1 |
a ND, not determined due to lack of activity. Means and 95% confidence limits.
The above results show that Tyr-181 is the site for VA oxidation by T. cervina LiP*. This residue (site-III) would also be involved in the oxidation of ABTS, although the Y181N variant retains some activity. The D146G mutation slightly affects VA oxidation, suggesting some contribution of Asp-146 to the catalytic activity. Substituting the His-170 side chain (in the H170S variant) does not modify VA oxidation. However, the H170S mutation affects ABTS oxidation, indicating that this residue (site-I) could be the second oxidation site for ABTS oxidation by T. cervina LiP* (whose existence was suggested by the residual activity of the Y181N variant).
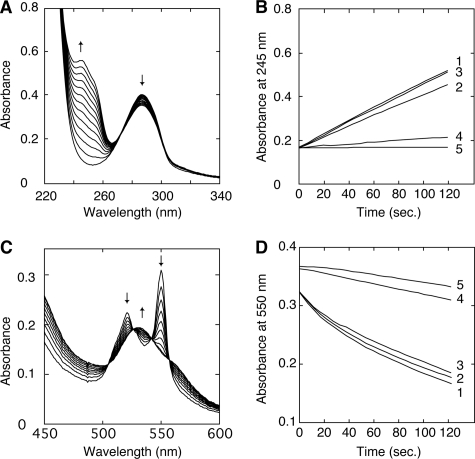
Some other LiP-type properties in T. cervina LiP* and its mutated variants were investigated using 1,4-DMB and Cc2+ as substrates. As shown in Fig. 3, A and C, formation of 1,4-benzoquinone and Cc3+, respectively, is shown by the spectral changes observed, including characteristic maxima. Fig. 3, B and D, show kinetic traces during oxidation of 1,4-DMB and Cc2+ by native LiP* and different variants, respectively. Only the Y181N variant (trace 4) drastically lost its oxidation activities for both substrates, whereas other mutations did not affect the activities significantly.
FIGURE 3.
Time course of 1,4-DMB and Cc2+ oxidation by T. cervina LiP* and mutated variants. A, spectral changes during 1,4-DMB (286 nm maximum) oxidation by LiP* forming 1,4-benzoquinone (254 nm maximum) (the spectra were acquired each 10 s). B, kinetic traces at 254 nm during 1,4-DMP oxidation by native LiP* (trace 1) and D146G (trace 2), H170S (trace 3), and Y181N (trace 4) variants, compared with a control without enzyme (trace 5). C, spectral changes during Cc2+ (520 and 550 nm maxima) oxidation by LiP* forming Cc3+ (528 nm maximum) (the spectra were acquired each 20 s). D, kinetic traces at 550 nm during Cc2+ oxidation by native LiP* (trace 1) and D146G (trace 2), H170S (trace 3), and Y181N (trace 4) variants, compared with a control without enzyme (trace 5).
Compound I Kinetics and Spectroscopy
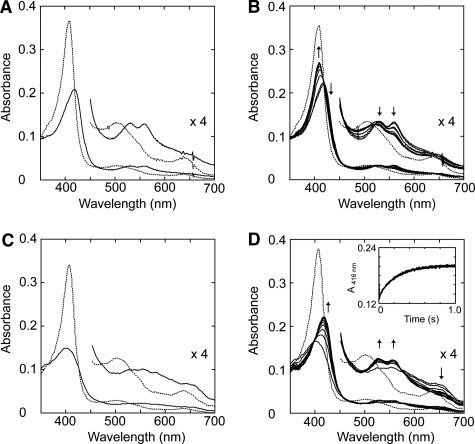
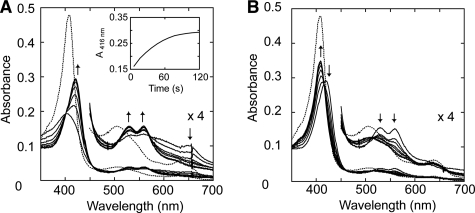
To investigate the mechanism of T. cervina LiP* catalysis, spectroscopic and kinetic studies on Compound I formation (and decay) were carried out. First, optimal conditions to form stable Compound I were studied by systematically changing the amount of H2O2 added (1–10 equivalents) and the pH (3.5–5.0). However, typical Compound I spectrum was not observed under any given condition with the diode array spectrophotometer (∼2 s for manual mixing of samples). Just after mixing, typical Compound II spectrum was obtained, with maxima at 419 (Soret band), 529 (β band), and 560 nm (α band) (Fig. 4A). In a few minutes, the spectrum was converted into resting-state spectrum (with an isosbestic point, between Compound II and resting state, at 416 nm) (Fig. 4B).
FIGURE 4.
Spectral changes of T. cervina LiP* after H2O2 addition measured with diode array spectrophotometer (A and B) and stopped-flow rapid scan equipment (C and D). A, spectra of 2.5 μm LiP* in 20 mm sodium succinate buffer (pH 4.0) containing 1 mm CaCl2 (resting state, dashed line), and 2 s after the addition of 3.75 μm H2O2 (Compound II-like spectrum; solid line). B, spectral changes observed during 2–120 s after the addition of H2O2 indicating self-reduction to the enzyme resting state (spectra acquired at 2 s, and then each 20 s). C, spectra of 19.2 μm LiP* in 20 mm sodium succinate buffer (pH 4.0) containing 1 mm CaCl2 (resting state, dashed line) and after 10 ms from the addition of 192 μm H2O2 (typical Compound I spectrum, solid line). D, spectral changes observed during 1000 ms after the addition of H2O2 indicating rapid decay of Compound I to Compound II (spectra acquired at 10 ms, and then each 100 ms) (inset, 416-nm trace showing Compound I decay). Details (×4 absorbance) of the 450–700 nm region are shown.
After peroxide oxidation of cytochrome c peroxidase, the radical is located in Trp-191 near the heme (as a protein radical), resulting in Compound II-like spectrum indicative of the oxo-ferryl heme (called compound ES) (34). To investigate the possibility of porphyrin π-cation radical migration in the peroxide-activated T. cervina LiP*, stopped-flow rapid scanning was used. As shown in Fig. 4C, after H2O2 addition (∼10 ms), LiP* is first converted into Compound I, with typical maxima at 401 nm (Soret), 527 nm (β), 554 nm (α), and 650 nm (CT1), and then rapidly (1 s) passes to Compound II spectrum (Fig. 4D), with an isosbestic point between Compounds I and II at 399 nm.
The D146G and H170S variants also exhibit the above spectral changes. However, the Y181N variant showed different behavior. Its Compound I spectrum (with maxima at 402, 534, 560, and 648 nm) was stable enough to be detected with the diode array spectrophotometer after manual mixing and was then spontaneously converted into Compound II (with maxima at 421, 529, and 560 nm) (Fig. 5A) and subsequently into the resting state (Fig. 5B). The different Compound I decay rates for LiP* and the Y181N variant are illustrated in the insets of Figs. 4D and 5A.
FIGURE 5.
Spectral changes of the Y181N variant after the addition of H2O2 measured with diode array spectrophotometer. A, spectrum of 3.3 μm Y181N in 20 mm sodium succinate buffer (pH 4.5) containing 1 mm CaCl2 (resting state, dashed line), and spectral changes of Y181N during the 2–120-s period after the addition of 4.9 μm H2O2 (solid lines), indicating Compound I self-reduction to Compound II (spectra acquired at 2 s, and then each 20 s) (inset, 416-nm trace showing Compound I decay). B, spectrum of resting Y181N (dashed line), and spectral changes of Y181N obtained during the 180–900-s period after H2O2 addition, indicating Compound II self-reduction to resting state (spectra acquired at 180 s, and then each 120 s). Details (×4 absorbance) of the 450–700 nm region are shown.
In Table 3, the kinetic constants for the formation and spontaneous decay of Compound I in native T. cervina LiP* and the three site-directed variants are shown. The second-order rate constant for Compound I formation in native LiP* is 3.6 × 106 s−1 · m−1, and those of the variants are in the same range. The first-order rate constant for Compound I decay in native LiP* is 4.35 s−1, and those of D146G and H170S variants are also similar. However, the constant for Y181N Compound I decay is 0.03 s−1, indicating that the Y181N Compound I is ∼150 times more stable than that of native LiP*, in agreement with the spectral observations.
TABLE 3.
Transient-state kinetic parameters for Compound I formation and its spontaneous decay in T. cervina LiP* and its site-directed variants D146G, H170S, and Y181N
| Compound I formation | Compound I decay | |
|---|---|---|
| × 106 s−1 · m−1 | s−1 | |
| LiP* | 3.6 ± 0.1 | 4.35 ± 0.12 |
| D146G | 2.9 ± 0.1 | 4.26 ± 0.15 |
| H170S | 3.5 ± 0.3 | 4.84 ± 0.25 |
| Y181N | 2.6 ± 0.2 | 0.03 ± 0.004 |
Means and 95% confidence limits.
EPR of Peroxide-activated LiP* and Y181N
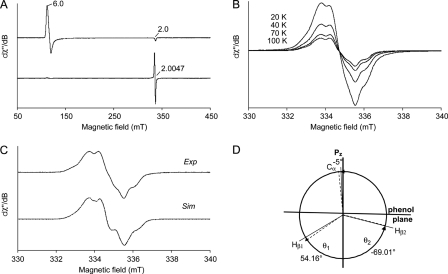
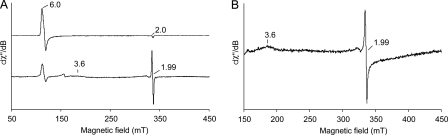
Low temperature (4 K) EPR was used to investigate further differences between the peroxide-activated LiP* and the Y181N variant, concerning heme and protein radicals. Spectra were recorded before and after the addition of H2O2 to the enzymes in the absence of reducing substrates. The spectra of native LiP* and Y181N are almost identical (Figs. 6A, top, and 7A, top, respectively) and dominated by the high-spin ferric signals (g⊥ = 6.0 and g‖ = 2.0) of resting peroxidases (35), which are consistent with the presence of CT1 bands in the electronic absorption spectra.
FIGURE 6.
Low temperature EPR spectra of T.cervina LiP*. A, spectra of LiP* resting state (top) indicating the high-spin ferric signals (g = 2.0 and 6.0), and its peroxide-activated form (Compound I) (bottom). B, effect of temperature (20, 40, 70, and 100 K) on the EPR signal in the peroxide-activated LiP*. C, high-resolution narrow scan spectrum of the protein radical in the peroxide-activated LiP* (Exp) paired with its simulation (Sim). D, schematic representation of the dihedral angles θ1(Hβ1), θ2(Hβ2), and θ3(Cα) of Tyr-181 in the LiP* crystal structure (dashed lines), together with those calculated from the EPR data (solid lines). EPR spectra were recorded under the following conditions: ν, 9.4 GHz; modulation amplitude, 1 (A) and 0.1 mT (B and C); microwave power, 2 (A) and 0.2 milliwatts (B and C); modulation frequency, 100 kHz; and temperature 4 (A) and 100 K (C).
FIGURE 7.
Low temperature EPR spectra of the Y181N variant. A, spectra of Y181N resting state (top) indicating the high-spin ferric signals (g = 2.0 and 6.0), and its peroxide-activated form (Compound I) (bottom). B, narrow scan EPR spectrum of the radical reported in A (bottom). EPR spectra were recorded under the following conditions: ν, 9.4 GHz; modulation amplitude, 1 (A) and 0.4 mT (B); microwave power, 2 (A) and 1 milliwatt (B); modulation frequency, 100 kHz; and temperature, 4 K.
However, the EPR spectra of peroxide-activated native LiP* and Y181N (Figs. 6A, bottom, and 7A, bottom, respectively) are different. After the addition of H2O2 to LiP*, the signals of high-spin ferric iron disappeared, indicating oxidation of heme iron FeIII to the ferryl state FeIV=O. At the same time, a free radical is formed, characterized by a giso = 2.0047, with a radical yield of 0.18 spin/heme (Fig. 6A, bottom). The new signal (giso = 2.0047) is detectable up to 100 K (Fig. 6B), suggesting a protein radical rather than a porphyrin radical, whose EPR signal would be hardly observable at temperatures over 30 K due to relaxation broadening (36). The signal was identified as corresponding to a tyrosyl radical, being superimposable to the simulated spectrum (Fig. 6C), as described below. Because the hydroxyl group of tyrosine is deprotonated simultaneously to its radical formation, the EPR spectrum is defined by the hyperfine interactions of the unpaired electron with the four protons of aromatic ring and the β-protons of the methylene group. The obtained singlet EPR signal was characterized by a hyperfine structure, which was poorly resolved but still detectable. The g tensor components (gx = 2.0080, gy = 2.0040, and gz = 2.0019 (estimated error 2 × 10−4)) and the hyperfine splitting constants for the β-protons of the side-chain methylene group (Ax(Hβ1) = 0.84 mT, Ay(Hβ1) = 0.62 mT, Az(Hβ1) = 0.90 mT, Ax(Hβ2) = 0.39 mT, Ay(Hβ2) = 0.27 mT, and Az(Hβ2) = 0.20 mT (estimated error ±0.05 mT)), obtained from the EPR spectral simulation, are consistent with the assignment to a tyrosyl radical (37).
Variability of the EPR spectrum of tyrosyl radicals in different protein environments is traditionally associated with the Cβ-C1 rotation of the phenolic group in tyrosine. According to the McConnell relation (20, 38), dihedral angles θ1(Hβ1) ∼ |51°| ± 5° and θ2(Hβ2) ∼ |69°| ± 5° were calculated from the isotropic part (Aiso) of the hyperfine tensors of the β-protons Hβ1 (Aβ1iso = 0.79 mT) and Hβ2 (Aβ2iso = 0.29 mT). These calculated data are in fair agreement with the corresponding dihedral angles of Tyr-181 (θ1 = 54.16° and θ2 = −69.01°) obtained from the LiP* crystal structure (Fig. 6D).
On the other hand, the Y181N variant after the addition of H2O2 shows a spectrum different from that of the peroxide-activated LiP* (Fig. 7A, bottom). This new spectrum, extending from g = 3.6 to g ∼ 1.99 (narrow scan reported in Fig. 7B), agrees well with that previously assigned to the porphyrin radical in P. chrysosporium LiP Compound I (36). The g tensor is characteristic of a heme in which the porphyrin π-cation radical is ferromagnetically coupled with an oxo-ferryl, Fe4+=O (S = 1) heme center (39–42). Moreover, the above mentioned temperature effect on porphyrin radical EPR spectra (36) was observed (see supplemental Fig. S1). Thus, the EPR results, together with the above spectroscopic and kinetic experiments, established that the peroxide-activated Y181N variant contains a porphyrin radical, in contrast to native LiP* that contains a tyrosyl radical, although further experiments might be advisable for definitive assignments.
DISCUSSION
The catalytic properties required for oxidation of lignin, including the ability to act on high redox potential and bulky molecules, are provided by several unique characteristics in the molecular architecture of ligninolytic peroxidases (5). Among them, oxidation sites on the protein surface, which collect electrons from substrates and deliver them to heme using different pathways, play a crucial role. Structure-function studies on P. chrysosporium LiP and P. eryngii VP have shown that a conserved tryptophan residue functions as their substrate oxidation site, initiating a long-range electron transfer pathway from the protein surface to the buried heme cofactor (16, 17). T. cervina LiP has been described as a unique peroxidase in which the catalytic tryptophan is absent, despite its LiP-type properties (22), and a tyrosine residue (Tyr-181) appears as a candidate for an alternative oxidation site (25). Although a catalytic role of a tyrosine residue had never been reported in any LiP or VP, different enzymes (including peroxidases, such as poplar peroxidase and the heme domain of animal cyclooxygenase) use tyrosines as redox-active residues (43, 44). These observations, together with homology modeling and chemical modification results (25, 26), suggested that Tyr-181 could function as the catalytic residue in T. cervina LiP. However, the detailed mechanism had remained unexplored till now.
The crystal structure of T. cervina LiP* maintains the consensus architecture of the heme cavity found in typical peroxidases and the two structural Ca2+ ions conserved in classes II and III of the plant, fungal, and bacterial peroxidases superfamily (7, 12). The heme cavity residues play roles as acid-base catalysts in the heterolysis of H2O2 for Compound I formation (at the distal side), as well as modulating the redox potential of the enzyme (at the proximal side). Thereby, it is expected that T. cervina LiP* will react with H2O2 in the same manner of other peroxidases. In fact, its apparent second-order rate constant for Compound I formation (k1app 3.6 × 106 s−1 m−1) is in the same range found for other superfamily members, such as P. chrysosporium MnP (12) and P. eryngii VP (17). In the crystal structure, three possible substrate oxidation sites exposed to the solvent were found. One of them would be located at the main heme access channel (involving Asp-146) enabling direct electron transfer to the heme δ-carbon. The other two sites correspond to surface residues, His-170 and Tyr-181, that would supply electrons to the heme, respectively, via: (i) long-range transfer to methyl-3; or (ii) direct transfer to the internal propionate.
Kinetic studies of site-directed variants show that the above sites work differently depending on the substrate, although Tyr-181 is the most crucial residue for catalysis in T. cervina LiP. Tyr-181 functions in oxidation of different types of substrates such as: (i) VA, the typical high redox potential (1.36 V) aromatic substrate of LiP and VP; and (ii) ABTS, a negatively charged aromatic disulfonate dye that is oxidized by a number of peroxidases due to its low redox potential (0.68 V). This aromatic residue is also responsible for other LiP-type catalytic properties of T. cervina LiP such as: (i) its ability to oxidize large molecules, which was shown here using Cc2+ (24); and (ii) the mechanism of one-electron oxidation of aromatic rings (to aryl cation radical intermediates), which was shown by the oxidation of 1,4-DMB to 1,4-benzoquinone, as described for P. chrysosporium LiP (45). Moreover, it is demonstrated that His-170 of T. cervina LiP, homologous to the catalytic tryptophan of other LiP and VP enzymes, is not involved in VA oxidation, as also found for the W164H variant of P. eryngii VP (16).
Detailed spectroscopic and transient-state kinetic studies of peroxide-activated T. cervina LiP*, including low temperature EPR, show that the porphyrin radical is unstable and rapidly moves to a tyrosine residue. This results in tyrosyl radical and oxo-ferryl heme formation, i.e. a compound ES-type intermediate (34), with the same behavior as the so-called Compound IB of the W164Y variant of P. eryngii VP (46). The theoretical dihedral angles for the tyrosyl radical estimated from the EPR data were nearly identical to those obtained for the Tyr-181 side chain in the T. cervina LiP* crystal structure, confirming radical location at this tyrosine residue. In contrast with that found for native LiP*, the porphyrin radical in the peroxide-activated Y181N variant was stable, indicating that electron transfer to the heme is blocked by the mutation. Taking into consideration the fact that the Y181N mutation completely removes the VA oxidation activity, it is concluded that Tyr-181 forms a catalytic radical by electron transfer to the heme in the peroxide-activated T. cervina LiP. This radical would function like the catalytic tryptophanyl radical formed in other LiP and VP enzymes, although the nature and length of the electron transfer pathways are different (being located at the opposite side of the peroxidase molecule).
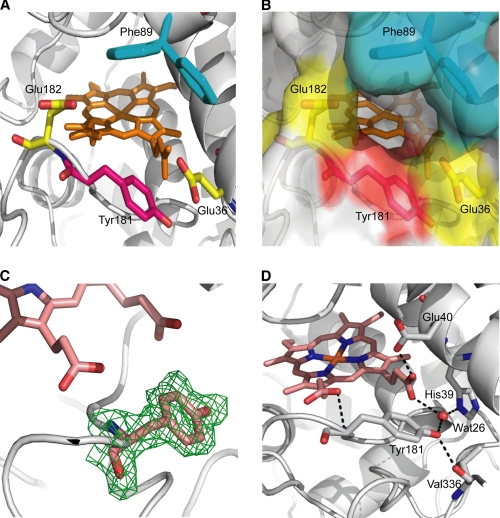
Studies on P. chrysosporium LiP and P. eryngii VP suggested a contribution of the protein environment around the catalytic tryptophan to substrate recognition and/or oxidation. An acidic environment was proposed to lower the local pH and provide a higher redox potential to the tryptophanyl radical (21), at the same time that it will favor binding of substrate cation radical intermediates that could act as enzyme-bound mediators (8, 47). On the other hand, an aromatic residue adjacent to Trp-171 in P. chrysosporium LiP was suggested to contribute to substrate binding by aromatic interactions (48). As shown in Fig. 8A, Tyr-181 of T. cervina LiP is surrounded by two acidic residues (Glu-36 and Glu-182) and Phe-89, which delimit the propionate access channel (Fig. 8B) and would play the same role of similar residues near the catalytic tryptophan of other LiP and VP enzymes, i.e. increasing the redox potential of the tyrosyl radical (49) and contributing to aromatic substrate binding. Moreover, the high mobility of the Phe-89 side chain, which is fully disordered in the crystal structure (the two most probable positions are shown in Fig. 8, A and B), could also be related to substrate binding.
FIGURE 8.
Tyr-181 microenvironment and interactions in T. cervina LiP* crystal structure (PDB entry 3Q3U). A, some amino acids forming the Tyr-181 (red carbons) microenvironment including two acidic residues (Glu-36 and Glu-182, yellow carbons) and one aromatic residue (Phe-89, blue carbons), whose side chain was not solved in the crystal structure due to its high mobility, but the positions with highest frequencies (62% in the right side, and 22% in the left side) were predicted with PyMOL (58). B, semitransparent solvent-accessible surface on the same residues included in A, showing the access channel to the heme propionates. C, a (2Fo − Fc) electron density map, contoured at the 1.0-s level, displaying the catalytic Tyr-181 residue. D, some interactions of Tyr-181 and neighbor residues.
Unambiguous identification of a tyrosine residue near the heme cofactor (occupying the 181 position of T. cervina LiP) is shown, for the first time in a ligninolytic peroxidase, by the (2Fo − Fc) electron density map included in Fig. 8C. The crystal structure of T. cervina LiP* also showed several specific interactions around Tyr-181 (Fig. 8D) potentially affecting substrate oxidation and/or subsequent electron transfer. The latter will be most probably produced through the internal propionate of heme followed by intraheme transfer to the porphyrin macrocycle in Compound I or to the iron center in Compound II. The above interactions include a strong hydrogen bond (2.40 Å) between the carboxylate group of Glu-40 and the internal heme propionate and several other bonds involving His-39, Tyr-181, and a water molecule (Wat-26). Electron transfer via the internal heme propionate has been suggested in other heme peroxidases including MnP and VP (14, 15) and predicted by mixed quantum mechanics and molecular mechanics calculations (50).
Additionally, the hydrogen bonds of the Tyr-181 hydroxyl group would assist the catalytic function of this aromatic residue. In the EPR spectrum of the tyrosyl radical from peroxide-activated T. cervina LiP*, the gx value with the acceptable variation (gx = 2.0078–2.0082) suggests the existence of a hydrogen bond with a length of 1.8–2.0 Å (51) because there is a clear relationship between the gx value and the hydrogen bond strength (52). The recent quantum mechanics and molecular mechanics calculations on the VP W164Y variant have demonstrated that the proton transfer to the nearby Glu-243, during tyrosine oxidation, proceeds barrierless, directly yielding the Tyr-164 neutral radical, but the variant is inactive because reorganization of hydrogen interactions (by water molecules and neighbor residues) precludes an easy recovery of the proton by the tyrosine residue (53). In the crystal structure of T. cervina LiP*, the phenolic hydroxyl group of Tyr-181 forms hydrogen bonds with the backbone carbonyl of Val-336 (2.72 Å) and with Wat-26 (2.74 Å) that forms two other H-bonds to His-39 (2.65 Å) and heme internal propionate (2.82 Å). The carboxylate of Glu-36, the acidic residue nearest to Tyr-181, is too far from the Tyr-181 hydroxyl group (4.08 Å) to be included in the proton transfer to form the phenoxy radical. However, the above interactions with the Tyr-181 phenolic hydroxyl could maintain the proton released from tyrosine forming an H-bond with the neutral radical formed, as suggested by the EPR results. This would contribute to peroxidase catalysis by retransferring the proton back to Tyr-181 after substrate oxidation.
The information currently available suggests a neutral (H-bonded) radical of Tyr-181 as the catalytic species in T. cervina LiP, as shown by the EPR spectra of peroxide-activated enzyme (Compound I). Low temperature EPR also revealed a neutral tryptophanyl radical in Compound I of VP (17, 20) and engineered P. chrysosporium LiP (21). However, in the latter LiP, a transient cation radical has been proposed as the real catalytic species (54), although no experimental evidence is available. The rationale for this proposal is the higher redox potential of the tryptophanyl cation radical (∼1.15 V) as compared with its neutral form (∼1.01 V) (55) that would facilitate lignin oxidation. Proton abstraction during phenol oxidation is generally simultaneous to electron abstraction, and this has been shown for tyrosyl radical formation in the W164Y VP variant (53). However, spectroscopic studies suggested that under acidic conditions, a cation-like radical (or H-bonded neutral radical) is formed at the high redox potential short-lived Tyr-161 of photosystem II (56, 57). A similar H-bonded tyrosyl radical seems to be formed at the T. cervina LiP Tyr-181, which has a very acidic environment and could contribute to oxidation of recalcitrant aromatics. The present study reports the first high redox potential peroxidase with a catalytic tyrosyl radical, as confirmed by crystallographic, spectroscopic, mutagenesis, and kinetic analyses, although further studies are required to fully elucidate its catalytic mechanism for oxidation of high redox potential substrates.
Supplementary Material
Acknowledgment
We thank Prof. H. Wariishi (Kyushu University, Japan) for the contribution to the initial molecular characterization of T. cervina LiP.
This work was supported by the Spanish projects BIO2008-01533 (RAPERO) and BFU2008-02595, the European projects BIORENEW (NMP2-CT-2006-026456) and PEROXICATS (KBBE-2010-4-265397), and the Italian Ministry of University and Research (MIUR) project 20072R7WWA.
The atomic coordinates and structure factors (code 3Q3U) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- LiP
- lignin peroxidase
- LiP*
- recombinant LiP
- MnP
- manganese peroxidase
- VP
- versatile peroxidase
- ABTS
- 2,2′-azino-bis(3-ethylbenzo-thiazoline-6-sulfonate)
- Cc3+
- ferricytochrome c
- Cc2+
- ferrocytochrome c
- 1,4-DMB
- 1,4-dimethoxybenzene
- MPD
- 2-methyl-2,4-pentanediol
- VA
- veratryl alcohol
- mT
- milliteslas.
REFERENCES
- 1. Brunow G., Lundquist K. (2010) in Lignin and Lignans: Advances in Chemistry (Heitner C., Dimmel D., Schmidt J. eds) pp. 267–299, CRC Press, Boca Raton [Google Scholar]
- 2. Martínez A. T., Ruiz-Dueñas F. J., Martínez M. J., Del Río J. C., Gutiérrez A. (2009) Curr. Opin. Biotechnol. 20, 348–357 [DOI] [PubMed] [Google Scholar]
- 3. Kirk T. K., Farrell R. L. (1987) Annu. Rev. Microbiol. 41, 465–505 [DOI] [PubMed] [Google Scholar]
- 4. Ruiz-Dueñas F. J., Martínez A. T. (2009) Microb. Biotechnol. 2, 164–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruiz-Dueñas F. J., Martínez A. T. (2010) in Biocatalysts Based on Heme Peroxidases (Torres E., Ayala M. eds) pp. 37–59, Springer-Verlag, Berlin [Google Scholar]
- 6. Hammel K. E., Cullen D. (2008) Curr. Opin. Plant Biol. 11, 349–355 [DOI] [PubMed] [Google Scholar]
- 7. Martínez A. T. (2002) Enzyme Microb. Technol. 30, 425–444 [Google Scholar]
- 8. Khindaria A., Yamazaki I., Aust S. D. (1996) Biochemistry 35, 6418–6424 [DOI] [PubMed] [Google Scholar]
- 9. Gold M. H., Youngs H. L., Gelpke M. D. (2000) Met. Ions Biol. Syst. 37, 559–586 [PubMed] [Google Scholar]
- 10. Kapich A. N., Korneichik T. V., Hatakka A., Hammel K. E. (2010) Enzyme Microb. Technol. 46, 136–140 [DOI] [PubMed] [Google Scholar]
- 11. Ruiz-Dueñas F. J., Morales M., García E., Miki Y., Martínez M. J., Martínez A. T. (2009) J. Exp. Bot. 60, 441–452 [DOI] [PubMed] [Google Scholar]
- 12. Dunford H. B. (1999) Heme Peroxidases, pp. 281–308, John Wiley & Sons, New York [Google Scholar]
- 13. Sundaramoorthy M., Kishi K., Gold M. H., Poulos T. L. (1997) J. Biol. Chem. 272, 17574–17580 [DOI] [PubMed] [Google Scholar]
- 14. Ruiz-Dueñas F. J., Morales M., Pérez-Boada M., Choinowski T., Martínez M. J., Piontek K., Martínez A. T. (2007) Biochemistry 46, 66–77 [DOI] [PubMed] [Google Scholar]
- 15. Smith A. T., Veitch N. C. (1998) Curr. Opin. Chem. Biol. 2, 269–278 [DOI] [PubMed] [Google Scholar]
- 16. Doyle W. A., Blodig W., Veitch N. C., Piontek K., Smith A. T. (1998) Biochemistry 37, 15097–15105 [DOI] [PubMed] [Google Scholar]
- 17. Pérez-Boada M., Ruiz-Dueñas F. J., Pogni R., Basosi R., Choinowski T., Martínez M. J., Piontek K., Martínez A. T. (2005) J. Mol. Biol. 354, 385–402 [DOI] [PubMed] [Google Scholar]
- 18. Mester T., Ambert-Balay K., Ciofi-Baffoni S., Banci L., Jones A. D., Tien M. (2001) J. Biol. Chem. 276, 22985–22990 [DOI] [PubMed] [Google Scholar]
- 19. Blodig W., Smith A. T., Winterhalter K., Piontek K. (1999) Arch. Biochem. Biophys. 370, 86–92 [DOI] [PubMed] [Google Scholar]
- 20. Pogni R., Baratto M. C., Teutloff C., Giansanti S., Ruiz-Dueñas F. J., Choinowski T., Piontek K., Martínez A. T., Lendzian F., Basosi R. (2006) J. Biol. Chem. 281, 9517–9526 [DOI] [PubMed] [Google Scholar]
- 21. Smith A. T., Doyle W. A., Dorlet P., Ivancich A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16084–16089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miki Y., Tanaka H., Nakamura M., Wariishi H. (2006) J. Fac. Agr. Kyushu Univ. 51, 99–104 [Google Scholar]
- 23. Kersten P. J., Kalyanaraman B., Hammel K. E., Reinhammar B., Kirk T. K. (1990) Biochem. J. 268, 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wariishi H., Sheng D., Gold M. H. (1994) Biochemistry 33, 5545–5552 [DOI] [PubMed] [Google Scholar]
- 25. Miki Y., Ichinose H., Wariishi H. (2010) FEMS Microbiol. Lett. 304, 39–46 [DOI] [PubMed] [Google Scholar]
- 26. Miki Y., Ichinose H., Wariishi H. (March 4, 2011) Biotechnol. Lett. 10.1007/s10529-011-0571-2 [DOI] [PubMed] [Google Scholar]
- 27. Miki Y., Morales M., Ruiz-Dueñas F. J., Martínez M. J., Wariishi H., Martínez A. T. (2009) Protein Expr. Purif. 68, 208–214 [DOI] [PubMed] [Google Scholar]
- 28. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 29. Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 30. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 31. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 32. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sivaraja M., Goodin D. B., Smith M., Hoffman B. M. (1989) Science 245, 738–740 [DOI] [PubMed] [Google Scholar]
- 35. Palmer G. (1983) in Iron Porphyrins: Part II (Lever A. B. P., Gray H. B. eds) pp. 43–88, Addison-Wesley, Reading, MA [Google Scholar]
- 36. Khindaria A., Aust S. D. (1996) Biochemistry 35, 13107–13111 [DOI] [PubMed] [Google Scholar]
- 37. Svistunenko D. A., Dunne J., Fryer M., Nicholls P., Reeder B. J., Wilson M. T., Bigotti M. G., Cutruzzolà F., Cooper C. E. (2002) Biophys. J. 83, 2845–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McConnell H. M., Robertson R. E. (1957) J. Phys. Chem. A 61, 1018 [Google Scholar]
- 39. Schulz C. E., Devaney P. W., Winkler H., Debrunner P. G., Doan N., Chiang R., Rutter R., Hager L. P. (1979) FEBS Lett. 103, 102–105 [DOI] [PubMed] [Google Scholar]
- 40. Benecky M. J., Frew J. E., Scowen N., Jones P., Hoffman B. M. (1993) Biochemistry 32, 11929–11933 [DOI] [PubMed] [Google Scholar]
- 41. Patterson W. R., Poulos T. L., Goodin D. B. (1995) Biochemistry 34, 4342–4345 [DOI] [PubMed] [Google Scholar]
- 42. Davydov R., Osborne R. L., Shanmugam M., Du J., Dawson J. H., Hoffman B. M. (2010) J. Am. Chem. Soc. 132, 14995–15004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sasaki S., Nonaka D., Wariishi H., Tsutsumi Y., Kondo R. (2008) Phytochemistry 69, 348–355 [DOI] [PubMed] [Google Scholar]
- 44. Smith W. L., Eling T. E., Kulmacz R. J., Marnett L. J., Tsai A. L. (1992) Biochemistry 31, 3–7 [DOI] [PubMed] [Google Scholar]
- 45. Kersten P. J., Tien M., Kalyanaraman B., Kirk T. K. (1985) J. Biol. Chem. 260, 2609–2612 [PubMed] [Google Scholar]
- 46. Ruiz-Dueñas F. J., Pogni R., Morales M., Giansanti S., Mate M. J., Romero A., Martínez M. J., Basosi R., Martínez A. T. (2009) J. Biol. Chem. 284, 7986–7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Choinowski T., Blodig W., Winterhalter K. H., Piontek K. (1999) J. Mol. Biol. 286, 809–827 [DOI] [PubMed] [Google Scholar]
- 48. Gelpke M. D. S., Lee J., Gold M. H. (2002) Biochemistry 41, 3498–3506 [DOI] [PubMed] [Google Scholar]
- 49. Tommos C., Skalicky J. J., Pilloud D. L., Wand A. J., Dutton P. L. (1999) Biochemistry 38, 9495–9507 [DOI] [PubMed] [Google Scholar]
- 50. Guallar V. (2008) J. Phys. Chem. B 112, 13460–13464 [DOI] [PubMed] [Google Scholar]
- 51. Bernini C., Sinicropi A., Basosi R., Pogni R. (2010) Appl. Magn. Res. 37, 279–288 [Google Scholar]
- 52. Un S., Gerez C., Elleingand E., Fontecave M. (2001) J. Am. Chem. Soc. 123, 3048–3054 [DOI] [PubMed] [Google Scholar]
- 53. Bernini C., Pogni R., Ruiz-Dueñas F. J., Martínez A. T., Basosi R., Sinicropi A. (2011) Phys. Chem. Chem. Phys. 13, 5078–5098 [DOI] [PubMed] [Google Scholar]
- 54. Piontek K., Smith A. T., Blodig W. (2001) Biochem. Soc. Trans. 29, 111–116 [DOI] [PubMed] [Google Scholar]
- 55. Stubbe J., van Der Donk W. A. (1998) Chem. Rev. 98, 705–762 [DOI] [PubMed] [Google Scholar]
- 56. Mino H., Kawamori A. (2008) Photosynth. Res. 98, 151–157 [DOI] [PubMed] [Google Scholar]
- 57. Barry B. A., Babcock G. T. (1988) Chemica Scripta 28A, 117–122 [Google Scholar]
- 58. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.