Abstract
The EDD (E3 identified by differential display) gene, first identified as a progestin-induced gene in T-47D breast cancer cells, encodes an E3 ubiquitin ligase with a HECT domain. It was reported that EDD is involved in the G2/M progression through ubiquitination of phospho-katanin p60. Previous study has also shown that EDD can act as a transcription cofactor independently of its E3 ligase activity. In this study, we uncover a new role for EDD during cell cycle progression in an E3 ligase-independent manner. We demonstrate that EDD can physically interact with p53 and that this interaction blocks the phosphorylation of p53 by ataxia telangiectasia mutated (ATM). Silencing of EDD induces phosphorylation of p53 at Ser15 and activates p53 target genes in fibroblasts and some transformed cells without activation of DNA damage response. The G1/S arrest induced by EDD depletion depends on p53. On the other hand, overexpression of EDD inhibits p53-Ser15 phosphorylation and suppresses the induction of p53 target genes during DNA damage, and this effect does not require its E3 ligase activity. Thus, through binding to p53, EDD actively inhibits p53 phosphorylation by ATM and plays a role in ensuring smooth G1/S progression.
Keywords: Cell Cycle, E3 Ubiquitin Ligase, Gene Regulation, p53, Phosphorylation Enzymes, ATM, EDD
Introduction
The tumor suppressor p53, one of the most important proteins in preventing human cancer, is a critical factor controlling cell cycle progression, particularly during the G1/S transition. Mutations of the p53 gene occur in at least half of all human cancers. Phosphorylation of p53 stabilizes p53 and hence induces both cell cycle arrest and apoptosis. Regulation of p53 phosphorylation is very complicated. There are three main kinases that are thought to be responsible for p53 phosphorylation at Ser15 residue under genotoxic stresses: ataxia telangiectasia mutated (ATM),3 ataxia telangiectasia and Rad3-related (ATR), and DNA-PK (DNA-dependent protein kinase) (1–3). ATM is a major player in phosphorylating p53 at Ser15 in response to ionizing radiation, and ATR plays a central role in response to UV light. The group of proteins phosphorylated by DNA-PK, including p53, is mainly involved in the non-homologous end joining DNA double strand break repair. However, all these three kinases can phosphorylate p53 at Ser15, and there is a lot of cross-talk between these three pathways. Besides Ser15, ATM can also stimulate Chk2 to phosphorylate p53 at Ser20, a site that is critical for the stability of p53. In addition to phosphorylation, other post-translational modifications such as acetylation, methylation, ubiquitination, and sumoylation regulate p53 as well (4). The status of these post-translational modifications determines whether p53 is active or not. Even in the cycling cells, there exist spontaneous pulses of p53, but without sustained active modifications, these p53 fail to induce p21 or cell cycle arrest (5). Therefore, p53 is controlled actively by both inhibitors and activators in a delicate manner so that cellular proliferation can proceed in unstressed condition, but upon encountering stresses, it can be quickly and effectively arrested. However, the full scope of these regulations remains to be further investigated.
EDD (E3 identified by differential display), originally isolated as a progestin-induced gene, encodes a 350-kDa E3 ubiquitin ligase containing a HECT (homologous to E6-AP carboxyl terminus) domain (6). Mutations of hyd (hyperplastic discs), the EDD homologue in Drosophila, lead to imaginal disc hyperplasia (7), suggesting involvement of EDD in cellular proliferation and differentiation. The phenotype seen in hyd mutant flies is due to activation of hedgehog and decapentaplegic expression (8). In mice, EDD plays an essential role in extraembryonic development as knock-out of EDD leads to embryonic lethality due to failed yolk sac and allantoic vascular development and defective chorioallantoic fusion (9).
In addition to a HECT domain, EDD also contains a poly(A)-binding protein C-terminal (PABC) domain near its carboxyl terminus, which also suggests a role in mRNA metabolism. EDD has been demonstrated to target Paip2 (poly(A)-binding protein (PABP))-interacting protein 2) and katanin for degradation (10, 11). On the other hand, interaction between EDD and adenomatous polyposis coli leads to up-regulation of adenomatous polyposis coli expression (12). EDD is also involved in transcriptional regulation and interacts with progesterone receptor as a coactivator (13). These E3 ligase-independent activities of EDD are further supported by a recent report that EDD enhances transactivation of smooth muscle-specific promoters by the myocardin family of proteins (14). Recent data from proteomics analysis also suggested that the EDD may be a substrate of ATM (15, 16). In addition, many other EDD-interacting proteins have been reported, including CIB (integrin-binding protein/DNA-dependent kinase-interacting protein) (13), TopBP1 (topoisomerase IIβ-binding protein) (17), Chk2 (18), and α4-phospho-protein (19). Thus, EDD appears to be a multifunctional protein involved in many intracellular processes. Interestingly, EDD mRNA and protein were found to be amplified or overexpressed in many types of cancer including breast and ovarian cancers, suggesting their involvement in tumorigenesis (20). Despite the accumulating knowledge about EDD, its involvement in normal cell cycle progression, its exact cellular functions, and corresponding molecular mechanism are yet to be fully explored.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK293, HEK293T, H1299, and human foreskin fibroblast cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (50 IU/ml), and streptomycin (50 μg/ml). HCT116 cells were grown in McCoy's 5A medium supplemented with 10% FBS, penicillin, and streptomycin. All cells were grown in a humidified incubator at 37 °C with 5% CO2 and 95% air. A standard calcium phosphate method was used for transfection of HEK293, HEK293T, and H1299 cells. HCT116 cells were transfected with electroporation with a Gene Pulser Xcell electroporation system (Bio-Rad).
Plasmid and Adenovirus Construction
All the p53-related constructs and recombinant adenovirus expressing control siRNA against GFP (Ad-siGFP) have been described previously (21). Mammalian expression pCMV-Tag2B vectors for FLAG-tagged EDD and FLAG-tagged EDD mutant (Cys-2768 → Ala) at the active site cysteine necessary for E3 ligase activity were gifts from Michelle Henderson and Charlie Watts (Garvan Institute of Medical Research). FLAG-EDD-N (a.a. 1–888) was obtained by digesting FLAG-EDD with SalI followed by self-ligation. FLAG-EDD-NM (a.a. 1–1876) was constructed by cloning FLAG-EDD EcoRI-digested fragment into pCMV-Tag2B vector. FLAG-EDD-C (a.a. 1876–2799) was cloned by moving the EcoRI/XhoI-digested fragment of FLAG-EDD into pCMV-Tag2A vector. FLAG-EDD-M (a.a. 888–1876) was obtained by cloning FLAG-EDD SalI/EcoRI-digested fragment into pCMV-Tag2C vector. To identify effective target sequences within EDD for shRNA, we initially picked four 19-nucleotide target sequences to clone into pSUPER vector. Among these four target sequences, target sequence 3 produced the best result in depleting EDD. Target sequence 5 was kindly provided by Michelle Henderson at Children's Cancer Institute Australia (18). The target sequence of siEDD#3 is 5′-GCGACTCTCCATGGTTTCT-3′. The target sequence of siEDD#5 is 5′-GCAGTGTTCCTGCCTTCTT-3′. The NotI/SalI fragments of the pSUPER-siEDD constructs containing H1 promoter and hairpin shRNA sequences were swapped to NotI/SalI sites of pShuttle, and the recombinant adenoviruses expressing siEDD#3 and siEDD#5 were prepared as described in the AdEasy system (22). All viruses were purified by CsCl banding.
Protein Purification
Escherichia coli strain DH5α transformed with either pGEX6P vector or pGEX6P-p53 was cultured in LB medium containing ampicillin at 37 °C to an A600 value of 0.6. GST fusion proteins were induced by 0.1 mm isopropyl-β-d-thiogalactopyranoside at 37 °C for 3 h; cells were then lysed in sodium chloride-Tris-EDTA (STE) buffer (100 mm NaCl, 1 mm EDTA) supplemented with lysozyme and protease inhibitors. After 15 min of treatment, the lysate was sonicated for 30 s and then purified using glutathione-Sepharose 4B (GE Healthcare) (23). Sepharose beads were washed four times with sodium chloride-Tris-EDTA (STE) buffer. The GST proteins were eluted out by reduced glutathione if needed. To prepare recombinant ATM proteins, 293T cells were transiently transfected with pBJ-HA-ATM or pBJ-HA-ATMkd using calcium phosphate, and HA-ATM kinase was purified from cell extracts using anti-HA beads as described (23). For purification of recombinant EDD protein, 293T cells were first transfected with FLAG-tagged EDD, and cellular extracts were prepared by lysing the cells with TNN buffer (50 mm Tris, 0.25 m NaCl, 5 mm EDTA, 0.5% Nonidet P-40) supplemented with 1 mm dithiothreitol, 1 mm NaF, 1 mm sodium orthovanadate, 20 nm microcystin, and a mixture of protease inhibitors including 10 μg of leupeptin/ml, 10 μg of aprotinin/ml, 10 μg of pepstatin/ml, 1 mm phenylmethylsulfonyl fluoride, 2 μg of antipain/ml, and 1 μg of chymostatin/ml. FLAG-tagged EDD was isolated on 0.4-ml anti-FLAG M2 agarose beads (Sigma). The beads were washed by TBS buffer four times and then eluted with 2 ml of TBS containing 0.1 mg/ml FLAG peptide (Sigma). Fractions containing EDD were desalted and concentrated by Microcon (Millipore) prior to storage in small aliquots at −80°.
Immunoprecipitation, Western Blot Analysis, and Immunocytochemical Staining
For immunoprecipitation, the transfected cells were harvested 24–48 h later with TNN buffer supplemented with 1 mm dithiothreitol, 1 mm NaF, 1 mm sodium orthovanadate, 20 nm microcystin, and a mixture of protease inhibitors as described above. Cells prepared for endogenous immunoprecipitation were directly lysed in TNN buffer. An aliquot of the cell lysates was saved for protein input control, and immunoprecipitation was carried out as described previously (24). The specific signals were detected with appropriate antibodies. The antibody specific to EDD (A300-573A) was purchased from Bethyl Laboratories. The monoclonal antibody (Ab-1) for p53 was purchased from Calbiochem.
Cells prepared for direct protein analysis were lysed in SDS lysis buffer (1% SDS, 60 mm Tris). Equal protein amounts were electrophoresed and blotted. DNA damage was induced by the addition of 0.3 μg/ml neocarzinostatin (NCS). Antibody against phospho-p53 (Ser15) (antibody number 9284) and phospho-p38 (9216) were purchased from Cell Signaling Technology. Anti-ATM pSer1981 (200-301-400) antibody was purchased from Rockland Immunochemicals. The monoclonal antibodies for p53 (DO1, PAb1801, PAb240, and PAb421) were gifts from Dr. Xinbin Chen. The antibody for β-actin was purchased from Sigma. The antibody for phospho-H2AX (Ser139) (JBW301) was purchased from Upstate Biotech Millipore. The antibodies for phospho-JNK (Cell Signaling Technology) and JNK (Santa Cruz Biotechnology Inc.) were gifts from Fannie Lin (UAB). The antibody for p38 (P71220) was purchased from BD Transduction Laboratories. The antibody for proliferating cell nuclear antigen (PC10) was purchased from Santa Cruz Biotechnology. The monoclonal antibody (Ab-3) for ATM was purchased from Calbiochem. For immunocytochemical staining, the cells were stained with mouse anti-p53 antibodies containing DO1, PAb1801, PAb240, and PAb421 (gifts from Dr. Xinbin Chen), a rabbit EDD antibody (A300-573A; Bethyl Laboratories), and fluorescein isothiocyanate- or Texas Red X-conjugated secondary antibody.
Luciferase Assays
The expression constructs (20 μg for FLAG-tagged EDD or FLAG-tagged EDD mutant (C2768A), 5 μg for pCMV-p53 or empty vector), the promoter plasmid (1 μg for pGL2-p21 promoter luciferase (25)), and 1 μg of pCMV-β-galactosidase plasmid were cotransfected in H1299 cells. Cells were harvested 48 h later in PBS; an aliquot was lysed in SDS lysis buffer for Western blotting, whereas the rest of the sample was lysed in reporter lysis buffer (Promega). Luciferase activity and β-galactosidase activity were measured according to manufacturer's instructions. Luciferase activity was normalized against β-galactosidase activity. All transient expression assays were performed in triplicate.
Real Time PCR
WI38 cells were infected with Ad-siGFP or Ad-siEDD. HCT116 cells were transfected with FLAG-tagged EDD, FLAG-tagged EDD mutant (C2768A), or an empty vector and then either left untreated or treated with NCS (300 ng/ml) for 2 h. RNA was extracted using TRIzol reagent (Invitrogen). Real time reverse transcription-PCR (RT-PCR) was performed in triplicate on an MX3005P thermal cycler (Stratagene) using SYBR Green dye to measure amplification and ROX as a reference dye. The detailed PCR conditions and the primer sequences of the p53 target genes have been described (21). Transcript levels were normalized with GAPDH levels, which were analyzed in parallel with test genes. Results were analyzed with MxPro 4.1 quantitative PCR software (Stratagene).
GST Pulldown Assay
GST fusion proteins were induced, lysed, and purified by the above method. Two micrograms of GST (as a control protein), p53, and p53-N terminus on Sepharose beads were nutated overnight at 4 °C with cellular lysates prepared from HEK293T cells and lysed with TNN buffer with protease inhibitors. Sepharose beads were washed five times with TNN buffer, eluted in SDS sample buffer, and subjected to SDS-PAGE and immunoblotting. Equal loading of GST proteins was assessed in parallel by SDS-PAGE followed by Coomassie Blue staining.
Bromodeoxyuridine Incorporation Assay
WI38 or HCT116 cells were plated on 24-well plates and then infected with adenovirus encoding the EDD siRNA or GFP siRNA. Forty-eight hours later, cells were labeled with 5-bromo-2′-deoxyuridine (BrdU) for 24 (WI38) or 8 h (HCT116), and the incorporated BrdU was detected with fluorescein isothiocyanate-conjugated anti-BrdU antibody following the manufacturer's instructions (BD Biosciences). Images were captured on a Zeiss fluorescence microscope (Axio Observer Inverted Microscope).
ATM Kinase Assay
ATM kinase assays were done in kinase buffer: 10 mm HEPES, pH 7.4, 50 mm potassium chloride, 10 mm magnesium chloride, 10 mm manganese chloride, 10% glycerol, 50 mm sodium chloride, 100 μm ATP, 1 mm dithiothreitol (DTT), 0.1 mm sodium orthovanadate, 20 nm microcystin, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 5 μg/ml pepstatin. Assays were performed in two stages. Substrate was first incubated alone or with purified EDD at 4 °C for 30 min. Then HA-ATM beads was added and incubated at 30 °C for 30 min.
RESULTS
Depletion of EDD Inhibited S-phase Entry
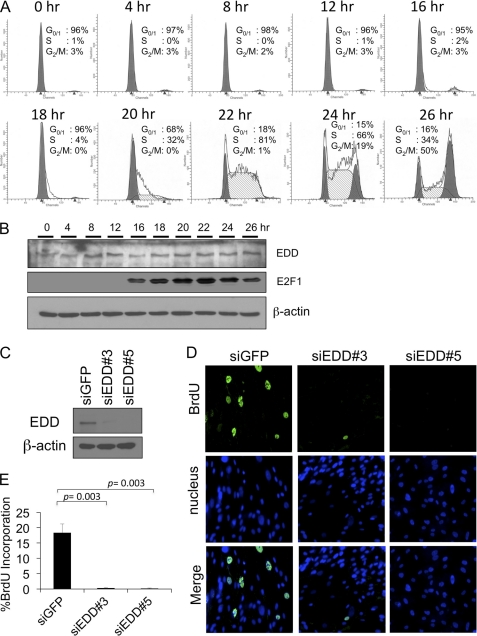
To investigate the possible involvement of EDD in the cell cycle, we first examined its expression during cell cycle progression. Primary human foreskin fibroblasts were synchronized by serum starvation for 48 h in medium containing 0.1% fetal bovine serum. Entry of cell cycle was induced by the addition of 20% fetal bovine serum as shown by propidium iodide staining (Fig. 1A) and the induction of E2F1 at G1/S entry and S-phase (Fig. 1B). We found that the expression of EDD remained relatively constant throughout the cell cycle (Fig. 1B). Previously, it was reported that EDD is required for G2/M progression through the regulation of katanin p60 (10). However, whether EDD is involved in progression of other cell cycle phases such as at the G1/S transition has not been explored. To test that, we depleted EDD in human fibroblast WI38 cells with recombinant adenoviruses expressing two different EDD shRNAs and determined their effects on the incorporation of BrdU. As shown in Fig. 1, C–E, depletion of EDD significantly inhibited BrdU incorporation, suggesting a role for EDD in the control of G1/S- or S-phases.
FIGURE 1.
EDD siRNA induces G1/S arrest in WI38 cells. A, primary human foreskin fibroblasts were brought to quiescence by serum starvation for 48 h and then stimulated by 20% FBS for indicated periods of time. The cells were fixed and analyzed with propidium iodide staining followed by flow cytometry analysis. B, cell lysates from synchronized human foreskin fibroblasts were subjected to Western blot analysis, and the immunoblot was probed with antibodies specific to EDD, E2F1, and β-actin, respectively. The E2F1 immunoblot serves as a control for cell cycle synchronization where the level of E2F1 appears at the G1/S transition. C, Western blot of EDD and β-actin level in WI38 infected with Ad-siEDD#3, Ad-siEDD#5, and Ad-siGFP. D and E, WI38 cells were infected with Ad-siGFP, Ad-siEDD#3, or Ad-siEDD#5 at a multiplicity of infection of 600. Forty-eight hours later, cells were labeled with BrdU for 24 h, and the incorporated BrdU was detected with fluorescein isothiocyanate-conjugated anti-BrdU antibody. The nuclei were stained with Hoechst 33258. D, representative images of 200× magnification. E, at least 300 nuclei were scored on each sample for BrdU incorporation by microscope, and the experiments were repeated three times. The graphs represent means ± standard errors. The p values are based on a paired two-tailed t test.
Depletion of EDD Induces p53 Phosphorylation at Ser15
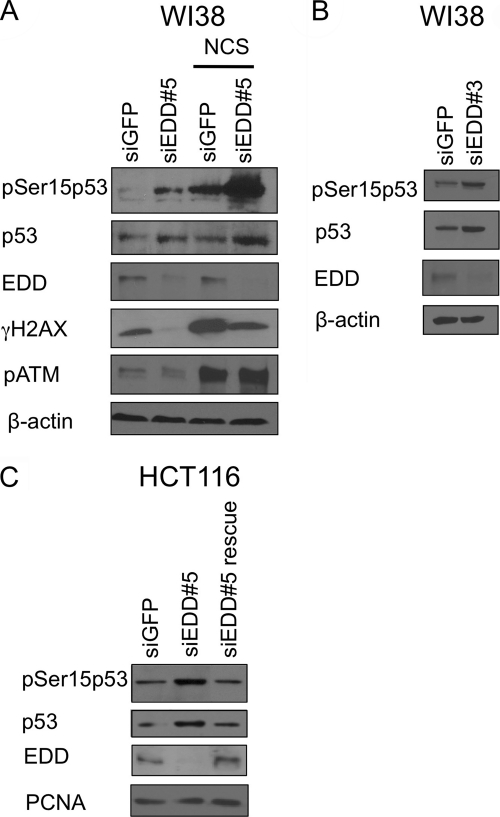
The tumor suppressor protein p53 plays a pivotal role in the regulation of G1/S checkpoint. Phosphorylation at Ser15 of p53 by ATM/ATR kinases is a key mechanism of cell cycle arrest at G1- or G1/S-phases (26, 27). Therefore, we explored the possibility that depletion of EDD could activate p53 Ser15 phosphorylation and block G1/S progression. To test this possibility, we examined the phosphorylation status of p53 Ser15 in EDD-depleted WI38 cells. We found that silencing of EDD with two different siRNAs induced p53 phosphorylation at Ser15 in normal growth condition (Fig. 2, A and B). The induction of Ser15 phosphorylation was further enhanced when the cells were treated with radiomimetic agent NCS (Fig. 2A). The induction of p53 phosphorylation by EDD siRNA was also seen in another cell line, HCT116, a colon cancer cell line (Fig. 2C). The observed effect was truly due to depletion of EDD protein because p53 Ser15 phosphorylation returned to its basal level when depletion of EDD was rescued (Fig. 2C).
FIGURE 2.
EDD siRNAs induce phosphorylation of p53 at Ser15. A, WI38 was infected with Ad-siEDD#5 and Ad-siGFP at a multiplicity of infection of 300. Three days later, cells were either left untreated or treated with NCS (300 ng/ml) for 2 h. Cell lysates were subjected to Western blotting analysis with antibodies specific to phosphorylated p53-Ser15, p53, phospho-H2AX, and EDD. B, Western blot of phosphorylated p53-Ser15, p53, and EDD in WI38 infected with another EDD siRNA virus Ad-siEDD#3 and Ad-siGFP for 3 days. C, HCT116 p53+/+ cells were infected by siRNA-expressing adenoviruses at a multiplicity of infection of 300. One day later, cells were transfected by either empty vector control or a FLAG-tagged EDD construct. Two days later, cells were harvested in TNN buffer. The cell lysates were analyzed by Western blotting. The proliferating cell nuclear antigen (PCNA) immunoblot serves as a protein loading control.
There are three possible scenarios for depletion of EDD to stimulate the phosphorylation of p53: 1) silencing of EDD could cause DNA damage and activate p53 indirectly; 2) EDD depletion could lead to cellular stress and therefore activate p53 indirectly; or 3) EDD could directly regulate phosphorylation of p53. To distinguish these possibilities, we examined the levels of Ser1981-phosphorylated ATM and phosphorylated histone 2AX (γH2AX), both considered markers for DNA damage. As shown in Fig. 2A, although NCS greatly induced phosphorylation of both ATM and H2AX, depletion of EDD did not induce ATM or H2AX phosphorylation, at the same time when it could induce p53 Ser15 phosphorylation. In fact, the γH2AX signal was decreased. These data strongly argue against the scenario that EDD depletion causes DNA damage. Because in the unirradiated cells expression of γH2AX is associated with DNA replication and its level is three times lower in G1-phase cells than in S/G2-phase cells (28), the drop of γH2AX level in EDD-depleted cells is consistent with inhibition of S-phase fraction in these cells, as shown in Fig. 1, D and E. Although p53 has been reported to be phosphorylated by stress kinases p38 or JNK at Ser15 residue upon certain stresses (29, 30), both p38 and JNK were not activated by EDD depletion (supplemental Fig. 1); thus, it is unlikely that p53 phosphorylation is induced by p38 or JNK during EDD depletion. Together, these data suggest that EDD directly regulates p53 phosphorylation.
A Role for EDD in the Regulation of p53 Target Gene Expression
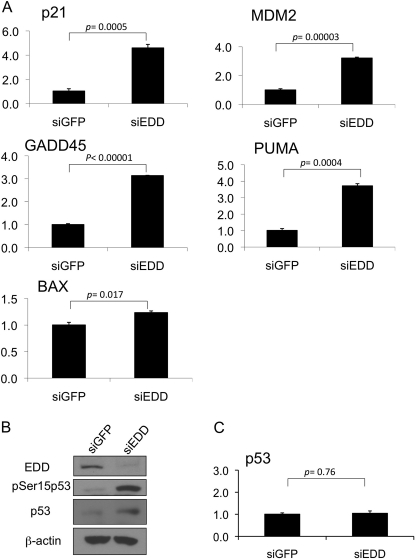
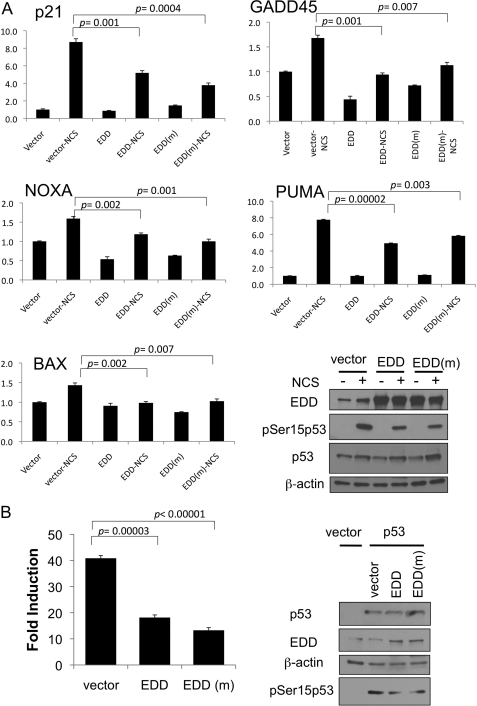
With the well established role for Ser15 phosphorylation in the transactivation function of p53 (31), regulation of Ser15 phosphorylation by EDD suggested that EDD controlled p53 transcriptional activity. We therefore examined the expression of several p53 target genes by real time quantitative RT-PCR in EDD-depleted WI38 cells. Indeed, concurrent with the induction of Ser15 phosphorylation, several of the p53 target genes such as p21, MDM2, GADD45, and PUMA were induced upon EDD depletion (Fig. 3, A and B). BAX was marginally induced. In this experiment, we also measured the levels of p53 transcripts in the control and EDD-depleted cells. The p53 transcript level was not induced by EDD depletion (Fig. 3C). This rules out the possibility that EDD depletion induced p53 transcript. Together with the results presented earlier, these data demonstrate a role for EDD in keeping p53 activity in check during unstressed condition.
FIGURE 3.
Silencing of EDD induces the expression of p53 target genes. A, WI38 cells were infected with Ad-siGFP or Ad-siEDD#5 at a multiplicity of infection of 600. Three days later, RNA was extracted, and quantitative real time PCR analysis was performed using primers specific for the selected p53 target genes or GAPDH as indicated. Results were normalized to GAPDH levels and are expressed relative to the expression of the genes in the siGFP control. The graphs represent means ± standard errors. The p values are based on a paired two-tailed t test. PUMA, p53 up-regulated modulator of apoptosis. B, an aliquot of cell lysates for panel A was analyzed by Western blotting with the indicated antibodies. C, quantitative real time PCR analysis was performed using primers specific for p53 in the same RNA extracted for panel A. Results were analyzed as described for panel A.
EDD Inhibits p53 Ser15 Phosphorylation, and This Function Does Not Require Its E3 Ligase Activity
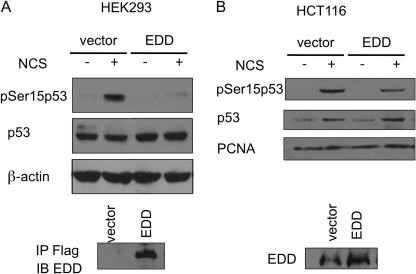
The induction of p53 Ser15 phosphorylation in the absence of DNA-damaging signaling response by EDD depletion suggests that EDD plays an active role in preventing phosphorylation of p53 at Ser15. To further test that, we overexpressed EDD in two cell lines, HEK293 and HCT116, and examined the effect on p53 Ser15 phosphorylation induced by radiomimetic NCS treatment. Indeed, overexpression of EDD inhibited NCS-induced p53 Ser15 phosphorylation in both cell lines (Fig. 4, A and B). It is worth noting that overexpression of EDD did not lead to a decrease in the p53 protein level, suggesting that despite EDD having an E3 ligase activity, it probably does not control p53 degradation. We also examined the effect on p53 target gene expression by EDD overexpression. HCT116 cells were transfected with an EDD expression vector and then treated with NCS to induce the expression of p53 target genes. In addition to the wild-type EDD, we also express an E3-deficient mutant EDD harboring C2768A mutation at the active site cysteine residue, which is necessary for E3 ligase activity in HECT domain proteins (13). This is to investigate whether the E3 ligase activity of EDD is required for p53 regulation. As shown in Fig. 5A, overexpression of EDD inhibited the induction of p53 target genes such as p21, GADD45, NOXA, PUMA, and BAX during NCS treatment. The C2768A mutant EDD was also capable of repressing the expression of p53 target genes. Consistent with these data, C2768A mutant EDD was equally efficient in inhibiting Ser15 phosphorylation (Fig. 5A, right low panels). Finally, we directly measured the p53 transcriptional activity by a p21 promoter-luciferase reporter assay in a p53-null cell line H1299 (21). Indeed, both wild-type and C2768A mutant EDD inhibited p53 transcriptional activity without significantly affecting the p53 protein level (Fig. 5B). This is also consistent with the immunoblotting result obtained from the same lysates as the reporter assay showing that both were able to repress Ser15 phosphorylation (Fig. 5B, right panels). The detectable phosphorylation of Ser15 in this transient transfection assay in the absence of DNA-damaging agents has been reported before (31, 32) and has been attributed to the DNA damage response triggered by transient transfection of DNA (31, 33, 34). Taken together, we conclude that EDD regulates p53-Ser15 phosphorylation and its transcriptional function, and this activity does not require the E3 ligase activity of EDD.
FIGURE 4.
EDD overexpression inhibits phosphorylation of p53 at Ser15 after DNA damage. A, HEK293 cells were transfected by electroporation with EDD plasmid or empty vector. Two days later, cells were either left untreated or treated by NCS for 1.5 h. Cell lysates were subjected to Western blotting analysis with the indicated antibodies. The FLAG-tagged EDD was immunoprecipitated (IP) with anti-FLAG beads and immunoblotted (IB) with EDD antibody (lower panel). B, HCT116 cells were transfected by electroporation with EDD plasmid or empty vector. Two days later, cells were either left untreated or treated by NCS for 2 h. Cell lysates were subjected to Western blotting analysis. PCNA, proliferating cell nuclear antigen.
FIGURE 5.
EDD overexpression inhibits the induction of p53 target genes after DNA damage. A, HCT116 cells were transfected with 20 μg of empty vector, EDD, or EDD mutant (EDD(m)) (C2768A). Two days later, cells were treated with NCS for 2 h. RNA was extracted, and quantitative real time PCR analysis was performed as described in the legend for Fig. 3. A portion of the cellular lysates was immunoblotted with antibody for p53 or EDD as indicated (right lower panels). PUMA, p53 up-regulated modulator of apoptosis. B, H1299 cells were transfected with the expression vectors for p53 and EDD or the EDD mutant (C2768A), along with a p53 activity reporter plasmid (a p21 promoter-luciferase plasmid) and pCMV-βgal. Luciferase activity of transfected p53 was determined as induction relative to that of empty vector control. Each sample was assayed in triplicate. A portion of the cellular lysates was immunoblotted with antibody for p53 or EDD (right panels). The graphs represent means ± standard errors. The p values are based on a paired two-tailed t test.
EDD Interacts with p53
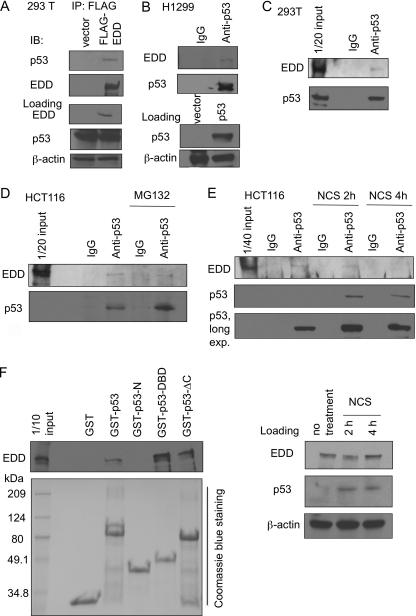
To determine the mechanism by which EDD regulates p53 phosphorylation, we tested whether EDD could interact with p53. When FLAG-EDD was overexpressed in 293T cells, endogenous p53 co-immunoprecipitated with FLAG-EDD (Fig. 6A). We also expressed p53 in a p53-null cell line H1299 and examined their interaction. As shown in Fig. 6B, endogenous EDD was co-immunoprecipitated with p53. We were also able to detect the interaction between EDD and p53, both at the endogenous levels, in 293T cells (Fig. 6C) and HCT116 cells (Fig. 6D). Consistent with the notion that EDD does not target p53 for proteasomal degradation, their interaction was not enhanced upon treatment by a proteasome inhibitor MG132 (Fig. 6D). We also examined their interaction after NCS treatment. Interestingly, although their interaction was still detectable after a 2-h treatment by NCS (Fig. 6E), considering the increase in the p53 protein after NCS treatment, it appears that their interaction is reduced after NCS treatment. The effect became more apparent upon longer treatment (4 h) of NCS. This result implies that EDD may play a role in preventing p53 activation primarily during unstressed condition and maintaining the normal progression of cell cycle. The distribution of EDD and p53 was also examined by immunofluorescence study. As shown in supplemental Fig. 2A, endogenous EDD in H1299 cells is localized diffusely in the nucleus and in the nuclear foci, whereas expressed GFP-p53 is found diffusely in the nucleus. This distribution pattern of EDD is also seen in HCT116 cells, where p53 is also seen diffusely within nucleus in a small population of untreated cells (5) and in more cells after a 1-h treatment of NCS (supplemental Fig. 2B). Although colocalization can occasionally be seen in some foci, their interaction most likely occurs in the nucleoplasm.
FIGURE 6.
Interaction between EDD and p53. A, HEK293T cells were first transfected with FLAG-EDD. FLAG-EDD was then immunoprecipitated (IP) from cell lysates, and the co-immunoprecipitated p53 was detected by immunoblotting (IB). B, lysates from H1299 that were transfected with p53 plasmid were immunoprecipitated with an anti-p53 antibody or a control mouse immunoglobulin G (IgG) antibody as indicated followed by immunoblotting. C, lysates from HEK293T cells were immunoprecipitated with an anti-p53 antibody or a control mouse IgG antibody followed by immunoblotting. One-twentieth of cell lysates before immunoprecipitation were analyzed by Western blotting and probed with p53 and EDD antibodies. D, HCT116 cells were left untreated or treated with MG132. Lysates were immunoprecipitated followed by immunoblotting. E, HCT116 cells were left untreated or treated with NCS for 2 or 4 h. Lysates were immunoprecipitated as described above followed by immunoblotting. F, GST pulldown assay was performed to test the interaction between EDD and purified GST-p53 or its domains: p53-N (N terminus; a.a. 1–70), p53-DBD (DNA binding domain; a.a. 94–292), and p53-ΔCT (lacking C terminus; a.a. 1–292). GST-p53 or its domains were purified as described under “Experimental Procedures” and incubated with HEK293T lysates, respectively.
Their interaction was also apparent when incubating EDD with purified GST-p53 protein and performing the glutathione S-transferase pulldown assay (Fig. 6F). Using this assay, we further investigated the interactions between EDD and several p53 deletional mutants (Fig. 6F). The N terminus of p53 (GST-p53-N, containing a.a. 1–70) did not interact with EDD. The DNA binding domain (DBD) of p53 (GST-p53-DBD) could interact with EDD. A deletion of the C terminus of p53 (GST-p53-ΔC) did not inhibit their interaction when compared with full-length GST-p53. Thus, we conclude that EDD mainly interacts with the DBD of p53.
EDD Regulates p53 Phosphorylation through ATM
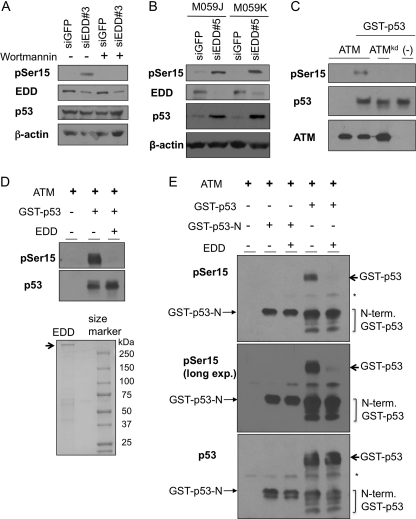
There are three kinases involved in the phosphorylation of p53 at Ser15: ATM, ATR, and DNA-PK. A phosphoinositide kinase-3 (PI3K) inhibitor wortmannin at a concentration of 20 μm completely blocked the phosphorylation of p53 induced by EDD siRNA (Fig. 7A). This concentration of wortmannin inhibits both ATM and DNA-PK, but not ATR in vivo (35). Accumulation of p53 phosphorylation was also observed in DNA-PK-deficient cells, M059J, as in the DNA-PK wild-type control cells M059K (Fig. 7B). These data suggest that EDD regulates ATM-mediated p53 phosphorylation. To directly test this possibility, we carried out an in vitro ATM kinase assay (23). In this assay, HA-ATM purified from 293T cells was able to phosphorylate GST-p53 at Ser15 in vitro, whereas a kinase-dead mutant HA-ATM (ATMkd) lost the activity (Fig. 7C). Incubation of purified EDD (Fig. 7D, lower panel) in this reaction greatly inhibited Ser15 phosphorylation by ATM. This effect most likely requires the binding between EDD and p53 because an N-terminal fragment of p53 (GST-p53-N), which could be effectively phosphorylated by ATM, did not bind to EDD (Fig. 6F), and its phosphorylation was not inhibited by EDD (Fig. 7E). Because GST was N-terminally tagged to p53, some partially degraded p53 proteins containing the N-terminal portions were retained in the glutathione beads and were co-eluted along with full-length protein during protein purification. Phosphorylation of these degradation products by ATM was also not inhibited by EDD, whereas in the same reaction mixture, EDD almost completely blocked the phosphorylation of full-length GST-p53. These data demonstrate that EDD, through binding to p53, prevents ATM from phosphorylating p53 at Ser15.
FIGURE 7.
EDD inhibits phosphorylation of p53 by ATM in vitro. A, WI38 cells infected with Ad-siGFP or Ad-siEDD#3 were left untreated or treated with wortmannin (20 μm). The cell lysates were harvest 1 h later and subjected to Western blot analysis. B, DNA-PK-deficient cell line M059J and its control M059K (DNA-PK wild type) were infected by Ad-siEDD#5 and Ad-siGFP. Three days later, cells were lysed, and p53 phosphorylation level at Ser15 was analyzed by immunoblotting. C, ATM and ATM kinase-dead mutant were immunoprecipitated from 293T cells, which were transfected with HA-ATM or HA-ATMkd construct. The ATM kinase assay was performed as described under “Experimental Procedures” and contained 200 nm GST-p53. Western blots were probed with an antibody against phospho-Ser15 of p53. D, 1 μg of EDD purified from 293T cells was incubated with 10 nm GST-p53 in 4 °C for 30 min first. Kinase assay was then performed as in C. E, 200 ng of EDD or 200 ng of BSA was incubated with 10 nm GST-p53 or GST-p53-N for 30 min. ATM kinase assay was then performed as in C. The top panel represents anti-pSer15-p53 immunoblot; the middle panel is a longer exposure (long exp.) of the same blot; and the low panel was probed with mixed p53 antibodies containing DO1, PAb1801, PAb240, and PAb421, which can recognize both GST-p53-N and GST-p53. * indicates nonspecific signals of IgG heavy chains. N-term GST-p53, N-terminal GST-p53.
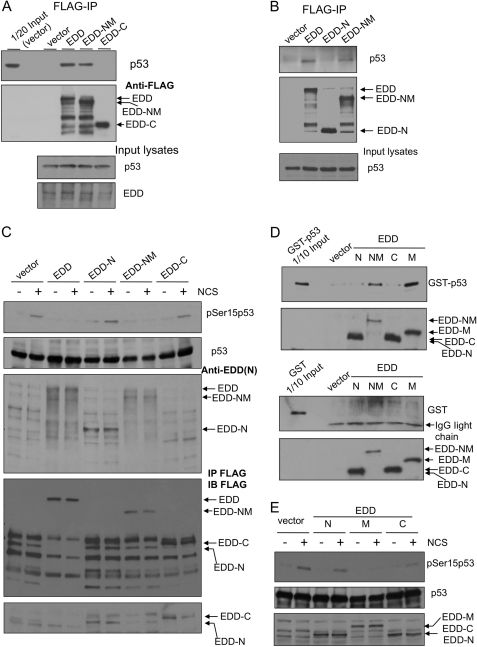
To further establish a role for the binding between EDD and p53 in repressing p53 Ser15 phosphorylation, we carried out domain mapping studies and identified the region of EDD that binds to p53 and represses its phosphorylation. We first examined the interaction between p53 and the N-terminal part (EDD-NM, a.a. 1–1876) or the C terminus (EDD-C, a.a. 1876–2799) of EDD. FLAG-EDD, FLAG-EDD-NM, and FLAG-EDD-C were expressed in HEK293 cells, and the endogenous p53 associated with the FLAG-tagged EDD proteins was isolated by immunoprecipitation with anti-FLAG beads. As shown in Fig. 8A, p53 interacts with EDD-NM, but not EDD-C. An EDD immunoblot (Fig. 8A, lower panel) shows a slightly high level of total EDD in FLAG-EDD-transfected cells when compared with vector-transfected cells, suggesting that the expression of FLAG-EDD was probably comparable with that of the endogenous EDD. We further excluded the N-terminal 888 a.a. of EDD as the responsible region for p53 binding because unlike EDD-NM, EDD-N (a.a. 1–888) failed to bind to p53 as shown by the co-immunoprecipitation assay (Fig. 8B). Next, we overexpressed these deletional EDD mutants in HEK293 cells and examined their activities in repressing p53 Ser15 phosphorylation upon NCS treatment, as in the experiment performed in Fig. 4A. Indeed, as shown in Fig. 8C, full-length EDD and a p53 binding-capable mutant EDD-NM repressed p53 phosphorylation, but the p53 binding-incapable mutants, EDD-N and EDD-C, failed to inhibit p53 Ser15 phosphorylation. To further narrow down the domain responsible for p53 binding and also to verify the binding in an in vitro assay, we incubated purified GST-p53 or GST protein with FLAG-EDD mutants: N, NM, C, and M (a.a. 888–1876), which were expressed and purified by anti-FLAG beads from HEK293 cells. Although GST did not bind to these EDD peptides, GST-p53 interacted only with EDD-NM and EDD-M (Fig. 8D). This result indicates that the region of a.a. 888–1876 in EDD is responsible for p53 binding. At last, we compared the activities of EDD-N, EDD-M, and EDD-C in repressing NCS-induced Ser15 phosphorylation of p53. Consistent with its ability to bind p53, EDD-M repressed p53 phosphorylation (Fig. 8E). Taken together, these data provide compelling evidence supporting a physiological role for EDD/p53 association in the regulation of p53 Ser15 phosphorylation.
FIGURE 8.
Mapping the interaction region of EDD and identifying its role in inhibiting phosphorylation of p53 at Ser15. A, HEK293 cells were transfected with an empty vector control, FLAG-EDD, or FLAG-tagged truncated mutants: EDD-NM (a.a. 1–1876) or EDD-C (C terminus; a.a. 1876–2799). FLAG-EDD and those mutants were immunoprecipitated (IP) from cell lysates, and the co-immunoprecipitated p53 was detected by immunoblotting. B, HEK293 cells were transfected with an empty vector control, FLAG-EDD, or FLAG-tagged truncated mutants: EDD-N (N terminus; a.a. 1–888) or EDD-NM. Lysates were immunoprecipitated with anti-FLAG beads followed by immunoblotting with p53 antibody. C, HEK293 cells were transfected by electroporation with an empty vector, FLAG-EDD, or FLAG-tagged truncated mutants: EDD-N, EDD-NM, or EDD-C. Two days later, cells were either left untreated or treated by NCS for 1 h. Cell lysates were subjected to Western blotting analysis with the indicated antibodies. The EDD antibody was raised against EDD N terminus and therefore do not recognize EDD-C. To show the expression of EDD-C, we immunoprecipitated these lysates with anti-FLAG beads and then immunoblotted (IB) with a FLAG antibody (the lower two panels). Because vector lanes had nonspecific bands (presumably heavy chain IgG) co-migrating with the EDD-N signal, we reprobed the membrane using a light chain specific secondary antibody (the bottom panel). D, HEK293 cells were transfected with FLAG-EDD or FLAG-tagged truncated mutants. The FLAG-tagged peptides were purified by anti-FLAG beads and then incubated with purified GST or GST-p53 (Fig. 6F), and the bound GST-p53 was pulled down by anti-FLAG beads. E, HEK293 cells were transfected by electroporation with an empty vector, FLAG-EDD, or truncated mutants: EDD-N, EDD-M, or EDD-C. Two days later, cells were either left untreated or treated by NCS for 1 h. Cell lysates were subjected to Western blotting analysis with the indicated antibodies. The FLAG-EDD mutants were identified by FLAG antibody.
The Role for EDD in G1/S Entry Is Mediated through p53
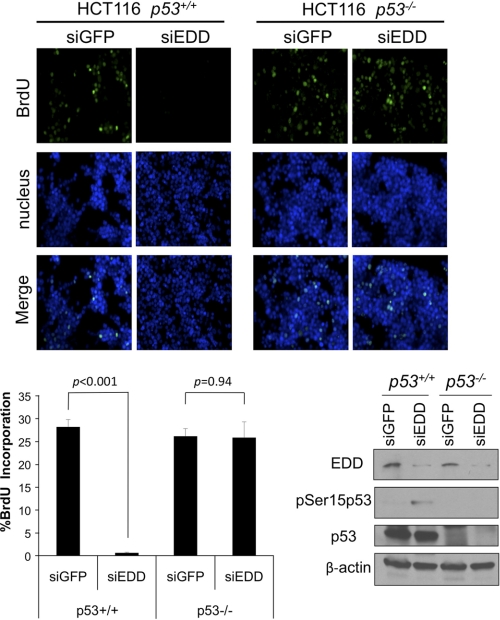
Given the key role of p53 in G1/S control and the regulation of p53 by EDD, we wanted to investigate whether EDD controls p53 for progression of G1/S entry. We infected a pair of isogenic p53+/+ and p53−/− HCT116 cells (36) with Ad-siEDD and examined the effect of EDD depletion on the S-phase fraction as measured by BrdU incorporation. As shown in Fig. 9, consistent with the result obtained in WI38 cells (Fig. 1, C–E), EDD knockdown inhibited the fraction of S-phase cells in p53 wild-type HCT116. This effect was obliterated in p53-null HCT116. Thus, we conclude that the effect of EDD depletion on G1/S entry is operated through p53.
FIGURE 9.
EDD siRNA induces G1/S arrest through p53. HCT116 p53+/+ or p53−/− cells were infected with Ad-siGFP or Ad-siEDD#3 at a multiplicity of infection of 600. Forty-eight hours later, cells were labeled with BrdU for 8 h, and the incorporated BrdU was detected with fluorescein isothiocyanate-conjugated anti-BrdU antibody. Upper panels are representative images of 200× magnification. In the left lower panel, the graphs represent means ± standard errors. At least 300 nuclei were scored on each sample for BrdU incorporation by microscope. The experiments were repeated three times. The p values are based on a paired two-tailed t test. Right lower panels, immunoblots of the lysates from this experiment.
DISCUSSION
With many important functions as the guardian of our genome, p53 is tightly controlled in many layers of regulation. Activation of p53 involves a complex regulatory network including at least phosphorylation and acetylation. It appears that none of the single modifications were able to fully recapitulate the in vivo dramatic activation of p53, and it has recently been suggested that the summation of these events leading to the release of p53 from repression factors such as MDM2 is the key step for p53 activation (37). Nonetheless, phosphorylation by ATM/ATR at Ser15 is considered to be an initial and important event for the activation of p53 in response to genotoxic stresses. On the other hand, phosphorylation of Ser15 may need to be reversed once the damaged DNA is repaired to resume the cellular proliferation. Wild-type p53-induced phosphatase 1 (Wip1, also named as protein phosphatase, Mg2+/Mn2+-dependent, 1D, or PPM1D) (38) and protein serine/threonine phosphatase-1 (PP-1) (39) have been identified to be responsible for the removal of Ser15 phosphorylation in p53. Here, we report another mode of regulation on this residue. By binding to the p53, EDD can directly block the activity of ATM in phosphorylating p53. With the constitutive expression of EDD throughout all phases of the cell cycle, it may function in policing p53 activity and constantly inhibiting the activation of p53 unless there are stressful stimuli. This regulation may be important for normal cellular proliferation because a constitutively active population of ATM kinase exists in normal cells even in the absence of exogenous damage (40, 41). Using quantitative time-lapse microscopy, spontaneous pulses of p53 with similar amplitude and duration as in sustained damage were detected in proliferating cells, probably derived from intrinsic DNA damage associated with normal growth (5). The activities of these pulses of p53 have been actively suppressed unless sustained severe damage changes the p53 modification state and releases p53 from suppression (5). Therefore, EDD may play an active repressive role to block p53 activity during normal growth. In the event of DNA damage, reduction of EDD and p53 binding and the accumulation of p53 protein and modifications might overwhelm the inhibitory effect by EDD. Although there are many proteins known to interact with p53 (42), to our knowledge, EDD is the only protein identified so far that directly inhibits p53 phosphorylation by ATM.
The interaction between EDD and p53 observed both inside the cells and in vitro suggests that EDD interacts with p53 and inhibits its phosphorylation. This mechanism is supported by the observation that EDD cannot inhibit ATM to phosphorylate an N-terminal fragment of p53, which does not bind EDD. The correlation between the ability of binding to p53 and that of inhibiting p53 phosphorylation among four EDD truncated mutants further strengthens this argument. We postulate that EDD might block the access of ATM to p53 through steric hindrance, given the fact that both EDD and ATM are near 350 kDa.
Recently, EDD was reported to regulate DNA damage checkpoints (44), and play a role during mitotic progression through its E3 ligase activity toward katanin (10). Here, we extend the involvement of EDD in normal cell cycle control to G1/S progression and provide evidence supporting a role of EDD in the regulation of p53 Ser15 phosphorylation. Although EDD contains a HECT domain with E3 ligase activity, an E3 ligase catalytic mutant EDD still suppresses p53 activity to the same degree as wild type. This is consistent with the notion that the binding of p53 by EDD somehow imposes physical hindrance to ATM. Furthermore, the interaction between the p53 and EDD is not enhanced by MG132, a proteasome inhibitor, implying that the proteasomal degradation pathway is not involved in the regulation. Other E3-independent functions of EDD are also identified. For example, EDD was reported to regulate progesterone receptor and myocardin through their physical interactions independently of its E3 ligase activity (13, 14). In these cases, EDD functions as a transcription coactivator with progesterone receptor and myocardin. It deserves further investigation to unveil the full scope of EDD activities.
Our findings may have implications in clinical cancer biology as well. EDD is frequently amplified and overexpressed in many types of cancer, including ovarian cancer, breast cancer, hepatocellular carcinoma, squamous cell carcinoma of the tongue, and metastatic melanoma (20). Furthermore, high nuclear EDD expression in serous ovarian carcinoma is associated with adverse prognosis, and depletion of EDD inhibits the growth of cisplatin-resistant ovarian cancer cells and partially restores their platinum sensitivity (43). It would be interesting to investigate whether the association of EDD overexpression with poor clinical outcome in cancer patients is related to its activity in suppressing p53 activation.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of wild-type and mutant EDD constructs from Drs. Michelle Henderson and Charlie Watts. We also thank Dr. Xinbin Chen for p53 antibodies and Bert Vogelstein for p53−/− and p53+/+ HCT116 cells.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1CA100857, RO1CA138641 (to W. C. L.), and ARRA 3 P30CA125123-03S5 and the Department of Defense Breast Cancer Research Program (Grant W81XWH-09-1-0338) (to W. C. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- ATM
- ataxia telangiectasia mutated
- ATR
- ataxia telangiectasia and Rad3-related
- DNA-PK
- DNA-dependent protein kinase
- EDD
- E3 identified by differential display
- HECT
- homologous to E6-AP carboxyl terminus
- NCS
- neocarzinostatin
- a.a.
- amino acids
- Chk2
- checkpoint kinase 2
- γH2AX
- phosphorylated histone 2AX
- DBD
- DNA binding domain.
REFERENCES
- 1. Appella E., Anderson C. W. (2001) Eur. J. Biochem. 268, 2764–2772 [DOI] [PubMed] [Google Scholar]
- 2. Lavin M. F., Kozlov S. (2007) Cell Cycle 6, 931–942 [DOI] [PubMed] [Google Scholar]
- 3. Okorokov A. L. (2003) Cell Cycle 2, 233–235 [PubMed] [Google Scholar]
- 4. Kruse J. P., Gu W. (2008) Cell 133, 930.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Loewer A., Batchelor E., Gaglia G., Lahav G. (2010) Cell 142, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Callaghan M. J., Russell A. J., Woollatt E., Sutherland G. R., Sutherland R. L., Watts C. K. (1998) Oncogene 17, 3479–3491 [DOI] [PubMed] [Google Scholar]
- 7. Mansfield E., Hersperger E., Biggs J., Shearn A. (1994) Dev. Biol. 165, 507–526 [DOI] [PubMed] [Google Scholar]
- 8. Lee J. D., Amanai K., Shearn A., Treisman J. E. (2002) Development 129, 5697–5706 [DOI] [PubMed] [Google Scholar]
- 9. Saunders D. N., Hird S. L., Withington S. L., Dunwoodie S. L., Henderson M. J., Biben C., Sutherland R. L., Ormandy C. J., Watts C. K. (2004) Mol. Cell Biol. 24, 7225–7234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maddika S., Chen J. (2009) Nat. Cell Biol. 11, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshida M., Yoshida K., Kozlov G., Lim N. S., De Crescenzo G., Pang Z., Berlanga J. J., Kahvejian A., Gehring K., Wing S. S., Sonenberg N. (2006) EMBO J. 25, 1934–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohshima R., Ohta T., Wu W., Koike A., Iwatani T., Henderson M., Watts C. K., Otsubo T. (2007) Genes Cells 12, 1339–1345 [DOI] [PubMed] [Google Scholar]
- 13. Henderson M. J., Russell A. J., Hird S., Muñoz M., Clancy J. L., Lehrbach G. M., Calanni S. T., Jans D. A., Sutherland R. L., Watts C. K. (2002) J. Biol. Chem. 277, 26468–26478 [DOI] [PubMed] [Google Scholar]
- 14. Hu G., Wang X., Saunders D. N., Henderson M., Russell A. J., Herring B. P., Zhou J. (2010) J. Biol. Chem. 285, 11800–11809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 16. Mu J. J., Wang Y., Luo H., Leng M., Zhang J., Yang T., Besusso D., Jung S. Y., Qin J. (2007) J. Biol. Chem. 282, 17330–17334 [DOI] [PubMed] [Google Scholar]
- 17. Honda Y., Tojo M., Matsuzaki K., Anan T., Matsumoto M., Ando M., Saya H., Nakao M. (2002) J. Biol. Chem. 277, 3599–3605 [DOI] [PubMed] [Google Scholar]
- 18. Henderson M. J., Munoz M. A., Saunders D. N., Clancy J. L., Russell A. J., Williams B., Pappin D., Khanna K. K., Jackson S. P., Sutherland R. L., Watts C. K. (2006) J. Biol. Chem. 281, 39990–40000 [DOI] [PubMed] [Google Scholar]
- 19. McDonald W. J., Sangster S. M., Moffat L. D., Henderson M. J., Too C. K. (2010) J. Cell Biochem. 110, 1123–1129 [DOI] [PubMed] [Google Scholar]
- 20. Clancy J. L., Henderson M. J., Russell A. J., Anderson D. W., Bova R. J., Campbell I. G., Choong D. Y., Macdonald G. A., Mann G. J., Nolan T., Brady G., Olopade O. I., Woollatt E., Davies M. J., Segara D., Hacker N. F., Henshall S. M., Sutherland R. L., Watts C. K. (2003) Oncogene 22, 5070–5081 [DOI] [PubMed] [Google Scholar]
- 21. Liu K., Bellam N., Lin H. Y., Wang B., Stockard C. R., Grizzle W. E., Lin W. C. (2009) Mol. Cell Biol. 29, 2673–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin W. C., Lin F. T., Nevins J. R. (2001) Genes Dev. 15, 1833–1844 [PMC free article] [PubMed] [Google Scholar]
- 24. Liu K., Lin F. T., Ruppert J. M., Lin W. C. (2003) Mol. Cell Biol. 23, 3287–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu K., Paik J. C., Wang B., Lin F. T., Lin W. C. (2006) EMBO J. 25, 4795–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Banin S., Moyal L., Shieh S., Taya Y., Anderson C. W., Chessa L., Smorodinsky N. I., Prives C., Reiss Y., Shiloh Y., Ziv Y. (1998) Science 281, 1674–1677 [DOI] [PubMed] [Google Scholar]
- 27. Canman C. E., Lim D. S., Cimprich K. A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M. B., Siliciano J. D. (1998) Science 281, 1677–1679 [DOI] [PubMed] [Google Scholar]
- 28. MacPhail S. H., Banáth J. P., Yu Y., Chu E., Olive P. L. (2003) Radiat. Res. 159, 759–767 [DOI] [PubMed] [Google Scholar]
- 29. Cheng W. H., Zheng X., Quimby F. R., Roneker C. A., Lei X. G. (2003) Biochem. J. 370, 927–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perfettini J. L., Castedo M., Nardacci R., Ciccosanti F., Boya P., Roumier T., Larochette N., Piacentini M., Kroemer G. (2005) J. Exp. Med. 201, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dumaz N., Meek D. W. (1999) EMBO J. 18, 7002–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dohoney K. M., Guillerm C., Whiteford C., Elbi C., Lambert P. F., Hager G. L., Brady J. N. (2004) Oncogene 23, 49–57 [DOI] [PubMed] [Google Scholar]
- 33. Huang L. C., Clarkin K. C., Wahl G. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4827–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Renzing J., Lane D. P. (1995) Oncogene 10, 1865–1868 [PubMed] [Google Scholar]
- 35. Sarkaria J. N., Tibbetts R. S., Busby E. C., Kennedy A. P., Hill D. E., Abraham R. T. (1998) Cancer Res. 58, 4375–4382 [PubMed] [Google Scholar]
- 36. Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. (1998) Science 282, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 37. Kruse J. P., Gu W. (2009) Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu X., Nannenga B., Donehower L. A. (2005) Genes Dev. 19, 1162–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li D. W., Liu J. P., Schmid P. C., Schlosser R., Feng H., Liu W. B., Yan Q., Gong L., Sun S. M., Deng M., Liu Y. (2006) Oncogene 25, 3006–3022 [DOI] [PubMed] [Google Scholar]
- 40. Bartkova J., Bakkenist C. J., Rajpert-De Meyts E., Skakkebaek N. E., Sehested M., Lukas J., Kastan M. B., Bartek J. (2005) Cell Cycle 4, 838–845 [DOI] [PubMed] [Google Scholar]
- 41. Tanaka T., Halicka H. D., Huang X., Traganos F., Darzynkiewicz Z. (2006) Cell Cycle 5, 1940–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vousden K. H., Prives C. (2009) Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 43. O'Brien P. M., Davies M. J., Scurry J. P., Smith A. N., Barton C. A., Henderson M. J., Saunders D. N., Gloss B. S., Patterson K. I., Clancy J. L., Heinzelmann-Schwarz V. A., Murali R., Scolyer R. A., Zeng Y., Williams E. D., Scurr L., Defazio A., Quinn D. I., Watts C. K., Hacker N. F., Henshall S. M., Sutherland R. L. (2008) Br. J. Cancer. 98, 1085–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Munoz M. A., Saunders D. N., Henderson M. J., Clancy J. L., Russell A. J., Lehrbach G., Musgrove E. A., Watts C. K., Sutherland R. L. (2007) Cell Cycle 6, 3070–3077 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.