Abstract
Biochemical characterization of intermediates involved in the assembly of the oxygen-evolving Photosystem II (PSII) complex is hampered by their low abundance in the membrane. Using the cyanobacterium Synechocystis sp. PCC 6803, we describe here the isolation of the CP47 and CP43 subunits, which, during biogenesis, attach to a reaction center assembly complex containing D1, D2, and cytochrome b559, with CP47 binding first. Our experimental approach involved a combination of His tagging, the use of a D1 deletion mutant that blocks PSII assembly at an early stage, and, in the case of CP47, the additional inactivation of the FtsH2 protease involved in degrading unassembled PSII proteins. Absorption spectroscopy and pigment analyses revealed that both CP47-His and CP43-His bind chlorophyll a and β-carotene. A comparison of the low temperature absorption and fluorescence spectra in the QY region for CP47-His and CP43-His with those for CP47 and CP43 isolated by fragmentation of spinach PSII core complexes confirmed that the spectroscopic properties are similar but not identical. The measured fluorescence quantum yield was generally lower for the proteins isolated from Synechocystis sp. PCC 6803, and a 1–3-nm blue shift and a 2-nm red shift of the 77 K emission maximum could be observed for CP47-His and CP43-His, respectively. Immunoblotting and mass spectrometry revealed the co-purification of PsbH, PsbL, and PsbT with CP47-His and of PsbK and Psb30/Ycf12 with CP43-His. Overall, our data support the view that CP47 and CP43 form preassembled pigment-protein complexes in vivo before their incorporation into the PSII complex.
Keywords: Membrane Biogenesis, Membrane Enzymes, Membrane Proteins, Photosynthesis, Protein Assembly, Photosystem II, Chlorophyll Proteins, Thylakoid Membrane
Introduction
Photosystem II (PSII)3 is the light-driven water:plastoquinone oxidoreductase of oxygenic photosynthesis, responsible for producing most of the oxygen in the atmosphere (1). It is located in the thylakoid membrane of chloroplasts and cyanobacteria and is a multisubunit lipoprotein complex composed of both intrinsic and extrinsic proteins. Crystal structures of dimeric PSII protein complexes isolated from the thermophilic cyanobacteria Thermosynechococcus elongatus (2–5) and Thermosynechococcus vulcanus (6, 7) have revealed the organization of the 20 subunits within each monomeric complex and the positions of the various cofactors. These include 35 chlorophyll (Chl) a molecules, two pheophytin a molecules, 12 carotenoids, two heme molecules, one non-heme iron, two calcium ions, two chloride ions, three plastoquinones, 25 lipids, and the Mn4Ca cluster, which catalyzes water oxidation (4).
We are interested in understanding how PSII is assembled from its individual components. Current models suggest a stepwise assembly in both cyanobacteria and chloroplasts involving distinct intermediates (8–10). However, as with other membrane protein complexes, detailed analysis of PSII assembly complexes is hindered by their low abundance in the membrane, and until now, early assembly intermediates of PSII have not been isolated and biochemically characterized.
Here, we describe the isolation of the CP47 and CP43 subunits from the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803). These PSII subunits each contain six transmembrane α-helices and, in the cyanobacterial PSII holoenzyme, bind Chl a (16 molecules in CP47 and 13 in CP43) and β-carotene (4). They lie on either side of the heterodimeric D1/D2 reaction center complex involved in light-induced transmembrane charge separation, and one of their roles is to act as an inner light-harvesting antenna system (11). CP43 is also involved in ligating the Mn4Ca cluster (3). CP47 and CP43 are tightly bound components of the larger PSII core complex, and relatively harsh treatments are required to remove CP47 and CP43 in vitro (12, 13).
It is known that the apopolypeptides of both CP47 (encoded by the psbB gene) and CP43 (encoded by psbC) are synthesized and inserted into the thylakoid membrane before their incorporation into the PSII complex of Synechocystis 6803 (8). However, it remains unclear whether they are able to bind pigment molecules in this “unassembled” state (10). To address this issue, we describe here a novel approach for the isolation and characterization of CP47 and CP43 complexes from Synechocystis 6803. Our results show that both CP47 and CP43 are likely to bind a largely complete set of pigments and are able to attach to neighboring low molecular mass (LMM) subunits before assembling into larger PSII complexes.
EXPERIMENTAL PROCEDURES
Cyanobacterial Strains and Growth Conditions
The glucose-tolerant strain of Synechocystis 6803 (14) and the psbA triple deletion strain, TD41 (15), were used in this work. For clarity, TD41 will be referred to here as ΔD1. Strains were grown in liquid BG-11 mineral medium and maintained on solid BG-11 plates containing 1.5% (w/v) agar, both containing 5 mm TES-KOH (pH 8.2) at a light intensity of 40 or 5 microeinstein m−2 s−1 of white fluorescent light, respectively, and at 29 °C. The medium was supplemented with 5 mm glucose and, where applicable, chloramphenicol (30 μg ml−1), erythromycin (10 μg ml−1), gentamycin (2 μg ml−1), kanamycin (50 μg ml−1), or spectinomycin (50 μg ml−1).
Construction of Mutants
To generate the His-tagged CP47 strain (ΔD1/CP47-His/ΔFtsH2), the gentamycin resistance cassette of plasmid pCP47His-tagGmR (16) was removed by BamHI digestion. After blunting the ends, an erythromycin resistance cassette was introduced to generate pCP47His-tagEryR. Ultimately, this plasmid was transformed into the ΔD1/ΔFtsH2 strain, which had been generated by transforming the ΔD1 mutant strain (15) with the plasmid used to produce the Syn0228GENT strain described previously (17). A control strain expressing His-tagged CP47 in a WT background (strain PSII-His) was generated by transforming the WT with pCP47His-tagEryR. To generate the His-tagged CP43 Synechocystis 6803 mutant strain (ΔD1/CP43-His), a His6-coding sequence was introduced at the 3′-end of the psbC gene (sll0851) by overlap extension PCR using the following primers: CP43+1000-Fw, 5′-ATATTTTCCCCTTCTTCGTAGGGGTGC-3′; CP43+1000-Rev, 5′-CTGCCATTAAAGAATTGGCTAAAGAAGCAGGTC-3′; CP43HT-Fw, 5′-CATCATCATCATCATCATTAGATTGAGACTTTTCTGATTTTGCAA-3′; and CP43HT-Rev, 5′-CTAGTAGTAGTAGTAGTAGTAGTCGAGGTCAGGCATGAACAA-3′. The overlap extension PCR product was cloned into the pGEMTeasy vector (Promega), and an erythromycin resistance cassette was introduced at an XmnI site 170 bp downstream of the stop codon of the gene. The genotypes of the mutants were verified by PCR analysis using gene-specific primers.
Isolation of CP47-His and CP43-His
CP47-His and CP43-His complexes were isolated from 10-liter cultures of the respective Synechocystis 6803 mutant strain that had been grown to stationary phase, concentrated using a cell harvester, and pelleted by centrifugation. Cells were then washed and finally resuspended in KPN buffer (40 mm potassium phosphate (pH 8.0) and 100 mm NaCl) containing EDTA-free Complete protease inhibitor (Roche) to a Chl a concentration of 1 mg ml−1. After two passages at 1250 p.s.i. through a prechilled French press cell (Amicon), unbroken cells were removed by centrifugation. Subsequently, crude membranes were pelleted by ultracentrifugation and resuspended in KPNG buffer (KPN buffer supplemented with 10% (v/v) glycerol). Solubilization of the sample was performed under gentle stirring for 30 min on ice after the addition of one-ninth of the sample volume of 10% (w/v) n-dodecyl β-d-maltoside (β-DM) in KPN buffer. Unsolubilized material was removed by ultracentrifugation, and the supernatant was incubated for 1 h at 4 °C on an end-over-end rotational wheel with nickel-iminodiacetic acid His affinity purification resin (Generon). After incubation, unbound material was removed, and the resin was washed with KPNGD buffer (KPNG buffer containing 0.04% (w/v) β-DM) until the flow-through became colorless (typically 40 column volumes). The resin was then washed with 10 column volumes each of KPNGD buffer containing increasing concentrations of imidazole (5, 10, and 20 mm). The His-tagged proteins were eluted using 10 and 5 column volumes of 50 and 100 mm imidazole in KPNGD buffer, respectively. These elution samples were then pooled and concentrated using 10-kDa molecular mass cutoff protein concentrators (Sartorius). In a second purification step, 500-μl aliquots containing ∼200 μg of Chl a were applied to a Superdex 200 FPLC column (GE Healthcare) operated at a flow rate of 1 ml min−1 with KPN buffer containing 0.04% (w/v) β-DM as the running buffer. The run was monitored at 280 nm using a Jasco MD-2015 Plus diode array detector, and 2-ml fractions were collected by a Frac-920 fraction collector (GE Healthcare). Selected fractions were pooled, supplemented with 10% (v/v) glycerol, and concentrated using 10-kDa molecular mass cutoff protein concentrators. Typically, the yield of CP47-His and CP43-His was ∼0.5–1.0 mg of chlorophyll/10-liter culture.
Isolation of His-tagged PSII Complexes
As a control, His-tagged PSII was purified from strain PSII-His using the same immobilized metal affinity chromatography purification protocol as used for CP43-His and CP47-His except that a 100-kDa molecular mass cutoff concentrator was used to concentrate samples. A 500-μl aliquot containing ∼200 μg of Chl a was then applied to 5 ml of TOYOPEARL 650S DEAE anion exchange chromatography resin (Anachem) in a second purification step. The anion exchange chromatography run was performed at a flow rate of 0.5 ml min−1 with KPN buffer containing 0.04% (w/v) β-DM as the running buffer. Initially, for the first 10 min, the running buffer also contained 5 mm MgSO4, and over the next 50 min, the concentration of MgSO4 was raised linearly to 200 mm. Fractions containing PSII were pooled, supplemented with glycerol to a final concentration of 10% (v/v) and concentrated using 100-kDa molecular mass cutoff protein concentrators.
Isolation of Spinach CP47 and CP43
Samples were prepared from larger PSII core complexes using the methods described previously (18, 19).
Protein Analysis
The Chl a content of samples was determined by extraction into methanol and absorption measurements at 666 and 750 nm (20). Protein samples were analyzed by Blue native (BN)-PAGE and SDS-PAGE as described previously (21). Unless stated otherwise, BN-8–12% (w/v) polyacrylamide and 18% (w/v) SDS-polyacrylamide gels containing 6 m urea were used. The resulting gels were either Coomassie Blue- or silver-stained (22) or electroblotted onto PVDF membrane using the iBlot system (Invitrogen) according to the manufacturer's instructions. Immunoblot analyses were performed using specific primary antibodies and a horseradish peroxidase-conjugated secondary antibody (GE Healthcare). Signals were visualized using a SuperSignal West Pico chemiluminescence kit (Pierce). The primary antibodies used in this study were as follows: anti-CP43 (directed against Chlamydomonas reinhardtii PsbC, serum P6, kindly provided by B. Diner), anti-CP47 (directed against barley PsbB (residues 380–394)), anti-D1 (directed against the C-terminal peptide) (23), anti-D2 (directed against the C-terminal peptide) (24), anti-His tag (Invitrogen), and anti-PsbH (from Synechocystis 6803) (25).
Reverse-phase HPLC Pigment Analysis
Pigments were extracted into 80% (v/v) acetone/water at 4 °C under dim light conditions, and the sample was centrifuged at maximum speed for 1 min in a microcentrifuge to pellet precipitated proteinaceous material before injection. The injection volume was 20 μl, and the pigments were resolved using a Gemini 5-μm C6-phenyl 110-Å column (Phenomenex). The following program was run at a flow rate of 1 ml min−1: 0–4 min at 100% system B, 4–20 min linear gradient from 100% system B to 40% system B and 60% system C, 20–24 min at 40% system B and 60% system C, 24–35 min at 100% system B, with system A being 65% (v/v) acetonitrile and 35% (v/v) water, system B being 90% (v/v) acetonitrile and 10% (v/v) water, and system C being 100% ethyl acetate. The run was monitored at 440 nm using a Jasco MD-2015 Plus diode array detector. Chl a and β-carotene quantification was performed after calibration with pigment standards (purchased from Sigma) of known concentrations.
Spectroscopic Analyses
CP43-His and CP47-His samples were diluted in buffer containing 20 mm BisTris-HCl (pH 6.5), 20 mm NaCl, 0.09% (w/v) β-DM, and 75% (v/v) glycerol. The samples for the absorption experiments were diluted to an absorbance of ∼0.5 cm−1 at the QY maxima, whereas for the fluorescence experiments, an absorbance if ≤0.1 cm−1 was used. The low temperature measurements were performed in an Utreks helium flow cryostat (5 K) or in an Oxford liquid nitrogen bath cryostat (77 K). Absorption spectra at 5 K were recorded in a home-built setup equipped with a 150-watt tungsten-halogen lamp, a monochromator, and a photodiode detector. The spectra were recorded using lock-in detection with 1-nm spectral resolution. Absorption spectra at 77 K were recorded in a PerkinElmer Lambda 40 UV-visible spectrophotometer with 1-nm spectral resolution. Fluorescence spectra at 77 K were recorded in a Jobin Yvon Fluorolog-3-11 fluorometer. The excitation wavelength was 488 nm (10-nm full-width half-maximum), and the spectral resolution was 1 nm. The fluorescence quantum yield was estimated by integrating the total fluorescence and comparing with the known value of the fluorescence quantum yield for higher plant CP43 at room temperature (19).
Mass Spectrometric Analysis
Low molecular mass proteins were identified using the methods described previously (26). For offline electrospray ionization MS, 20 μl of purified CP47-His and CP43-His complexes, respectively, were precipitated overnight in 80% (v/v) acetone/water at −20 °C. After centrifugation at 13,000 × g for 10 min, the supernatant was discarded, and the pellet was air-dried. The dry pellet was dissolved in solution containing 70% (v/v) acetone, 19% (v/v) water, 10% (v/v) 2-propanol, and 1% (v/v) formic acid. Dissolved proteins were directly applied to a nanospray emitter. Mass spectra were obtained using a Waters Q-Tof Premier spectrometer equipped with a nanoelectrospray ionization source. For scanning LMM proteins, mass spectra at a mass range of 800–2500 m/z were acquired. Data were recorded at a capillary voltage of 0.8 kV and a cone voltage of 37 V. After MS data collection at a rate of 1 s/scan, scans were averaged. After acquisition of fragment ion spectra at a collision energy between 26 and 40 eV, data were analyzed using MassLynx/BioLynx 4.1 software. Sequence tags obtained from the fragment spectra were used for similarity search (EMBL-EBI Data Bank). The samples were analyzed in several repetitions.
RESULTS
Isolation of CP47-His and CP43-His from Synechocystis 6803
The low abundance of PSII assembly intermediates in WT Synechocystis 6803 is a major barrier to their isolation. We have adopted a 3-fold approach to aid purification of unassembled CP47 and CP43 complexes. First, we incorporated His6-tags at the C terminus of each protein to allow purification by immobilized metal affinity chromatography. Previous work has already shown that the presence of His-tags at these positions does not prevent assembly of active PSII (27, 28). Second, we blocked assembly of the PSII holoenzyme using a parental strain, ΔD1, which is unable to make the D1 reaction center subunit (8, 15), and finally, in the case of CP47, we also inactivated the membrane-bound FtsH2 protease (Slr0228), which leads to elevated levels of unassembled PSII subunits, including the CP47 apopolypeptide, in the ΔD1 strain (17). Construction and validation of the ΔD1/CP43-His and ΔD1/CP47-His/ΔFtsH2 strains are described under “Experimental Procedures.”
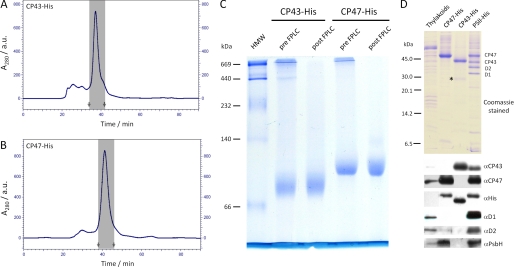
The CP43-His and CP47-His complexes were successfully purified from detergent-solubilized thylakoid membranes by immobilized metal affinity chromatography (supplemental Fig. 1). Small amounts of residual high molecular mass contaminants were removed by a subsequent size exclusion chromatography step (Fig. 1, A and B). Both CP47-His and CP43-His were largely monodisperse as assessed by BN-PAGE (Fig. 1C).
FIGURE 1.
Isolation and protein analysis of CP47-His and CP43-His isolated from mutants of Synechocystis 6803 blocked at an early stage of PSII assembly. CP43-His (A) and CP47-His (B) were purified by Ni2+ affinity chromatography (supplemental Fig. 1), followed by FPLC size exclusion chromatography. Shown are elution profiles of the FPLC size exclusion runs monitored at 280 nm, with the fractions that were later pooled marked by gray areas (start and stop indicated by arrows). a.u., absorbance units. C, CP43-His and CP47-His samples before (pre-FPLC) and after (post-FPLC) purification by FPLC size exclusion chromatography were analyzed on a BN-8–12% (w/v) polyacrylamide linear gradient gel. Samples corresponding to 0.5 μg of Chl a were loaded per lane. A high molecular weight marker (HMW; GE Healthcare) was used to calibrate the gel. D, Coomassie Blue-stained 18% (w/v) SDS-polyacrylamide gel of the isolated proteins and immunoblot analyses with the indicated antibodies. A low molecular mass marker (GE Healthcare) was used to calibrate the gel. WT thylakoid membranes (0.5 μg of Chl a) and final samples of CP47-His and CP43-His (1 μg of Chl a) and PSII-His (1 μg of Chl a) were loaded on the gels. A minor degradation product of CP43-His is indicated by the asterisk.
Protein Composition
SDS-PAGE combined with immunoblot analyses confirmed the purity of each preparation and the presence of His-tagged CP47 and CP43 (Fig. 1D). In the case of CP47-His, we were also able to detect the presence of PsbH, a neighboring low molecular mass subunit in the PSII holoenzyme (Fig. 1D). Co-purification of PsbH and CP47-His was further demonstrated by two-dimensional BN-PAGE (supplemental Fig. 2). Together, these data confirm earlier conclusions that CP47 and PsbH are capable of forming a complex at an early stage in PSII assembly (25). Interestingly, despite the absence of D1, the D2 reaction center subunit, which interacts with CP47 in the holoenzyme, was also detected in the CP47-His complex, albeit at substoichiometric levels (Fig. 1D). Analysis by mass spectrometry confirmed the presence of PsbH and also identified PsbL and PsbT (Table 1). In the case of CP43-His, low levels of a CP43 degradation product were detected by immunoblotting (designated by an asterisk in Fig. 1D) (data not shown), and the PsbK and Psb30/Ycf12 subunits were identified by mass spectrometry (Table 1).
TABLE 1.
LMM proteins detected in CP43-His and CP47-His complexes by mass spectrometry
| Gene | Accession number | Detected signals (charge state) | De novo sequenced protein fragment | Total sequence coverage | |
|---|---|---|---|---|---|
| % | |||||
| CP47-His complex | |||||
| PsbH | P14835 | 998.56 ([M + 6H]6+) | PVMGVFMALFLVFL | 38 | |
| 1001.57 ([M + 6H]6+) | PVMGVFMALFLVFL | ||||
| 1003.85([M + 6H]6+) | MGVFMALFLVFLL | ||||
| 1004.70 ([M + 6H]6+) | TPVMGVFMALFLVF | ||||
| 1006.99 ([M + 6H]6+) | VPGWGTTPVMGVFMA | ||||
| 1171.16 ([M + 5H]5+) | TTPVMGVFMALFLVFLLII | ||||
| 1397.41 ([M + 5H]5+) | GTTPVMGVFMALFLVFL | ||||
| PsbL | Q55354 | 1118.64 ([M + 4H]4+) | LLVAVLGILFSSYF | 36 | |
| 1246.45 ([M + 3H]3+) | LLLVAVLGI | ||||
| PsbT | P74787 | 892.53 ([M + 4H]4+) | LVLTMALAVLF | 52 | |
| 1189.33 ([M + 3H]3+) | SVAYILVLT | ||||
| CP43-His complex | |||||
| PsbK | P73676 | 1050.15 ([M + 4H]4+) | YQIFDPLVDVL | 24 | |
| 1055.64 ([M + 4H]4+) | DPLVDVL | ||||
| Psb30/Ycf12 | Q55438 | 1202.76 ([M + 3H]3+) | IVLAG | 13 | |
Pigment Composition
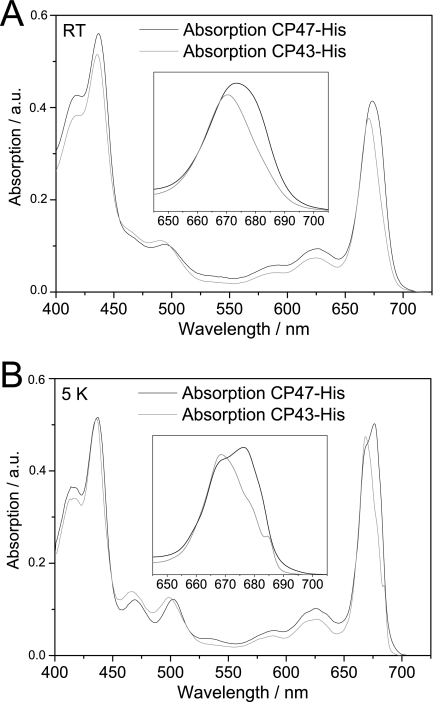
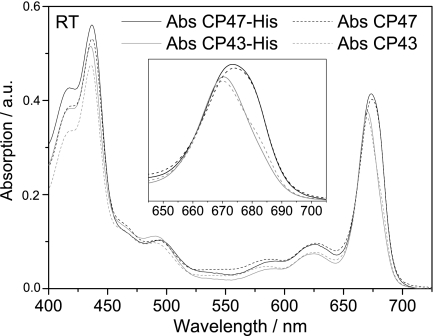
A notable feature of both the CP47-His and CP43-His complexes was the presence of bound pigment. Reverse-phase HPLC pigment analyses confirmed the presence of both Chl a and β-carotene in CP47-His and CP43-His complexes (supplemental Fig. 3), with Chl/carotenoid ratios of ∼7.7 for CP47-His and 5.0 for CP43-His. The wavelengths of maximum absorption in the red region (λmax) measured at room temperature were 674 nm for CP47-His and 671 nm for CP43-His (Fig. 2A), close to the values reported for CP47 and CP43 isolated by fragmentation of larger complexes (13). The relative amplitudes of the Soret and carotenoid absorption bands in the room temperature absorption spectra for CP47-His and CP43-His were also quite similar to their spinach counterparts (Fig. 3), consistent with similar Chl/carotenoid ratios in each type of complex.
FIGURE 2.
CP43 and CP47 absorption spectra at room temperature (A) and 5 K (B). The spectra were normalized to the Chl content in the QY region from 645 to 710 nm assuming 13 Chl molecules in CP43 and 16 Chl molecules in CP47 (3, 5, 40). The insets show an enlargement of the absorption in the QY region. RT, room temperature; a.u., absorbance units.
FIGURE 3.
Comparison of the room temperature absorption spectra of CP43-His and CP47-His isolated from Synechocystis 6803 and CP43 and CP47 isolated from spinach. RT, room temperature; Abs, absorbance; a.u., absorbance units.
Absorption at 5 K and Comparison with Spinach
To assess the optical properties of the pigments bound to CP47-His and CP43-His in more detail, we measured low temperature absorption spectra at 5 K and compared them with those of the widely studied CP47 and CP43 complexes isolated by detachment from larger spinach PSII core complexes (Fig. 2B) (29–32). In the Chl Soret region, the spectral shape is almost identical for both inner antenna complexes, with two bands at 416 and 436 nm identical in value to their spinach counterparts. The carotenoid absorption bands for CP47-His are at 469 and 502 nm (467 and 502 nm for spinach CP47), and those for CP43-His are at 467 and 499 nm (462 and 497 nm for spinach CP43). Fig. 2B (inset) shows an enlargement of the Chl QY absorption region. For CP47-His, the absorption spectrum showed two bands at 668 and 676 nm and a shoulder around 683 nm. The absorption red tail extends to 700 nm, suggesting the presence of a spectral component in the red edge of the spectrum (also observed for spinach at 690 nm). Overall, when comparing CP47-His from Synechocystis 6803 with spinach CP47, the spectral shapes show some differences in positions and relative amplitude, such as stronger and weaker absorptions for the cyanobacterial inner antenna protein around 667 nm and in the red edge, respectively. For CP43-His, the absorption spectra show a broad maximum at 668.5 nm, two shoulders at 673 and 679 nm, and a narrow band at 684.5 nm. The absorption spectra of the spinach CP43 inner antenna (19) display a very similar spectral shape, the main difference between the two organisms being the 2.5-nm red shift of the narrow band at 684.5 nm in Synechocystis 6803 with respect to the 682 nm narrow band in spinach.
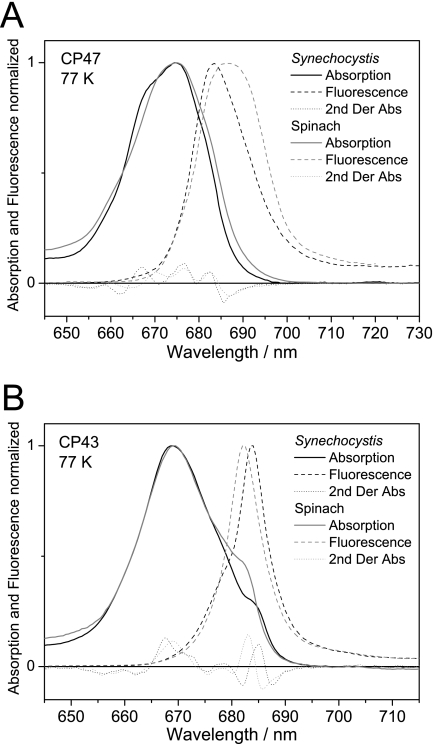
Absorption and Fluorescence at 77 K and Comparison with Spinach
Fig. 4 shows the absorption and fluorescence spectra at 77 K normalized to a value of 1 at the absorption and fluorescence maxima for CP47 (Fig. 4A) and CP43 (Fig. 4B) from Synechocystis 6803 and spinach to facilitate the comparison of their respective spectral shapes. The second derivatives of the absorption (multiplied by −4) are displayed to identify the spectral components present in the spectra. For CP47, the second derivatives of the absorption spectra show differences at wavelengths shorter than 675 nm, with maxima at 658.5 and 667 nm for Synechocystis 6803 and 660 and 670.5 nm for spinach. At wavelengths longer than 675 nm, the peak positions at 676.5 and 682.5 nm are the same. However, the red edge is less pronounced in cyanobacterial CP47-His, which suggests that, although the 682.5 nm band is at the same position in both organisms, this band is narrower in Synechocystis 6803. This could be an indication of a more “defined” protein environment or a lower degree of electron-phonon coupling (coupling of the electronic transitions to protein lattice vibrations, which induce broadening of the absorption) in CP47-His. The fluorescence spectrum also shows a narrower and blue-shifted spectral distribution in Synechocystis 6803 with respect to spinach, which indicates that the 682.5 nm pigment(s) are emitting states with large contributions to the overall fluorescence spectrum and that the red chlorophyll (peaking around 690 nm in spinach) gives a smaller contribution. However, a “red” chlorophyll is probably also present in Synechocystis 6803, in view of the shoulder at around 690 nm in the 77 K emission spectrum. For CP43, the second derivatives confirm that the narrow red band is shifted from 682 nm in spinach to 684.5 nm in Synechocystis 6803 and that the other absorption bands peak at the same wavelengths in both organisms. The 2-nm red shift is also observed in the fluorescence spectra (which peak at 682 nm in spinach and 684 nm in Synechocystis 6803), indicating that the pigment(s) giving rise to the narrow absorption bands are states with large contributions to the 77 K steady-state emission spectra (19). In contrast to spinach, the 77 K emission spectrum of CP43-His from Synechocystis 6803 reveals a shoulder at about 679 nm, which is probably related to the feature peaking at about 678 nm in the second derivative of the absorption spectrum. CP43 from spinach shows an absorption band peaking at the same wavelength, but in the emission, this feature is not observed, most likely as a result of overlap with the emission from the 682 nm band. The efficiency of excitation energy transfer to the reaction center was assessed by calculating the fluorescence quantum yield at room temperature. The calculated fluorescence quantum yield values for the Synechocystis 6803 inner antenna proteins are both lower than those for spinach (∼50 and 75% for CP47-His and CP43-His, respectively).
FIGURE 4.
Comparison of the low temperature absorption and fluorescence spectra in the QY band region between the CP47 and CP43 inner antenna proteins of Synechocystis 6803 and spinach. Absorption and fluorescence spectra for CP47-His (A) and CP43-His (B) from Synechocystis 6803 and for CP47 and CP43 from spinach were recorded at 77 K and normalized to 1 at the absorption and fluorescence maxima. Second derivatives of absorption spectra (2nd Der Abs) are also shown.
DISCUSSION
Although the structure of the cyanobacterial PSII holoenzyme is now known to atomic resolution (6), there is still much to learn about how the complex is assembled and then repaired following irreversible damage (reviewed in Ref. 10). Here, we have addressed a very basic question: at what stage can chlorophyll and carotenoid molecules be inserted into the inner antenna proteins CP47 and CP43 of PSII? To address this, we have generated His-tagged strains of Synechocystis 6803 to allow rapid isolation of unassembled CP47-His and CP43-His protein complexes from the membranes of a ΔD1 deletion strain. In the case of CP47, we also exploited the observation that the levels of unassembled CP47 increase from 10 to 20% of WT levels to 100% of WT levels when the FtsH2 thylakoid protease is inactivated (17). Thylakoid FtsH proteases appear to play an important role in removing unassembled protein subunits (17). Consequently, the use of FtsH mutants might be a useful strategy to increase levels of other unstable complexes or assembly complexes in the thylakoid membrane in both cyanobacteria and chloroplasts.
Our results indicate that the isolated His-tagged CP47 and CP43 complexes contain a very similar set of chlorophyll and carotenoid pigments compared with spinach CP47 and CP43 isolated by fragmentation of larger core complexes. Evidence comes from the close similarity in spectral shape and relative amplitudes in the absorption spectra in the visible range from 400 to 750 nm between the Synechocystis 6803 complexes and spinach PSII core complexes (Fig. 3) as well as similarities in low temperature absorption and fluorescence spectra (Figs. 2 and 4). Thus, we can conclude that the insertion of pigment into CP47 and CP43 in Synechocystis 6803 is able to occur before their incorporation into a larger PSII complex, at least in the PSII assembly mutants studied here. Previous work has shown that the availability of Chl a is important for the synthesis and accumulation of full-length CP47 apopolypeptide (33, 34) and that absence of carotenoid leads to impaired synthesis of CP47 and especially of CP43 (35). Also, accumulation of CP47 and CP43 is severely impaired in site-directed mutants lacking an appropriate amino acid ligand to chlorophyll (36, 37). These data therefore suggest that binding of pigment starts to occur at an early stage in the assembly of CP47 and CP43 in the WT, perhaps even cotranslationally during insertion of the apopolypeptides into the membrane to help stabilize the complex. A close synchronization between pigment binding and the synthesis and folding of CP47 and CP43 would also help to minimize potential toxic effects of free chlorophyll in the membrane (38). Whether all or only a fraction of the chlorophyll molecules bind to CP47 and CP43 at this early stage of PSII assembly in the WT is unclear. How chlorophyll and carotenoid are presented to the CP47 and CP43 apopolypeptides is also currently unknown, although there is some suggestion that enzymes involved in chlorophyll biosynthesis might be docked onto the thylakoid membrane (39).
The precise Chl/carotenoid ratio to be expected for CP47-His and CP43-His is still not clear. In the most recent structure, 35 Chl a molecules are present per PSII monomer, with 16 molecules bound to CP47 and 13 to CP43 (4). However, the assignment of carotenoids within the PSII structural models has proved to be more difficult. With improved resolution, the number has increased from 7 (3) to 11 (5) and most recently to 12 molecules (4). analysis by Müh et al. (40) concluded that five carotenoids are associated with CP47 and four with CP43. However, many of these carotenoids are located at the interfaces between subunits, such as between CP43 and PsbK/PsbZ (41), and at the interface of the two monomers between CP47 and D1-PsbT (40). The Chl/carotenoid ratios determined here (7.7 for CP47-His and 5.0 for CP43-His) are much greater than those predicted by Müh et al. (40) (3.2 for CP47 and 3.25 for CP43) and might reflect absence of carotenoid in the smaller assembly complexes described here and/or loss of carotenoid during purification.
Comparison of the low temperature absorption and fluorescence spectra of the inner antenna proteins isolated from Synechocystis 6803 and spinach shows that the spectral distribution of absorption bands is similar but not identical in both organisms. Regarding the functionality of the antenna complexes with respect to their efficiency of excitation energy transfer to the reaction center, the fluorescence quantum yields for CP47-His and CP43-His were calculated and found to be generally lower than those for their spinach counterparts. The differences may be of several origins. One explanation might derive from the fact that the Synechocystis 6803 antenna complexes are assembly complexes isolated at a stage preceding their assembly into the core complex (not isolated from the core complex, as in the case of spinach). This implies that the energy absorbed by the antenna protein pigments is not “meant” to be used by the reaction center for photochemistry and that there might therefore be an intrinsic energy dissipation pathway, possibly mediated by LMM PSII subunits. Alternatively, the assembly complexes could be in a protein conformation that is not favorable for efficient excitation energy transfer (because it is not needed), which might change to an optimized conformation once the complexes are integrated into the core complex. Another possibility is that the differences may arise from variations in the macrostructure of the inner antenna systems between the two organisms and sequence differences between them. In spinach, the core antennas play a role in transferring excitation energy absorbed by other peripheral antennas: the minor antennas CP24, CP26, and CP29 and the main light-harvesting complex II. In Synechocystis 6803, a role as an “intermediary” in excitation energy transfer could be bypassed by the presence of the dynamic phycobilisomes, which can funnel excitation energy directly to the reaction center. Therefore, the requirement of highly efficient excitation energy transfer would be less of a determinant in Synechocystis 6803 than in spinach. We note that these possibilities are not mutually exclusive and that we cannot conclude from the available data which possibility or possibilities are the right one(s). To shed light on these issues, it will be necessary to perform further comparative spectroscopic studies between assembly complexes and complexes isolated from the core complex.
An additional novel aspect of our work is the detection of LMM subunits in the isolated CP43-His and CP47-His complexes. Although our data show that these subunits are capable of binding to unassembled CP47 and CP43, additional evidence is required to confirm that such complexes also form in the WT strain and are not merely “dead-end” complexes that only accumulate in assembly-defective mutants. For instance, we have shown here that PsbH is capable of forming a complex with CP47-His (Fig. 1 and supplemental Fig. 2). Low levels of a CP47-PsbH complex have also been detected in WT thylakoids (25), and accumulation of full-length unassembled CP47 apopolypeptide is impaired in a PsbH-null mutant (25). Together, these data suggest a stabilizing effect of PsbH on CP47 accumulation during the early steps of PSII assembly in the WT (25). The small chlorophyll-a/b-binding-like protein ScpD can also bind to CP47 in the vicinity of PsbH (42), but ScpD is not expressed under the illumination conditions used here (data not shown), which would explain its absence in the CP47-His complex. Overall, the currently available data suggest that the low level of unassembled CP47 found in the WT thylakoid membrane exists predominantly as a CP47-PsbH complex and can include ScpD under high light conditions. Importantly, we were able to detect the co-purification of PsbT and PsbL with CP47-His. These LMM subunits are located next to CP47 in the PSII holoenzyme and lie at the interface of the two monomers in the dimeric complex. Whether PsbT and PsbL are present at stoichiometric levels in the CP47-His complex and attach to CP47 early in assembly in the WT is presently unclear.
CP43 has been detected in two distinct complexes (CP43a and CP43b) in solubilized thylakoid membrane extracts of Synechocystis 6803, so it is likely that CP43 also binds LMM subunits at an early stage of PSII assembly in the WT (8). Based on the PSII crystal structures, three LMM subunits might form a stable complex with CP43: PsbK, PsbZ, and Psb30/Ycf12 (3–5). Using mass spectrometry, we have been able to identify PsbK in the CP43-His complex. A tight association between PsbK and CP43 has previously been documented in fragmentation studies of PSII core complexes (39), and the early binding of PsbK to CP43 during assembly has been inferred from analysis of Chlamydomonas PSII mutants (43). In addition, we have identified a LMM polypeptide in the His-CP43 complex containing a short sequence match with Psb30/Ycf12 (Table 1). Hence, we assign Psb30/Ycf12 as an additional component of the CP43-His complex. Whether PsbZ is also a component of the CP43-His complex remains to be clarified.
In conclusion, we have described the first isolation and characterization of unassembled CP47 and CP43 subunits from the thylakoid membrane. Our experimental results indicate that CP47 and CP43 are capable of forming preassembled pigment-protein complexes containing neighboring LMM subunits found in the holoenzyme. On the basis of our data here and earlier results, we propose a modular assembly of PSII in which D1-PsbI (44), D2-cytochrome b559 (8), CP47-PsbH-PsbL-PsbT, and CP43-PsbK-Psb30 subcomplexes, the latter possibly including PsbZ, are combined together to form first a PSII reaction center-like complex, then the RC47 complex (a PSII core complex lacking CP43), and finally a monomeric core complex (reviewed in Ref. 10). Subsequent formation of oxygen-evolving PSII would require light-driven assembly of the Mn4Ca cluster and attachment of the extrinsic proteins.
Supplementary Material
This work was supported by Engineering and Physical Sciences Research Council (EPSRC) Grant EP/F002070X/1 and in part by Czech Academy of Science Institutional Research Concept AV0Z50200510 and Grant Project IAA400200801.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- PSII
- Photosystem II
- Chl
- chlorophyll
- LMM
- low molecular mass
- TES
- N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid
- β-DM
- n-dodecyl β-d-maltoside
- BN
- Blue native
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Wydrzynski T. J., Satoh K. (eds) (2005) in Photosystem II: The Light-driven Water:Plastoquinone Oxidoreductase, Springer, Dordrecht, The Netherlands [Google Scholar]
- 2. Zouni A., Witt H. T., Kern J., Fromme P., Krauss N., Saenger W., Orth P. (2001) Nature 409, 739–743 [DOI] [PubMed] [Google Scholar]
- 3. Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. (2004) Science 303, 1831–1838 [DOI] [PubMed] [Google Scholar]
- 4. Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009) Nat. Struct. Mol. Biol. 16, 334–342 [DOI] [PubMed] [Google Scholar]
- 5. Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2005) Nature 438, 1040–1044 [DOI] [PubMed] [Google Scholar]
- 6. Shen J. R., Umena Y., Kawakami K., Kamiya N. (2010) The 15th International Congress of Photosynthesis, Beijing, August 22–27, 2010, Crystal Structure of Oxygen Evolving Photosystem II at Atomic Resolution, Abstr. PS6.5, Chinese Academy of Sciences, Beijing, China [Google Scholar]
- 7. Kamiya N., Shen J. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 98–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Komenda J., Reisinger V., Müller B. C., Dobáková M., Granvogl B., Eichacker L. A. (2004) J. Biol. Chem. 279, 48620–48629 [DOI] [PubMed] [Google Scholar]
- 9. Rokka A., Suorsa M., Saleem A., Battchikova N., Aro E. M. (2005) Biochem. J. 388, 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nixon P. J., Michoux F., Yu J., Boehm M., Komenda J. (2010) Ann. Bot. 106, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bricker T. M., Frankel L. K. (2002) Photosynth. Res. 72, 131–146 [DOI] [PubMed] [Google Scholar]
- 12. Ghanotakis D. F., de Paula J. C., Demetriou D. M., Bowlby N. R., Petersen J., Babcock G. T., Yocum C. F. (1989) Biochim. Biophys. Acta 974, 44–53 [DOI] [PubMed] [Google Scholar]
- 13. Alfonso M., Montoya G., Cases R., Rodríguez R., Picorel R. (1994) Biochemistry 33, 10494–10500 [DOI] [PubMed] [Google Scholar]
- 14. Williams J. G. (1988) Methods Enzymol. 167, 766–778 [Google Scholar]
- 15. Nixon P. J., Trost J. T., Diner B. A. (1992) Biochemistry 31, 10859–10871 [DOI] [PubMed] [Google Scholar]
- 16. Debus R. J., Campbell K. A., Gregor W., Li Z. L., Burnap R. L., Britt R. D. (2001) Biochemistry 40, 3690–3699 [DOI] [PubMed] [Google Scholar]
- 17. Komenda J., Barker M., Kuviková S., de Vries R., Mullineaux C. W., Tichy M., Nixon P. J. (2006) J. Biol. Chem. 281, 1145–1151 [DOI] [PubMed] [Google Scholar]
- 18. Groot M. L., Peterman E. J., van Stokkum I. H., Dekker J. P., van Grondelle R. (1995) Biophys. J. 68, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Groot M. L., Frese R. N., de Weerd F. L., Bromek K., Pettersson A., Peterman E. J., van Stokkum I. H., van Grondelle R., Dekker J. P. (1999) Biophys. J. 77, 3328–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Komenda J., Barber J. (1995) Biochemistry 34, 9625–9631 [DOI] [PubMed] [Google Scholar]
- 21. Boehm M., Nield J., Zhang P., Aro E. M., Komenda J., Nixon P. J. (2009) J. Bacteriol. 191, 6425–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blum H., Beier H., Gross H. J. (1987) Electrophoresis 8, 93–99 [Google Scholar]
- 23. Nixon P. J., Komenda J., Barber J., Deak Z., Vass I., Diner B. A. (1995) J. Biol. Chem. 270, 14919–14927 [DOI] [PubMed] [Google Scholar]
- 24. Andronis C., Kruse O., Deák Z., Vass I., Diner B. A., Nixon P. J. (1998) Plant Physiol. 117, 515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Komenda J., Tichý M., Eichacker L. A. (2005) Plant Cell Physiol. 46, 1477–1483 [DOI] [PubMed] [Google Scholar]
- 26. Granvogl B., Zoryan M., Plöscher M., Eichacker L. A. (2008) Anal. Biochem. 383, 279–288 [DOI] [PubMed] [Google Scholar]
- 27. Sugiura M., Inoue Y. (1999) Plant Cell Physiol. 40, 1219–1231 [DOI] [PubMed] [Google Scholar]
- 28. Bricker T. M., Morvant J., Masri N., Sutton H. M., Frankel L. K. (1998) Biochim. Biophys. Acta 1409, 50–57 [DOI] [PubMed] [Google Scholar]
- 29. Van Dorssen R. J., Breton J., Plijter J. J., Satoh K., Van Gorkom H. J., Amesz J. (1987) Biochim. Biophys. Acta 893, 267–274 [Google Scholar]
- 30. Hughes J. L., Picorel R., Seibert M., Krausz E. (2006) Biochemistry 45, 12345–12357 [DOI] [PubMed] [Google Scholar]
- 31. Reppert M., Zazubovich V., Dang N. C., Seibert M., Jankowiak R. (2008) J. Phys. Chem. B 112, 9934–9947 [DOI] [PubMed] [Google Scholar]
- 32. Neupane B., Dang N. C., Acharya K., Reppert M., Zazubovich V., Picorel R., Seibert M., Jankowiak R. (2010) J. Am. Chem. Soc. 132, 4214–4229 [DOI] [PubMed] [Google Scholar]
- 33. Sobotka R., Dühring U., Komenda J., Peter E., Gardian Z., Tichy M., Grimm B., Wilde A. (2008) J. Biol. Chem. 283, 25794–25802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sobotka R., Komenda J., Bumba L., Tichy M. (2005) J. Biol. Chem. 280, 31595–31602 [DOI] [PubMed] [Google Scholar]
- 35. Sozer O., Komenda J., Ughy B., Domonkos I., Laczkó-Dobos H., Malec P., Gombos Z., Kis M. (2010) Plant Cell Physiol. 51, 823–835 [DOI] [PubMed] [Google Scholar]
- 36. Shen G., Vermaas W. F. (1994) Biochemistry 33, 7379–7388 [DOI] [PubMed] [Google Scholar]
- 37. Manna P., Vermaas W. (1997) Eur. J. Biochem. 247, 666–672 [DOI] [PubMed] [Google Scholar]
- 38. Krieger-Liszkay A., Fufezan C., Trebst A. (2008) Photosynth. Res. 98, 551–564 [DOI] [PubMed] [Google Scholar]
- 39. Schottkowski M., Ratke J., Oster U., Nowaczyk M., Nickelsen J. (2009) Mol. Plant 2, 1289–1297 [DOI] [PubMed] [Google Scholar]
- 40. Müh F., Renger T., Zouni A. (2008) Plant Physiol. Biochem. 46, 238–264 [DOI] [PubMed] [Google Scholar]
- 41. Iwata S., Barber J. (2004) Curr. Opin. Struct. Biol. 14, 447–453 [DOI] [PubMed] [Google Scholar]
- 42. Promnares K., Komenda J., Bumba L., Nebesarova J., Vacha F., Tichy M. (2006) J. Biol. Chem. 281, 32705–32713 [DOI] [PubMed] [Google Scholar]
- 43. Sugimoto I., Takahashi Y. (2003) J. Biol. Chem. 278, 45004–45010 [DOI] [PubMed] [Google Scholar]
- 44. Dobáková M., Tichy M., Komenda J. (2007) Plant Physiol. 145, 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.