Abstract
The role of signaling pathways in the regulation of cellular iron metabolism is becoming increasingly recognized. Iron chelation is used for the treatment of iron overload but also as a potential strategy for cancer therapy, because iron depletion results in cell cycle arrest and apoptosis. This study examined potential signaling pathways affected by iron depletion induced by desferrioxamine (DFO) or di-2-pyridylketone-4,4-dimethyl-3-thiosemicarbazone (Dp44mT). Both chelators affected multiple molecules in the mitogen-activated protein kinase (MAPK) pathway, including a number of dual specificity phosphatases that directly de-phosphorylate MAPKs. Examination of the phosphorylation of major MAPKs revealed that DFO and Dp44mT markedly increased phosphorylation of stress-activated protein kinases, JNK and p38, without significantly affecting the extracellular signal-regulated kinase (ERK). Redox-inactive DFO-iron complexes did not affect phosphorylation of JNK or p38, whereas the redox-active Dp44mT-iron complex significantly increased the phosphorylation of these kinases similarly to Dp44mT alone. Iron or N-acetylcysteine supplementation reversed Dp44mT-induced up-regulation of phospho-JNK, but only iron was able to reverse the effect of DFO on JNK. Both iron chelators significantly reduced ASK1-thioredoxin complex formation, resulting in the increased phosphorylation of ASK1, which activates the JNK and p38 pathways. Thus, dissociation of ASK1 could serve as an important signal for the phosphorylation of JNK and p38 activation observed after iron chelation. Phosphorylation of JNK and p38 likely play an important role in mediating the cell cycle arrest and apoptosis induced by iron depletion.
Keywords: Iron, Iron Metabolism, Metals, Protein Metal Ion Interaction, Transport Metals, Iron, Iron Metabolism, Metals, Transport Metals
Introduction
Complex signaling pathways play crucial functions in many cellular processes, and recently, the role of signal transduction pathways in iron metabolism has been described (1, 2). For instance, one of these include the recently established role for the bone morphogenetic protein-Smad signaling pathway in the regulation of hepcidin, the hormone of iron metabolism (3).
Iron is critical for cellular proliferation, and its depletion results in cell cycle arrest and apoptosis (4–6). As a result, iron chelation is a promising strategy for cancer therapy (7, 8). A well known iron chelator that is clinically used for the treatment of iron-overload disease is desferrioxamine (DFO)3 (Fig. 1A) (7, 8). It has also been demonstrated that DFO possesses anti-proliferative activity, with the chelator having entered clinical trials for cancer treatment (7, 9, 10). However, the efficacy of DFO is limited by its low lipophilicity and membrane permeability (11, 12), and hence, other iron chelators with improved lipophilicity have been developed (8). One such chelator is di-2-pyridylketone-4,4-dimethyl-3-thiosemicarbazone (Dp44mT) (Fig. 1A), which showed marked membrane permeability and pronounced anti-cancer activity against multiple human and rodent tumor xenograft models (13, 14).
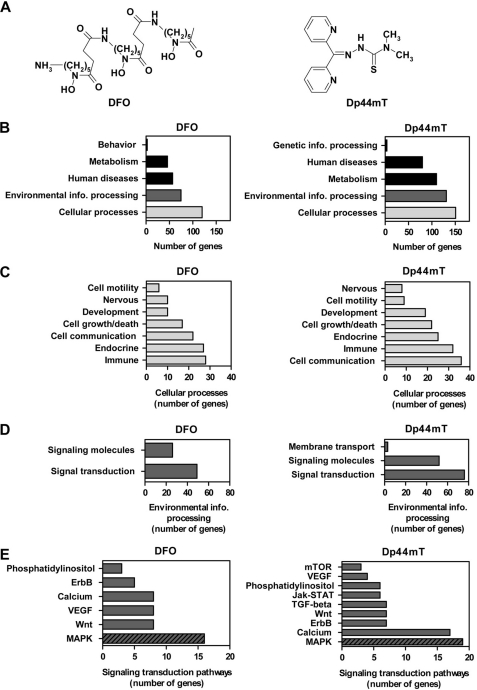
FIGURE 1.
Structures of the chelators used and microarray data showing that DFO and Dp44mT significantly alter the expression of a large number of genes involved in the MAPK signaling transduction pathway. A, chemical structures of DFO and Dp44mT. B, functional pathway annotation of genes significantly altered (p < 0.05) by DFO (250 μm) or Dp44mT (25 μm) after a 24 h/37 °C incubation of DMS-53 lung cancer cells as assessed by a human genome gene array (Affymetrix U133 plus 2.0 array). Only genes with a greater than log2 value of 1.0 as compared with the control were included in the analysis. Genes in the category of cellular processes (C) and environmental information processing (D) were further sub-categorized according to their functions. E, various signaling transduction pathways affected. Lists of genes were assessed using DAVID. Pathway annotation was obtained through the public data base, KEGG pathway mapping.
The cytotoxicity of iron chelators can be attributed to their ability to cause cell cycle arrest and apoptosis due to iron depletion. DFO mainly arrests cells at the G1/S (15–17) phase of the cycle and has been shown to induce apoptosis in multiple cell types (16, 18). Inhibition of cell cycle progression is also reflected by the effect of DFO on cell cycle-related molecules, particularly decreasing the protein levels of cyclin D1 and to lesser extent, cyclin A and B1 (19, 20). The expression of cyclin-dependent kinase 2 (cdk2) is also decreased (19). In addition, DFO also affects gene expression mediated by transcription factors, such as hypoxia-inducible factor-1α (HIF-1α) and p53 (21).
The chelator, Dp44mT has a much higher anti-proliferative efficacy than DFO and was shown to reduce progression of cells through the G1/S and G2/M phases of the cell cycle (22). Moreover, Dp44mT was shown to mediate the mitochondrial apoptotic pathway by significantly increasing caspases-3, -8, and -9 protein levels and activities (13). Tumor xenografts taken from Dp44mT-treated mice demonstrated increased apoptosis and up-regulation of the tumor growth and metastasis suppressor, N-myc downstream-regulated gene-1 (Ndrg1) (13, 14). Recently, Dp44mT was also shown to selectively inhibit topoisomerase IIα (23). Furthermore, unlike DFO, the iron complexes of Dp44mT can cause reactive oxygen species (ROS) generation, which may mediate cytotoxicity (13).
Hence, the anti-cancer activity of iron chelators can be elicited through multiple molecular targets, and the cellular signaling pathways affected by iron depletion remain unclear. Thus, we examined the molecular targets of these iron chelators and assessed potential pathways leading to their cytotoxic mechanisms. We performed gene array expression screening after incubation of cells with DFO or Dp44mT and observed that a number of signaling pathways were affected after analysis using bioinformatics resources. Among these pathways was the mitogen-activated protein kinase (MAPK) pathway, which showed the highest number of genes that were modulated by these agents.
The MAPKs are evolutionarily conserved kinases that link extracellular signals to regulate diverse cellular processes, including cell growth, proliferation, differentiation, migration, and apoptosis (24, 25). This pathway consists of a core of three protein kinases, including the extracellular signal-regulated kinases 1 and 2 (ERK1/2), c-Jun amino-terminal kinase (JNK), and p38 kinases (24, 25). Generally, JNK and p38 have anti-proliferative and pro-apoptotic roles, although ERK1/2 is primarily involved in the proliferative response (24, 26).
In this study, we demonstrate that iron depletion mediated by chelators increases the phosphorylation of JNK and p38 MAPKs, as well as the phosphorylation of their downstream targets, namely p53 and ATF-2. Activation of JNK and p38 was, at least in part, mediated by the apoptosis signal-regulating kinase 1 (ASK1), a MAP3K, which in turn is activated as a result of dissociation from its complexation with thioredoxin (Trx) after iron chelation. These results are important for understanding the role of cellular signaling in the response to iron depletion.
EXPERIMENTAL PROCEDURES
Chelators
Dp44mT was synthesized and characterized using standard procedures (27, 28). DFO was obtained from Novartis (Basel, Switzerland).
Cell Culture
The human DMS-53 small cell lung carcinoma, SK-N-MC, neuroepithelioma, and SK-Mel-28 melanoma cell lines were obtained from the American Type Culture Collection (Manassas, VA). The cells were grown in RPMI 1640 medium by standard procedures, as described previously (29). Cultures were used when cells were at ∼80% confluence.
RNA Isolation and Semi-quantitative RT-PCR
Total RNA was isolated using TRIzol® reagent (Invitrogen). Reverse transcriptase-PCR (RT-PCR) was performed using the primers listed in Table 1, as per standard procedures (30, 31). RT-PCR was optimized to be semi-quantitative and in the log phase of amplification. β-Actin was used as a loading control, and its expression was unaltered in all of the treatment conditions used.
TABLE 1.
List of primers for amplification of human mRNA
F indicates forward, and R indicates reverse.
| Pair no. | Primer name | Accession no. | Oligonucleotide (5′ to 3′) sequence | Product size |
|---|---|---|---|---|
| bp | ||||
| 1 | hCACNA1E | NM_000721 | F-TCTGCCATCTACTTCATTGTG | 762 |
| R-CTCCTTGCCTTCTGTCCT | ||||
| 2 | hDDIT3 | NM_004083 | F-GCGTATGTGGGATTGAG | 573 |
| R-GCTGGAAGCCTGGTAT | ||||
| 3 | hNFATC4 | NM_004554 | F-GACTGGCTCCAACTTCCTG | 565 |
| R-CCCTCCATACGGGTCACTA | ||||
| 4 | hNFKB2 | NM_001077494 | F-GAAGACCTTGCTGCTAAATGC | 399 |
| R-TGCTCCACTGACTGGCTCCC | ||||
| 5 | hPLA2G3 | NM_015715 | F-CCCATGTCATTCTACAACCA | 537 |
| R-CTAACTTCCCACCTCCTAC | ||||
| 6 | hTAOK2 | NM_016151 | F-AGGTGCGGTTCTTACAGA | 327 |
| R-TGCCCACGAAGGAGTT | ||||
| 7 | hTGFB2 | NM_001135599 | F-CTTTCTACAGACCCTACTTCA | 642 |
| R-GTATCCATTTCCACCCTA | ||||
| 8 | hGADD45B | NM_015675 | F-CCAGCTACTGCGAAGAAAG | 554 |
| R-CTCCCAAGTCCCAAAGTGT | ||||
| 9 | hMAPK8 | NM_002752 | F-TTGACAGACGACGATG | 645 |
| R-GACGCCTTATGTAGTGA | ||||
| 10 | hMAPK14 | NM_001315 | F-CCGAGCCAGTCCAAAA | 464 |
| R-AGGTGCCCGAGCGTTAC | ||||
| 11 | hMAPK1 | NM_002745 | F-CGTCACTCGGGTCGTA | 506 |
| R-CAACCTGCTGCTCAAC | ||||
| 12 | hDUSP1 | NM_004417 | F-TCCCGAATGTGCTGAGTT | 552 |
| R-CTGGAGGAAGGGTGTTT | ||||
| 13 | hDUSP5 | NM_004419 | F-TGGTCAGGGAAGGGGTAG | 563 |
| R-GCCGTCGCTGTTAGAGGA | ||||
| 14 | hDUSP10 | NM_007207 | F-CACCGTTGTTTAGGTCTTCC | 477 |
| R-TACCTCAGCCCATCCC | ||||
| 15 | hDUSP16 | NM_030640 | F-GCCGTCGCTGTTAGAGGA | 563 |
| R-TGGTCAGGGAAGGGGTAG | ||||
| 16 | hNFKB1 | NM_003998 | F-TCTTCACTGGGCTGGA | 853 |
| R-CGCTTGGGTAACTCTG |
Microarray Processing and Data Analysis
RNA isolation and microarray processing were performed, as described previously (21, 31). The Affymetrix Human Genome U133 Plus 2.0 430 2.0 array (Affymetrix, Santa Clara, CA) was used, which assesses >47,000 transcripts from over 38,500 genes.
Initial data analysis for the conversion of images to raw expression values was performed with Affymetrix Expression ConsoleTM version 1.1. Three separate arrays were carried out for each group. Absolute expression signals as well as the mean probe level fold changes were expressed as SigLogRatio, i.e. log2 of the fold change. To be considered as modulated by the iron chelators, the intensity value threshold was set at a log2 value of 1 increase or decrease compared with the control, with significance at p < 0.05, as determined by Student's t test. Definitive evidence of differential expression was validated using RT-PCR assessment using three independent RNA samples. Pathways were annotated and classified through the DAVID (david.abcc.ncifcrf.gov; accessed March, 2008) and KEGG pathway mapping databases (accessed March, 2008). The complete array dataset can be accessed via the Gene Expression Omnibus (www.ncbi.nlm.nih.gov), using GSM662881–GSM662886.
Protein Extraction and Western Blotting
Protein extraction and Western blotting were performed using well established procedures (21, 31). Rabbit anti-human phospho-specific and nonphospho-specific ERK1/2 (catalog no. 9910 and 4695), JNK (catalog no. 4668 and 9252), p38 (catalog no. 9215 and 9212), p53 (catalog no. 9919), ATF-2 (catalog no. 9920), and ASK1 (catalog no. 3761 and 3762) antibodies (Cell Signaling Technology, Danvers, MA) were incubated at 1:1000–1:2500 dilutions. Rabbit anti-human Trx1 antibody (catalog no. 2429, Cell Signaling Technology) was used at a 1:1000 dilution. Mouse monoclonal anti-human TfR1 (Invitrogen; catalog no. 136800) was incubated at 1:1000. The secondary antibodies employed were anti-rabbit and anti-mouse (Sigma), each at a dilution of 1:10,000. As an internal control for protein loading, membranes were also probed for β-actin.
MAPK Phospho-antibody Array
A phospho-specific MAPK antibody array (Full Moon Biosystems, Sunnyvale, CA) was utilized as described previously (32). This antibody array contained 185 characterized phospho-specific antibodies for proteins in the MAPK pathway and also antibodies for the paired nonphosphorylated targets to determine the relative level of phosphorylation. For data analysis, background signals were removed from all measurements. A ratio was calculated to measure the extent of protein phosphorylation. Results from quadruplicate samples were averaged.
Measurement of Glutathione and Oxidized Glutathione
Intracellular GSH and oxidized GSH (GSSG) were determined using the GSH/GSSG ratio assay kit (catalog no. 371757; Calbiochem) according to the manufacturer's instructions. Briefly, cells were seeded in 100-mm dishes for experimental treatments. After a 24 h/37 °C incubation with the chelators, the cells were washed with ice-cold PBS and lysed in 50 μl of PBS by three freeze-thaw cycles. The lysates were then acidified with 5% metaphosphoric acid, and the supernatant was separated by centrifugation at 10,000 × g for 10 min at 4 °C.
Immunoprecipitation
Immunoprecipitation was performed using Dynabeads® protein G following the manufacturer's procedure (Invitrogen). Briefly, cells were washed with ice-cold PBS and lysed using Nonidet P-40 lysis buffer (Invitrogen) containing protease and phosphatase mixture inhibitors (Roche Diagnostics). Protein (1 mg) was incubated for 2 h/4 °C with 5 μg of monoclonal mouse anti-human thioredoxin antibody (Abcam, catalog no. ab16845). The mixture was added to 50 μl of Dynabeads® protein G (Invitrogen) and allowed to incubate overnight. The beads were washed four times, resuspended in SDS-loading buffer, and incubated at 70 °C/10 min. The supernatant was separated on 4–12% BisTris gel (Invitrogen). ASK1 and Trx1 were detected using the rabbit anti-human antibodies described above (Cell Signaling Technology) at a 1:1000 dilution.
Densitometry
Densitometric analysis of band intensities obtained from RT-PCR and Western blotting experiments were carried out using Quantity One software (Bio-Rad). The relative intensities of target bands were normalized using the relative β-actin loading control.
Statistical Analysis
Data are expressed as means ± S.E. of at least three experiments. Experimental data were compared using Student's t test and considered statistically significant when p < 0.05.
RESULTS
Chelators Modulate the MAPK Signaling Pathway
To characterize potential molecular pathways affected by DFO and Dp44mT, we performed a whole genome microarray screen after DMS-53 lung carcinoma cells were incubated with DFO (250 μm) or Dp44mT (25 μm) over 24 h/37 °C. These chelator concentrations and this incubation period were used because we previously demonstrated that under these conditions the chelators modulate iron-responsive genes (e.g. transferrin receptor 1 (TfR1) (21, 30)). The higher concentration of DFO was implemented due to its limited ability to permeate membranes (33, 34). Dp44mT was utilized at a lower concentration because this ligand shows high permeability and demonstrates marked iron chelation efficacy and anti-proliferative activity (13, 14). Initially, the DMS-53 cell type was chosen for these studies because Dp44mT showed potent anti-tumor efficacy against this cell type in vitro and in vivo (14).
Incubation of cells with Dp44mT resulted in the alteration in expression of 1548 genes, whereas DFO affected 731 genes. The majority of affected genes was either of unknown function or uncharacterized molecular interaction. Consequently, 291 (Dp44mT) and 160 (DFO) genes were successfully annotated into known functional categories using the data base, DAVID.
We identified “cellular processes” and “environmental information processing” as the major categories affected by the chelators, although to a lesser extent “human diseases” and “metabolism” were also affected (Fig. 1B). Cellular processes can be further subcategorized into multiple processes, with the majority of these being involved in “cell communication,” “immune,” and “endocrine” functions with greater than 20 genes being affected in each of these categories (Fig. 1C). In the category of environmental information processing, most of the affected genes were related to “signaling transduction pathways,” with 49 and 76 genes altered after incubation with DFO or Dp44mT, respectively (Fig. 1D). Considering the large number of genes affected in this category, the effect of DFO or Dp44mT on signaling pathways was further examined. Both chelators affected the expression of genes involved in phosphatidylinositol, ErbB, calcium, vascular endothelial growth factor (VEGF), Wnt, and MAPK signaling (Fig. 1E). The alteration of VEGF signaling is in agreement with the well known effect of iron depletion on the up-regulation of VEGF expression (35, 36). Incubation of cells with Dp44mT also affected the expression of the mTOR, Jak-STAT, and TGF-β-signaling pathways, although DFO did not. For both chelators, the signaling pathway that contained the highest number of affected genes was the MAPK signaling system (DFO, 16 genes, and Dp44mT, 19 genes; Fig. 1E). These results highlighted the potential role of MAPK signaling in the mechanism of action of these chelators, and thus, the MAPK signaling transduction pathway was then further examined.
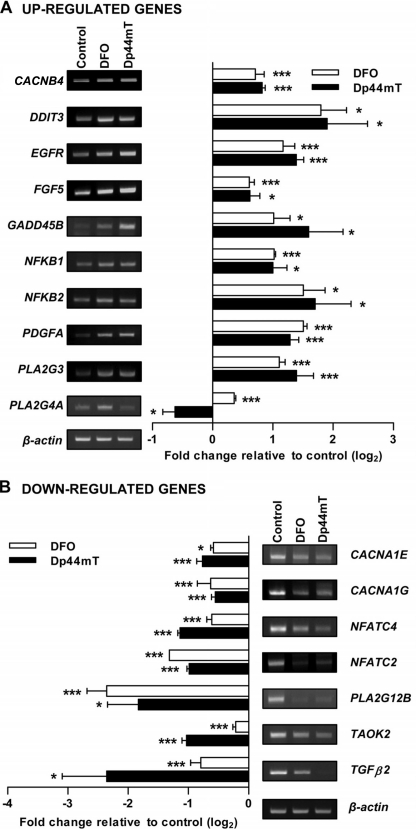
The expression of MAPK-related genes identified from the array (Table 2) was confirmed via RT-PCR using the primers listed in Table 1. The RT-PCR validation confirmed the majority (21 genes) of the results identified from the array (Figs. 2 and 3A), although some genes either showed unchanged expression or were not reliably detected. Genes up-regulated by both chelators included CACNB4, DDIT3, EGFR, FGF5, GADD45B, NFKB1, NFKB2, PDGFA, and PLA2G3 (Fig. 2A). Interestingly, the expression of PLA2G4A was up-regulated after incubation with DFO but was down-regulated after Dp44mT. The genes down-regulated by both chelators included CACNA1E, CACNA1G, NFATC4, NFATC2, PLA2G12B, TAOK2, and TGFβ2 (Fig. 2B).
TABLE 2.
Genes in MAPK signaling transduction pathway affected by DFO or Dp44mT
| Affymetrix ID | Pubmed ID | Gene name | Gene abbreviation | Fold change (log2)a |
|---|---|---|---|---|
| Altered gene expression due to DFO | ||||
| 209383_at | BC003637 | DNA damage-inducible transcript 3 | DDIT3 | 2.81 |
| 201041_s_at | NM_004417 | Dual specificity phosphatase 1 | DUSP1 | 2.42 |
| 201044_x_at | AA530892 | Dual specificity phosphatase 1 | DUSP1 | 2.83 |
| 221563_at | N36770 | Dual specificity phosphatase 10 | DUSP10 | 2.15 |
| 215501_s_at | AK022513 | Dual specificity phosphatase 10 | DUSP10 | 2.76 |
| 209457_at | U16996 | Dual specificity phosphatase 5 | DUSP5 | 3.05 |
| 211485_s_at | AF211188 | Fibroblast growth factor 18 | FGF18 | 1.47 |
| 231382_at | AI798863 | Fibroblast growth factor 18 | FGF18 | 1.84 |
| 210310_s_at | AB016517 | Fibroblast growth factor 5 | FGF5 | 1.10 |
| 207574_s_at | NM_015675 | Growth arrest and DNA damage-inducible, β | GADD45B | 4.25 |
| 209305_s_at | AF078077 | Growth arrest and DNA damage-inducible, β | GADD45B | 4.10 |
| 224542_s_at | U43342 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | NFATC2 | −1.15 |
| 205897_at | NM_004554 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 | NFATC4 | −1.02 |
| 209239_at | M55643 | Nuclear factor of κ light polypeptide gene enhancer in B-cells 1 (p105) | NFKB1 | 1.05 |
| 209636_at | BC002844 | Nuclear factor of κ light polypeptide gene enhancer in B-cells 2 (p49/p100) | NFKB2 | 1.54 |
| 224411_at | AF349540 | Phospholipase A2, group xiib | PLA2G12B | −1.24 |
| 220780_at | NM_015715 | Phospholipase A2, group iii | PLA2G3 | 1.79 |
| 210145_at | M68874 | Phospholipase A2, group iva (cytosolic, calcium-dependent) | PLA2G4A | 1.10 |
| 1555940_a_at | BG198711 | Protein kinase C, α | PRKCA | −1.16 |
| 209908_s_at | BF061658 | Transforming growth factor, β2 | TGFB2 | −1.00 |
| 1561967_at | AI688132 | Transforming growth factor, β2 | TGFB2 | −1.01 |
| Altered gene expression due to Dp44mT | ||||
| 244256_at | AI912770 | Calcium channel, voltage-dependent, α1e subunit | CACNA1E | −2.26 |
| 210380_s_at | AF126966 | Calcium channel, voltage-dependent, α1g subunit | CACNA1G | −3.83 |
| 207693_at | NM_000726 | Calcium channel, voltage-dependent, β4 subunit | CACNB4 | 1.10 |
| 243244_at | H22005 | Calcium channel, voltage-dependent, β4 subunit | CACNB4 | 2.14 |
| 209383_at | BC003637 | DNA damage-inducible transcript 3 | DDIT3 | 3.10 |
| 215501_s_at | GenBank | Dual specificity phosphatase 10 | DUSP10 | 2.29 |
| 221563_at | N36770 | Dual specificity phosphatase 10 | DUSP10 | 1.73 |
| 1558740_s_at | R30807 | Dual specificity phosphatase 16 | DUSP16 | 2.24 |
| 236511_at | AI798976 | Dual specificity phosphatase 16 | DUSP16 | 2.55 |
| 209457_at | U16996 | Dual specificity phosphatase 5 | DUSP5 | 3.14 |
| 211550_at | AF125253 | Epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | EGFR | 1.31 |
| 207574_s_at | NM_015675 | Growth arrest and DNA damage-inducible, β | GADD45B | 4.18 |
| 209305_s_at | AF078077 | growth arrest and DNA damage-inducible, β | GADD45B | 4.16 |
| 238769_at | AW450572 | MAPK kinase kinase kinase kinase 4 | MAP4K4 | −1.79 |
| 238623_at | AI633559 | MAPK kinase kinase kinase 4 | MAPK3K4 | −3.15 |
| 1563367_at | BG704389 | Microtubule-associated protein τ | MAPT | −2.44 |
| 206814_at | NM_002506 | Nerve growth factor, β polypeptide | NGFB | −1.29 |
| 1556568_a_at | N46436 | Nemo-like kinase | NLK | −1.79 |
| 216867_s_at | X03795 | Platelet-derived growth factor α polypeptide | PDGFA | 1.63 |
| 220780_at | NM_015715 | Phospholipase A2, group iii | PLA2G3 | 2.89 |
| 217253_at | L37198 | Stathmin 1/oncoprotein 18 | STMN1 | −2.20 |
| 204986_s_at | NM_016151 | Tao kinase 2 | TAOK2 | −1.44 |
| 209909_s_at | M19154 | Transforming growth factor, β2 | TGFB2 | −4.67 |
| 220407_s_at | NM_003238 | Transforming growth factor, β2 | TGFB2 | −3.98 |
| 228121_at | AU145950 | Transforming growth factor, β2 | TGFB2 | −4.63 |
a Data are relative to control.
FIGURE 2.
Validation of MAPK pathway genes identified in the microarray. RT-PCR confirmation of genes that are part of the MAPK pathway from the microarray analysis as described in Fig. 1 and detailed in Table 2. Shown are the genes that were up-regulated (A) or down-regulated (B) by either DFO (250 μm) or Dp44mT (25 μm) after a 24 h/37 °C incubation with DMS-53 cells. It should be noted that semi-quantitative RT-PCR may not necessarily reflect direct fold change in gene expression, but it was used to validate the up- and down-regulated expression observed in the microarray. RT-PCR panels and densitometric analysis are from a typical experiment of three performed or the mean ± S.E. of at least three independent experiments, respectively. *, p < 0.05; ***, p < 0.001.
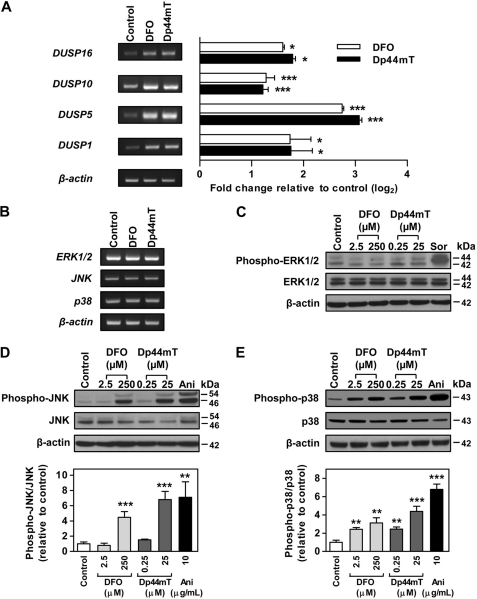
FIGURE 3.
DFO and Dp44mT increase the expression of MAPK phosphatases and the phosphorylation of JNK and p38. mRNA expression of MAPK phosphatases, dual specificity phosphatases (DUSP) (identified from the microarray) (A), and MAPKs, ERK1/2, JNK, and p38 (B) in DMS-53 cells after incubation with DFO (250 μm) or Dp44mT (25 μm) over 24 h/37 °C. C–E, effect of DFO (2.5 and 250 μm) or Dp44mT (0.25 and 25 μm) after a 24 h/37 °C incubation of DMS-53 cells on the protein (phosphorylated and nonphosphorylated) expression of MAPKs as follows. C, ERK1/2 (phospho-Thr-202/Tyr-204); D, JNK (phospho-Thr-183/Tyr-185); and E, p38 (phospho-Thr-180/Tyr-182). Experiments were performed using Western blotting implementing rabbit anti-human phospho-specific and nonphospho-specific antibodies for ERK1/2, JNK, or p38. Sorbitol (Sor; 500 mm) and anisomycin (Ani; 10 μg/ml) were included as positive controls. It should be noted that semi-quantitative RT-PCR may not necessary reflect direct fold change in gene expression but was used to validate the up- and down-regulated expression observed in the microarray. Densitometry of phospho- and nonphospho-JNK includes both bands. RT-PCR and Western blot panels are typical experiments from three performed, and the densitometry is the mean ± S.E. of at least three experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
The gene expression changes confirmed above encompassed both upstream and downstream targets of the MAPK signaling pathway. In particular, the upstream molecules include the following: growth factor-related genes (EGFR, FGF5, and TGFβ2) and genes encoding voltage-dependent calcium channels (CACNB4, CACNA1G, and CACNA1E). The downstream gene targets include the following: NFKBs, phospholipase A2 (PLA2G3, PLA2G4A, and PLA2G12B), and NFATCs. Both chelators increased expression of GADD genes, including DDIT3 (also known as GADD153) and GADD45B, as consistent with a previous report from our laboratory that also found increased GADD45 expression after iron chelation (34). GADD45B is implicated in the activation of JNK and p38 MAPK (37), whereas GADD153 is downstream and is activated by the p38 pathway (38). Together, this gene array study revealed that the MAPK pathway was affected by DFO and Dp44mT. Considering the critical roles of this pathway in cell proliferation and apoptosis (24, 25), we examined the effects of the chelators on the central MAPKs.
Chelators Increase the Phosphorylation of JNK and p38 MAPKs
In addition to the genes above, the array also identified genes corresponding to MAPK phosphatases (Table 2), which are also known as dual specificity phosphatases. These enzymes dephosphorylate threonine and tyrosine residues on MAPKs (39). The gene array and RT-PCR showed that the dual specificity phosphatases were significantly (p < 0.05) up-regulated to a similar extent by DFO and Dp44mT (Fig. 3A). These were DUSP5 (2.9-fold) > DUSP1 (1.7-fold) > DUSP16 (1.7-fold) and DUSP10 (1.3-fold), as determined from RT-PCR. Recently, other phosphatases have also been shown to be regulated by iron levels resulting in protein hyperphosphorylation in macrophages (40).
Although the mRNA expression of MAPK pathway-related genes was altered, the gene array did not reveal significant changes in the mRNA expression of major MAPKs, namely ERK1/2, JNK, and p38. Indeed, using RT-PCR, no significant alteration in the mRNA expression of these latter kinases was observed after a 24 h incubation with either DFO or Dp44mT at the same concentrations used in the gene array studies (Fig. 3B). However, this was not unexpected, as the activation of MAPKs is governed by their protein phosphorylation state (41), and so the latter was then examined.
Western blot analysis after incubation with DFO (2.5 or 250 μm) or Dp44mT (0.25 or 25 μm) demonstrated that the expression of phospho-ERK1/2 and ERK1/2 protein levels was not significantly (p > 0.05) altered (Fig. 3C). As a positive control, incubation of cells with sorbitol (500 mm) was used (42), and this significantly (p < 0.001) increased phospho-ERK1/2 expression. In contrast to ERK1/2, the phosphorylation of the other MAPKs, JNK and p38, was significantly affected by incubation of the cells with chelators (Fig. 3, D and E). Phosphorylation of JNK (JNK1 and JNK2/3 at 46 and 54 kDa, respectively (43)) was markedly and significantly (p < 0.001) increased after incubation with the highest concentrations of DFO or Dp44mT, as reflected by the ratio of phospho-JNK/JNK (Fig. 3D). The expression of nonphosphorylated JNK1 (46 kDa (43)) was much stronger in this cell type than the nonphosphorylated JNK2/3 (54 kDa (43)) in the control and treatment groups (Fig. 3D). Incubation with anisomycin (10 μg/ml) was also included as a positive control, as it is known to increase phosphorylation of JNK and p38 (44).
The ratio of phospho-p38 to nonphosphorylated p38 was also significantly (p < 0.01) increased even after incubation with the lower concentrations of DFO (2.5 μm) or Dp44mT (0.25 μm) (Fig. 3E). Nonphosphorylated p38 expression was not significantly altered after incubation with either of the chelators. Hence, these results showed that both chelators increased the phosphorylation of JNK and p38 MAPKs but not ERK1/2. Similar results were also observed in the SK-N-MC neuroepithelioma and SK-Mel-28 melanoma cell types (data not shown).
Chelators Affect the Phosphorylation of Other Molecules in MAPK Signaling
Because protein phosphorylation is crucial in MAPK signaling (24, 41, 45), we used a phospho-specific antibody array that focuses on MAPK signaling molecules to determine whether DFO or Dp44mT affected other MAPK-related targets. These studies were performed using DMS-53 lung carcinoma cells under the same conditions as the gene array (i.e. 250 μm DFO or 25 μm Dp44mT over a 24 h incubation). Because JNK and p38 were phosphorylated (Fig. 3, D and E), based on these Western results we were able to calculate a ratio between phosphorylated and nonphosphorylated forms and deduce a cutoff ratio for phosphorylation changes in the antibody array that were likely to be significant. Using a cutoff ratio of 1.1, we identified 19 and 12 alterations in the expression of phosphorylated proteins after incubation with DFO or Dp44mT, respectively (Table 3).
TABLE 3.
Alteration of protein phosphorylation in the MAPK signaling pathway due to DFO or Dp44mT
| Molecule | Phospho-site | Ratioa | Effects of phosphorylation on biological processes (phospho-site) |
|---|---|---|---|
| Increase in phosphorylation due to DFO | |||
| ATF2 | Thr-71 or -53 | 1.14 | Regulates cell cycle, cell growth, and transcription |
| c-Kit | Tyr-721 | 1.16 | Regulates cell adhesion |
| FAK | Tyr-925 | 1.12 | Regulates apoptosis, cell differentiation, and transcription |
| Histone H3.1 | Ser-10 | 1.18 | Unknown |
| HSP27 | Ser-78 | 1.12 | Cytoskeletal reorganization, regulates apoptosis, and cell cycle |
| HSP27 | Ser-82 | 1.16 | Cytoskeletal reorganization and regulates apoptosis |
| HSP27 | Ser-15 | 1.19 | Cytoskeletal reorganization and regulates apoptosis |
| IRS-1 | Ser-312 | 1.12 | Unknown |
| p38 MAPK | Tyr-182 | 1.23 | Cytoskeletal reorganization, regulates apoptosis, cell adhesion, cell cycle, motility; dual phosphorylation on Thr-180 and Tyr-182, which activates the enzyme |
| p53 | Ser-315 | 1.10 | Regulates apoptosis, cell cycle, cell growth, and transcription |
| p53 | Ser-37 | 1.11 | Regulates apoptosis and transcription |
| p53 | Ser-33 | 1.12 | Regulates apoptosis, cell growth, and transcription |
| p53 | Ser-46 | 1.13 | Regulates apoptosis, cell growth, and transcription |
| p53 | Ser-15 | 1.14 | Regulates apoptosis, cell cycle, cell growth, and transcription |
| p53 | Thr-18 | 1.15 | Regulates transcription |
| p53 | Ser-9 | 1.15 | Regulates apoptosis, cell cycle, and transcription |
| SAPK/JNK | Thr-183 | 1.17 | Regulates apoptosis, cell adhesion, and transcription; dual phosphorylation on Thr-183 and Tyr-185, which activates the enzyme |
| SAPK/JNK | Tyr-185 | 1.20 | Regulates apoptosis, cell adhesion, and transcription; dual phosphorylation on Thr-183 and Tyr-185, which activates the enzyme |
| Tau | Ser-356 | 1.14 | Unknown |
| Increase in phosphorylation due to Dp44mT | |||
| Histone H3.1 | Ser-10 | 1.18 | Unknown |
| HSP27 | Ser-15 | 1.10 | Cytoskeletal reorganization and regulates apoptosis |
| HSP27 | Ser-78 | 1.10 | Cytoskeletal reorganization, regulates apoptosis, and cell cycle |
| HSP27 | Ser-82 | 1.15 | Cytoskeletal reorganization and regulates apoptosis |
| p38 MAPK | Tyr-182 | 1.12 | Cytoskeletal reorganization, regulates apoptosis, cell adhesion, cell cycle, and cell motility; dual phosphorylation on Thr-180 and Tyr-182, which activates the enzyme |
| p53 | Ser-37 | 1.10 | Regulates apoptosis, cell growth, and transcription |
| p53 | Thr-18 | 1.11 | Regulates transcription |
| p53 | Ser-15 | 1.12 | Regulates apoptosis, cell cycle, cell growth, and transcription |
| p53 | Ser-9 | 1.13 | Regulates apoptosis, cell growth, and transcription |
| SAPK/JNK | Thr-183 | 1.14 | Regulates apoptosis, cell adhesion, transcription; dual phosphorylation on Thr-183 and Tyr-185, which activates the enzyme |
| SAPK/JNK | Tyr-185 | 1.26 | Regulates apoptosis, cell adhesion, transcription; dual phosphorylation on Thr-183 and Tyr-185; which activates the enzyme |
| Tau | Ser-396 | 1.14 | Unknown |
a Data are relative to control.
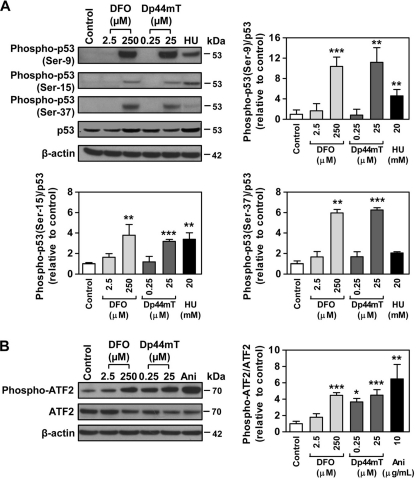
Consistent with the results presented in Fig. 3, D and E, there was increased expression of phosphorylated JNK (Thr-183 and Tyr-185) and p38 (Tyr-182) after incubation of cells with either chelator (Table 3). In addition, the antibody array identified multiple downstream targets of JNK and p38, which showed an increase in phosphorylation after incubation with DFO or Dp44mT (Table 3). Both chelators induced phosphorylation of p53 (Ser-9, Ser-15, Ser-37, and Thr-18), histone H3.1 (Ser-10), HSP27 (Ser-15, Ser-78, and Ser-82), and Tau (Ser-356 for DFO and Ser-396 for Dp44mT). Furthermore, incubation with DFO increased phosphorylation of p53 at Ser-315, Ser-33, and Ser-46 and also ATF-2 at Thr-71 or Thr-53. A previous study also showed that incubation with DFO increased phosphorylation of p53 at Ser-15 and Ser-20 (46).
Some of these alterations in phosphorylation (i.e. p53; Ser-9, -15, and -37 and ATF-2; Thr-71 or -53) were confirmed using Western blot analysis employing specific antibodies for the different phosphorylation sites as shown in Fig. 4. Similar to the antibody array studies, the Western analysis showed that DFO (250 μm) and Dp44mT (25 μm) significantly (p < 0.01–0.001) increased the ratio of phospho-p53(Ser-9)/p53, phospho-p53(Ser-15)/p53, and phospho-p53(Ser-37)/p53 (Fig. 4A). The ratio of phospho-ATF2 (Thr-71 or -53)/ATF2 was also significantly (p < 0.001) increased after incubation with DFO (250 μm) or Dp44mT (25 μm; Fig. 4B). However, the antibody array did not identify the increase of phospho-ATF2 after incubation with Dp44mT, because it was below the cutoff ratio used. Incubation with hydroxyurea or anisomycin served as positive controls, as they increase the phosphorylation of p53 and ATF-2, respectively (47). Although hydroxyurea significantly (p < 0.01) increased the ratio of phospho-p53 (Ser-9)/p53 and (Ser-15)/p53, it did not have any significant effect on the ratio of phospho-p53 (Ser 37)/p53 (Fig. 4A).
FIGURE 4.
Iron chelators increase phosphorylation of p53 and ATF2 that are downstream targets of JNK and p38 kinases. A, p53 at multiple phosphorylation sites; B, ATF2 (Thr-71) that was significantly increased upon incubation of DMS-53 cells with DFO (250 μm) or Dp44mT (25 μm) over 24 h/37 °C. These targets were identified using the phospho-MAPK antibody array (Table 3) and validated here by Western blotting. The panels and the densitometric analysis are typical experiments from three performed or mean ± S.E. of at least three experiments, respectively. *, p < 0.05; **, p < 0.01; ***, p < 0.001. HU, hydroxyurea; Ani, anisomycin.
JNK and p38 kinases play an important role in the activation of p53 via phosphorylation (48). In particular, phosphorylation of human p53 at Ser-15 and Ser-37 by both these kinases has been identified in response to stress stimuli (48), although other well known kinases, especially ataxia telangiectasia mutated, can also phosphorylate p53 at multiple sites upon DNA damage (48). Hence, these data suggested that a number of immediate targets of JNK and p38, such as p53 and ATF2, were phosphorylated after incubation with chelators, indicating the activation of these kinases. These results further demonstrated the effect of iron chelation on the MAPK pathway.
Iron Complexes of Dp44mT Increase Phosphorylation of JNK and p38 Kinases
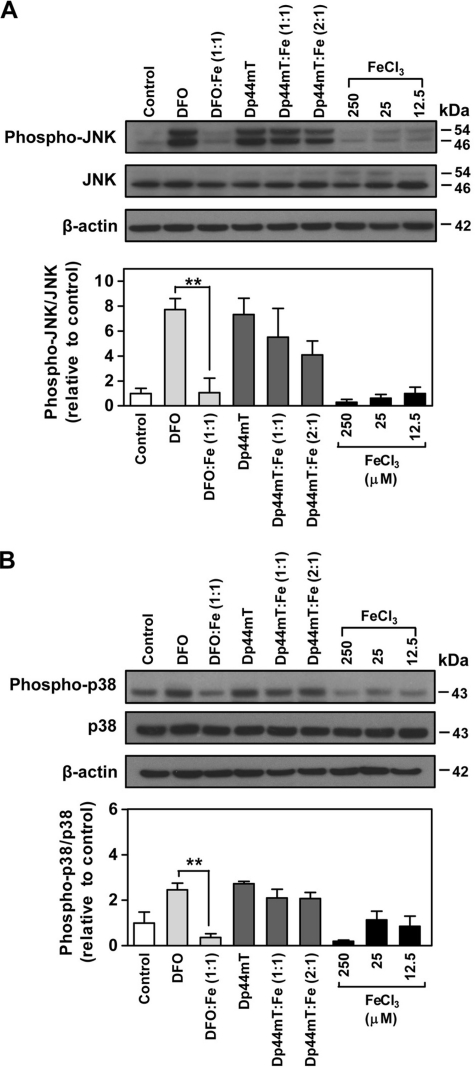
To understand the importance of iron complexation in the DFO- and Dp44mT-mediated phosphorylation of JNK and p38, DMS-53 cells were incubated for 24 h with the iron complexes of these ligands. These iron complexes were generated by pre-complexation of the chelator with iron(III) (in the form of FeCl3). The iron complex of DFO was prepared in a 1:1 ligand/iron molar ratio at 250 μm, because it is a hexadentate ligand, whereas Dp44mT was examined at 1:1 and 2:1 ligand/iron molar ratios (25 and 12.5 μm), as it is tridentate (8). Media containing the relevant concentrations of iron alone (i.e. 12.5, 2,5 and 250 μm) were also included as controls and showed that incubation of cells with iron alone (as FeCl3) had no significant effect on the phosphorylation of JNK or p38 (Fig. 5, A and B).
FIGURE 5.
Effect of chelator-iron complexes on the phosphorylation of JNK and p38. The expression of JNK (phospho-Thr-183/Tyr-185) (A) and p38 (phospho-Thr-180/Tyr-182) (B) in DMS-53 cells after incubation with either chelator alone (250 μm DFO or 25 μm Dp44mT) or chelator-iron(III) complexes at the same concentrations for 24 h/37 °C. The hexadentate DFO-iron complex was examined at a 1:1 molar ratio (chelator-iron(III) ratio; 250 μm). Tridentate Dp44mT was assessed at 1:1 molar ratio (chelator/iron(III) ratio; 25 μm) or 2:1 molar ratio (chelator/iron(III) ratio; 25:12.5 μm). The iron(III) complexes were pre-formed using ferric chloride (FeCl3). FeCl3 at 12.5, 25, or 250 μm was included as a relevant control as it is a component of the complexes. Western blotting was performed using rabbit anti-human phospho- and nonphospho-specific antibodies for JNK or p38. Densitometry of phospho- and nonphospho-JNK includes both bands. The panels and the densitometric analysis are a typical experiment from three performed or mean ± S.E. of at least three experiments, respectively. **, p < 0.01.
After incubation of cells with the DFO-iron complex, the ratio of phospho-JNK/JNK was significantly (p < 0.01) lower than that found for DFO, resulting in JNK phosphorylation that was similar to the control (Fig. 5A). In contrast, JNK phosphorylation induced by the Dp44mT-iron (1:1 and 2:1) complexes remained at a level that was not significantly different than that of Dp44mT alone. Similarly, studies examining phosphorylation of p38 showed that the DFO-iron complex significantly (p < 0.01) abolished the DFO-mediated increase in the ratio of phospho-p38/p38 (Fig. 5B). Conversely, the 1:1 or 2:1 Dp44mT-iron complexes caused a significant (p < 0.01) increase in the ratio of phospho-p38/p38 when compared with the control, and this was not significantly different than the effect of Dp44mT alone (Fig. 5B).
These results indicate that the ability of DFO to bind intracellular iron is crucial to mediate phosphorylation of JNK and p38, as its redox-inert iron complex (8) (which cannot bind intracellular iron) did not increase phosphorylation of these kinases. However, Dp44mT-iron complexes were still able to induce phosphorylation of JNK and p38 kinases. This is likely due to the pronounced redox activity of Dp44mT-iron complexes that induce ROS generation (13, 27, 49). Hence, in addition to Dp44mT-binding iron, its redox-active iron complex is also able to induce phosphorylation of JNK and p38, probably through the formation of ROS. In fact, various pro-oxidants such as H2O2 have been shown to increase JNK and p38 phosphorylation (50). It is important to note that the redox-inert DFO-iron complex possesses no anti-proliferative activity, whereas the Dp44mT-iron complexes exert anti-proliferative activity that is similar to that of Dp44mT alone (27). Hence, the anti-proliferative activity of the chelator-iron complexes or the chelator itself can be correlated with their ability to induce JNK and p38 phosphorylation that is known to have anti-proliferative and pro-apoptotic roles (23, 25).
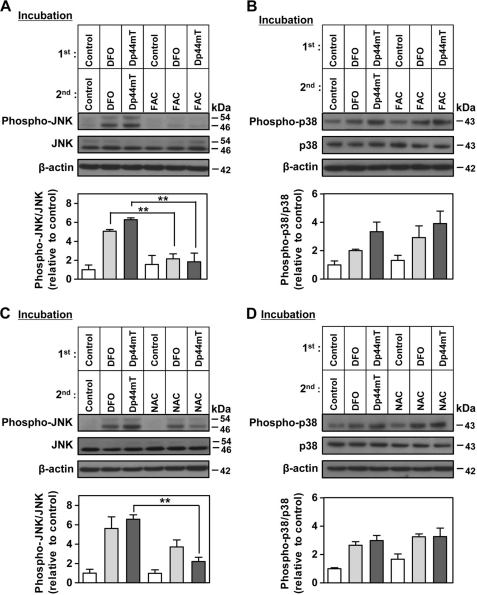
Iron Supplementation Restores Chelator-induced Phosphorylation of JNK but Not p38
To assess if intracellular iron depletion by DFO and Dp44mT mediated the increase of JNK and p38 phosphorylation, we examined the effect of cellular iron reconstitution using an established protocol implementing incubation with ferric ammonium citrate ([FAC] = 100 μg/ml) (30, 34, 51). In these studies, DMS-53 cells were first incubated with control media, Dp44mT (25 μm), or DFO (250 μm) for 24 h/37 °C and then re-incubated with FAC for a further 24 h/37 °C (Fig. 6, A and B). As a positive control for the regulation of cellular iron levels by chelators and FAC, the expression of the well characterized iron-regulated molecule, TfR1, was also assessed (30, 34). As expected, TfR1 expression was significantly (p < 0.001) increased 3–4-fold after incubation with DFO (250 μm) or Dp44mT (25 μm), indicating iron depletion. The chelator-mediated increase in the TfR1 level was then significantly (p < 0.001) decreased by half after supplementation of iron using FAC (100 μg/ml; data not shown), as we previously showed using this protocol (30, 34).
FIGURE 6.
Effect of iron or NAC supplementation on the DFO- or Dp44mT-induced phosphorylation of JNK and p38. The expression of JNK (phospho-Thr-183/Tyr-185) (A) and p38 (phospho-Thr-180/Tyr-182) (B) after DMS-53 cells were incubated (i.e. first incubation) with either DFO (250 μm) or Dp44mT (25 μm) for 24 h/37 °C and then re-incubated (i.e. second incubation) with either ferric ammonium citrate (100 μg/ml), DFO (250 μm), or Dp44mT (25 μm) for another 24 h/37 °C. The expressions of JNK (phospho-Thr-183/Tyr-185) (C) and p38 (phospho-Thr-180/Tyr-182) (D) after DMS-53 cells were incubated the same way as in A but using NAC (5 mm) in the second incubation rather than FAC. Western blotting was performed using anti-rabbit phospho- and nonphospho-specific antibodies for JNK or p38. Densitometry of phospho- and nonphospho-JNK includes both bands. The panels and the densitometric analysis are either a typical experiment from three performed or the mean ± S.E. of at least three experiments, respectively. **, p < 0.01.
Cells incubated with DFO (250 μm) or Dp44mT (25 μm) throughout the experiment showed an increase in the phospho-JNK/JNK ratio relative to cells incubated with control medium alone (Fig. 6A), which is consistent with the results in Fig. 3D. When cells were incubated in control medium for 24 h and then incubated with FAC for 24 h, there was no significant change in the phospho-JNK/JNK ratio as compared with cells incubated with control medium throughout (Fig. 6A). These data showed that incubation of control cells with FAC alone did not affect JNK phosphorylation as the cells were already iron-replete. However, re-incubation of cells with FAC after the incubation with DFO or Dp44mT significantly (p < 0.01) reduced the ratio of phospho-JNK/JNK to control levels (Fig. 6A). These results demonstrated that the addition of iron to chelator-treated cells resulted in diminished JNK phosphorylation and therefore demonstrated that DFO- or Dp44mT-mediated JNK phosphorylation was due to cellular iron depletion. Interestingly, when similar iron reconstitution experiments were performed to study the increased p38 phosphorylation after incubation with chelators, a different response was observed (Fig. 6B). In this case, the increase in the phospho-p38/p38 ratio after DFO or Dp44mT treatment was not reduced after re-incubation with FAC. This could be interpreted that iron-repletion under the conditions used did not rescue the increase in p38 phosphorylation (Fig. 6B), unlike the effect observed for phospho-JNK (Fig. 6A). Nevertheless, the ability of DFO to bind intracellular iron was important for its capacity to phosphorylate p38, because its iron complex did not increase phospho-p38 (Fig. 5B). These results highlight that the chelation-mediated increase in phospho-JNK and phospho-p38 responds differently to reconstitution of cellular iron levels. Indeed, the initial stimulus of iron chelation leads to an increase of phospho-p38 levels that were not restored by iron supplementation.
N-Acetylcysteine Supplementation Reduces the Effect of Dp44mT-induced Phosphorylation of JNK
Because the Dp44mT-iron complex is redox-active and increases ROS production in cells (13) and also elevates phosphorylation of JNK and p38 (Fig. 5, A and B), it was important to assess whether this latter effect was due to ROS generation. To assess this, a similar approach to the FAC re-incubation studies performed in Fig. 6, A and B, was employed, but using NAC (Fig. 6, C and D). This anti-oxidant increases intracellular glutathione levels that can quench ROS (52). Indeed, experiments in this study demonstrated that an incubation for 24 h/37 °C with NAC (5 mm) significantly (p < 0.001) increased cellular GSH levels by 151 + 5% (three experiments) relative to the control (100%; data not shown).
When cells were re-incubated with NAC after incubation with Dp44mT, a significant (p < 0.01) reduction of the phospho-JNK/JNK ratio was observed relative to cells re-incubated with Dp44mT (Fig. 6C). In contrast, re-incubation with NAC did not significantly reduce DFO-mediated phosphorylation of JNK. As a control, re-incubation with NAC alone after the initial incubation with control medium did not affect JNK phosphorylation relative to control medium. Similar to the studies examining phosphorylation of p38 after re-incubation with FAC (Fig. 6B), re-incubation with NAC after incubation with either DFO or Dp44mT did not decrease the phospho-p38/p38 ratio to control levels (Fig. 6D). Collectively, these results showed that Dp44mT-mediated JNK phosphorylation is mediated by both iron depletion and cellular ROS induction, whereas the response of phospho-p38 to the chelators was more complex.
Chelator-induced Dissociation of ASK1-Thioredoxin Complex and Phosphorylation of ASK1
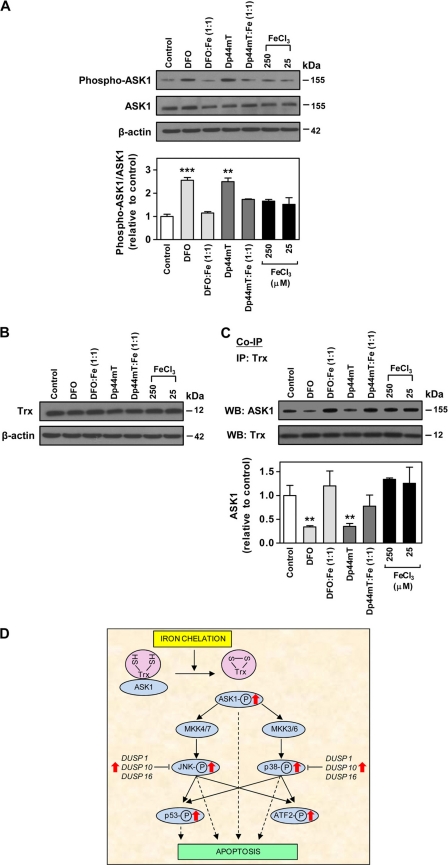
The mechanism involved in the increased phosphorylation of JNK and p38 observed above after incubation of cells with chelators was important to elucidate. A pathway known to induce phosphorylation of JNK and p38 has been shown to occur via ASK1 through activation of the MAPKK 4/7-JNK- and the MAPKK 3/6-p38-signaling cascade (45). In healthy cells, ASK1 forms an inactive complex with Trx, thus inhibiting the activation of this kinase (50, 53). When oxidation of Trx cysteine residues occur, the ASK1-Trx complex dissociates (50, 53), leading to the phosphorylation and activation of ASK1 (50, 53). Our previous studies have shown that DFO and Dp44mT increase Trx oxidation, as these chelators reduce the activity of thioredoxin reductase (64). Hence, we hypothesized that phosphorylation of JNK and p38 after cellular iron depletion could be mediated by ASK1, as the kinase becomes phosphorylated as a result of its dissociation from the ASK1-Trx complex (50, 53).
To assess the role of ASK1, we first examined the level of ASK1 phosphorylation after incubation of DMS-53 cells with DFO (250 μm) or Dp44mT (25 μm) over 24 h/37 °C. The results showed that DFO or Dp44mT significantly (p < 0.01–0.001) increased the phospho-ASK1/ASK1 ratio to 2.55 ± 0.16- and 2.50 ± 0.16-fold of the control, respectively (Fig. 7A). Iron complexes (1:1 ligand/metal ratio) of DFO and Dp44mT were also tested. Iron alone at the relevant concentration (25 or 250 μm) used for the formation of the chelator complexes did not have any significant effect on ASK1 phosphorylation relative to control. In addition, the DFO-iron complexes did not increase ASK1 phosphorylation relative to the control, although the Dp44mT-iron complex caused a slight but not significant increase in the phospho-ASK1/ASK1 ratio. Thus, as per the effect of these chelator-iron complexes on JNK and p38 (Fig. 5, A and B), these results showed that the iron-binding property of the chelators was important for the increase in ASK1 phosphorylation.
FIGURE 7.
Effect of DFO and Dp44mT on phospho-ASK1 and ASK1-Trx complex. A, expression of phospho-ASK1 (Ser-83) and ASK1 in DMS-53 cells after incubation with either chelator alone (250 μm DFO or 25 μm Dp44mT) or chelator-iron(III) complexes at 1:1 (chelator/iron) molar ratio as determined by Western blotting using phospho- and nonphospho-specific antibodies for ASK1. B, expression of Trx in DMS-53 cells after incubation as in A. C, co-immunoprecipitation (Co-IP) examining the ASK1-Trx interaction as determined in DMS-53 cells after incubation as in A using mouse anti-human Trx antibody. The levels of ASK1 and Trx in the immunoprecipitates (IP) were determined using rabbit anti-human ASK1 and Trx antibodies. The panels and the densitometric analysis are either a typical experiment from three performed or the mean ± S.E. of at least three experiments, respectively. **, p < 0.01; ***, p < 0.001. WB, Western blot. D, schematic summary of the effect of iron chelation on the MAPK signaling transduction pathway. The chelators, DFO and Dp44mT increase phosphorylation of ASK1 because of the dissociation of the ASK1-Trx complex that is caused by the oxidation of Trx.4 This results in elevation of phospho-ASK1, which can modulate signaling cascades to increase phosphorylation of JNK and p38. A number of dual specificity phosphatases (DUSPs), including DUSP1, DUSP10, and DUSP16, which directly de-phosphorylate JNK and p38, were also increased by the chelators. This latter effect could be a counteractive response to the increased phosphorylation of JNK and p38. Hyperphosphorylation of p53 at multiple sites was also prominent as it is directly targeted by the JNK and p38 pathways. JNK, p38, ASK1, and p53 are pro-apoptotic molecules, and their increased phosphorylation could serve as important signals leading to apoptosis after iron depletion mediated by chelators.
Considering that ASK1 forms a complex with Trx (50, 53), the expression of the latter was assessed after incubation with chelators. The ligands DFO, Dp44mT, or their 1:1 iron complexes did not affect the protein expression of Trx in cells (Fig. 7B). To assess if the increase in phospho-ASK1 expression was due to dissociation of ASK1-Trx, we performed co-immunoprecipitation to examine the levels of the ASK1-Trx complex after incubation of DMS-53 cells with DFO (250 μm) or Dp44mT (25 μm) over 24 h/37 °C. We observed that incubation with DFO or Dp44mT significantly (p < 0.01) reduced the ASK1-thioredoxin complex to 34 ± 3 and 35 ± 5% (n = 4) of the control, respectively (Fig. 7C). Furthermore, the iron complexes (1:1 ratio) of both chelators were not able to significantly reduce the level of the ASK1-Trx complex relative to the control. These results are consistent with the studies on the phosphorylation of ASK1 (Fig. 7A). To ensure equal amounts of thioredoxin were successfully immunoprecipitated from the cell lysates, Western analysis was performed, and the Trx level was found to be equal (Fig. 7C). Similar data were also obtained using the SK-N-MC and SK-Mel-28 cell types (data not shown). Therefore, these results demonstrated that increased phosphorylation of JNK and p38 after iron chelation was, at least in part, mediated by the dissociation of ASK1-Trx and thus the activation of ASK1.
DISCUSSION
It is increasingly recognized that iron plays roles in cell signaling pathways (1, 2). Because iron chelators induce cell cycle arrest and apoptosis (4, 6, 22), it was important to examine the molecular signaling pathways and mechanisms leading to these processes. In this study, we examined the MAPK pathway because of the high number of genes altered in this pathway upon incubation of cells with DFO or Dp44mT. Indeed, our investigation demonstrated that iron depletion mediated by chelators increases the phosphorylation of JNK and the p38 MAPKs, as well as the phosphorylation of the downstream targets of JNK and p38, namely p53 and ATF-2 (48, 54, 55). The increased phosphorylation of JNK and p38 could be mediated by ASK1 that was activated as a result of dissociation from its complex with Trx after iron chelation.
For the first time, our results demonstrate that DFO and Dp44mT increase phosphorylation of ASK1 as the ASK1-Trx complex dissociates as a consequence of Trx oxidation after iron chelation, as summarized in Fig. 7D. Hence, elevated phospho-ASK1 could mediate a signaling cascade leading to increased phosphorylation of the JNK and p38 MAPK pathway. In addition, the expression of a number of genes in the MAPK pathway was also affected by DFO and Dp44mT, and these include DUSP1, DUSP10, and DUSP16, which serve as phosphatases that de-phosphorylate JNK and p38. The increased mRNA expression of these phosphatases can be speculated to be a counteractive response to the up-regulation of phospho-JNK and p38. Hyper-phosphorylation of p53 at multiple sites was also prominent after incubation with DFO or Dp44mT and could be mediated by the JNK and p38 pathways.
The JNK and p38 pathways are largely tumor suppressive considering their role in apoptosis and cell cycle control (24, 56). Thus, the chelator-mediated phosphorylation of these molecules may be important upstream mediators of apoptosis. Indeed, the selective p38 inhibitor, SB203580, has been shown to suppress DFO-induced apoptosis (57, 58). Furthermore, these stress-activated protein kinases have also been implicated in cell cycle regulation (56). Of relevance, p38 is proposed to induce G1/S arrest through a variety of mechanisms, including direct activation of p53 phosphorylation, direct stabilization of p21WAF1/CIP1, and decreased cyclin D1 protein stability (56). JNK is also known to increase stability of p21WAF1/CIP1 and phosphorylate p53 (56). Thus, activation of p38 and JNK may also play a role in iron chelation-induced cell cycle arrest. In fact, we observed increased phosphorylation of p53 at multiple sites (Fig. 4A). Previously, iron chelation-induced activation of p53 was shown to occur through HIF-1α-dependent and -independent mechanisms (59). Thus, activation of JNK and p38 after incubation with chelators could also lead to p53 activation, because p53 is the immediate downstream target of these kinases (56).
Incubation of cells with DFO or Dp44mT also increased ASK1 phosphorylation (Fig. 7A), which plays a role as an apoptosis-signaling molecule in the MAPK pathway. ASK1 executes apoptosis mainly via mitochondrion-dependent caspase activation (60). Overexpression of wild type ASK1 induces cytochrome c release from the mitochondrion, which activates caspase-3 and -9 (60). Our previous studies have shown that Dp44mT-induced tumor cell apoptosis was mediated through the release of mitochondrial cytochrome c to the cytosol and activation of caspase-3, -8, and -9 (13). Hence, phosphorylation of ASK1 after iron chelation can either promote activation of JNK and p38 leading to apoptosis and/or directly result in the apoptosis cascade.
As demonstrated by iron supplementation experiments (Fig. 6A), DFO and Dp44mT-induced cellular iron depletion was an important factor that activated JNK. These results highlight the sensitivity of the JNK-signaling pathway toward cellular iron levels. Furthermore, previous investigations have also shown that JNK signaling can stimulate ferritin degradation and thus the level of cellular iron pools (1). These results indicate an important association between iron metabolism and the JNK-signaling pathway. It should be noted that previous studies have examined the effect of DFO on MAPKs and demonstrated increased phospho-ERK1/2 levels after short incubations of up to 12 h (61, 62), which may be related to an early response against stress. Clearly, these latter results are in contrast to the increased levels of phospho-JNK and -p38 observed in this study after 24 h, where the anti-proliferative effects of these pathways are probably involved in the activity of chelators against tumor cells.
In comparison with DFO, Dp44mT-induced JNK activation is mediated via iron depletion and the ability of its iron complex to redox cycle (49), thus promoting ROS generation (13). This is shown by the observation that the Dp44mT-iron complex increased JNK phosphorylation, unlike the redox-inert DFO-iron complex (Fig. 5A) (8). In addition, supplementation of cells with GSH by incubation with NAC rescued the effect of Dp44mT on increasing JNK phosphorylation, although NAC had no effect on the ability of DFO to induce this effect (Fig. 6C). Considering these results, it is of interest that the JNK-signaling pathway is ROS-sensitive (24). However, unlike JNK and p38, it remains unclear why the ASK1-Trx complex and ASK1 phosphorylation were not significantly affected by the redox-active Dp44mT-iron complex (27, 49), because the ASK1-Trx complex is also known to be a sensor of redox state (50). Potentially, this could be explained by the differential access of these complexes into various cellular compartments, therefore affecting the response of different molecules.
Considering this study, it is of interest to note that blocking the p38 pathway using the inhibitor, SB202190, resulted in reduced total iron uptake in erythroid cells, highlighting the potential significance of this pathway in affecting iron metabolism (63). Indeed, the alterations in p38 phosphorylation observed in this study after manipulation of cellular iron status indicate that this signaling pathway has potential roles in regulating molecules involved in iron metabolism.
In conclusion, this investigation shows for the first time that iron chelation can modulate the ASK1-Trx-signaling system, which activates the MAPK signaling pathway by increasing phosphorylation of JNK and p38 kinases. DFO-induced JNK phosphorylation is solely reliant on the ability of this chelator to deplete iron, whereas the effect of Dp44mT can be explained by both its ability to deplete iron and to generate ROS. Up-regulation of the MAPK signaling pathway may play an important role in mediating the cell cycle arrest and apoptosis induced by iron depletion.
Acknowledgments
We acknowledge the critical comments on the manuscript prior to submission by Dr. Katie Dixon, Dr. Patric Jansson, Dr. Zaklina Kovacevic, Dr. Darius Lane, Dr. David Lovejoy, Dr. Helena Mangs, and Angelica Merlot of the Iron Metabolism and Chelation Program. Dr. Yohan Suryo Rahmanto of the Department of Pathology, University of Sydney, is also thanked for critical input.
This work was supported in part by a Senior Principal Research Fellowship, Project Grants from the National Health and Medical Research Council of Australia, and a Discovery Grant from the Australian Research Council (to D. R. R.).
- DFO
- desferrioxamine
- Dp44mT
- di-2-pyridylketone-4,4-dimethyl-3 thiosemicarbazone
- ROS
- reactive oxygen species
- MAP3K
- MAPK kinase kinase
- Trx
- thioredoxin
- FAC
- ferric ammonium citrate
- NAC
- N-acetylcysteine
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Antosiewicz J., Ziolkowski W., Kaczor J. J., Herman-Antosiewicz A. (2007) Free Radic. Biol. Med. 43, 265–270 [DOI] [PubMed] [Google Scholar]
- 2. Deb S., Johnson E. E., Robalinho-Teixeira R. L., Wessling-Resnick M. (2009) Biometals 22, 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Babitt J. L., Huang F. W., Xia Y., Sidis Y., Andrews N. C., Lin H. Y. (2007) J. Clin. Invest. 117, 1933–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Le N. T., Richardson D. R. (2002) Biochim. Biophys. Acta 1603, 31–46 [DOI] [PubMed] [Google Scholar]
- 5. Brodie C., Siriwardana G., Lucas J., Schleicher R., Terada N., Szepesi A., Gelfand E., Seligman P. (1993) Cancer Res. 53, 3968–3975 [PubMed] [Google Scholar]
- 6. Hileti D., Panayiotidis P., Hoffbrand A. V. (1995) Br. J. Haematol. 89, 181–187 [DOI] [PubMed] [Google Scholar]
- 7. Buss J. L., Torti F. M., Torti S. V. (2003) Curr. Med. Chem. 10, 1021–1034 [DOI] [PubMed] [Google Scholar]
- 8. Kalinowski D. S., Richardson D. R. (2005) Pharmacol. Rev. 57, 547–583 [DOI] [PubMed] [Google Scholar]
- 9. Donfrancesco A., Deb G., Dominici C., Pileggi D., Castello M. A., Helson L. (1990) Cancer Res. 50, 4929–4930 [PubMed] [Google Scholar]
- 10. Blatt J., Stitely S. (1987) Cancer Res. 47, 1749–1750 [PubMed] [Google Scholar]
- 11. Estrov Z., Tawa A., Wang X. H., Dubé I. D., Sulh H., Cohen A., Gelfand E. W., Freedman M. H. (1987) Blood 69, 757–761 [PubMed] [Google Scholar]
- 12. Selig R. A., Madafiglio J., Haber M., Norris M. D., White L., Stewart B. W. (1993) Anticancer Res. 13, 721–725 [PubMed] [Google Scholar]
- 13. Yuan J., Lovejoy D. B., Richardson D. R. (2004) Blood 104, 1450–1458 [DOI] [PubMed] [Google Scholar]
- 14. Whitnall M., Howard J., Ponka P., Richardson D. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14901–14906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kulp K. S., Green S. L., Vulliet P. R. (1996) Exp. Cell Res. 229, 60–68 [DOI] [PubMed] [Google Scholar]
- 16. Simonart T., Degraef C., Andrei G., Mosselmans R., Hermans P., Van Vooren J. P., Noel J. C., Boelaert J. R., Snoeck R., Heenen M. (2000) J. Invest. Dermatol. 115, 893–900 [DOI] [PubMed] [Google Scholar]
- 17. Lucas J. J., Szepesi A., Domenico J., Takase K., Tordai A., Terada N., Gelfand E. W. (1995) Blood 86, 2268–2280 [PubMed] [Google Scholar]
- 18. Fukuchi K., Tomoyasu S., Tsuruoka N., Gomi K. (1994) FEBS Lett. 350, 139–142 [DOI] [PubMed] [Google Scholar]
- 19. Gao J., Richardson D. R. (2001) Blood 98, 842–850 [DOI] [PubMed] [Google Scholar]
- 20. Nurtjahja-Tjendraputra E., Fu D., Phang J. M., Richardson D. R. (2007) Blood 109, 4045–4054 [DOI] [PubMed] [Google Scholar]
- 21. Saletta F., Rahmanto Y. S., Noulsri E., Richardson D. R. (2010) Mol. Pharmacol. 77, 443–458 [DOI] [PubMed] [Google Scholar]
- 22. Noulsri E., Richardson D. R., Lerdwana S., Fucharoen S., Yamagishi T., Kalinowski D. S., Pattanapanyasat K. (2009) Am. J. Hematol. 84, 170–176 [DOI] [PubMed] [Google Scholar]
- 23. Rao V. A., Klein S. R., Agama K. K., Toyoda E., Adachi N., Pommier Y., Shacter E. B. (2009) Cancer Res. 69, 948–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dhillon A. S., Hagan S., Rath O., Kolch W. (2007) Oncogene 26, 3279–3290 [DOI] [PubMed] [Google Scholar]
- 25. Raman M., Chen W., Cobb M. H. (2007) Oncogene 26, 3100–3112 [DOI] [PubMed] [Google Scholar]
- 26. Lewis T. S., Shapiro P. S., Ahn N. G. (1998) Adv. Cancer Res. 74, 49–139 [DOI] [PubMed] [Google Scholar]
- 27. Richardson D. R., Sharpe P. C., Lovejoy D. B., Senaratne D., Kalinowski D. S., Islam M., Bernhardt P. V. (2006) J. Med. Chem. 49, 6510–6521 [DOI] [PubMed] [Google Scholar]
- 28. Kalinowski D. S., Yu Y., Sharpe P. C., Islam M., Liao Y. T., Lovejoy D. B., Kumar N., Bernhardt P. V., Richardson D. R. (2007) J. Med. Chem. 50, 3716–3729 [DOI] [PubMed] [Google Scholar]
- 29. Richardson D. R., Baker E. (1990) Biochim. Biophys. Acta 1053, 1–12 [DOI] [PubMed] [Google Scholar]
- 30. Le N. T., Richardson D. R. (2004) Blood 104, 2967–2975 [DOI] [PubMed] [Google Scholar]
- 31. Suryo Rahmanto Y., Dunn L. L., Richardson D. R. (2007) Carcinogenesis 28, 2172–2183 [DOI] [PubMed] [Google Scholar]
- 32. Zhong D., Liu X., Khuri F. R., Sun S. Y., Vertino P. M., Zhou W. (2008) Cancer Res. 68, 7270–7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Richardson D., Ponka P., Baker E. (1994) Cancer Res. 54, 685–689 [PubMed] [Google Scholar]
- 34. Darnell G., Richardson D. R. (1999) Blood 94, 781–792 [PubMed] [Google Scholar]
- 35. Beerepoot L. V., Shima D. T., Kuroki M., Yeo K. T., Voest E. E. (1996) Cancer Res. 56, 3747–3751 [PubMed] [Google Scholar]
- 36. Richardson D. R. (2005) Curr. Med. Chem. 12, 2711–2729 [DOI] [PubMed] [Google Scholar]
- 37. Takekawa M., Saito H. (1998) Cell 95, 521–530 [DOI] [PubMed] [Google Scholar]
- 38. Wang X. Z., Ron D. (1996) Science 272, 1347–1349 [DOI] [PubMed] [Google Scholar]
- 39. Jeffrey K. L., Camps M., Rommel C., Mackay C. R. (2007) Nat. Rev. Drug Discov. 6, 391–403 [DOI] [PubMed] [Google Scholar]
- 40. Gomez M. A., Alisaraie L., Shio M. T., Berghuis A. M., Lebrun C., Gautier-Luneau I., Olivier M. (2010) J. Biol. Chem. 285, 24620–24628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bardwell L., Shah K. (2006) Methods 40, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abe J., Kusuhara M., Ulevitch R. J., Berk B. C., Lee J. D. (1996) J. Biol. Chem. 271, 16586–16590 [DOI] [PubMed] [Google Scholar]
- 43. Park Y. C., Ye H., Hsia C., Segal D., Rich R. L., Liou H. C., Myszka D. G., Wu H. (2000) Cell 101, 777–787 [DOI] [PubMed] [Google Scholar]
- 44. Cano E., Hazzalin C. A., Mahadevan L. C. (1994) Mol. Cell. Biol. 14, 7352–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cuevas B. D., Abell A. N., Johnson G. L. (2007) Oncogene 26, 3159–3171 [DOI] [PubMed] [Google Scholar]
- 46. Ashcroft M., Taya Y., Vousden K. H. (2000) Mol. Cell. Biol. 20, 3224–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Clerk A., Sugden P. H. (1997) Biochem. J. 325, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu G. S. (2004) Cancer Biol. Ther. 3, 156–161 [DOI] [PubMed] [Google Scholar]
- 49. Jansson P. J., Hawkins C. L., Lovejoy D. B., Richardson D. R. (2010) J. Inorg. Biochem. 104, 1224–1228 [DOI] [PubMed] [Google Scholar]
- 50. Matsuzawa A., Ichijo H. (2008) Biochim. Biophys. Acta 1780, 1325–1336 [DOI] [PubMed] [Google Scholar]
- 51. Fu D., Richardson D. R. (2007) Blood 110, 752–761 [DOI] [PubMed] [Google Scholar]
- 52. Schafer F. Q., Buettner G. R. (2001) Free Radic. Biol. Med. 30, 1191–1212 [DOI] [PubMed] [Google Scholar]
- 53. Saitoh M., Nishitoh H., Fujii M., Takeda K., Tobiume K., Sawada Y., Kawabata M., Miyazono K., Ichijo H. (1998) EMBO J. 17, 2596–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gupta S., Campbell D., Dérijard B., Davis R. J. (1995) Science 267, 389–393 [DOI] [PubMed] [Google Scholar]
- 55. Zhu T., Lobie P. E. (2000) J. Biol. Chem. 275, 2103–2114 [DOI] [PubMed] [Google Scholar]
- 56. Wilkinson M. G., Millar J. B. (2000) FASEB J. 14, 2147–2157 [DOI] [PubMed] [Google Scholar]
- 57. Choi S. C., Kim B. S., Song M. Y., Choi E. Y., Oh H. M., Lyou J. H., Han W. C., Moon H. B., Kim T. H., Oh J. M., Chung H. T., Jun C. D. (2003) Free Radic. Biol. Med. 35, 1171–1184 [DOI] [PubMed] [Google Scholar]
- 58. Kim B. S., Yoon K. H., Oh H. M., Choi E. Y., Kim S. W., Han W. C., Kim E. A., Choi S. C., Kim T. H., Yun K. J., Kim E. C., Lyou J. H., Nah Y. H., Chung H. T., Cha Y. N., Jun C. D. (2002) Cell. Immunol. 220, 96–106 [DOI] [PubMed] [Google Scholar]
- 59. Wenger R. H., Camenisch G., Desbaillets I., Chilov D., Gassmann M. (1998) Cancer Res. 58, 5678–5680 [PubMed] [Google Scholar]
- 60. Matsuzawa A., Ichijo H. (2001) J. Biochem. 130, 1–8 [DOI] [PubMed] [Google Scholar]
- 61. Kim B. M., Chung H. W. (2008) Toxicol. Appl. Pharmacol. 228, 24–31 [DOI] [PubMed] [Google Scholar]
- 62. Seo G. S., Lee S. H., Choi S. C., Choi E. Y., Oh H. M., Choi E. J., Park D. S., Kim S. W., Kim T. H., Nah Y. H., Kim S., Kim S. H., You S. H., Jun C. D. (2006) Free Radic. Biol. Med. 40, 1502–1512 [DOI] [PubMed] [Google Scholar]
- 63. Mardini L., Gasiorek J., Derjuga A., Carrière L., Schranzhofer M., Paw B. H., Ponka P., Blank V. (2010) Biochem. J. 432, 145–151 [DOI] [PubMed] [Google Scholar]
- 64. Yu Y., Suryo Rahmanto Y., Hawkins C. L., Richardson D. R. (2011) Mol. Pharmacol., in press [DOI] [PubMed] [Google Scholar]