Abstract
EfeB/YcdB is a member of the dye-decolorizing peroxidase (DyP) protein family. A recent study has shown that this protein can extract iron from heme without breaking the tetrapyrrole ring. We report the crystal structure of EfeB from Escherichia coli O157 bound to heme at 1.95 Å resolution. The EfeB monomer contains two domains. The heme molecule is located in a large hydrophobic pocket in the C-terminal domain. A long loop connecting the two domains extensively interacts with the heme, which is a distinctive structural feature of EfeB homologues. A large tunnel formed by this loop and the β-sheet of C-terminal domain provides a potential cofactor/substrate binding site. Biochemical data show that the production of protoporphyrin IX (PPIX) is closely related to the peroxidation activity. The mutant D235N keeps nearly the same activity of guaiacol peroxidase as the wild-type protein, whereas the corresponding mutation in the classic DyP protein family completely abolished the peroxidation activity. These results suggest that EfeB is a unique member of the DyP protein family. In addition, dramatically enhanced fluorescence excitation and emission of EfeB-PPIX was observed, implying this protein may be used as a red color fluorescence marker.
Keywords: Crystal Structure, Heme, Iron Metabolism, Peroxidase, Porphyrin, EfeB, PPIX, Heme Deferrochelatase
Introduction
Iron is an essential element for most living organisms (1). The ability to acquire enough iron from an environment with little free iron in mammalian hosts is a key feature of most successful pathogens (2). Secretion and utilization of iron-sequestering compounds known as siderophores are an iron-uptake strategy employed by pathogenic bacteria (3–4).
Heme is the most abundant iron-containing compound in vertebrates. The binding affinity of iron with the tetrapyrrole ring is extremely high, and there is no evidence that any siderophore can retrieve iron from the heme. To make full use of the rich source of iron in the host, pathogenic bacteria have developed diverse mechanisms for heme uptake and iron release. In general, the heme utilization system consists of three parts: a heme receptor that scavenges the heme molecule, a specific ABC transporter that delivers heme into cytosol, and enzymes that release iron from the heme tetrapyrrole ring (5–7). It has long been observed that heme transport and utilization is closely related to the virulence of pathogenic Escherichia coli strains (8–12).
The enzymes involved in iron release have drawn significant attention for a long time because of a sophisticated catalytic mechanism of iron discharge (13–16). Most identified enzymes release iron through cleavage of the tetrapyrrole ring (6, 17). These enzymes can be divided into two subfamilies according to their three-dimensional structures and reaction products. The first group of enzymes exist as monomers and belong to orthologs of the mammalian heme oxygenase, which is characterized by production of biliverdin, carbon monoxide (CO), and iron (7, 18–19). The second group exists as a functional dimer and also degrades the heme molecule with CO and iron release. However, biliverdin was not identified from resulting products (20–21).
Recently, EfeB/YcdB and YfeX from Escherichia coli were reported to be enzymes catalyzing the release of iron from heme (22). Biochemical studies showed that YfeX is located in the cytoplasm. Surprisingly, EfeB was found to be a Tat-dependent translated hemoprotein (22–24). The supposition that EfeB and YfeX have deferrochelation activity was supported by the accumulation of protoporphyrin IX (PPIX)2 in cells overexpressing these two proteins (22). It was proposed that this kind of enzyme has an advantage over previously identified enzymes in that EfeB and YfeX release iron without the production of CO, an antibacterial gas. Homologues of EfeB and YfeX are widespread in the bacterial kingdom, indicating that this group of enzyme is crucial for bacterial survival and propagation. Because EfeB and YfeX do not exist in higher eukaryotic organisms, they may serve as latent targets of antimicrobial drugs.
Although the exact catalytic mechanism of EfeB remains largely unknown, the involvement of this protein in bacterial iron acquisition is not in doubt. Interestingly, a genomic study showed that the expression level of EfeB increased in response to both iron deficiency and low pH (25–28). Moreover, it was found that EfeB/YcdB had modest guaiacol peroxidase activity at acidic pH values (23, 29). The bi-functional property of EfeB makes it a puzzling protein and raises a question about the correlation between two seemingly unrelated functions.
To gain further insight into the mechanism underlying the complicated enzymatic activities, we overexpressed EfeB from E. coli (strain O157) and determined the crystal structure of its complex with heme at 1.95 Å resolution. The structure shows that EfeB exists as homodimers with each monomer containing a conserved heme binding site. A long loop designated as the switch loop, which contains residues 222–243 and connects the N-terminal and C-terminal domains, is a distinctive structural feature of EfeB. Biochemical data of the correlation between the production of PPIX and the peroxidation activity of EfeB strongly suggest that this protein is an outsider from the DyP peroxidase family. In addition, dramatically enhanced fluorescence excitation and emission of EfeB-PPIX were observed, implying this protein may be used as a red color fluorescence marker.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Site-directed Mutagenesis
The efeB gene was amplified from genomic DNA of E. coli O157 and was subcloned into the pET21b vector (Novagen) between NdeI and XhoI restriction cutting sites. N-terminal Tat signal sequence (residues 1–35) of EfeB was removed during cloning resulting in a construct of 36–423-His tag fusion for protein expression. Based on the structure of the EfeB-heme complex and sequence conservation analysis, 16 efeB mutants (Q68W, G223F, P227F, D235A, D235N, D235W, G236F, N239W, H329A, L346W, R347E G349F G349L, S351Q, L366W, F368W) were produced using a two-step PCR strategy.
BL21 (DE3) cells carrying the plasmid of the recombinant EfeB protein were grown in L Broth media supplemented with 100 μg/ml ampicillin at 37 °C. Once an A600 of 1.0 was reached, the incubation temperature was decreased to 15 °C. Protein expression was induced by adding IPTG to a final concentration of 0.12 mm. The cells were harvested by centrifugation after 12 h of expression. The red color as well as the fluorescent characteristic of the cell indicates the accumulation of PPIX (22). The pellet was resuspended in lysis buffer (25 mm Tris-HCl, pH 8.0, 200 mm NaCl, and 1 mm PMSF), and lysed by sonication. The cell lysate was incubated with hemin at a final concentration of 50 μm at room temperature for 30 min and then centrifuged for 45 min at 28,500 × g. The soluble fractions containing the EfeB-heme complex were loaded onto a Ni-chelating Sepharose (GE Healthcare) affinity column equilibrated with lysis buffer. The affinity column was extensively washed with lysis buffer. All proteins were eluted by elution buffer (25 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 250 mm imidazole). Alone, the red color (no fluorescence) of the elution indicated that heme binds to EfeB. The elution was then loaded onto an ion exchange column (Source 15Q, GE Healthcare) and was eluted using a linear 150-ml gradient of 0–0.5 m NaCl. Finally, the EfeB-heme complex was purified using size exclusion (Superdex-200, GE Healthcare) chromatography. Fractions were pooled according to protein purity analyzed by SDS-PAGE, and the final protein concentration was 10 mg/ml. All mutant proteins were purified using the same procedure.
The Se-Met-labeled EfeB-heme complex was expressed using E. coli grown in Se-Met-containing medium using the metabolic inhibition pathway, and it was purified using the same procedure as that of the EfeB-heme complex. To purify apo-EfeB that was used for titration experiments, the cell lysate was directly purified using the protocol of EfeB-heme complex without incubation with hemin. The colorless fractions of apo-EfeB were collected in each step of the purification. The EfeB-PPIX complex was purified following the protocol of the EfeB-heme complex, except that the cell lysate was incubated with PPIX.
Crystallization and Data Collection
Both heme-bound EfeB and heme-bound Se-EfeB were crystallized using the hanging drop diffusion method at 20 °C by mixing equal volumes of protein complex with reservoir solution containing 0.2 m NH4Cl and 20% PEG3350. Crystals appeared after about 1 week and reached full size in 2 weeks. Both native and anomalous diffraction data were collected at Shanghai Synchrotron Radiation facility (SSRF) beamline BL17u1. The best data were obtained from a crystal of heme-bound Se-EfeB. The crystal belongs to space group P21 and contains two dimers per asymmetric unit. The unit cell dimensions are a = 50.64 Å, b = 155.568 Å, c = 97.374 Å, and β = 94.696°. To prevent radiation damage, crystals were equilibrated in a cryoprotectant buffer containing 15% glycerol (v/v) plus reservoir buffer and then flash frozen in a 100 K nitrogen stream. Data sets were processed using the HKL2000 software suite (30).
Structure Determination
The EfeB structure was solved by single anomalous dispersion (SAD) phasing. Twenty Se sites were found using the program SOLVE (31). Initial single anomalous dispersion phases were then improved, and the chain was automatically traced using the program RESOLVE (32). The atomic model was built using COOT (33) and refined using PHENIX (34). 2Fo-Fc and Fo-Fc electron density maps show that each protein molecule binds to one heme molecule in a large hydrophobic pocket. Data collection and structure refinement statistics are summarized in Table 1. All the molecular graphics figures were generated using PyMol.
TABLE 1.
Statistics of crystallographic analysis
| Data collection | |
| Space group | P21 |
| Unit cell (Å) | a = 50.640 b = 155.568 c = 97.374 β = 94.70 |
| Resolution (Å) | 50-1.95 |
| Completeness | 96.1 (92.4)a |
| Redundancy | 6.5 (5.2) |
| I/σ(I) | 20.4 (3.6) |
| Rsymb(%) | 10.3 (44.8) |
| Refinement | |
| Resolution | 50-1.95 |
| Rwork/Rfreec(%) | 17.03/20.57 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.106 |
| Ramachandran plot (%)d | |
| Most favored (%) | 91.7 |
| Additionally allowed (%) | 7.8 |
| Generously allowed | 0.5 |
| Disallowed (%) | 0.0 |
a Values in parentheses are for reflections in the highest resolution shell.
b Rsym = ΣhklΣi|I(hkl)i − <I(hkl)>/ΣhklΣI<I(hkl)i> over i observations.
c Value of Rfree for 1.91% of randomly selected reflections excluded from refinement.
d As defined in PROCHECK.
Isothermal Titration Calorimetry
ITC was employed to measure the binding affinities of wild-type EfeB with heme and PPIX using ITC200. All titrations were carried out at pH 8.0 in 20 mm Bicine buffer (35). Hemin was dissolved in a solution of 0.1 m NaOH and then diluted to 200 μm with the Bicine buffer. The pH was then adjusted with hydrochloric acid. Heme solutions were freshly prepared, centrifuged, and then filtrated just before the titration experiments. Titrations were carried out at 25 °C. 200 μm hemin was titrated against 20 μm protein with a 0.4-μl injection followed by 19 × 2 μl injection using a syringe rotating at 400 rpm. The interval between successive injections was 2 min. As control, heme was titrated with Bicine buffer.
Fluorescence Spectroscopy Tests Production of PPIX
Bacteria were grown aerobically in LB medium at 37 °C. The incubation temperature was reduced to 15 °C at an A600 of 1.0. IPTG was then added to a final concentration of 0.12 mm. After 3 h of induction, cells were harvested by centrifugation for 15 min at 5200 × g at 4 °C. The bacterial pellets were resuspended in Bug Buster buffer (Novagen) at a concentration of 1 g of bacterial wet weight in 5 ml of buffer (100 A600/ml). The mixture was incubated on a shaking table for 30 min at room temperature to lyse the cells. Then it was centrifuged again for 15 min at 28,500 × g at 4 °C. The supernatants were scanned with specific excitation wavelength and emission wavelengths. PPIX and EfeB-PPIX complex were also characterized by fluorescence spectroscopy.
Peroxidase Activity
The guaiacol peroxidase activity of EfeB and its mutants were determined using the method described in the previous report (23), with the concentration of H2O2 reduced to 1 mm. In addition, the peroxidase activity was also defined using 10 mm catechol as a substrate in the reaction system containing 1 mm H2O2. The oxidation of catechol substrate was measured in the 480 nm (36).
RESULTS
EfeB Binds PPIX and Enhances Its Fluorescence Intensity
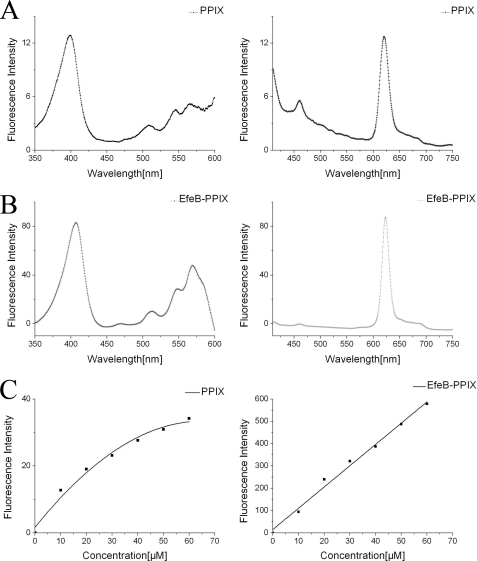
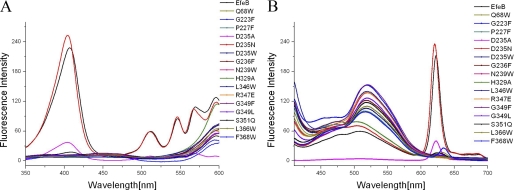
Fluorescence spectroscopy showed that PPIX as the resulting product of EfeB has a distinctive excitation wavelength at 398.5 nm and emission wavelength at 620 nm (Fig. 1A). However, for the EfeB-PPIX complex, the excitation wavelength shifts to 407.5 nm, and the emission wavelength shifts to 622.5 nm (Fig. 1B) because of the interaction of PPIX with protein EfeB. Surprisingly, the fluorescence intensity of free PPIX increased nonlinearly with a rising concentration of PPIX. This may be caused by its low solubility and self-aggregation (37–38) and may affect the PPIX concentration determination based on fluorescence spectroscopy. The EfeB-PPIX complex has a much higher fluorescence intensity compared with that of free PPIX at a corresponding concentration. And more importantly, the fluorescence intensity of EfeB-PPIX increased linearly as the concentration rises (Fig. 1C), indicating that the concentration of PPIX could be determined according to the fluorescence intensity of the EfeB-PPIX complex. Additionally, fluorescence scans for the cell lysate overexpressing EfeB also showed the same fluorescent characteristics as the purified EfeB-PPIX complex, indicating that the fluorescence scans for cell lysates overexpressing wild-type protein or its mutants at the excitation wavelength of 407.5 nm and the emission wavelength of 622.5 nm can be used to detect the output of PPIX.
FIGURE 1.
Fluorescence characteristic of free PPIX and EfeB-PPIX complex. The initial concentration of PPIX is 2 mm and was diluted to different concentrations from 10 to 60 μm using Tris-HCl buffer (10 mm Tris-HCl, pH 8.0, 100 mm NaCl). Apo-EfeB was diluted to different concentrations from 10 to 60 μm using the same buffer, and then PPIX was added to the apo-EfeB solution to a final concentration ranging from 10 to 60 μm and was mixed and measured. A, excitation and emission wavelength scan for free PPIX of 10 μm, showing that the distinctive excitation wavelength is 398 nm and emission wavelength is 620 nm. B, excitation and emission wavelength scan for EfeB-PPIX complex of 10 μm, showing that the excitation wavelength is 407.5 nm and emission wavelength is 622.5 nm. C, fluorescence intensity of free PPIX increased nonlinearly as concentration rises. However, the fluorescence intensity of EfeB-PPIX complex increased linearly with rising concentrations.
Overall Structure of EfeB Bound to Heme
The final model of the EfeB-heme complex contains four protein molecules in the asymmetric unit, termed A, B, C, and D. Each monomer includes residues 48–422 and unambiguously binds to a heme molecule according to the electron density map. No electron density is observed for residues 36–47. Four oxygen molecules and 1158 water molecules are included in the model according to the electron density map and stereochemistry.
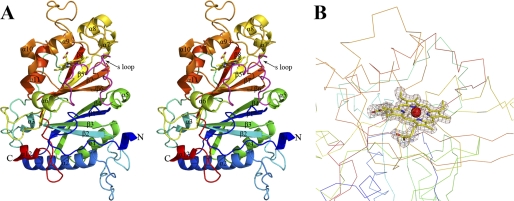
The EfeB monomer shows a ferredoxin-like fold and consists of 12 α-helices and 8 β-strands. The protein can be divided into an N-terminal domain (residues 36–221) and a C-terminal domain (residues 244–422). These two domains are connected by a 22-amino acid-long loop (residues 222–243). Each domain is an α+β fold and contains an anti-paralleled β-sheet composed of four β-strands. These two β-sheets pack against with each other in a β-barrel structure pattern and have an angle of 70°. The β-barrel structure is sandwiched by five α-helices from the N-terminal domain and seven α-helices from the C-terminal domain. This resembles the characteristic of the DyP-type peroxidase protein family (39–42) (Fig. 2).
FIGURE 2.
Stereoview of the structure of heme-bound EfeB monomer and electron density for the heme moiety. A, rainbow-colored scheme representation of the EfeB-heme complex. 12 α-helices and 8 β-strands are labeled. The heme molecule is shown as a stick. The S loop represents a switch loop and is colored in magenta. B, difference electron density map (Fo-Fc) calculated at 1.95 Å resolution using phase from the final model with heme and waters omitted and contoured at 1.5σ reveals an electron density consistent with the heme molecule within molecule A.
Dimerization of EfeB
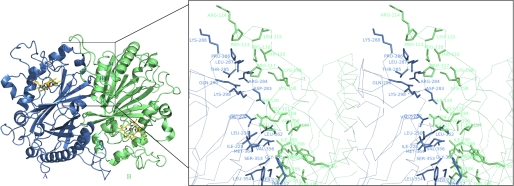
The four EfeB molecules in the asymmetric unit interact head to tail forming two homodimers (AB and CD) with approximate 2-fold symmetry. Superposition of the two monomers from each dimer gives an r.m.s. deviation of less than 0.3 Å for all corresponding Cα atoms. The dimer interface is predominantly hydrophobic and can be divided into two identical subinterfaces. Each subinterface consists of α4, β4, and a loop between α2 and β2 of the N-terminal domain from one monomer pack against β6, β7, α7, a loop between α7 and α8, a loop between α8 and α9 of the C-terminal domain and loop between α5 and α6 from the other monomer (Fig. 3). The total buried surface of the EfeB monomer upon dimerization is 2163 Å2, about 12.69% of the solvent accessible surface area of 17045.2 Å2. The existence of the EfeB dimer in solution was tested by gel filtration on a superdex 200 column. BSA and VibB-(1–215) fragment were used as standards. Existing as monomer in solution, BSA has the molecular mass of 67 kDa and gives the elution volume of 14.1 ml. The molecular mass of VibB-(1–215) is 23.5 kDa. It exists as very stable dimer (47 kDa) in solution and gives the elution volume of 14.8 ml. Furthermore, the elution volume was 14.9 ml for the apo-EfeB and 14.2 ml for the heme bound EfeB (supplemental Fig. S1). These results indicated that the apo-EfeB mainly exists as a monomer in solution, whereas the heme-bound EfeB may be in the equilibrium of monomer and dimer.
FIGURE 3.
Structure of the homodimer and stereoview of the dimer interface. The homodimer is shown in schematic representation; molecule A is in blue and molecule B is in green. Two heme molecules are shown in stick representation. The zoomed region shows the secondary structures contributing to the dimer formation. The residues that form the interface are shown in sticks with molecule A in blue and molecule B in green.
Structure of the Active Site
The electron density map revealed unambiguous binding of a heme molecule to the C-terminal domain of each monomer. The C-terminal domain of EfeB is mainly loosely packed resulting into a large hydrophobic cavity for heme binding. This cavity is sandwiched by the β-sheet and α-helices 7–11 and has a volume much bigger than a heme molecule. To stabilize the substrate, the flexible loop (residues 222–243) that connects the N- and C-terminal domain roughly occupies the superfluous space. This long loop does not make extensive contacts with other parts of the protein except for the interaction with heme. Therefore, this loop may flip out or flip in to facilitate the turnover of substrates and products. Hereafter it is called the “switch loop” (Fig. 2). Because the switch loop is also involved in the interaction between the two monomers of the dimer, it is not surprising that heme binding can stabilize the switch loop and facilitate dimerization.
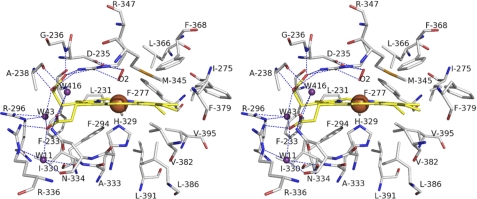
The heme molecule is positioned in the active site with the porphyrin ring deeply buried in the hydrophobic pocket. The porphyrin ring makes extensive interactions with hydrophobic residues Leu231, Phe233, Ala238, Ile275, Phe277, Phe294, Ile330, Ala333, Met345, Leu366, Pro368, Phe379, Val382, Leu386, Leu391, and Val395. Two propionic acid moieties adopt outward orientation. The propionic acid moiety on pyrrole A bends nearly 90° and is stabilized by a hydrogen bond network between the carboxyl group, side chain of Arg347, amino group of Gly236, amino group of Ala238, and a water molecule (W43). Arg347 also forms a hydrogen bond with a nearby residue Asp235. W43 is hydrogen bonded by the side chain of Arg296 and the carboxyl group of the propionic acid moiety on pyrrole D. The second propionic acid moiety is also stabilized by a hydrogen bond network consisting of its carboxyl group, side chain of Arg336, amide group of Asn334, and three water molecules (W11, W43, and W416). These two hydrogen bond networks incorporate with each other through the water molecule W43 (Fig. 4).
FIGURE 4.
Stereoview of heme binding site. The heme carbons are yellow sticks with iron atoms shown as brown spheres. Residues contributing to heme binding are shown as sticks with carbons in gray. The red stick represents the oxygen molecule and acts as the distal ligand. The water molecules are depicted as violet-purple spheres. The hydrogen bonds are depicted as dashed lines.
It has been proved that the heme iron in the EfeB-heme complex exists in the ferric form and in a high-spin configuration (23). Our data also showed that the ferric iron in the purified EfeB-heme complex cannot bind oxygen molecule (see supplemental Fig. S2). However, the crystal structure of heme-bound EfeB shows that the heme iron atom (Fe (III)) is hexacoordinated in the plane of the porphyrin. The average distance between the Fe (III) and pyrrole nitrogens is 2.12 Å. 2Fo-Fc and Fo-Fc electron density maps display a diatomic molecule acting as the distal ligand. Because the heme iron atom can be photoreduced to the ferrous state because of the prolonged x-ray exposure (43–45), it was most likely in the ferrous form and coordinated by an oxygen molecule. The bond length between the oxygen molecule and heme iron is 2.35 Å. His329 acts as the proximal axial ligand. The distance between the heme iron and the Nϵ atom of the imidazole ring is 2.14 Å. The Nδ atom of the imidazole ring is hydrogen bonded by the carboxyl group of Glu389 with a bond length of 2.72 Å, forming a distinct Fe-His-Glu triad. This resembles the distinctive characteristic of the Fe-His-Asp triad, which is a widespread motif at the active site of many metalloenzymes. It is believed that the character of histidine as a metal ligand can be modulated by glutamic acid (46).
An ITC experiment was performed to quantify the interaction between EfeB and heme. The result showed that heme binds to EfeB at a stoichiometry of 1:1 with a dissociation constant (Kd) of 2.96 (±0.43) × 10−8 m (supplemental Fig. S3A). The EfeB-PPIX complex can also be purified from cell lysate containing EfeB and additive PPIX. However, the ITC run for PPIX and EfeB does not provide a significant result because of its low solubility and self-stacking (37–38). Titrating heme into the EfeB-PPIX complex gives a Kd of 2.14 (±0.5) × 10−7 m (supplemental Fig. S3B). It is expected based on the protein and ITC results that PPIX occupies the same binding site as heme.
Structural comparisons of our structure with previously published structures of DyP family peroxidases indicate that the heme molecules adopt an identical conformation, and the residues interacting with heme are highly conserved (39–40). Based on the previous study of DyP family peroxidases, it can be expected that Asp235 and Arg347 may also function as proton acceptor and charge stabilizer, respectively. To test if these residues in the present structure are essential for the activity of EfeB, three mutants D235N, H329A, and R347E were produced, and PPIX production of each mutant was tested through a fluorescence scan. No PPIX production was observed in the supernatants from cell extracts containing overexpressed H329A and R347E. However, mutant D235N produced the same amount of PPIX as the wild-type EfeB (Fig. 5).
FIGURE 5.
PPIX production of EfeB mutants tested by fluorescence scan of cell lysates of bacteria overexpressing mutants. Native EfeB was used as a positive control. A, excitation wavelength scan for mutants and native EfeB. The obvious excitation wavelength at 407.5 nm was detected for EfeB. No distinctive excitation wavelength was observed for these mutants except for D235N,D235A and S351Q, which kept about 102, 17, and 8% fluorescence intensity of native EfeB, respectively. B, emission wavelength scan for mutants and native EfeB. The distinct emission wavelength at 622.5 nm was observed for EfeB, no emission wavelength was detected for these mutants except for D235N,D235A and S351Q, which kept about 102, 17, and 8% fluorescence intensity of native EfeB, respectively.
Binding Site for the Potential Cofactor
Although the cell overexpressing the EfeB produces a large amount of PPIX, the purified EfeB protein does not show any observable catalytic activity. These results imply that this reaction requires an unidentified cofactor missing from the purified protein (22). On the other hand, significant guaiacol peroxidase activity of EfeB has been reported. So far, the physiological substrate has not been identified. Whether the physiological substrate also plays the role of the cofactor is still unknown. The cofactor/substrate should interact with the heme molecule and act as a proton donor during the reaction that removes iron from heme. Thus, the binding site for the physiological cofactor/substrate should be observed close to the heme binding site.
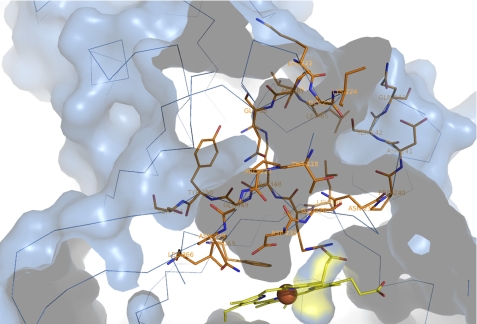
Indeed, the structure of heme-bound EfeB reveals the presence of a large amphiphilic tunnel spanning from the surface of the protein molecule to the heme molecule. The tunnel in EfeB is primarily formed by 14 residues (Lys222, Gly223, Lys224, Thr226, Pro227, Ile228, Asn229, Asp235, Gly236, Asn239, Pro240, Asp241, Ser242, and Gln243) from the switch loop, 6 residues (Leu346, Arg347, Arg348, Gly349, Tyr350, and Ser351) from β6, 2 residues (Leu366 and Phe368) from β7 and 4 residues (Leu65, Thr66, Pro67, and Gln68) from the loop between α1 and β1 (Fig. 6). The diameter of the tunnel is about 4.5 Å, which is much smaller than the size of a heme molecule. Thus, it is unlikely that this tunnel functions as a passageway for the entrance of the heme molecule. The residues forming the amphiphilic tunnel are divided into two groups. Considering the flexibility of the switch loop, the residues from the switch loop may change their positions and orientations upon unknown cofactor binding. The other residues from β6, β7, and the loop between α1 and β1 could be more stabile because they have extensive interactions with other parts of the protein. It can be seen that this tunnel is constructed by a fixed and a mobile wall.
FIGURE 6.
The unidentified cofactor binding tunnel. The protein is shown in surface representation with a cross-sectioned slab removed to allow viewing of the internal ligand-binding tunnel. 26 residues involved in the formation of the tunnel are shown as sticks with carbons in orange.
Structure-aided sequence alignment shows that the residues making up the tunnel are highly conserved in EfeB orthologs across different bacterial species (Fig. 7). The importance of these residues is further confirmed by the mutagenesis study. Twelve mutants were produced: Q68W, G223F, P227F, D235W, G236F, N239W, L346W, G349F G349L, S351Q, L366W, and F368W. All mutants have a bigger side chain, which was supposed to block the entrance of the physiological cofactor/substrate into the tunnel. Soluble expression was obtained for all mutants. Nevertheless, no distinctive excitation and emission wavelength of PPIX was observed through the fluorescence scan of the cell lysate except for S351Q, which gave a little fluorescence intensity of about 8% of the control (Fig. 5). The red color and the distinctive Soret peak of the purified mutant proteins represent the characteristic of a heme-binding protein. This strongly suggested that all these mutants nearly lose the ability to produce PPIX, the product in the enzymatic reaction of removing iron from heme. Results from these experiments support the postulate that the tunnel works as a passageway for the entrance of the physiological cofactor/substrate involved in the enzymatic reaction, rather than the entrance for a heme molecule.
FIGURE 7.
Sequence alignment of EfeB and its homologous proteins. Residues that make up the tunnel are labeled with solid squares at the top, and residues involved in stabilization of heme molecule are marked in solid triangles at the bottom. The proximal ligand His329 is in dark gray and the switch loop is highlighted using a gray line.
Correlation between PPIX Production and Peroxidase Activity
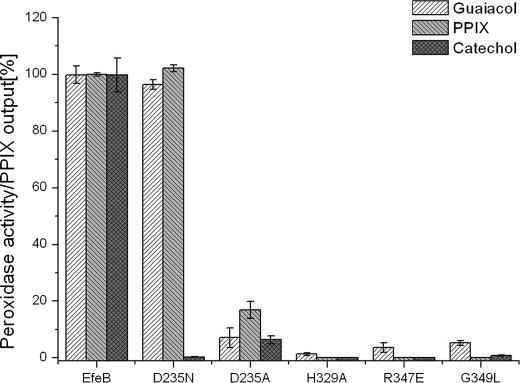
A previous study has indicated the bi-functional property of EfeB/YcdB. To elucidate the correlation between PPIX production and the guaiacol peroxidase activity, peroxidase activity of five mutants (D235A, D235N, H329A, R347E, G349L), which have effects on PPIX production, was measured. These results showed that all mutants that lost PPIX production exhibit decreased guaiacol peroxidase activity to varying degrees. Once again, the mutant D235N was surprising, remaining with almost the same guaiacol peroxidase activity as the wild-type protein (Fig. 8). In addition, the incubation of this mutant D235N with H2O2 generated major spectral changes, which is the same as the addition of H2O2 to wild-type protein (see supplemental Fig. S4). These results are inconsistent with the previous study on DyP in that the corresponding residue Asp171 acts as a proton acceptor and is essential for the enzymatic activity (39). The opposite effects produced by this mutant in EfeB and DyP suggest that EfeB may also show divergent mechanisms with different substrates.
FIGURE 8.
Correlation of peroxidase activity and the PPIX production. Both guaiacol and catechol were used as substrates to determine the peroxidase activities of EfeB and its mutants. PPIX production was measured using fluorescence spectroscopy. The peroxidase activity and PPIX production of native EfeB were set to 100%.
To address this question, we tested whether the guaiacol analog catechol can be oxidized by hydrogen peroxide through EfeB catalysis. This activity was confirmed by an absorption peak at 480 nm when catechol was substituted for guaiacol as the substrate of EfeB. Whether the mutants have the same effects on catechol peroxidase activity as guaiacol peroxidase activity were then tested. With this, the mutant D235N almost completely lost peroxidase activity on catechol. It is expected that the residue Asp235 may play the role of proton acceptor in this reaction. The mutagenesis study strongly suggests that the DyP peroxidase family may adopt divergent mechanisms on different substrates.
DISCUSSION
Although the exact catalytic mechanism of EfeB is controversial, the involvement of EfeB in iron acquisition is well established (25, 28–29). Because iron acquisition is required for pathogenic bacterial survival and virulence, EfeB may serve as latent targets for antimicrobial drugs. The accumulation of PPIX in the cells overexpressing EfeB strongly suggests that this protein is a deferrochelatase. However, it may be argued that this may be due to the sequestration of heme by EfeB, because the synthesis of PPIX is generally regulated by intracellular heme concentration (47). In this regard, even the loss-of-function mutation H329A can be interpreted as a loss-of-binding capacity of heme. Therefore, this mutagenesis study of the residues in the active site and the potential cofactor/substrate binding tunnel adds weight to the previous argument, as these mutants can still bind heme but lose the ability to produce PPIX in cells.
Significantly, EfeB was also reported as a DyP family member with modest guaiacol peroxidase activity (23). The present work shows that EfeB can also use catechol as a substrate. The bi-functional property makes EfeB a puzzling protein attractive to biochemists. Structural comparison of DyP family members indicates the high conservation of the active site (39–40). The residues corresponding to His329, Asp235, and Arg347 are His308, Asp171, and Arg329 in the protein DyP (PDB code 2D3Q). Sugano et al. (39) reported that the activity of the D171N mutant drastically decreased in relation to that of wild-type DyP. This suggested that Asp171 and Arg329 functioned, respectively, as the proton acceptor and charge stabilizer. However, in the present mutagenesis study, mutant D235N did not affect the PPIX production or the guaiacol peroxidase activity, while completely losing catechol peroxidase activity. Moreover, the addition of H2O2 to the D235N mutant generated major spectral changes, and this is also inconsistent with a previous study on DyP where no spectral change was observed when H2O2 was incubated with the mutant D171N.
These data imply that PPIX production and peroxidase activity are closely related to each other. The mechanism of the DyP family peroxidase could be much more complicated than what was expected. The residue Asp235 may play divergent roles in different enzymatic processes. Because identification of the physical substrate of any DyP peroxidase family has been unsuccessful until now, further effort is needed to determine the catalytic mechanism of these kinds of proteins.
Notably, a fluorescent spectrum study of EfeB-PPIX may provide a new method to detect PPIX and measure its concentration in the cell lysate with typical excitation wavelengths at 407.5 nm and emission wavelengths at 622.5 nm. In addition, EfeB and its isoenzymes may be used as a red color fluorescence marker.
Currently, the cofactor involved in removing iron from heme has not been identified; thus, the detailed mechanism for the cofactor extracting iron from heme is still unknown. The unidentified cofactor may function as proton donors in the iron removing process. This supposition was supported by the phenomenon that iron can be removed from heme by treatment with both HCl and glacial acetic acid under a reducing agent such as ferrous sulfate (48–49). Reduction of ferric iron to ferrous iron may be necessary because iron (III) is very difficult to be released from a tetrapyrrole ring. In addition, the tetrapyrrole ring could be distorted during the enzymatic reaction because it happens during the process of metal insertion catalyzed by ferrochelatase (50).
The present study provides a high resolution structure of EfeB/YcdB in complex with heme. EfeB shows a unique heme binding motif highly conserved in this protein family. The switch loop connecting the N-terminal and C-terminal domains could be a distinctive feature of this kind of protein and control the turnover of substrates. During preparation of this manuscript, the structures of apo-EfeB and EfeB in complex with PPIX were released in the Protein Data Bank (PDB code: 2WX6 for apo-EfeB, and PDB code: 2WX7 for EfeB-PPIX). EfeB also exists as a dimer in these two structures. PPIX occupies the same binding site as heme does. Structural comparison shows that EfeB adopts quite similar conformations under these three conditions. A major conformational change appears in the switch loop region. This is coincident with the present postulate that the switch loop is flexible and comprises the mobile wall of the large tunnel for unknown cofactor/substrate binding.
Accession Codes
The atomic coordinates and structure factors have been deposited in the Protein Data Bank with the Accession Code 3O72, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ.
Supplementary Material
Acknowledgments
The genomic DNA of E. coli O157 is a gift from Dr. Jianling Jin. We thank Dr. Pingwei Li for critical reading of this manuscript. We thank the staff at beamline BL17u1 at the Shanghai Synchrotron Radiation facility for support with data collection and Dr. Edward C. Mignot, Shandong University, for linguistic advice.
This work was supported by the State Key Laboratory of Microbial Technology, Shandong University and Grant 2006AA02A324 from the Hi-Tech Research and Development Program of China.
The atomic coordinates and structure factors (code 3O72) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- PPIX
- protoporphyrin IX
- hemin
- ferriprotoporphyrin IX
- ITC
- isothermal titration calorimetry
- Tat system
- twin-arginine translocation system
- Se-Met
- selenomethionine
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- r.m.s. deviation
- root mean square deviation
- DyP
- dye-decolorizing peroxidase.
REFERENCES
- 1. Andrews S. C., Robinson A. K., Rodríguez-Quiñones F. (2003) FEMS Microbiol. Rev. 27, 215–237 [DOI] [PubMed] [Google Scholar]
- 2. Litwin C. M., Calderwood S. B. (1993) Clin. Microbiol. Rev. 6, 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Neilands J. B. (1995) J. Biol. Chem. 270, 26723–26726 [DOI] [PubMed] [Google Scholar]
- 4. Crosa J. H., Walsh C. T. (2002) Microbiol. Mol. Biol. Rev. 66, 223–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stojiljkovic I., Perkins-Balding D. (2002) DNA Cell Biol. 21, 281–295 [DOI] [PubMed] [Google Scholar]
- 6. Tong Y., Guo M. (2009) Arch. Biochem. Biophys. 481, 1–15 144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frankenberg-Dinkel N. (2004) Antioxid. Redox Signal 6, 825–834 [DOI] [PubMed] [Google Scholar]
- 8. Torres A. G., Redford P., Welch R. A., Payne S. M. (2001) Infect. Immun. 69, 6179–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Law D., Kelly J. (1995) Infect. Immun. 63, 700–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wiles T. J., Kulesus R. R., Mulvey M. A. (2008) Exp. Mol. Pathol. 85, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagy G., Dobrindt U., Schneider G., Khan A. S., Hacker J., Emödy L. (2002) Infect Immun 70, 4406–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bonacorsi S. P., Clermont O., Tinsley C., Le Gall I., Beaudoin J. C., Elion J., Nassif X., Bingen E. (2000) Infect. Immun. 68, 2096–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Montellano P. R. (2000) Curr. Opin. Chem. Biol. 4, 221–227 [DOI] [PubMed] [Google Scholar]
- 14. Yoshida T., Migita C. T. (2000) J. Inorg. Biochem. 82, 33–41 [DOI] [PubMed] [Google Scholar]
- 15. Kumar D., de Visser S. P., Shaik S. (2005) J. Am. Chem. Soc. 127, 8204–8213 [DOI] [PubMed] [Google Scholar]
- 16. Matsui T., Iwasaki M., Sugiyama R., Unno M., Ikeda-Saito M. (2010) Inorg. Chem. 49, 3602–3609 [DOI] [PubMed] [Google Scholar]
- 17. Wilks A. (2002) Antioxid. Redox Signal 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 18. Hirotsu S., Chu G. C., Unno M., Lee D. S., Yoshida T., Park S. Y., Shiro Y., Ikeda-Saito M. (2004) J. Biol. Chem. 279, 11937–11947 [DOI] [PubMed] [Google Scholar]
- 19. Schuller D. J., Zhu W., Stojiljkovic I., Wilks A., Poulos T. L. (2001) Biochemistry 40, 11552–11558 [DOI] [PubMed] [Google Scholar]
- 20. Lee W. C., Reniere M. L., Skaar E. P., Murphy M. E. (2008) J. Biol. Chem. 283, 30957–30963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu R., Skaar E. P., Zhang R., Joachimiak G., Gornicki P., Schneewind O., Joachimiak A. (2005) J. Biol. Chem. 280, 2840–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Létoffé S., Heuck G., Delepelaire P., Lange N., Wandersman C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11719–11724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sturm A., Schierhorn A., Lindenstrauss U., Lilie H., Brüser T. (2006) J. Biol. Chem. 281, 13972–13978 [DOI] [PubMed] [Google Scholar]
- 24. Müller M., Klösgen R. B. (2005) Mol. Membr. Biol. 22, 113–121 [DOI] [PubMed] [Google Scholar]
- 25. Cao J., Woodhall M. R., Alvarez J., Cartron M. L., Andrews S. C. (2007) Mol. Microbiol. 65, 857–875 [DOI] [PubMed] [Google Scholar]
- 26. Stancik L. M., Stancik D. M., Schmidt B., Barnhart D. M., Yoncheva Y. N., Slonczewski J. L. (2002) J. Bacteriol. 184, 4246–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maurer L. M., Yohannes E., Bondurant S. S., Radmacher M., Slonczewski J. L. (2005) J. Bacteriol. 187, 304–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rajasekaran M. B., Nilapwar S., Andrews S. C., Watson K. A. (2010) Biometals 23, 1–17 [DOI] [PubMed] [Google Scholar]
- 29. Cartron M. L., Mitchell S. A., Woodhall M. R., Andrews S. C., Watson K. A. (2007) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 37–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31. Terwilliger T. C., Fahrner R., Eisenberg D. (1999) Acta Crystallogr. Sect. D, 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Terwilliger T. C. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 35. Robinson C. R., Liu Y., Thomson J. A., Sturtevant J. M., Sligar S. G. (1997) Biochemistry 36, 16141–16146 [DOI] [PubMed] [Google Scholar]
- 36. Lasovsky J., Hrbac J., Sichertova D., Bednar P. (2007) Luminescence 22, 501–506 [DOI] [PubMed] [Google Scholar]
- 37. Scolaro L. M., Castriciano M., Romeo A., Patane S., Cefali E., Allegrini M. (2002) J. Phys. Chem B 106, 2453–2459 [Google Scholar]
- 38. Margalit R., Shaklai N., Cohen S. (1983) Biochem. J. 209, 547–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sugano Y., Muramatsu R., Ichiyanagi A., Sato T., Shoda M. (2007) J. Biol. Chem. 282, 36652–36658 [DOI] [PubMed] [Google Scholar]
- 40. Zubieta C., Krishna S. S., Kapoor M., Kozbial P., McMullan D., Axelrod H. L., Miller M. D., Abdubek P., Ambing E., Astakhova T., Carlton D., Chiu H. J., Clayton T., Deller M. C., Duan L., Elsliger M. A., Feuerhelm J., Grzechnik S. K., Hale J., Hampton E., Han G. W., Jaroszewski L., Jin K. K., Klock H. E., Knuth M. W., Kumar A., Marciano D., Morse A. T., Nigoghossian E., Okach L., Oommachen S., Reyes R., Rife C. L., Schimmel P., van den Bedem H., Weekes D., White A., Xu Q., Hodgson K. O., Wooley J., Deacon A. M., Godzik A., Lesley S. A., Wilson I. A. (2007) Proteins 69, 223–233 [DOI] [PubMed] [Google Scholar]
- 41. Welinder K. G., Mauro J. M., Nørskov-Lauritsen L. (1992) Biochem. Soc. Trans 20, 337–340 [DOI] [PubMed] [Google Scholar]
- 42. Banci L. (1997) J. Biotechnol. 53, 253–263 [DOI] [PubMed] [Google Scholar]
- 43. Podust L. M., Ioanoviciu A., Ortiz de Montellano P. R. (2008) Biochemistry 47, 12523–12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arcovito A., Moschetti T., D'Angelo P., Mancini G., Vallone B., Brunori M., Della Longa S. (2008) Arch. Biochem.. Biophys. 475, 7–13 [DOI] [PubMed] [Google Scholar]
- 45. Beitlich T., Kühnel K., Schulze-Briese C., Shoeman R. L., Schlichting I. (2007) J. Synchrotron Radiat. 14, 11–23 [DOI] [PubMed] [Google Scholar]
- 46. Goodin D. B., McRee D. E. (1993) Biochemistry 32, 3313–3324 [PubMed] [Google Scholar]
- 47. McNicholas P. M., Javor G., Darie S., Gunsalus R. P. (1997) FEMS Microbiol. Lett. 146, 143–148 [DOI] [PubMed] [Google Scholar]
- 48. van den Berg J. W., Koole-Lesuis R., Edixhoven-Bosdijk A., Brouwers N. (1988) Clin. Chem. 34, 2125–2126 [PubMed] [Google Scholar]
- 49. Schwartz S., Dahl J., Ellefson M., Ahlquist D. (1983) Clin. Chem. 29, 2061–2067 [PubMed] [Google Scholar]
- 50. Karlberg T., Hansson M. D., Yengo R. K., Johansson R., Thorvaldsen H. O., Ferreira G. C., Hansson M., Al-Karadaghi S. (2008) J. Mol. Biol. 378, 1074–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.