Abstract
We report that the activation level of AMP-dependent protein kinase AMPK is elevated in cancer cell lines as a hallmark of their transformed state. In OVCAR3 and A431 cells, c-Src signals through protein kinase Cα, phospholipase Cγ, and LKB1 to AMPK. AMPK controls internal ribosome entry site (IRES) dependent translation in these cells. We suggest that AMPK activation via PKC might be a general mechanism to regulate IRES-dependent translation in cancer cells.
Keywords: AMP Kinase, Phospholipase C, Protein Kinase C (PKC), Signal Transduction, Src, Cancer
Introduction
The nonreceptor tyrosine kinase c-Src was the first proto-oncogene to be discovered in the vertebrate genome. c-Src is overexpressed and activated in many human cancers, including lung, skin, colon, breast, and ovarian malignancies. The elevated protein levels and catalytic activity have been linked to the development of cancer and progression to metastases. However, despite the fact that c-Src is one of the oldest and most investigated proto-oncogenes, the precise functions of c-Src in cancer remain unclear. In addition to increasing cell proliferation, a key role of c-Src in cancer seems to be to promote invasion and motility, functions that might contribute to tumor progression (for review, see Refs. 1–3).
Previously, we had shown that AMPK2 activation appeared to result from progressive activation of c-Src (4). Recently published data also demonstrate that AMPK activation can be induced by c-Src, independently of the AMP/ATP ratio in endothelial cells (5, 6).
The AMP-activated protein kinase (AMPK) maintains the balance between ATP production and consumption in eukaryotic cells (for review, see Refs. 7, 8). Mammalian AMPK is sensitive to the cellular AMP/ATP ratio and is activated by metabolic stresses such as hypoxia and glucose deprivation. AMPK is also modulated by cytokines that regulate whole body energy balance, including leptin and interleukin-6 (for review, see Refs. 7, 8).
AMPK is a conserved heterotrimeric protein comprised of a catalytic (α1 or α2) subunit and two regulatory (β1 or β2 and γ1, γ2, or γ3) subunits. Following binding of AMP to the γ subunit, AMPK is activated by LKB1, which phosphorylates AMPK at a conserved threonine residue (Thr-172) on the α subunit. This mechanism allows the system to act as a sensor of cellular energy status (for review, see Refs. 7, 8).
Upon activation, AMPK switches on catabolic pathways that generate ATP, such as the uptake and metabolism of glucose and fatty acids. AMPK also switches off anabolic pathways that consume ATP, but are not necessary for survival, such as the synthesis of fatty acids, glycogen, and proteins. Thus, AMPK plays a dual role in cancer. On the one hand, AMPK activates glycolysis and angiogenesis, promoting cell survival (7–10). On the other hand, active AMPK also switches off anabolic processes, including protein synthesis, through the inhibition of mTOR (11).
Previously, we found that AMPK activity increases during HPV16 transformation of human keratinocytes (4). Therefore, we were intrigued to examine the status of AMPK in cancer cells. Here, we report that AMPK activity is high in several known cancer cell lines. We find that AMPK activation is regulated through c-Src, PLCγ, PKCα, and LKB1 in some of these cancer cell lines.
EXPERIMENTAL PROCEDURES
Materials
All tissue culture reagents were purchased from Biological Industries Bet-Haemek Ltd., Israel. Compound C, 5-aminoimidazole-4-carboxamide riboside (AICAR), bisindolymaleimide I (BIS1), and rapamycin were from Calbiochem. PP1 was purchased from Synthose. U73122 was from Sigma. siRNA against c-Src, PKCα, and LKB1 and nontargeting siRNA, siRNA Universal Buffer, and DharmaFECT1 were from Dharmacon. GST-PKCα and TPA were from Cell Signaling Technology. Lipofectamine 2000 reagent was purchased from Invitrogen. Reporter Lysis Buffer and the dual-luciferase kit were from Promega.
Cell Culture
A431 (human epithelial carcinoma), OVCAR3 (ovarian cancer), HeLa (cervical cancer), HT-29 (colon cancer), SKOV3 (ovarian cancer), T-24 (bladder cancer), PC3 (prostate cancer), NIH 3T3 and SrcNIH (NIH 3T3 transformed with active Src (Y529F) (12)) cells were grown in DMEM supplemented with 10% fetal calf serum (FCS). MCF7 (breast cancer) cells were grown in RPMI 1640 medium supplemented with 10% FCS. All media were supplemented with 100 units/ml penicillin and 100 mg/ml streptomycin, and all cells were grown in a humidified atmosphere containing 5% CO2 at 37 °C.
siRNA Transfection
A431 and OVCAR3 cells were transfected using DharmaFECT1 according to the manufacturer's protocol. In brief, for a 6-well plate 2 μm siRNAs in 1 × siRNA buffer were diluted into serum-free medium and incubated for 5 min at room temperature. 5 μl of DharmaFECT1 was diluted with 195 μl of serum-free medium, and after 5 min at room temperature the reagent was incubated with 2 μm siRNA for 20 min at room temperature. 1.6 ml of DMEM supplemented with 10% FCS was added to the siRNA·DharmaFECT1 complex and dropped onto the cells. 96 h after transfection, cells were harvested.
Drug Treatment
At 60–70% confluence, cells were starved of serum for 2 h. The medium was replaced with medium lacking serum containing PP1, BIS1, rapamycin, AICAR, compound C, or U73122 and the cells were incubated further, as indicated.
Protein Determination
The protein content of cell lysates generated by nondenaturing detergents was quantified using the micro-Bradford assay (13) with the Zor-Selinger modifications (14). Protein determination for denatured lysates was performed using the bound Coomassie Blue method: samples (5 μl) and BSA standards (1–10 μg) were spotted onto numbered 3MM filter paper strips (0.5 × 1.5 cm; Whatman). Filter papers were stained with Coomassie Brilliant Blue G-250 (2.5 g/liter in 40% methanol, 10% acetic acid; Bio-Rad), and unbound dye was washed with destaining solution (20% methanol, 7% acetic acid). Bound Coomassie was eluted by incubating the filters in 1 ml of 3% SDS at 37 °C for 1 h with shaking in a 24-well plate. 200-μl samples were transferred to a 96-well plate, and the absorbance of the eluted solution was measured at 595 nm in an ELISA reader.
Western Blot Analysis
Cells were washed with PBS (50 mm NaH2PO, 50 mm Na2HPO4, 0.77 m NaCl), and denatured cell lysates were prepared by scraping the cells in the presence of Laemmli sample buffer (40% glycerol, 0.2 m Tris, pH 6.8, 20% β-mercaptoethanol, 12% SDS, and bromphenol blue) and boiling the samples for 10 min. Aliquots of cell extracts containing the same amounts of protein were resolved by SDS-PAGE and electroblotted onto nitrocellulose membranes (Sartorius). The membranes were blocked with TBS-T (10 mm Tris-HCl, pH 7.5, 50 mm NaCl, and 0.2% Tween 20) containing 5% low fat milk, incubated with primary antibodies overnight at 4 °C, and then with HRP-conjugated secondary antibodies (Jackson Immunoresearch) for 45 min at room temperature. After each incubation the membranes were thoroughly washed with TBS-T (four times for 3 min). Immunoreactive bands were visualized using enhanced chemiluminescence (ECL). For densitometric analysis, several exposures were made, and only subsaturation exposures were analyzed. Densitometry of immunoblots was performed with ImageJ 1.61 (National Institutes of Health) or EZQuant software.
When needed, blots were stripped of antibodies by using Restore Plus Western blot stripping buffer (Pierce), according to the manufacturer's instructions, or by incubating in 2% SDS, 10 mm β-mercaptoethanol, 62.5 mm Tris-HCl, pH 6.8, at 55 °C for 20 min. After stripping, the blots were washed with TBST, blocked with 5% milk in TBST, and reprobed. The following antibodies were used: anti-ACC (catalogue no. 3602, dilution 1:1,000), anti-phospho-ACC-S79 (catalog 3661, 1:1,000), anti-AMPKα (2532, 1:1,000), anti-phospho-AMPKα-T172 (2531, 1:1,000), anti-phospho-c-Src-Y416 (2101, 1:1,000), anti-LKB1 (3050, 1:1,000), anti-phospho-LKB1-S428 (3051, 1:1,000), anti-PKCα (2056, 1:1,000), anti-phospho-PKCα-T638 (9375, 1:1,000), anti-PKCζ (9372, 1:1,000), anti-phospho-PKCζ-T410/403 (9378, 1:1,000), anti-PLCγ (2822, 1:1,000), anti-phospho-PLCγ-Y783 (2821, 1:1,000), anti-Hif-1α (3716, 1:1,000), anti-GAPDH (2118, 1:1,000) from Cell Signaling Technology; anti-ERK2 (sc-7383, 1:1,000), from Santa Cruz Biotechnology.
DNA Transfection and Drug Treatment
A431 and OVCAR3 cells were transfected with the bicistronic reporter construct (1 μg of DNA/well). 48 h after transfection the cells were treated for 4 h with AICAR or compound C (CC). After 52 h, cell lysates were prepared and assayed for firefly luciferase and Renilla luciferase activity. The bicistronic translation reporter plasmids (pEF-FFL-IRES-SPL and pEF-SPL-IRES-FFL) were kindly provided by Prof. R. Fukunaga (Osaka, Japan) (15). The control bicistronic reporter plasmid (pRF) was kindly provided by Prof. A.E. Willis (Nottingham, UK) (16). The bicistronic reporter plasmid containing the 5′-UTR of HIF-1α and the pStemRhifF plasmid, which contains a hairpin before the SPL cistron, were kindly provided by Prof. G. J. Goodall (Adelaide, Australia) (17).
Luciferase Assay
Cells were transfected with the appropriate reporter plasmids, as above. At the time indicated, cells were harvested, and protein extracts were prepared by lysis with Reporter Lysis Buffer. FFL and SPL activities were assayed with a dual-luciferase kit using a Luminoskan Ascent (Thermo Labsystem) luminometer. For the bicistronic constructs containing the 5′-UTR of HIF-1α, FFL activity was normalized to SPL activity, to correct for transfection efficiencies.
Purification of Recombinant GST-LKB1 Expressed in HEK293T Cells
GST-tagged LKB1 was transfected and expressed in HEK293 cells and purified on glutathione-Sepharose as described previously (18). The pEBG-2T-GST-LKB1 construct was kindly provided by Prof. D. G. Hardie (Dundee, Scotland, UK) (18).
Coomassie Gels
Gels were stained with GelCode blue stain reagent (Pierce), according to the manufacturer's instructions. The amount of purified protein was estimated using a BSA standard curve, which was loaded on the same gel.
In Vitro Kinase Assay
Increasing amounts of GST-LKB1 (10, 20, 40, and 80 ng) were incubated for 30 min at 30 °C with 60 mm HEPES-NaOH, pH 7.5, 3 mm MgCl2, 3 mm MnCl2, 3 μm sodium orthovanadate, 1.2 mm DTT, 20 μm ATP, and 1 μCi of [γ-32P]ATP in the presence or absence of active GST-PKCα (20 ng/reaction). The reaction was terminated by the addition of SDS sample buffer and the proteins resolved by SDS-PAGE. Proteins were transferred to a nitrocellulose membrane, and the membrane was subjected to autoradiography. Afterward, membranes were blocked with 5% low fat milk in TBST and reacted with anti-LKB1, anti-phospho-LKB1-S428 and anti-PKCα.
Immunoprecipitation
For co-immunoprecipitation, the cells were lysed by scraping at 4 °C with lysis buffer containing 50 mm HEPES, pH 7.5, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 150 mm NaCl, 1 mm sodium orthovanadate, 10 mm β-glycerolphosphate, 50 mm sodium fluoride, 5 mm sodium pyrophosphate, 10 μg/ml soybean trypsin inhibitor, 10 μg/ml leupeptin, 1 μg/ml aprotinin, 313 μg/ml benzamidine, and 0.2 mm AEBSF. Lysates were centrifuged at 19,000 × g for 10 min. Lysates comprising 0.5–1 mg of protein were used to immunoprecipitate c-Src and PLCγ1 from A431 and OVCAR3 cells. Antibodies against c-Src (mAb 327 or sc-208; Santa Cruz) or PLCγ1 (2822 from Cell Signaling or M156 from Abcam) were coupled to protein G-Sepharose (mAb 327) or A-Sepharose (sc-208, 2822 and M156) (Amersham Biosciences). 60 μl of protein A or G was coupled to 250 μl of α-c-Src (mAb 327) antibody (in medium) or to 2 μg of α-c-Src (sc-208) antibody by incubating them for 2 h at 4 °C. 60 μl of protein A was coupled to 1:50 diluted α-PLCγ (2822) antibody or 20 μl of α-PLCγ (M156) antibody by incubating them for 2 h at 4 °C. After coupling, the beads were washed five times with PBS to remove excess antibody. Lysates were incubated with coupled beads overnight at 4 °C on a rotating platform. Immunocomplexes were washed once with lysis buffer and five times with PBS, sample buffer was added to the beads, and the samples were boiled for 10 min before SDS-PAGE separation.
RESULTS
Activity of AMPK Is Elevated in Cancer Cell Lines
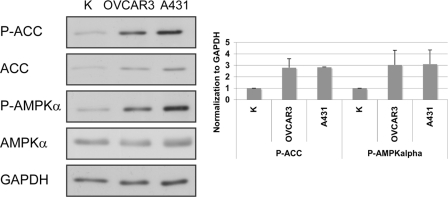
To characterize AMPK regulation in transformed cells, we examined AMPK activity in a set of cancer cell lines, including A431, OVCAR3, HeLa, HT-29, MCF7, SKOV-3, T-24, and PC3 cells. We compared these cells with primary keratinocytes, which represent nontransformed cells of epithelial origin. The levels of the activated (phosphorylated) form of AMPK (Thr-172), as well as the phosphorylation of its direct substrate, ACC (Ser-79), were elevated in A431, OVCAR3, HeLa, T-24, MCF7, and PC-3 cells, compared with K cells, but not in SKOV-3 and HT-29 cells (Fig. 1 and supplemental Fig. S1). In this study we conducted more detailed experiments on two of the cell lines: OVCAR3 and A431.
FIGURE 1.
AMPK is more active in A431 and OVCAR3 cells than in primary keratinocytes. Phosphorylation of AMPK on Thr-172 indicates constitutive activation in OVCAR3 and A431 cells. P-AMPKα and P-ACC levels were normalized to GAPDH levels. The graph shows the calculated averages ± S.D. from three independent experiments. In the cell lines, the direct substrate of AMPK, ACC, was also highly phosphorylated on Ser-79 (P-ACC).
AMPK Is Regulated by c-Src in OVCAR3 and A431 Cells
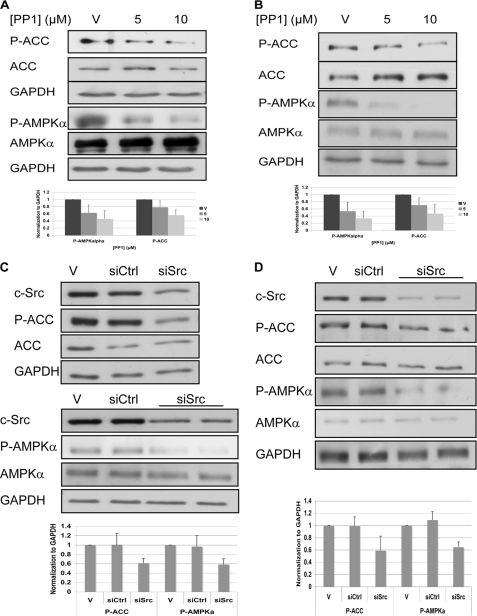
c-Src is known to be activated in OVCAR3 and A431 cells (19, 20). Therefore, we decided to investigate whether c-Src regulates AMPK activity in these cell lines. Inhibition of c-Src by its inhibitor, PP1 (21), led to reduced activation of AMPK (as indicated by P-AMPKα) and reduced phosphorylation of its direct substrate, ACC, in both OVCAR3 and A431 cells (Fig. 2, A and B). Furthermore, siRNA-mediated depletion of c-Src using siRNA also resulted in reduced activity of AMPK in both cell lines (Fig. 2, C and D).
FIGURE 2.
AMPK is regulated by c-Src in OVCAR3 and A431 cells. A and B, cells were starved for serum and after 2 h were treated with PP1 to inhibit c-Src. After 4 h cells were lysed and analyzed by Western blotting. C and D, cells were transfected with siRNA against c-Src. After 96 h cells were lysed and analyzed by Western blotting. A and C, A431 cells; B and D, OVCAR3 cells. Representative Western blot analysis using the indicated antibodies is shown. GAPDH served as a gel loading control. V, vehicle; siCtrl, nontargeting siRNA; siSrc, siRNA against c-Src. P-AMPKα and P-ACC levels were normalized to GAPDH levels (bottom). The graphs show the calculated averages ± S.D. from four (A and B) or three (C and D) independent experiments.
c-Src Regulates AMPK through PKCα
Because previous studies demonstrated that the activation of AMPK is PKCζ-dependent (22, 23), we tested whether PKCζ mediates AMPK activation by c-Src. Interestingly, the inhibition of PKC by BIS1 significantly attenuated AMPK activity (supplemental Fig. S2, A and B). Nevertheless, PP1 treatment had no effect on the phosphorylation of PKCζ (Thr-410/Thr-403) (supplemental Fig. S2, C and D). Therefore, PKCζ is not the intermediate involved in the activation of AMPK by c-Src in these cells. However, the finding that BIS1 treatment led to AMPK inhibition (supplemental Fig. S2, A and B) implied that other PKC isoforms may be involved in the regulation of AMPK by c-Src.
A connection between c-Src and PKCα had been reported previously in breast cancer cells (24, 25). We therefore examined the effect of Src inhibition on PKCα in our cells. Treatment of A431 and OVCAR3 cells with the Src inhibitor, PP1, led to a reduction in the phosphorylated (activated) form of PKCα (Thr-638) (18), in both A431 and OVCAR3 cells (supplemental Fig. S3, A and B). Furthermore, siRNA against c-Src also led to a reduction in PKCα activation (supplemental Fig. S3, C and D). These results confirm that c-Src regulates PKCα in these cell lines.
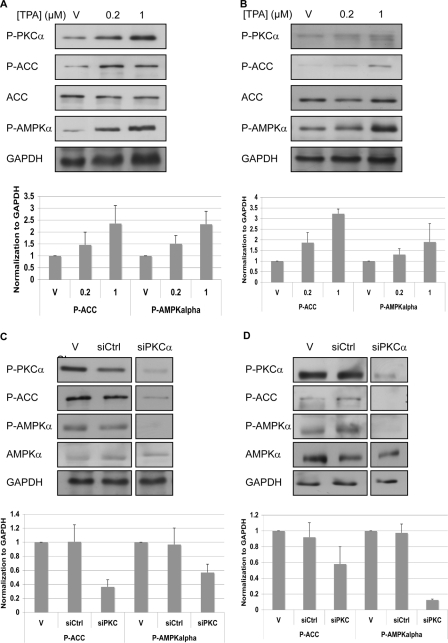
The phorbol ester, TPA, is known to activate PKC. Treatment of OVCAR3 and A431 cells with TPA resulted in increased phosphorylation of PKCα (Thr-638), as well as of AMPKα (Thr-172) and ACC (Ser-79) (Fig. 3, A and B). Furthermore, siRNA against PKCα led to reduced activity of AMPK in A431 and OVCAR3 cells (Fig. 3, C and D).
FIGURE 3.
c-Src regulates AMPK through PKC. A and B, cells were treated with TPA to activate PKC. After 1 h cells were lysed and analyzed by Western blotting. C and D, cells were transfected with siRNA against PKCα. After 96 h cells were lysed and analyzed by Western blotting. The strips were from a single blot. The figure shows where the scans were cut for presentation purposes. A and C, A431 cells. B and D, OVCAR3 cells. Representative Western blot analysis using the indicated antibodies is shown. GAPDH served as a gel loading control. P-AMPKα and P-ACC levels were normalized to GAPDH levels (bottom). The graphs show the calculated averages ± S.D. from four independent experiments (A–D). V, vehicle; siCtrl, nontargeting siRNA; siPKCα, siRNA against PKCα.
Although the up-regulation of PKCα by c-Src has been shown previously, the mechanism of this activation remains unknown (25). Recently, it was found that c-Src can be co-immunoprecipitated with PKCα from breast cancer cell lines (25). Therefore, we tested whether such an association occurs in OVCAR3 and A431 cells. c-Src was immunoprecipitated with two different c-Src-specific antibodies, but Western blot analysis with c-Src and PKCα antibodies failed to show an interaction between PKCα and c-Src (data not shown). Thus, c-Src regulates PKCα in OVCAR3 and A431 cells, in an indirect manner.
c-Src Does Not Regulate PKCα through PDK1
The initial step in the activation of PKC can be executed by PDK1 (26, 27). Furthermore, it has been reported that the phosphorylation of PDK1 by c-Src is required for PDK1 activation (28, 29). Therefore, we explored whether PDK1 mediates PKCα activation by c-Src in A431 and OVCAR3 cells. Depletion of PDK1 by siRNA in these cells had a minor effect, if any, on the activity of PKCα (supplemental Fig. S4). Therefore, we conclude that PDK1 is not a major player in transmitting the signal from Src to PKCα.
c-Src Regulates PKCα through PLCγ
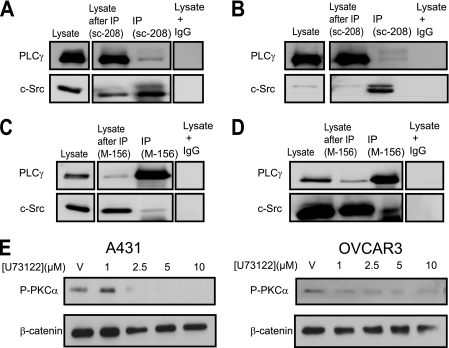
Regulation of PLCγ1 by c-Src has been reported previously (30–32), and regulation of PKC isoforms by PLCγ1 has long been known (for review, see Ref. 33). Therefore, we examined whether Src signaled to PKCα via PLCγ. Following immunoprecipitation of c-Src from A431 and OVCAR3 lysates, we observed a weak association between c-Src and PLCγ1 (Fig. 4, A and B). The reciprocal co-immunoprecipitation with anti-PLCγ1 antibody confirmed these results (Fig. 4, C and D). Interestingly, an antibody against an epitope in the N terminus of PLCγ1 precipitated c-Src (Fig. 4, C and D), whereas an antibody against an epitope in the C terminus failed (supplemental Fig. S5).
FIGURE 4.
A–D, c-Src interacts with PLCγ1. Protein lysates from A431 (A and C) or OVCAR3 cells (B and D) were immunoprecipitated with c-Src-specific antibodies (sc-208, which recognizes an epitope in the C terminus) or control IgG (A and B) or PLCγ1-specific antibody (M-156, which recognizes an epitope in the N terminus) or control IgG (C and D). The immunoprecipitates were blotted with anti-Src and anti-PLCγ1. The strips were from a single blot. The figure shows where the scans were cut for presentation purposes. E, PLCγ activates PKCα. A431 and OVCAR3 cells were treated for 6 h with U73122 at the concentrations shown. All experiments were performed at least two times. Representative blots are shown.
In light of these results, we speculated that PLCγ1 mediates the activation of PKCα by c-Src. Inhibition of c-Src by PP1 or its depletion by siRNA led to a reduction in the phosphorylation of PLCγ1 at Tyr-783 (supplemental Fig. S6). Phosphorylation is one of the key mechanisms that regulate the activity of PLCγ1, and phosphorylation at Tyr-783 was reported to activate PLCγ1 (34). We also note that in SrcNIH cells, which are NIH 3T3 cells transformed with active Src (Y529F) (12), the levels of phosphorylated PLCγ1 were higher than in the parental NIH 3T3 cell line. Moreover, PP1 strongly inhibited the phosphorylation of PLCγ1 at Tyr-783, in SrcNIH cells (supplemental Fig. S7).
We next inhibited PLCγ using U73122 (35, 36). This led to a decrease in the phosphorylation of PKCα (Fig. 4E). Thus, the signal from c-Src to AMPK is transmitted via PLCγ and PKCα.
Activation of AMPK by c-Src Is Mediated through PKCα and LKB1
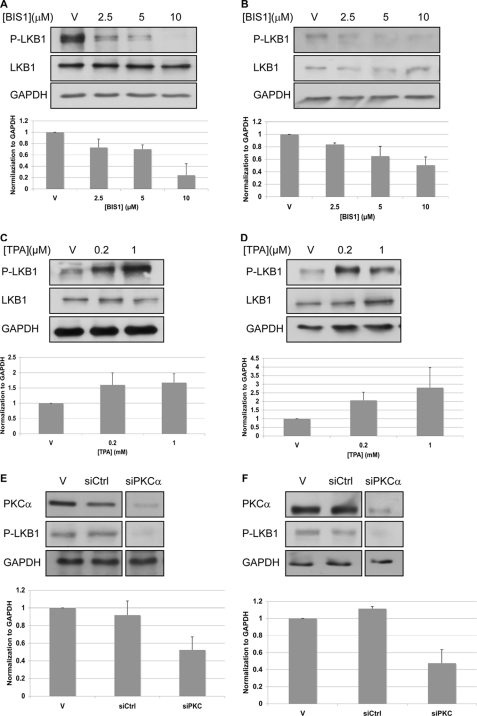
Bioinformatics analysis suggested that AMPKα might be a direct substrate of PKCα. However, co-immunoprecipitation failed to detect an interaction between AMPKα and PKCα in OVCAR3 or A431 cells (data not shown). Previously, it had been shown that PKCζ activates AMPK through the serine/threonine kinase, LKB1. Following the phosphorylation of LKB1 at Ser-428, LKB1 is translocated from the nucleus to the cytosol, where it associates with AMPK and activates it (22, 23). Therefore, we examined whether LKB1 also mediates the activation of AMPK by PKCα. We found that the inhibition of PKC by BIS1 caused a reduction in the phosphorylation of LKB1 at Ser-428 (Fig. 5, A and B), whereas the activation of PKCα by TPA led to increased phosphorylation of LKB at Ser-428 (Fig. 5, C and D). Furthermore, the depletion of PKCα by siRNA led to a marked reduction in the phosphorylation of LKB1. The inhibition of c-Src by PP1 or the depletion of c-Src by siRNA led to a reduction in the phosphorylation of LKB1 (supplemental Fig. S8).
FIGURE 5.
Activation of AMPK is mediated through PKCα and LKB1. A and B, cells were starved for serum and after 2 h were treated with BIS1. After 4 h cells were lysed and analyzed by Western blotting. C and D, cells were treated with TPA to activate PKC. After 1 h cells were lysed and analyzed by Western blotting. E and F, cells were transfected with siRNA against PKCα. After 96 h cells were lysed and analyzed by Western blotting. The strips were from a single blot. The figure shows where the scans were cut for presentation purposes. A, C, and E, A431 cells. B, D, and F, OVCAR3 cells. Representative Western blot analysis using the indicated antibodies is shown. GAPDH served as a gel loading control. A–F, P-LKB1 levels were normalized to GAPDH levels (bottom). The graphs show the calculated averages ± S.D. from two independent experiments.
To examine whether PKCα directly phosphorylates LKB1, we performed an in vitro kinase assay using purified GST-LKB1 and active GST-PKCα, in the presence of MgATP and [γ-32P]ATP and then separated the reaction mixes by SDS-PAGE and transferred them to nitrocellulose. Autoradiography of the blot showed that in the absence of active PKCα, GST-LKB1 was not significantly phosphorylated. However, in the presence of active PKCα, [γ-32P]ATP was incorporated into GST-LKB1, and the degree of incorporated [γ-32P]ATP was proportional to the amount of GST-LKB1 in the assay. Furthermore, Western blot analysis using anti-phospho-LKB1 antibody showed an increase in Ser-428 phosphorylation of LKB1 (supplemental Fig. S9A). Thus, PKCα activates LKB1 in OVCAR3 and A431 cells.
Inhibition of LKB1 using siRNA led to reduced phosphorylation of AMPK and ACC, but not of PKCα, indicating that LKB1 transmits the signal from PKCα to AMPK (supplemental Fig. S8E). Inhibition of Src using PP1 led to decreased phosphorylation of PLCγ, PKCα, LKB1, AMPK, and ACC (supplemental Fig. S9B). In light of these results, we conclude that c-Src regulates AMPK via PLCγ, PKCα, and LKB1 in OVCAR3 and A431 cells.
Involvement of AMPK in the Regulation of IRES-dependent Translation
AMPK plays a central role in the regulation of protein synthesis. It inhibits mTOR via the phosphorylation and activation of tuberous sclerosis complex 2 (TSC2). TSC2 is a subunit of the TSC1/TSC2 (hamartin/tuberin) complex which negatively regulates mTOR signaling. Thus, AMPK mediates the inhibition of cap-dependent translation (11, 37).
Previously, we showed that in the course of cell transformation a switch from cap to IRES-dependent translation occurred. This switch corresponded with the progressive activation of AMPK during transformation (4). Here, we examined whether AMPK can divert translation toward IRES-dependent translation in OVCAR3 and A431 cells. To this end we used a luciferase reporter system that utilizes reciprocal bicistronic plasmids from which both cap and IRES-dependent translation can be measured (15). In both cell lines, the activation of AMPK by AICAR (11) led to an increase in IRES-dependent translation (supplemental Fig. S10, A, B, E, and F) and a reduction in cap-dependent translation (supplemental Fig. S10, C, D, G, and H). Correspondingly, the addition of the AMPK inhibitor, compound C (38), led to a decline in the IRES fraction. Interestingly, compound C treatment also reduced cap-dependent translation (supplemental Fig. S10, C, D, G, and H). To verify that we were indeed measuring IRES-dependent translation from the HIF 5′-UTR, and not read-through translation, we used the pStemRHifF plasmid (Fig. S10I), in which cap-dependent translation is strongly inhibited. The ratio of firefly to Renilla luciferase activity was even higher from pStemRhifF than from pRhifF, indicating that cap-dependent translation of the Renilla luciferase was repressed and the firefly luciferase was read from the IRES site in the 5′-UTR (17).
Hif-1α is directly involved in cell survival and was reported to undergo protein translation by both cap and IRES-dependent mechanisms (17). It has been reported that c-Src regulates the synthesis of Hif-1α protein (39). Furthermore, it has been documented that AMPK regulates Hif-1α via a signaling pathway that is independent of the PI3- kinase/PKB and MAPK pathways (40). In view of these results, we hypothesized that through AMPK, c-Src might regulate the IRES-dependent protein synthesis of Hif-1α in A431 and OVCAR3 cells.
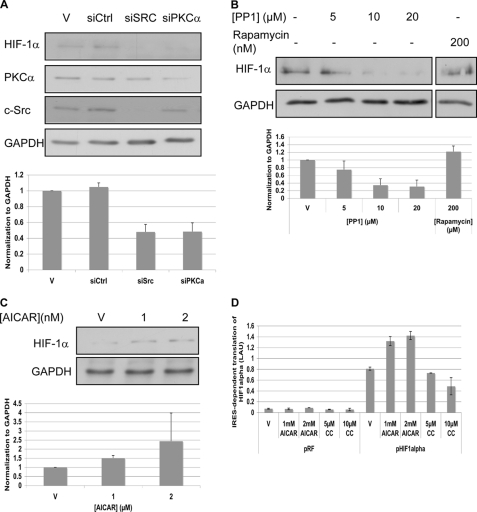
The depletion of c-Src or of PKCα using siRNA, as well as PP1 treatment, led to a reduction in the level of Hif-1α in OVCAR3 cells (Fig. 6, A and B). We note that Hif-1α could not be detected on Western blots from A431 cells.
FIGURE 6.
IRES- versus cap-dependent translation. A, OVCAR3 cells were transfected with siRNA against c-Src or PKCα. After 96 h cells were lysed and analyzed by Western blotting. B, OVCAR3 cells were starved for serum and after 2 h were treated with PP1 or rapamycin. After 4 h cells were lysed and analyzed by Western blotting. The strips were from a single blot. The figure shows where the scans were cut for presentation purposes. A and B, Hif-1α levels were normalized to GAPDH levels (bottom). The graphs show the calculated averages ± S.D. from two independent experiments, whereas the blots show one representative experiment. C, OVCAR3 cells were starved for serum and after 2 h were treated with AICAR. After 4 h cells were lysed and analyzed by Western blotting. Hif-1α levels were normalized to GAPDH levels (bottom). A representative blot is shown. The graphs show the calculated averages ± S.D. from three independent experiments. D, OVCAR3 cells were transfected with the HIF-1α bicistronic construct. Activation of AMPK by AICAR or its inhibition by compound C had an effect on the FFL/SPL ratio. Error bars represent the S.D. of duplicate samples. V, vehicle. The experiment was performed two times.
Rapamycin, a selective inhibitor of mTOR, and hence of cap-related translation, had no effect on the accumulation of Hif-1α in OVCAR3 cells (Fig. 6B). Furthermore, the activation of AMPK by AICAR treatment led to increased accumulation of Hif-1α (Fig. 6C). These findings were corroborated by the demonstration that IRES-dependent translation from the 5′-UTRs of HIF1α increased under AICAR treatment, whereas compound C treatment led to reduced translation from the 5′-UTR of HIF1α (Fig. 6D).
Regulation of AMPK by c-Src Is Cell Type-specific
The regulation of AMPK by c-Src was seen in a number of cell lines, including L-HF1, BP-HF1 (4), A431, and OVCAR3 cells. However, in HeLa and MCF7 cells, the inhibition of c-Src by PP1 had no effect on AMPK activity (supplemental Fig. S11, A and B), suggesting that the c-Src → PKCα → AMPK module is cell type- (or tissue)-specific. Even in these cells, however, the inhibition of PKC by BIS1 treatment led to a reduction in the phosphorylation of AMPK and ACC (supplemental Fig. S11, C and D), suggesting that this part of the signaling module may be widespread but that the upstream elements and PKC isotypes may vary.
DISCUSSION
In this study, we delineate a signaling pathway from the oncoprotein c-Src to AMPK. AMPK, a Ser/Thr protein kinase, serves as an energy sensor in all eukaryotic cells (7, 8). We observed an increase in the activation of AMPK in a number of cancer cell lines, including OVCAR3 and A431 cells (Fig. 1 and supplemental Fig. S1). The increased activity of AMPK in A431 and OVCAR3 cells is c-Src-dependent, as inhibition of c-Src resulted in decreased activity of AMPK (Fig. 2). In primary keratinocytes, we saw no evidence of a connection between c-Src and AMPK (supplemental Fig. S12).
A signaling pathway leading to the activation of AMPKα by c-Src has not been previously established in cancer cells. Here, we have found that in OVCAR3 and A431 cells, c-Src regulates AMPKα through PKCα, PLCγ, and LKB1. Although PKC can be activated by PDK1, and PDK1 has been reported to be regulated by c-Src (28), we were unable to find evidence for PDK1-dependent activity of PKCα in OVCAR3 and A431 cells (supplemental Fig. S4). However, PDK1 was not completely abolished by the siRNA in this assay, and the remaining protein might have been sufficient to induce PKCα activation.
Another possible connection between c-Src and PKCα is through PLCγ. It has long been known that PLCγ regulates the activity of PKC isoforms (for review, see Ref. 33). Furthermore, a connection between PLCγ and c-Src was postulated previously (30–32). Here we have shown, using siRNA techniques and pharmacological tools, that c-Src regulates PLCγ in A431 and OVCAR3 cells (supplemental Fig. S6). Furthermore, we monitored a weak physical association between c-Src and PLCγ1 (Fig. 4). It is interesting to note that only an antibody that targets an epitope in the N terminus of PLCγ could precipitate c-Src (Fig. 4, C and D), whereas an antibody against an epitope in the C terminus failed to achieve this (supplemental Fig. S5). The C-epitope antibody may interfere with the interaction of c-Src and PLCγ1. The finding that the SH3 domain of PLCγ1 is in close proximity to the C terminus supports this hypothesis (41).
The existence of a link between c-Src and PLCγ1 is strongly supported by the finding that the activity of PLCγ1 is higher in SrcNIH cells than in the parental NIH 3T3 cell line. Moreover, we found that the Src inhibitor, PP1, reduces Tyr-783 phosphorylation on PLCγ1 in a dose-dependent manner (supplemental Fig. S7). Hence, in OVCAR3 and A431 cells, c-Src signals to PKCα via PLCγ.
LKB1 is the major upstream kinase of AMPK (for review, see Refs. 7, 8), therefore we looked for a connection between PKCα and LKB1. We found that the phosphorylation of LKB1 by PKCα leads to activation of AMPK (Fig. 5). Additionally, in a cell-free assay, PKCα directly phosphorylated LKB1 at Ser-428 (supplemental Fig. S9). In light of these results, we conclude that AMPK is regulated by c-Src through PKCα, PLCγ1, and LKB1 in OVCAR3 and A431 cells.
We speculate that this signaling pathway, in which Src or other tyrosine kinases activate AMPK via PKC, might be common to a number of cell types. It would be interesting to investigate whether this pathway is activated in tumor tissue. It has been recently shown that in endothelial cells AMPK is activated by c-Src, via the PI3-kinase pathway (22, 23). Even though c-Src does not regulate AMPKα in HeLa and MCF7 cells (supplemental Fig. S11, A and B), we did find reduced activity of AMPK when PKC was inhibited in these cells (supplemental Fig. S11, C and D). We speculate that not only c-Src, but also other tyrosine kinases or receptor tyrosine kinases might be involved in the regulation of AMPK via PKC. Furthermore, not only PKCα, but also other PKC isoforms, might be involved in the regulation of AMPK. As mentioned above, it has been reported previously that AMPK is PKCζ-dependent in heart and skeletal muscle (22, 23). This dependence was not seen in A431 and OVCAR3 cells, but might be relevant to other cell lines. Further, we note that increased AMPK activity was monitored in HeLa cells compared with primary keratinocytes (supplemental Fig. S1), although it is well known that HeLa cells are LKB1-deficient (42).
AMPK is activated under stress, enabling cells to survive. Cancer cells are subjected to significant stress, due to the accumulation of chromosomal aberrations and damaged proteins, as well as to their rapid rates of proliferation and the consequent metabolic stress. Although LKB1 is generally considered to be a tumor suppressor, the role of AMPK in cancer is unclear. It has been suggested that AMPK is essential for tumors to adapt to metabolic stress (43 and references therein). The activation of AMPK has been reported to decrease cap-dependent translation (11). Here, we have shown that AMPK also plays a role in the regulation of IRES-dependent translation. We found that IRES-dependent translation was up-regulated when AMPK was activated and down-regulated when AMPK was inhibited (supplemental Fig. S10, A, B, E, and F). As expected, following AMPK activation, there was a reduction in cap-dependent translation (supplemental Fig. S10, C, D, G, and H). Surprisingly, AMPK inhibition by compound C also reduced cap-dependent translation (supplemental Fig. S10, C, D, G, and H), possibly due to a toxic effect of compound C on A431 and OVCAR3 cells. The activation of AMPK led to the increased accumulation of Hif-1α, whereas rapamycin treatment (which inhibits cap-dependent translation) had no effect on the accumulation of this protein in OVCAR3 cells (Fig. 6). Thus, activation of AMPK by c-Src leads to induced IRES-dependent translation in OVCAR3 cells.
In summary, we have shown that AMPK is regulated by c-Src through PKCα, PLCγ, and LKB1 in OVCAR3 and A431 cells. Furthermore, our results imply a correlation between AMPK activation and IRES-dependent translation. We suggest that activation of AMPK by c-Src may signal the cell to save energy, to allow cell survival under stress.
Supplementary Material
Acknowledgments
We thank D. G. Hardie, R. Fukunaga, A. E. Willis, and G. J. Goodall for plasmids and helpful advice.
This work was supported by the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum and the Israel Ministry of Science and Technology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S12.
- AMPK
- AMP-dependent protein kinase
- AICAR
- 5-aminoimidazole-4-carboxamide riboside
- BIS1
- bisindolymaleimide I
- FFL
- firefly luciferase
- IRES
- internal ribosome entry site
- mTOR
- mammalian target of rapamycin
- PLCγ
- phospholipase Cγ
- SPL
- sea pansy (Renilla) luciferase
- TPA
- 12-O-tetradecanoylphorbol-13-acetate.
REFERENCES
- 1. Martin G. S. (2001) Nat. Rev. Mol. Cell Biol. 2, 467–475 [DOI] [PubMed] [Google Scholar]
- 2. Martin G. S. (2004) Oncogene 23, 7910–7917 [DOI] [PubMed] [Google Scholar]
- 3. Yeatman T. J. (2004) Nat. Rev. Cancer 4, 470–480 [DOI] [PubMed] [Google Scholar]
- 4. Mizrachy-Schwartz S., Kravchenko-Balasha N., Ben-Bassat H., Klein S., Levitzki A. (2007) PLoS One 2, e628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zou M. H., Hou X. Y., Shi C. M., Kirkpatick S., Liu F., Goldman M. H., Cohen R. A. (2003) J. Biol. Chem. 278, 34003–34010 [DOI] [PubMed] [Google Scholar]
- 6. Zou M. H., Kirkpatrick S. S., Davis B. J., Nelson J. S., Wiles W. G., 4th, Schlattner U., Neumann D., Brownlee M., Freeman M. B., Goldman M. H. (2004) J. Biol. Chem. 279, 43940–43951 [DOI] [PubMed] [Google Scholar]
- 7. Hardie D. G. (2008) Int. J. Obes. 32, S7–S12 [DOI] [PubMed] [Google Scholar]
- 8. Hardie D. G. (2007) Nat. Rev. Mol. Cell Biol. 8, 774–785 [DOI] [PubMed] [Google Scholar]
- 9. Ashrafian H. (2006) Lancet 367, 618–621 [DOI] [PubMed] [Google Scholar]
- 10. Hardie D. G., Scott J. W., Pan D. A., Hudson E. R. (2003) FEBS Lett. 546, 113–120 [DOI] [PubMed] [Google Scholar]
- 11. Hahn-Windgassen A., Nogueira V., Chen C. C., Skeen J. E., Sonenberg N., Hay N. (2005) J. Biol. Chem. 280, 32081–32089 [DOI] [PubMed] [Google Scholar]
- 12. Lin P. H., Shenoy S., Galitski T., Shalloway D. (1995) Oncogene 10, 401–405 [PubMed] [Google Scholar]
- 13. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 14. Zor T., Selinger Z. (1996) Anal. Biochem. 236, 302–308 [DOI] [PubMed] [Google Scholar]
- 15. Ueda T., Watanabe-Fukunaga R., Fukuyama H., Nagata S., Fukunaga R. (2004) Mol. Cell. Biol. 24, 6539–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stoneley M., Paulin F. E., Le Quesne J. P., Chappell S. A., Willis A. E. (1998) Oncogene 16, 423–428 [DOI] [PubMed] [Google Scholar]
- 17. Lang K. J., Kappel A., Goodall G. J. (2002) Mol. Biol. Cell 13, 1792–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boudeau J., Baas A. F., Deak M., Morrice N. A., Kieloch A., Schutkowski M., Prescott A. R., Clevers H. C., Alessi D. R. (2003) EMBO J. 22, 5102–5114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wiener J. R., Windham T. C., Estrella V. C., Parikh N. U., Thall P. F., Deavers M. T., Bast R. C., Mills G. B., Gallick G. E. (2003) Gynecol. Oncol 88, 73–79 [DOI] [PubMed] [Google Scholar]
- 20. Osherov N., Levitzki A. (1994) Eur. J. Biochem. 225, 1047–1053 [DOI] [PubMed] [Google Scholar]
- 21. Hanke J. H., Gardner J. P., Dow R. L., Changelian P. S., Brissette W. H., Weringer E. J., Pollok B. A., Connelly P. A. (1996) J. Biol. Chem. 271, 695–701 [DOI] [PubMed] [Google Scholar]
- 22. Xie Z., Dong Y., Zhang M., Cui M. Z., Cohen R. A., Riek U., Neumann D., Schlattner U., Zou M. H. (2006) J. Biol. Chem. 281, 6366–6375 [DOI] [PubMed] [Google Scholar]
- 23. Xie Z., Dong Y., Scholz R., Neumann D., Zou M. H. (2008) Circulation 117, 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Longo M., Brama M., Marino M., Bernardini S., Korach K. S., Wetsel W. C., Scandurra R., Faraggiana T., Spera G., Baron R., Teti A., Migliaccio S. (2004) Bone 34, 100–111 [DOI] [PubMed] [Google Scholar]
- 25. Tan M., Li P., Sun M., Yin G., Yu D. (2006) Oncogene 25, 3286–3295 [DOI] [PubMed] [Google Scholar]
- 26. Dutil E. M., Toker A., Newton A. C. (1998) Curr. Biol. 8, 1366–1375 [DOI] [PubMed] [Google Scholar]
- 27. Le Good J. A., Ziegler W. H., Parekh D. B., Alessi D. R., Cohen P., Parker P. J. (1998) Science 281, 2042–2045 [DOI] [PubMed] [Google Scholar]
- 28. Taniyama Y., Weber D. S., Rocic P., Hilenski L., Akers M. L., Park J., Hemmings B. A., Alexander R. W., Griendling K. K. (2003) Mol. Cell. Biol. 23, 8019–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang K. J., Shin S., Piao L., Shin E., Li Y., Park K. A., Byun H. S., Won M., Hong J., Kweon G. R., Hur G. M., Seok J. H., Chun T., Brazil D. P., Hemmings B. A., Park J. (2008) J. Biol. Chem. 283, 1480–1491 [DOI] [PubMed] [Google Scholar]
- 30. Shah K., Vincent F. (2005) Mol. Biol. Cell 16, 5418–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nozawa H., Howell G., Suzuki S., Zhang Q., Qi Y., Klein-Seetharaman J., Wells A., Grandis J. R., Thomas S. M. (2008) Clin. Cancer Res. 14, 4336–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang J., Taba Y., Pang J., Yin G., Yan C., Berk B. C. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martiny-Baron G., Fabbro D. (2007) Pharmacol. Res. 55, 477–486 [DOI] [PubMed] [Google Scholar]
- 34. Kim H. K., Kim J. W., Zilberstein A., Margolis B., Kim J. G., Schlessinger J., Rhee S. G. (1991) Cell 65, 435–441 [DOI] [PubMed] [Google Scholar]
- 35. Kassis J., Moellinger J., Lo H., Greenberg N. M., Kim H. G., Wells A. (1999) Clin. Cancer Res. 5, 2251–2260 [PubMed] [Google Scholar]
- 36. Kayali A. G., Eichhorn J., Haruta T., Morris A. J., Nelson J. G., Vollenweider P., Olefsky J. M., Webster N. J. (1998) J. Biol. Chem. 273, 13808–13818 [DOI] [PubMed] [Google Scholar]
- 37. Dowling R. J., Zakikhani M., Fantus I. G., Pollak M., Sonenberg N. (2007) Cancer Res. 67, 10804–10812 [DOI] [PubMed] [Google Scholar]
- 38. Kim E. K., Miller I., Aja S., Landree L. E., Pinn M., McFadden J., Kuhajda F. P., Moran T. H., Ronnett G. V. (2004) J. Biol. Chem. 279, 19970–19976 [DOI] [PubMed] [Google Scholar]
- 39. Karni R., Dor Y., Keshet E., Meyuhas O., Levitzki A. (2002) J. Biol. Chem. 277, 42919–42925 [DOI] [PubMed] [Google Scholar]
- 40. Lee M., Hwang J. T., Lee H. J., Jung S. N., Kang I., Chi S. G., Kim S. S., Ha J. (2003) J. Biol. Chem. 278, 39653–39661 [DOI] [PubMed] [Google Scholar]
- 41. Rebecchi M. J., Pentyala S. N. (2000) Physiol. Rev. 80, 1291–1335 [DOI] [PubMed] [Google Scholar]
- 42. Hawley S. A., Boudeau J., Reid J. L., Mustard K. J., Udd L., Mäkelä T. P., Alessi D. R., Hardie D. G. (2003) J. Biol. 2, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones R. G., Thompson C. B. (2009) Genes Dev. 23, 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.