Abstract
Cytosine arabinoside (ara-C) is irreversibly deaminated by cytidine deaminase (CDD) to a nontoxic metabolite. A common polymorphism, A79C, in CDD changes a lysine residue to glutamine resulting in decreased enzyme activity. We determined CDD A79C genotypes for 457 children with AML treated on CCG 2941 and 2961 and analyzed the impact of CDD genotype on therapy outcomes. Post-Induction treatment related mortality (TRM) was significantly elevated in children with the CC genotype (5 year TRM 17 ± 13% CC vs 7 ± 4% AA, 5 ± 4% AC, p= 0.05). This was more notable in children who received IDA-FLAG (ara-C= 7590 mg/m2) (5 year TRM 24 ± 21% CC vs 6 ± 6% AA, 6 ± 7% AC, p=0.07) as consolidation therapy compared to IDA-DCTER (ara-C= 800 mg/m2) (5 year TRM 15 ± 20% CC vs 8 ± 6% AA, 4 ± 6% AC; p=0.29). Relapse-free survival was non-significantly increased in children with the CC genotype treated with IDA-FLAG (76 ± 20% CC vs 59 ± 12% AA and 55 ± 14% AC; p= 0.40). These data indicate that children with a low activity CDD genotype are at increased risk of treatment-related mortality with Ara-C based therapy for AML.
Introduction
Ara-C (1-B-D-arabinofuranosylcytosine), an arabinose containing analogue of the pyrimidine nucleoside deoxycytidine, is an active chemotherapeutic agent for hematological malignancies, particularly acute myeloid leukemia (AML) (1 –3). Ara-C forms the backbone of both induction and consolidation therapy for AML, given at standard or high doses (4). The cytotoxic effect of ara-C requires transport into the cells followed by metabolic activation. Intracellular transport of ara-C, when administered in standard doses, is mediated by the human equilibrative transporter hENT1 (5,6). At high doses, ara-C diffuses into the cell at a rate that exceeds that of pump-mediated transport (5,6). Inside the cell, ara-C is phosphorlylated to its active triphosphate form (ara-CTP) through the sequential action of deoxycytidine kinase (dCK), deoxycytidylate kinase and nucleoside diphosphate kinase (7–9). Phosphorylation by dCK is the rate-limiting step in this process. Ara-CTP is incorporated into DNA and inhibits DNA synthesis in a competitive fashion (10,11). Cytidine deaminase (CDD) is a pyrimidine salvage pathway enzyme that catalyzes the hydrolytic deamination of cytidine and deoxycytidine to their corresponding uracil nucleosides. CDD deaminates ara-C, resulting in the formation of its inactive metabolite 1-B-D-arabinofuranosyluracil (12–14). Several studies have suggested that increased levels of CDD may play an important role in the development of resistance to ara-C.
There is a non-synonymous single nucleotide polymorphism (SNP) in the CDD gene, which results in an amino acid change from lysine to glutamine at position 27. In-vitro studies have shown that the variant glutamine allele (C genotype) has reduced activity compared to the wild-type lysine allele (A genotype) (15, 16). We hypothesized that the children treated with ara-C for AML with the variant CC genotype would have inferior outcomes with increased toxicity compared to other genotypes. In this study we demonstrate increased treatment-related mortality in children with a homozygous CC genotype.
Patients and Methods
Patients
The study population consisted of 457 children from birth to 21 years with de novo AML treated on Children's Cancer Group (CCG) protocols 2941 and 2961 between 1995 and 2002. Children with therapy related AML were excluded from this analysis. Clinical data, including age, sex, white blood cell (wbc) count at diagnosis, race, presence of chloroma, presence of CNS disease, immunophenotype and cytogenetic abnormalities were collected prospectively. Patients with Down Syndrome were not eligible for these trials. Cases were classified on the basis of criteria established and revised by the French-American-British (FAB) Cooperative Study Group by central pathology review. All FAB categories except acute promyelocytic leukemia (APL-AML M3) were eligible for enrollment and were treated on the same chemotherapy regimens. All patients or legal guardians consented to enrollment on the therapeutic studies after approval of the study by the Institutional Review Board (IRB) of each participating institution, and to the submission of samples for biological studies. Informed consent was provided in accordance with the Declaration of Helsinki. The genotyping study and analysis were approved by the IRB of Cincinnati Children's Hospital and Medical Center.
Blood samples obtained from 205 healthy blood donors served as controls to estimate the genotype frequencies in the normal population. One hundred and four were Caucasian and 101 were African-American.
Chemotherapy Treatment Regimen
CCG-2961 was a phase III randomized trial for patients age <21 years with previously untreated acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS) conducted between August 1996 and December 2002. CCG-2941 was a feasibility pilot study for the successor CCG-2961 trial and had a similar study design. All patients received intensively timed induction therapy with IDA-DCTER (idarubicin, dexamethasone, cytarabine, thioguanine, etoposide and daunomycin) given on days 0 to 3 followed by DCTER (dexamethasone, ara-C, thioguanine, etoposide and daunomycin) given on days 10 to 13. The total dose of ara-C during induction (course 1) was 1600 mg/m2. On recovery of white blood cell and platelet counts, patients were randomly assigned to receive consolidation (course 2) therapy with Regimen A, which was a second course of DCTER (total dose of ara-C 1600 mg/m2) or Regimen B which was IDA-FLAG (idarubicin, fludarabine, ara-C (total dose of ara-C 7590 mg/m2) and G-CSF). Intrathecal ara-C was used for central nervous system (CNS) prophylaxis. Following consolidation, patients with a matched related donor proceeded to allogeneic marrow transplant with ablative conditioning (busulfan and cytoxan). Those without a related donor received intensification with high-dose ara-C (total dose of ara-C 24,000 mg/m2) and L-asparaginase and additional intrathecal ara-C. Patients who did not receive an allogeneic transplant were further randomized to receive immune modulation with Interleukin-2 or standard follow-up care (17, 18). Treatment related mortality was unacceptably high in children enrolled in the study between 1996 and 1999 and the study was suspended at this time and amended to recommend a uniform approach to infection prevention, including requirements for mandatory hospitalization during periods of expected neutropenia, use of broad spectrum antibiotics, including vancomycin for fever, anti-fungal therapy for fever beyond 72 hours, immunoglobulin replacement and avoidance of corticosteroids as anti-emetics (17,18).
Genotyping
DNA was extracted from diagnostic marrow samples using standard methods and normalized to 10 ng/ml. Genotyping for the CDD A79C (NCBI rs 2072671) polymorphism was performed using a fluorescence based allelic discrimination assay (Taqman, Applied Biosystems, Foster City, CA). Gene-specific polymerase chain reaction (PCR) primers and fluorogenic probes for allelic discrimination were supplied by Applied Biosystems (Taqman Drug Metabolism Genotyping Assays). PCR cycling reactions were performed in 96-well micro titer plates in a GeneAmp PCR System 9700 (Perkin-Elmer) according to the conditions specified by the manufacturer. Results were analyzed by the automated TaqMan allelic discrimination assay using sequence detection system 1.2.3 software (ABI TaqMan 7300, Applied Biosystems). Genotyping results were duplicated in 10% of samples; concordance between repeats was 100%.
Genomic DNA was extracted from peripheral blood samples from our control population using QIAamp blood DNA isolation kits (Qiagen Sciences, Maryland, USA) as per the manufacturer's protocol. Genotyping was performed as described for the patient population.
Statistical Analysis
Data obtained from CCG-2941 through April 14, 2005 and from CCG-2961 through April 26, 2006 were used for analyses. The Kaplan-Meier method was used to calculate estimates of overall survival (OS), event-free survival (EFS) and disease-free survival (DFS) (19). Estimates are reported with their Greenwood standard errors (20). Differences in these estimates were tested for significance using the log-rank statistic (21). Cumulative incidence estimates were used to determine treatment-related mortality (TRM) and relapse-free survival (RFS). Differences between TRM and RFS estimates were tested for significance using Gray's test (22). Patients lost to follow-up were censored at their date of last known contact or at a cutoff 6 months prior to April 2005 or April 2006 for CCG-2941 and CCG-2961, respectively, in order to prevent deaths and relapses being reported sooner than ongoing follow-up. The significance of observed differences in proportions was tested using the Chi-squared test and Fisher's exact test when data were sparse.
Definitions
Overall survival (OS) is defined as time from study entry to death from any cause. Event-free survival (EFS) is defined as time from study entry to failure at the end of two courses, relapse or death from any cause. Disease-free survival (DFS) is defined as time from remission to failure at the end of two courses, relapse or death from any cause. Treatment related mortality (TRM) is assessed at three time points i.e. study entry, end of course one or end of course two and defined as the time from either study entry, from the end of one course of therapy or from the end of two courses of therapy to death from causes other than progressive disease where failures at the end of two courses, relapses and deaths from progressive disease were competing events. Relapse-free survival (RFS) is defined as the time from the end of one or two courses of therapy to death from progressive disease, failure at the end of two courses, or relapse where deaths from causes other than progressive disease were competing events.
Results
Genotype frequencies and demographics for cases are summarized in Table I. CDD genotypes varied significantly by ethnicity with the CDD variant C allele being more frequent in white children (AC+CC 80%) compared with black (AC+CC 3%), Hispanic (AC+CC 13%), Asian children (AC+CC 1%), and other races (AC+CC 3%), p<0.001. Genotype frequencies did not differ significantly by gender. No significant difference was observed with respect to other known AML prognostic factors, eg wbc at presentation, age or cytogenetic abnormality among the CDD genotype groups. Genotype frequencies were in Hardy Weinberg equilibrium and did not differ from those seen in healthy race-matched controls (data not shown).
Table I.
Demographic characteristics by CDD A79C genotype
| AA | AC | CC | AC+CC | AA vs AC+CC | |||||
|---|---|---|---|---|---|---|---|---|---|
| Characteristic | N | % | N | % | N | % | N | % | P |
| Total | 229 | 179 | 49 | 228 | |||||
| Age: median & range (yrs) | 10.5 | (0.15 – 20.9) | 10.0 | (0.01 – 19.8) | 9.6 | (0.13 – 18.4) | 10.0 | (0.01 – 19.8) | 0.61 |
| WBC: median & range Study | 18,900 | (300 – 860,000) | 19,600 | (1,000 – 600,000) | 25,500 | (1,500 – 324,000) | 20,800 | (1,000 – 600,000) | 0.61 |
| CCG-2941 | 13 | 6 | 18 | 10 | 7 | 14 | 25 | 11 | 0.06 |
| CCG-2961 | 216 | 94 | 161 | 90 | 42 | 86 | 203 | 89 | |
| Gender | |||||||||
| Male | 130 | 57 | 96 | 54 | 25 | 51 | 121 | 53 | 0.48 |
| Female | 99 | 43 | 83 | 46 | 24 | 49 | 107 | 47 | |
| Race | |||||||||
| White | 129 | 57 | 142 | 79 | 41 | 84 | 183 | 80 | <0.001 |
| Black | 33 | 14 | 5 | 3 | 1 | 2 | 6 | 3 | <0.001 |
| Hispanic | 42 | 18 | 23 | 13 | 7 | 14 | 30 | 13 | 0.16 |
| Asian | 11 | 5 | 3 | 2 | 0 | 0 | 3 | 1 | 0.06 |
| Other | 13 | 6 | 6 | 3 | 0 | 0 | 6 | 3 | 0.16 |
| Unknown | 1 | 0 | 0 | 0 | |||||
| FAB subtype | |||||||||
| M0 | 8 | 4 | 13 | 7 | 4 | 8 | 17 | 8 | 0.10 |
| M1 | 32 | 14 | 35 | 20 | 4 | 8 | 39 | 17 | 0.43 |
| M2 | 67 | 29 | 48 | 27 | 16 | 33 | 64 | 28 | 0.86 |
| M4 | 63 | 28 | 45 | 25 | 11 | 22 | 0.54 | ||
| 56 | 25 | ||||||||
| M5 | 40 | 17 | 29 | 16 | 8 | 16 | 37 | 16 | 0.82 |
| M6 | 7 | 3 | 2 | 1 | 1 | 2 | 3 | 1 | 0.34 |
| M7 | 7 | 3 | 4 | 2 | 5 | 10 | 9 | 4 | 0.79 |
| De novo (NOS) | 5 | 2 | 3 | 2 | 0 | 0 | 3 | 1 | 0.72 |
| Cytogenetics | |||||||||
| Normal | 32 | 23 | 24 | 22 | 10 | 34 | 34 | 25 | 0.92 |
| t(8;21) | 27 | 20 | 16 | 15 | 3 | 10 | 19 | 14 | 0.25 |
| Abn 16 | 14 | 10 | 9 | 8 | 4 | 14 | 13 | 9 | 0.98 |
| Abn 11 | 27 | 20 | 25 | 23 | 5 | 17 | 30 | 22 | 0.79 |
| t(6;9) | 1 | 1 | 4 | 4 | 0 | 0 | 4 | 3 | 0.37 |
| −7/7- | 4 | 3 | 4 | 4 | 0 | 0 | 4 | 3 | 1.000 |
| −5/5- | 2 | 2 | 1 | 1 | 1 | 3 | 2 | 1 | 1.000 |
| +8 | 7 | 5 | 9 | 8 | 2 | 7 | 11 | 8 | 0.47 |
| +21 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 1 | 0.50 |
| Pseudodiploid | 16 | 12 | 9 | 8 | 4 | 14 | 13 | 9 | 0.68 |
| Hyperdiploid | 5 | 4 | 3 | 3 | 0 | 0 | 3 | 2 | 0.50 |
| Hypodiploid | 2 | 2 | 3 | 3 | 0 | 0 | 3 | 2 | 1.000 |
| Unknown | 92 | 40 | 70 | 39 | 20 | 41 | 90 | 39 | |
| Response at end of first course | |||||||||
| Remission | 196 | 87 | 147 | 87 | 45 | 92 | 192 | 88 | 0.87 |
| Failure | 19 | 8 | 10 | 6 | 2 | 4 | 12 | 6 | 0.31 |
| Death | 10 | 4 | 12 | 7 | 2 | 4 | 14 | 6 | 0.48 |
| Inevaluable | 4 | 10 | 0 | 10 | |||||
From study entry
Overall survival (OS) from study entry did not differ significantly among the CDD genotype groups (5 yr OS 52 ± 7% for AA, 46 ± 8% for AC, 55 ± 15% for CC; p=0.56). Similarly, we observed no significant difference in EFS (5 year EFS 40 ± 7% AA, 37 ± 7% AC, 41 ± 15% CC; p=0.90) nor treatment related mortality (TRM) from study entry (5 year TRM 15 ± 5% AA vs 18 ± 6% AC vs 20 ± 12% CC; p=0.59.)
From end of induction
Overall survival (OS) from the end of one course of therapy for patients in remission did not differ significantly among the CDD genotype groups (5 yr OS 58 ± 8% for AA, 55 ± 9% for AC, 59 ± 16% for CC; p=0.91) as well as DFS (5 yr OS 48 ± 8% for AA, 45 ± 9% for AC, 45 ± 16% for CC; p=0.86) and RFS (5 yr RFS 60 ± 8% for AA, 58 ± 9% for AC, 65 ± 15% for CC; p=0.90). Treatment related mortality from end of course one, when only low doses of ara-C were used, was not statistically significant by CDD genotype (5 year TRM 12 ± 5% AA vs 13 ± 6% AC vs 20 ± 12% CC; p=0.55), and remission induction rate was similar in all groups (87% AA vs 87% AC vs 92% CC; p= 0.63).
From end of course 2 (post randomization)
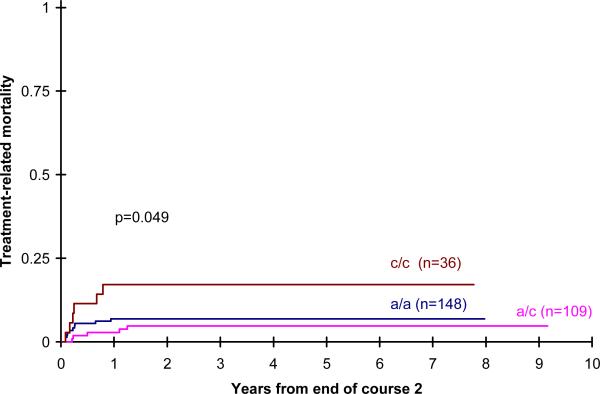
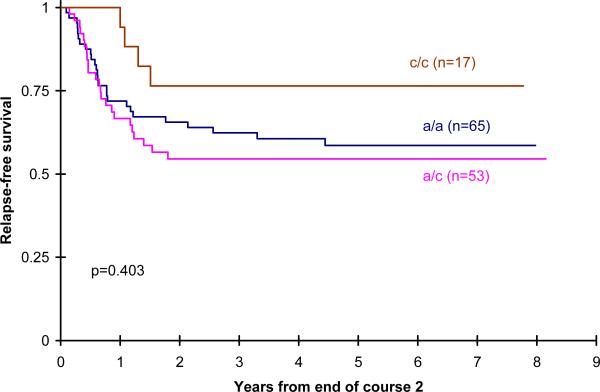
After the second course of treatment, however, when larger doses of ara-C were used, there was a significant increase in the cumulative incidence of treatment related mortality in children with the CC genotype compared with AA and AC genotypes (5 year TRM 7 ± 4% AA vs 5 ± 4% AC vs 17 ± 13% CC; p= 0.049; p=0.05) (Figure1). The impact of CDD genotype on TRM was more notable in children randomized to IDA-FLAG consolidation receiving a higher cumulative dose of ara-C (9190 mg/m2)(5 year TRM 6 ± 6% AA vs 6 ± 7% AC vs 24 ± 21% CC;p=0.06) than those randomized to DCTER, receiving a lower cumulative dose of ara-C (3200mg/m2) (5 year TRM 8 ± 6% AA vs 4 ± 6% AC vs 15 ± 20% CC; p=0.39). These outcomes are summarized in Table II. RFS was increased in children with the CC genotype treated with IDA-FLAG but this was not statistically significant, perhaps because numbers of cases were small (59 ± 12% AA vs 55 ± 14% AC vs 76 ± 20% CC; p= 0.40) (Figure 2). However OS (5 yr OS 62 ± 8% for AA, 62 ± 8% for AC, 62 ± 16% for CC; p=0.97) and DFS (5 yr DFS 51 ± 8% for AA, 52 ± 10% for AC, 51 ± 17% for CC; p=0.93) from the end of two courses was not statistically significant by genotype.
Figure 1.
Post induction treament related mortality by CDD genotype
Table II.
Treatment-related mortality by therapy received
| CDD Genotype | TRM from study entry N= | Post induction TRM N= | Post randomization TRM N=293 | IDA-DCTER Total Cytarabine 3200 mg/m2 N=137 | IDA-FLAG Total Cytarabine 9190 mg/m2 N=135 |
|---|---|---|---|---|---|
| AA | 15 ±5% | 12 ±5% | 6.9% ± 4.2% | 8.2% ± 6.4% | 6.3% ± 6.1% |
| AC | 18 ±6% | 13 ±6% | 4.8% ± 4.2% | 4.3% ± 6.0% | 5.9% ± 6.7% |
| CC | 20 ±12% | 20 ±12% | 17.1% ± 12.7% | 15.4% ± 20% | 23.5% ± 20.6% |
| P-value | 0.59 | 0.55 | 0.05 | 0.39 | 0.06 |
Figure 2.
Post randomization RFS by CDD genotype for IDA-FLAG patients
When non-hematological toxicities were examined, overall toxicity was not significantly different in the three genotype groups (AA 64%, AC 66%, CC 59%, AA vs AC vs CC, p = 0.675) though patients with the variant CC genotype were more likely to report mucositis post-randomization (AA 21%, AC 32%, CC 34%, p = 0.04). In attempt to examine specific Ara-C toxicities we looked at cerebellar and clinical pulmonary toxicity. The number of cases of cerebellar toxicity was small (5 in course 1 and 6 in course 2) and no differences were seen by genotype. Similarly, the overall incidence of pulmonary toxicity was low and not different by genotype.
Discussion
In this study, we found that children with a low activity CC genotype were at an increased risk of treatment related mortality when treated with ara-C based therapy for AML. Analysis of post-induction outcomes showed that this effect was more notable in the group of patients randomized to receive IDA-FLAG therapy where the total dose of ara-C was considerably higher (9190 mg/m2) as compared to the IDA-DCTER arm (3200 mg/m2). We expected that increased in vivo exposure to ara-C would result in a reduced relapse rate. In our dataset, RFS was improved by 17% in children with the homozygous low activity CC genotype compared with other genotypes, although this did not achieve statistical significance, possibly due to limited sample size. The balance of increased TRM but reduced relapse resulted in equivalent overall survival in children with the CC genotype.
Several in vitro studies have indicated a role for increased CDD enzyme activity in the development of resistance to ara-C (23, 24). Yusa et al demonstrated that chronically HIV-1 infected H-9 cells showed 21 fold resistance to ara-C owing to the induction of cytidine deaminase activity (23). This conclusion was supported by the observation that ara-C resistance could be reversed by using 3,4,5,6-tetrahydrouridine, an inhibitor of cytidine deaminase (25, 26). Similar observations were reported in erythroleukemia cell lines, AML cell lines and mouse tumors (25, 27–30). More definitive evidence of the crucial role CDD plays in ara-C metabolism comes from the demonstration of ara-C resistance in murine fibroblasts that constitutively overexpress the human CDD gene (31,32). In fact, retroviral transduction of CDD in human hematopoeitic progenitor cells is being explored as a cytoprotective strategy in conjunction with dose-intensive cytarabine therapy in the animal model with promising preliminary results (33, 34).
Attempts to study the relationship between CDD activity in primary human AML blasts maintained in short term culture and clinical drug response have yielded inconsistent results. Smyth et al found no correlation between the levels of ara-C deamination and remission induction in their series of 29 adult AML patients(35). Mejer and Nygaard demonstrated that while CDD activity was lower in AML cells compared to normal leukocytes, CDD activity did not predict response to ara-C based therapy in their patient population of 17 adults (28). Other authors have reported that increased CDD activity correlates both with clinical drug resistance and duration of remission (24,36–38). Some have reported a non-significant trend for treatment failure with increased CDD activity (29). Jahns-Streubel and colleagues found pre-therapeutic CDD levels to be the most sensitive predictor of blast clearance in their patient subset of 36 adults with de novo AML treated with TAD-9 induction therapy (39). In their patient population, CDD activity was also predictive of remission duration. Schroder et al corroborated these findings by demonstrating that median CDD activity was higher in patients with refractory and recurrent disease (40). They assessed both quantitative CDD mRNA expression and enzyme activity.
CDD is not the only gene that may modify Ara-C metabolism and response to therapy. Pharmacogenetic variants in genes such as hENT1 that transports Ara-C across the cell membrane, deoxycytidine kinase (CdR kinase) that is the rate limiting step in Ara-CTP formation, pyrimidine nucleotidase 1 that opposes the action of deoxycytidine kinase, and UMP/CMP kinase that catalyses the conversion of Ara-CMP to Ara-DMP could also modify outcomes and future studies are needed to examine this possibility (6, 41–43).
The strength of our study is the large sample size, representing children treated on a uniform clinical treatment protocol with long and complete follow-up. A weakness of our study is the lack of direct measurement of CDD activity in the children, in whom we implied CDD activity from genotype. While Kirch et al have demonstrated reduced enzyme activity with the A79C polymorphism using in vitro deamination of ara-C (15), it is possible that additional polymorphisms within the CDD gene also influence enzyme activity, or that variants in other enzymes important in ara-C metabolism play a major role. In the patients treated on CCG 2961, increased mortality due to infections was noted in the patients on the IDA-FLAG arm raising the question whether Fludarabine contributed to additional immunosuppression in this subset (41). This may be a confounding factor when assessing post-randomization TRM in the patients who received IDA-FLAG i.e. higher dose of ara-C.
This emphasizes the importance of replicating these data in independent datasets, and in clinical protocols using different doses of ara-C to determine the significance of this polymorphism.
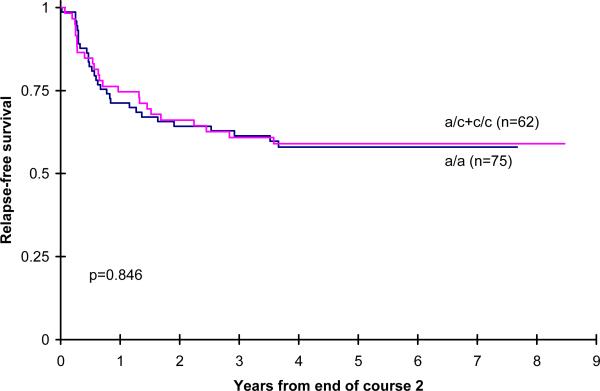
Figure 3.
Post randomization RFS by CDD genotype for patients receiving IDA-DCTER
References
- (1).Beard ME, Fairley GH. Acute leukemia in adults. Semin Hematol. 1974 Jan;11(1):5–24. [PubMed] [Google Scholar]
- (2).Ellison RR, Holland JF, Weil M, Jacquillat C, Boiron M, Bernard J, Sawitsky A, Rosner F, Gussoff B, Silver RT, Karanas A, Cuttner J, Spurr CL, Hayes DM, Blom J, Leone LA, Haurani F, Kyle R, Hutchison JL, Forcier RJ, Moon JH. Arabinosyl cytosine: a useful agent in the treatment of acute leukemia in adults. Blood. 1968 Oct;32(4):507–23. [PubMed] [Google Scholar]
- (3).Keating MJ, McCredie KB, Bodey GP, Smith TL, Gehan E, Freireich EJ. Improved prospects for long-term survival in adults with acute myelogenous leukemia. JAMA. 1982 Nov 19;248(19):2481–6. [PubMed] [Google Scholar]
- (4).Bodey GP, Freireich EJ, Monto RW, Hewlett JS. Cytosine arabinoside (NSC-63878) therapy for acute leukemia in adults. Cancer Chemother Rep. 1969 Feb;53(1):59–66. [PubMed] [Google Scholar]
- (5).Wiley JS, Jones SP, Sawyer WH. Cytosine arabinoside transport by human leukaemic cells. Eur J Cabcer Clin Oncol. 1983;19:1067–1074. doi: 10.1016/0277-5379(83)90029-9. [DOI] [PubMed] [Google Scholar]
- (6).Galmarini CM, Thomas X, Calvo F, Rousselot P, Rabilloud M, El Jaffari A, et al. In vivo mechanisms of resistence to cytarabine in acute myeloid leukemia. Br J Haematol. 2002;117:860–868. doi: 10.1046/j.1365-2141.2002.03538.x. [DOI] [PubMed] [Google Scholar]
- (7).Kessel D, Hall TC, Rosenthal D. Uptake and phosphorylation of cytosine arabinoside by normal and leukemic human blood cells in vitro. Cancer Res. 1969 Feb;29(2):459–63. [PubMed] [Google Scholar]
- (8).Chou TC, Arlin Z, Clarkson BD, Phillips FS. Metabolism of 1-beta-D-arabinofuranosylcytosine in human leukemic cells. Cancer Res. 1977 Oct;37(10):3561–70. [PubMed] [Google Scholar]
- (9).Schrecker AW, Goldin A. Antitumor effect and mode of action of 1-beta-D-arabinofuranosylcytosine 5'-phosphate in leukemia L1210. Cancer Res. 1968 Apr;28(4):802–3. [PubMed] [Google Scholar]
- (10).Furth JJ, Cohen SS. Inhibition of mammalian DNA polymerase by the 5'-triphosphate of 1-beta-d-arabinofuranosylcytosine and the 5'-triphosphate of 9-beta-d-arabinofuranoxyladenine. Cancer Res. 1968 Oct;28(10):2061–7. [PubMed] [Google Scholar]
- (11).Graham FL, Whitmore GF. Studies in mouse L-cells on the incorporation of 1-beta-D-arabinofuranosylcytosine into DNA and on inhibition of DNA polymerase by 1-beta-D-arabinofuranosylcytosine 5'-triphosphate. Cancer Res. 1970 Nov;30(11):2636–44. [PubMed] [Google Scholar]
- (12).Chabner BA, Johns DG, Coleman CN, Drake JC, Evans WH. Purification and properties of cytidine deaminase from normal and leukemic granulocytes. J Clin Invest. 1974 Mar;53(3):922–31. doi: 10.1172/JCI107633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Cacciamani T, Vita A, Cristalli G, Vincenzetti S, Natalini P, Ruggieri S, Amici A, Magni G. Purification of human cytidine deaminase: molecular and enzymatic characterization and inhibition by synthetic pyrimidine analogs. Arch Biochem Biophys. 1991 Nov 1;290(2):285–92. doi: 10.1016/0003-9861(91)90543-r. [DOI] [PubMed] [Google Scholar]
- (14).Betts L, Xiang S, Short SA, Wolfenden R, Carter CW., Jr. Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. J Mol Biol. 1994 Jan 14;235(2):635–56. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- (15).Kirch HC, Schröder J, Hoppe H, Esche H, Seeber S, Schütte J. Recombinant gene products of two natural variants of the human cytidine deaminase gene confer different deamination rates of cytarabine in vitro. Exp Hematol. 1998 May;26(5):421–5. [PubMed] [Google Scholar]
- (16).Gilbert JA, Salavaggione OE, Ji Y, Pelleymounter LL, Eckloff BW, Wieben ED, Ames MM, Weinshilboum RM. Gemcitabine pharmacogenomics: cytidine deaminase and deoxycytidylate deaminase gene resequencing and functional genomics. Clin Cancer Res. 2006 Mar 15;12(6):1794–803. doi: 10.1158/1078-0432.CCR-05-1969. [DOI] [PubMed] [Google Scholar]
- (17).Lange BJ, Dinndorf P, Smith FO, Arndt C, Barnard D, Feig S, et al. Pilot study of idarubicin-based intensive-timing induction therapy for children with previously untreated acute myeloid leukemia: Children's Cancer Group Study 2941. J Clin Oncol. 2004 Jan 1;22(1):150–6. doi: 10.1200/JCO.2004.04.016. [DOI] [PubMed] [Google Scholar]
- (18).Lange BJ, Smith FO, Feusner J, Barnard DR, Dinndorf P, Feig S, Heerema NA, Arndt C, Arceci RJ, Seibel N, Weiman M, Dusenbery K, Shannon K, Luna-Fineman S, Gerbing RB, Alonzo TA. Outcomes in CCG-2961, a Children's Oncology Group phase 3 trial for untreated pediatric acute myeloid leukemia: a report from the children's oncology group. Blood. 2008 Feb 1;111(3):1044–53. doi: 10.1182/blood-2007-04-084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Kaplan E, Meier P, Kaplan E, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53(282):457–481. [Google Scholar]
- (20).Greenwood M. Report on Public Health and Medical Subjects. His Majesty's Stationery Office. 33; London England: 1926. The natural duration of cancer. [Google Scholar]
- (21).Kalbfleisch J, Prentice R. The Statistical Analysis of Failure Time Data. John Wiley; New York: 1980. [Google Scholar]
- (22).Gray R. A Class of K-sample tests for comparing the cumulative incidence of a competing risk. Annals of Statistics. 1988;16:1141–1154. [Google Scholar]
- (23).Yusa K, Oh-hara T, Tsukahara S, Tsuruo T. Human immunodeficiency virus type 1 induces 1-beta-D-arabinofuranosylcytosine resistance in human H9 cell line. J Biol Chem. 1992 Aug 25;267(24):16848–50. [PubMed] [Google Scholar]
- (24).Steuart CD, Burke PJ. Cytidine deaminase and the development of resistance to arabinosyl cytosine. Nat New Biol. 1971 Sep 22;233(38):109–10. doi: 10.1038/newbio233109a0. [DOI] [PubMed] [Google Scholar]
- (25).Kreis W, Hession C, Soricelli A, Scully K. Combinations of tetrahydrouridine and cytosine arabinoside in mouse tumors. Cancer Treat Rep. 1977 Oct;61(7):1355–64. [PubMed] [Google Scholar]
- (26).Riva C, Barra Y, Carcassonne Y, Cano JP, Rustum Y. Effect of tetrahydrouridine on metabolism and transport of 1-beta-D-arabinofuranosylcytosine in human cells. Chemotherapy. 1992;38(5):358–66. doi: 10.1159/000239026. [DOI] [PubMed] [Google Scholar]
- (27).Honma Y, Onozuka Y, Okabe-Kado J, Kasukabe T, Hozumi M. Hemin enhances the sensitivity of erythroleukemia cells to 1-beta-D-arabinofuranosylcytosine by both activation of deoxycytidine kinase and reduction of cytidine deaminase activity. Cancer Res. 1991 Sep 1;51(17):4535–8. [PubMed] [Google Scholar]
- (28).Mejer J, Nygaard P. Cytosine arabinoside phosphorylation and deamination in acute myeloblastic leukemia cells. Leuk Res. 1978;2(2):127–31. [Google Scholar]
- (29).Tattersall MH, Ganeshaguru K, Hoffbrand AV. Mechanisms of resistance of human acute leukaemia cells to cytosine arabinoside. Br J Haematol. 1974 May;27(1):39–46. doi: 10.1111/j.1365-2141.1974.tb06772.x. [DOI] [PubMed] [Google Scholar]
- (30).Furner RL, Mellett LB, Herren TC. Influence of tetrahydrouridine on the phosphorylation of 1-beta-D-arabinofuranosyl-cytosine (ara-C) by enzymes from solid tumors in vitro. J Pharmacol Exp Ther. 1975 Jul;194(1):103–10. [PubMed] [Google Scholar]
- (31).Momparler RL, Laliberte J, Eliopoulos N, Beausejour C, Cournoyer D. Transfection of murine fibroblast cells with human cytidine deaminase cDNA confers resistance to cytosine arabinoside. Anticancer Drugs. 1996 May;7(3):266–74. doi: 10.1097/00001813-199605000-00005. [DOI] [PubMed] [Google Scholar]
- (32).Momparler RL, Eliopoulos N, Bovenzi V, Letourneau S, Greenbaum M, Cournoyer D. Resistance to cytosine arabinoside by retrovirally mediated gene transfer of human cytidine deaminase into murine fibroblast and hematopoietic cells. Cancer Gene Ther. 1996 Sep-Oct;3(5):331–8. [PubMed] [Google Scholar]
- (33).Bardenheuer W, Lehmberg K, Rattmann I, Brueckner A, Schneider A, Sorg UR, Seeber S, Moritz T, Flasshove M. Resistance to cytarabine and gemcitabine and in vitro selection of transduced cells after retroviral expression of cytidine deaminase in human hematopoietic progenitor cells. Leukemia. 2005 Dec;19(12):2281–8. doi: 10.1038/sj.leu.2403977. [DOI] [PubMed] [Google Scholar]
- (34).Rattmann I, Kleff V, Sorg UR, Bardenheuer W, Brueckner A, Hilger RA, Opalka B, Seeber S, Flasshove M, Moritz T. Gene transfer of cytidine deaminase protects myelopoiesis from cytidine analogs in an in vivo murine transplant model. Blood. 2006 Nov 1;108(9):2965–71. doi: 10.1182/blood-2006-03-011734. Epub 2006 Jul 11. [DOI] [PubMed] [Google Scholar]
- (35).Smyth JF, Robins AB, Leese CL. The metabolism of Cytosine Arabinoside as a predictive test for clinical response to the drug in Acute Myeloid Leukemia. Europ J Cancer. 1976;12:567–573. doi: 10.1016/0014-2964(76)90164-x. [DOI] [PubMed] [Google Scholar]
- (36).Preisler HD, Rustum Y, Priore RL. Relationship between leukemic cell retention of cytosine arabinoside triphosphate and the duration of remission in patients with acute non-lymphocytic leukemia. Eur J Cancer Clin Oncol. 1985 Jan;21(1):23–30. doi: 10.1016/0277-5379(85)90196-8. [DOI] [PubMed] [Google Scholar]
- (37).Rustum YM, Preisler HD. Correlation between leukemic cell retention of 1-beta-D-arabinofuranosylcytosine 5'-triphosphate and response to therapy. Cancer Res. 1979 Jan;39(1):42–9. [PubMed] [Google Scholar]
- (38).Colly LP, Peters WG, Richel D, Arentsen-Honders MW, Starrenburg CW, Willemze R. Deoxycytidine kinase and deoxycytidine deaminase values correspond closely to clinical response to cytosine arabinoside remission induction therapy in patients with acute myelogenous leukemia. Semin Oncol. 1987 Jun;14(2 Suppl 1):257–61. [PubMed] [Google Scholar]
- (39).Jahns-Streubel G, Reuter C, Auf der Landwehr U, Unterhalt M, Schleyer E, Wörmann B, Büchner T, Hiddemann W. Activity of thymidine kinase and of polymerase alpha as well as activity and gene expression of deoxycytidine deaminase in leukemic blasts are correlated with clinical response in the setting of granulocyte-macrophage colony-stimulating factor-based priming before and during TAD-9 induction therapy in acute myeloid leukemia. Blood. 1997 Sep 1;90(5):1968–76. [PubMed] [Google Scholar]
- (40).Schröder JK, Kirch C, Seeber S, Schütte J. Structural and functional analysis of the cytidine deaminase gene in patients with acute myeloid leukaemia. Br J Haematol. 1998 Dec;103(4):1096–103. doi: 10.1046/j.1365-2141.1998.01084.x. [DOI] [PubMed] [Google Scholar]
- (41).Hubeek I, Stam RW, Peters GJ, Broekhuizen R, Meijerink JP, van Wering ER, Gibson BE, Creutzig U, Zwaan CM, Cloos J, Kuik DJ, Pieters R, Kaspers GJ. The human equilibrative nucleoside transporter 1 mediates in vitro cytarabine sensitivity in childhood acute myeloid leukaemia. Br J Cancer. 2005 Dec 12;93(12):1388–94. doi: 10.1038/sj.bjc.6602881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Stam RW, den Boer ML, Meijerink JP, Ebus ME, Peters GJ, Noordhuis P, Janka-Schaub GE, Armstrong SA, Korsmeyer SJ, Pieters R. Differential mRNA expression of Ara-C-metabolizing enzymes explains Ara-C sensitivity in MLL gene-rearranged infant acute lymphoblastic leukemia. Blood. 2003 Feb 15;101(4):1270–6. doi: 10.1182/blood-2002-05-1600. [DOI] [PubMed] [Google Scholar]
- (43).Galmarini CM, Mackey JR, Dumontet C. Nucleoside analogues: mechanisms of drug resistance and reversal strategies. Leukemia. 2001 Jun;15(6):875–90. doi: 10.1038/sj.leu.2402114. [DOI] [PubMed] [Google Scholar]