Abstract
Objective
Human 8-oxoguanine glycosylase 1 (OGG1) excises oxidatively damaged promutagenic base 8-oxoguanine, a lesion previously observed in a rat model of type 2 diabetes (T2DM). The objective of the present study is to determine whether genetic variation in OGG1 is associated with type 2 diabetes (T2DM) in a Mexican American cohort.
Methods
Ten SNPs including two tagging SNPs (rs1052133, rs2072668) across the OGG1 gene region were selected from the Hapmap database and genotyped in the entire cohort (n = 670; 29% diabetes; 39 families) by TaqMan assay. Association analyses between the SNPs and T2DM were performed using the measured genotype approach as implemented in the program SOLAR.
Results
Of the ten SNPs genotyped, only five were polymorphic. The minor allele frequencies of these 5 SNPs ranged from 1–38%. Of the SNPs examined for association, the Ser(326)Cys (rs1052133) exhibited significant association with T2DM (p = 0.016) after accounting for age and sex effects. Another intronic variant (rs2072668), which was in strong linkage disequilibrium (r2 = 0.96) with Ser(326)Cys also exhibited significant association with T2DM (p = 0.031).
Conclusions
These results suggest for the first time that the variants in OGG1 could influence diabetes risk in these Mexican American families and support a role for alterations of OGG1 in the pathogenesis of T2DM.
Key Words: Mexican Americans, OGG1, Type 2 diabetes, Ser326Cys polymorphism, Association, Tagging SNPs, rs1052133
Introduction
Reactive oxygen species (ROS) are known to cause DNA strand breaks and base modifications in nuclear and mitochondrial DNA which in turn play a key role in the cellular dysfunction and apoptosis that are critical to the pathogenesis of many diseases including diabetes and its complications. Accumulating evidence indicates that ROS mediated oxidative DNA damage plays a major role in β-cell dysfunction and loss that contributes to the development of type 2 diabetes (T2DM) and its complications [1]. Among the various oxidatively modified DNA bases generated by ROS, the 7,8-hydroxy-2'-deoxy-guanosine (8-OHdG) is the most abundant and potent mutagenic base as it can mispair with adenine residues instead of the usual cytosine residues, therefore leading to an increased frequency of G:C to T:A transversion mutations that contribute to tumorogensis and carcinogenesis [2]. The repair of 8-OHdG is therefore crucial to maintain the functional integrity of cellular DNA. The enzyme that recognizes and excises 8-OHdG is 8-oxo-guanine glycosylase 1 (OGG1). OGG1 gene maps to human chromosome 3p26.1, a region that frequently shows loss of heterozygosity in several human cancers [3].
Genetic alterations in OGG1 are thought to influence the development of oxidative stress and thus contribute to the pathophysiology of many diseases including cancer. While many sequence variants within the OGG1 gene have been identified, the main focus has been on the Ser(326)-Cys variant, since several epidemiological studies have associated the Ser(326)Cys polymorphism with many types of cancer including kidney, colon and lung cancer [4]. Carriers of Cys/Cys were found to have lower OGG1 activity and impaired ability to repair 8-OHdG than the carriers of Ser/Ser allele, thus contributing to the cancer risk [5]. Recently, the Ser(326)Cys variant was reported to be associated with decreased insulin sensitivity in subjects with normal glucose tolerance suggesting that genetic alterations in OGG1 may contribute to insulin resistance and potentially T2DM [6]. However, the association between OGG1 polymorphisms and T2DM has not yet been explored.
Therefore, the objective of the present study is to examine whether OGG1 polymorphisms are associated with T2DM. To address this issue, we used data from the San Antonio Family Diabetes/Gallbladder Study (SAFDGS), which represents a cohort of Mexican American population at high risk for T2DM and its complications.
Methods
Patient Information and Phenotypes
The SAFDGS family member recruitment and data collection procedures were reported previously [7]. Briefly, probands were low-income Mexican Americans with T2DM, and all 1st, 2nd, and 3rd degree relatives of probands were invited to participate in the study. A variety of metabolic, hemodynamic, anthropometrie, and demographic variables were collected from about 700 individuals drawn from 39 large Mexican American families. Blood samples were obtained after a 12-hour fast for measurement of various phenotypes including glucose, total cholesterol, triglycerides, and HDL cholesterol, and they were collected again 2 h after a standardized oral glucose load to measure plasma glucose. Diabetes status was defined by the 1999 criteria of the World Health Organization (i.e., fasting glucose levels ≥126 mg/dl and/or 2-hour glucose levels ≥ 200 mg/dl). Participants who did not meet these criteria but reported to be under treatment with either oral antidiabetic agents or insulin and who gave a history of diabetes were also considered to have T2DM. The Institutional Review Board of the University of Texas Health Science Center at San Antonio approved all procedures, and all subjects gave informed consent.
SNP Selection and Genotyping
Tagging SNPs for OGG1 gene were selected from the Hapmap database. Only two tagging SNPs (rs1052133, rs2072668) were identified based on the pair-wise tagging (r2 ≥ 0.80, MAF ≥ 0.05) using genotype data from the unrelated Hapmap CEU individuals. To assure adequate coverage of the locus, 8 SNPs (MAF <1–13%) covering the locus (rs125701, rs1801126, rs3219001, rs3219007, rs1805373, rs3219012, rs1052140, rs2304277) were also selected for analyses from the SNP database (www.ncbi.nlm.nih. gov/SNP). All the 10 SNPs were genotyped in the study participants (n = 670,39 families) by TaqMan assay (Applied Biosystems, Calif, USA). Allelic discrimination PCR was carried out on a GeneAmp PCR system 9700 (Applied Biosystems), and fluorescent signals were detected on an ABI PRISM 7700 sequence detector (Applied Biosystems).
Statistical Analysis
The genotypic data were checked for Mendelian inconsistencies using INFER, the subroutine of the program PEDSYS. All polymorphisms were tested for Hardy-Weinberg Equilibrium (HWE). Allele frequencies were estimated using maximum likelihood methods, which accounted for pedigree structure. Linkage disequilibrium (LD) between SNPs was estimated using the r2 values. We performed association analysis in our complex pedigree-based data using the measured genotype approach (MGA) within the variance components (VC) analytical framework [8]. In this approach, VCs are modeled as random effects (e.g. additive genetic effects and random environmental effects), whereas the effects of measured covariates such as age and sex are modeled as fixed effects on the trait mean. The marker genotypes were incorporated in the mean effects model as a measured covariate, assuming additivity of allelic effects [8, 9]. The effect of this measured genotype (i.e., association parameter) together with other covariate effects (e.g., age and sex) and VCs was estimated by maximum likelihood methods. The hypothesis of no association is tested by comparing the likelihood of a model in which the effect of the measured genotype is estimated with a model where the effect of the measured genotype was fixed at zero. Twice the difference in the log-likelihoods of these models yields a test statistic that is asymptotically distributed, approximating a χ2 distribution with one degree of freedom. A p value ≤ 0.05 is considered significant. Prior to performing MGA, the quantitative transmission disequilibrium test (QTDT) was used to examine hidden population stratification [10]. All statistical methods described above were implemented in the computer program SOLAR [8].
Results and Discussion
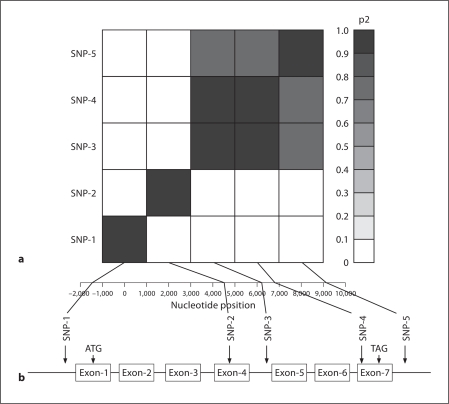
Table 1 shows the demographic, clinical, and biochemical characteristics for the subjects genotyped. In the total sample, 29, 28 and 14% of the subjects had hypertension, T2DM and albuminuria, respectively. To examine the association between the genetic variants in OGG1 and T2DM, ten SNPs including two tagging SNPs were selected from the SNP database and genotyped in the study participants. Of the SNPs genotyped, only five were polymorphic (rs105701, rs1805373, rs2072668, rs1052133, rs2304277) in this cohort. The location, alíele frequencies, and association results of the genotyped SNPs are presented in table 2. The minor allele frequency of the SNPs varied from 1% (rs1805373) to 38% (rs2072688). Genotypic data were consistent with the Hardy-Weinberg Equilibrium expectations, and there was no evidence for hidden population stratification in the data as tested by QTDT. Before performing association analysis, we estimated the pairwise LD (r2) between all the 5 SNPs and found that the pairwise LD ranged from 0 to 0.96 (fig. 1). As can be seen from figure 1, the relatively high pairwise LDs (r2 > 0.8) were found for the following SNPs pairs: rs2072668 (SNP-3), rs1052133 (SNP-4) [r2 = 0.96]; rs2072668 (SNP-3), rs2304277 (SNP-5) [r2 = 0.88]; rs1052133 (SNP-4), rs2304277 (SNP-5) [r2 = 0.87]. Association analysis in our family data was carried out using the measured genotype approach within the variance components analytical framework implemented in SOLAR [8]. Of the SNPs examined for association, rs1052133 (SNP-4) [p = 0.016] and rs2072668 (SNP-3) [p = 0.031] exhibited significant association with T2DM after adjusting for age and sex effects (table 2), although, as noted above, these SNPs are in strong LD.
Table 1.
Clinical characteristics of the genotyped SAFDGS participants used for the present studya
| Variables | |
|---|---|
| Females | 61% |
| Type 2 diabetes | 28% |
| Age, years | 44.8 ± 16.2 |
| Systolic blood pressure, mm Hg | 127.1 ± 16.8 |
| Diastolic blood pressure, mm Hg | 70.3 ± 9.6 |
| Body mass index | 30.9 ± 7.0 |
| Total cholesterol, mg/dl | 194.0 ± 37.9 |
| High density lipoprotein-cholesterol, mg/dl | 46.0 ± 12.0 |
| Triglycerides (log transformed) | 4.9 ± 0.6 |
| Albumin to Creatinine Ratio (log transformed) | 2.4 ± 0.8 |
Sample size varies from 610 (cholesterol) to 670 (age).
Table 2.
Association analysis between the OGG1 polymorphisms and T2DM in SAFDGS
| SNPs | SNP_ID | Positions bp | Gene location | Major/minor allele, % | p valuesa |
|---|---|---|---|---|---|
| SNP-1 | rs125701 | −1,493 | Promoter | G (93)/A (7) | 0.379 |
| SNP-2 | rs1805373 | 4,537 | Gln(229) Arg | G (99)/A(l) | 0.100 |
| SNP-3 | rs2072668 | 6,169 | Intron 4 | C (62)/G (38) | 0.031 |
| SNP-4 | rs1052133 | 6,802 | Ser(326)Cys | C (64)/G (36) | 0.016 |
| SNP-5 | rs2304277 | 9,109 | 3'UTR | G (63)/A (37) | 0.135 |
Adjusted for age and sex terms.
Fig. 1.
a Linkage disequilibrium (LD) between SNP pairs within the OGG1 gene. SNPs are labeled on the y-axis, and the locations (bp) within the gene are shown on the X-axis. Pairwise LD is estimated using r2 values and depicted in the figure by the color intensity of the shaded box, as shown in the legend. The diagonal represents a comparison of eachSNP against itself (i.e., r2 = 1.0). b Schematic diagram of human OGG1 gene structure on 3p26.1 and the location of the polymorphisms genotyped in SAFDGS. The exons of the OGG1 are represented by box and the intron by a thin line. SNPs 1–5 locations in the gene structure and their LD pattern are indicated by the vertical line/arrow.
In reference to Ser(326)Cys polymorphism (i.e., rs1052133), the individuals with Ser326 and Ser(326)Cys genotypes are more likely to be at risk for diabetes, with relative risks of 1.99 (95% CI: 1.48–2.57) and 1.44 (95% CI: 1.09–1.86), respectively. Given the LD patterns among the examined SNPs, we obtained a p value after adjusting for multiple testing, using an effective number of SNPs calculated following the method of Li and Ji [11]. For a statistical significance threshold of ≤0.05, the required experiment-wide significance threshold, using the effective number of SNPs (which was 4) to adjust for multiple testing, was ∼0.013. Our nominal p value of 0.016 associated with rs1052133, which was uncorrected, thus approximates the p value corrected for multiple testing. In addition, it is worth noting that this study (n = 670) is powered at 80% to detect a SNP (at an experiment-wide significance of 0.05) that accounts for 1.7% of variation in T2DM phenotype. However, the observed additive variation attributable to rs1052133 is 2.7%, which adds confidence to the observed association with T2DM.
DNA lesions caused by ROS and other free radicals have been implicated in the etiology of many diseases including diabetes [1]. One of the oxidatively induced pro mutagenic bases is 8-OHdG. Diabetic patients have higher concentrations of 8-OHdG in urine [12], blood cells [13], muscle [14], pancreas [15, 16] and kidney [17]. In addition, poor glycaemic control is associated with high urinary 8-OHdG excretion [13]. A rise in 8-OHdG has been correlated with HbAlc levels indicating that increased 8-OHdG may contribute to the progression of diabetic microvascular complications [18]. In summary, 80HdG has been considered a potential biomarker of ox-idative stress and oxidative DNA damage in diabetes. OGG1 is the key component responsible for the removal of 8-OHdG from oxidatively damaged DNA. Alteration of OGG1 enzyme activity due to genetic variation in OGG1 gene may influence the development of oxidative stress and oxidative DNA damage and thus may contribute to the risk for T2DM and its complications. The purpose of the present study is therefore to explore the hypothesis that genetic alterations in OGG1 may confer risk for T2DM. To our knowledge, no study has thus far examined the relationship between the OGG1 variants and diabetes risk in Mexican Americans.
Of the genetic variants of OGG1 examined for association, the Ser(326)Cys variant, which is in strong LD with rs2072668 (r2 = 0.96), exhibited significant association with T2DM after accounting for age and sex effects. Although, the exact mechanism by which the Ser(326)-Cys variant contributes to T2DM is not known, it is conceivable that Ser(326)Cys may impair the activity of OGG1 to repair 8-OHdG in T2DM. 8-OHdG in turn would promote an increase in G:C → TA transversions, DNA damage and loss of pancreatic β-cell, thereby contributing to the development of T2DM. In support of this notion, a significant association has recently been reported between Ser(326)Cys variant of OGG1 and type 2 diabetes [19] and decreased insulin sensitivity in patients with normal glucose tolerance test [6]. Further confirmatory studies of this genetic association and/or functional studies are required to support the observed association between the Ser(326)Cys variant and T2DM.
In conclusion, this study assessed common genetic variations in OGG1 in relation to T2DM risk in a Mexican American population. The evidence presented here supports the potential role of OGG1 variants in conferring susceptibility to T2DM, although the magnitude of the genetic effect appears to be small. Validation of these findings in larger studies and in studies of other populations is needed. The data presented here would also provide a basis for other investigators to explore OGG1 as a candidate gene for development of T2DM.
Acknowledgements
This study was supported by a Grant-in-Aid from the American Heart Association, American Society of Nephrology, and Satellite HealthCare (FT.), and Faculty Development Award from the American Diabetes Association and New Investigator Award from South Texas Veterans Healthcare System (S.L.H.), and the George O'Brien Kidney Research Center (F.T, S.L.H., H.E.A.) and the National Institute of Diabetes, Digestive and Kidney Diseases (R.D.; DK53889, D.M.L.; DK42273, and DK47482). We warmly thank the families from the SAFDGS for their kind generosity and cooperation in taking part in this study.
References
- 1.Hadjivassiliou V, Green MH, James RF, Swift SM, Clayton HA, Green IC. Insulin secretion, DNA damage, and apoptosis in human and rat islets of Langerhans following exposure to nitric oxide, peroxynitrite, and cytokines. Nitric Oxide. 1998;2:429–441. doi: 10.1006/niox.1998.0203. [DOI] [PubMed] [Google Scholar]
- 2.Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature. 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- 3.Roldán-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc Natl Acad Sci USA. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss JM, Goode EL, Ladiges WC, Ulrich CM. Polymorphic variation in hOGG1 and risk of cancer: a review of the functional and epidemiologic literature. Mol Carcinog. 2005;42:127–141. doi: 10.1002/mc.20067. [DOI] [PubMed] [Google Scholar]
- 5.Lee AJ, Hodges NJ, Chipman JK. Interindividual variability in response to sodium dichromate-induced oxidative DNA damage: role of the Ser326Cys polymorphism in the DNA-repair protein of 8-oxo-7,8-dihydro-2′-deoxyguanosine DNA glycosylase 1. Cancer Epidemiol Biomarkers Prev. 2005;14:497–505. doi: 10.1158/1055-9965.EPI-04-0295. [DOI] [PubMed] [Google Scholar]
- 6.Wang CL, Hsieh MC, Hsin SC, Lin HY, Lin KD, Lo CS, Chen ZH, Shin SJ. The hOGG1 Ser326Cys gene polymorphism is associated with decreased insulin sensitivity in subjects with normal glucose tolerance. J Hum Genet. 2006;51:124–128. doi: 10.1007/s10038-005-0335-8. [DOI] [PubMed] [Google Scholar]
- 7.Puppala S, Dodd GD, Fowler S, Arya R, Schneider J, Farook VS, Granato R, Dyer TD, Almasy L, Jenkinson CP, Diehl AK, Stern MP, Blangero J, Duggirala R. A genomewide search finds major susceptibility loci for gallbladder disease on chromosome 1 in Mexican Americans. Am J Hum Genet. 2006;78:377–392. doi: 10.1086/500274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Almasy L, Blangero J. Exploring positional candidate genes: linkage conditional on measured genotype. Behav Genet. 2004;34:173–177. doi: 10.1023/B:BEGE.0000013731.03827.69. [DOI] [PubMed] [Google Scholar]
- 9.Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986;50:181–194. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 10.Abecasis GR, Cookson WO, Cardon LR. Pedigree tests of transmission disequilibrium. Eur J Hum Genet. 2000;8:545–551. doi: 10.1038/sj.ejhg.5200494. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 12.Malin R, Rantalaiho V, Huang XH, Wirta O, Pasternack A, Leinonen JS, Alho H, Jokela H, Koivula T, Tanaka T, Okada K, Ochi H, Toyokuni S, Lehtimäki T. Association between M/L55-polymorphism of paraoxonase enzyme and oxidative DNA damage in patients with type 2 diabetes mellitus and in control subjects. Hum Genet. 1999;105:179–180. doi: 10.1007/s004399900074. [DOI] [PubMed] [Google Scholar]
- 13.Dandona P, Thusu K, Cook S, Snyder B, Makowski J, Armstrong D, Nicotera T. Oxidative damage to DNA in diabetes mellitus. Lancet. 1996;347:444–445. doi: 10.1016/s0140-6736(96)90013-6. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki S, Hinokio Y, Komatu K, Ohtomo M, Onoda M, Hirai S, Hirai M, Hirai A, Chiba M, Kasuga S, Akai H, Toyota T. Oxidative damage to mitochondrial DNA and its relationship to diabetic complications. Diabetes Res Clin Pract. 1999;45:161–168. doi: 10.1016/s0168-8227(99)00046-7. [DOI] [PubMed] [Google Scholar]
- 15.Sakuraba H, Mizukami H, Yagihashi N, Wada R, Hanyu C, Yagihashi S. Reduced beta-cell mass and expression of oxidative stress-related DNA damage in the islet of Japanese Type II diabetic patients. Diabetologia. 2002;45:85–96. doi: 10.1007/s125-002-8248-z. [DOI] [PubMed] [Google Scholar]
- 16.Ku YP, Jin M, Kim KH, Ahn YJ, Yoon SP, You HJ, Chang IY. Immunolocalization of 8-OHdG and OGG1 in pancreatic islets of streptozotocin-induced diabetic rats. Acta Histochem. 2009;111:138–144. doi: 10.1016/j.acthis.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Ha H, Kim C, Son Y, Chung MH, Kim KH. DNA damage in the kidneys of diabetic rats exhibiting microalbuminuria. Free Radic Biol Med. 1994;16:271–274. doi: 10.1016/0891-5849(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 18.Leinonen J, Lehtimäki T, Toyokuni S, Okada K, Tanaka T, Hiai H, Ochi H, Laippala P, Rantalaiho V, Wirta O, Pasternack A, Alho H. New biomarker evidence of oxidative DNA damage in patients with non-insulin-dependent diabetes mellitus. FEBS Lett. 1997;417:150–152. doi: 10.1016/s0014-5793(97)01273-8. [DOI] [PubMed] [Google Scholar]
- 19.Daimon M, Oizumi T, Toriyama S, Karasawa S, Jimbu Y, Wada K, Kameda W, Susa S, Muramatsu M, Kubota I, Kawata S, Kato T. Association of the Ser326Cys polymorphism in the OGG1 gene with type 2 DM. Biochem Biophys Res Commun. 2009;386:26–29. doi: 10.1016/j.bbrc.2009.05.119. [DOI] [PubMed] [Google Scholar]