Abstract
Summary
Stem cells have the ability to self-renew, and give rise to one or more differentiated cell types. Embryonic stem cells can differentiate into all cell types of the body and have unlimited self-renewal capacity. Somatic stem cells are found in many adult tissues. They have an extensive but finite lifespan and can differentiate into a more restricted range of cell types. Increasing evidence indicates that the multilineage differentiation ability of stem cells is defined by the potential for expression of developmentally regulated transcription factors and of lineage specification genes. Gene expression, or as emphasized here, the potential for gene expression, is largely controlled by epigenetic modifications of DNA (DNA methylation) and chromatin (such as post-translational histone modifications) in the regulatory regions of specific genes. Epigenetic modifications can also influence the timing of DNA replication. We highlight here how mechanisms by which genes are poised for transcription in undifferentiated stem cells are being uncovered through the mapping of DNA methylation profiles on differentiation-regulated promoters and at the genome-wide level, histone modifications, and transcription factor binding. Epigenetic marks on developmentally regulated and lineage specification genes in stem cells seem to define a state of pluripotency.
Key Words: Chromatin, Differentiation, DNA methylation, Embryonic stem cell, Epigenetics, Mesenchymal stem cell
Abstract
Zusammenfassung
Epigenetische Basis für das Differenzierungspotential von mesenchymalen und embryonalen Stammzellen
Stammzellen haben die Fähigkeit zur Selbsterneuerung und bilden die Ausgangsbasis für einen oder mehrere differenzierte Zelltypen. Embryonale Stammzellen können in alle Zelltypen des Körpers differenzieren und haben eine unbegrenzte Selbsterneuerungskapazität. Somatische Stammzellen finden sich in vielen adulten Geweben. Sie haben eine lange, aber dennoch begrenzte Lebensdauer und können nur in ein bestimmtes Spektrum von Zelltypen differenzieren. Dafür dass die Multilineage-Differen-zierungsfähigkeit der Stammzellen bestimmt wird durch deren Potential, entwicklungsgemäß regulierte Transkriptionsfaktoren und Abstammungsspezifikationsgene zu exprimieren, gibt es einen zunehmende Zahl von Belegen. Genexpression bzw. das Potential zur Genexpression, wie es hier betont wird, wird hauptsächlich durch epigenetische Modifikationen der DNA (DNA-Methylierung) des Chromatins (z.B. posttranslationale Histonmodifikationen) in den Regulatorregionen von speziellen Genen gesteuert. Epigenetische Modifikationen können auch das Timing der DNA-Replikation beeinflussen. Der Fokus dieser Übersicht liegt darauf, wie Mechanismen, durch die Gene zur Transkription in undifferenzierte Stammzellen ausbalanciert werden, erkannt werden durch das Mapping von DNA-Methylierungsprofilen an differenzierungsregulierenden Promotern und auf genomweiter Ebene, Histonmodifikationen und Transkriptionsfaktorbindung. Epigenetische Marker auf entwicklungsgemäß regulierten und Abstammungsspezifikationsgenen in Stammzellen scheinen den Grad der Pluripotenz zu definieren.
Introduction
Stem cells are defined by their ability to self-renew and to give rise to at least one more differentiated cell type. Embryonic stem cells (ESCs), in vitro derivatives of the inner cell mass of blastocysts, retain the ability of the inner cell mass to differentiate into all cell types of the body, and acquire unlimited self-renewal potential. For these reasons, and due to their perceived use in regenerative medicine, human ESCs (hESCs) have received considerable attention since their derivation 10 years ago [1]. Multiple extracellular factors are required for the establishment and maintenance of pluripotency in ESCs, and these have been reviewed elsewhere [2, 3]. The multilineage differentiation ability of ESCs is defined by the potential for expression of lineage specification genes. This review analyzes the mechanisms by which these genes are poised for transcription. Stem cells have in recent years also been identified in many adult organs. Stromal stem cells, present in a variety of mesenchymal tissues, are also being scrutinized due to their potential use in autologous cell replacement therapy [4, 5]. In contrast to ESCs, mesenchymal stem cells (MSCs) seem to be restricted to forming preferentially mesodermal cell types such as adipocytes, osteocytes, and chondrocytes. However, rare subsets of MSCs identified in bone marrow seem to have the ability to form cells types of all 3 germ layers, and have challenged the limited differentiation potential of somatic stem cells [6]. An abundant source of MSCs are adipose tissue-derived stem cells (ASCs) isolated from liposuction material [7, 8]. Like bone marrow-derived MSCs, ASCs can differentiate into mesodermal cell types; however, recent findings suggest a limited differentiation ability even within mesodermal lineages [9, 10]. So, although MSCs retain the ability of express various lineage-specific genes upon differentiation, this potential is more restricted than in ESCs.
The potential for gene expression in stem cells is regulated by epigenetic processes that confer a specific chromatin configuration on gene regulatory regions and on coding sequences. Epigenetic mechanisms refer to heritable modifications on DNA and chromatin that do not affect DNA sequence. The best characterized epigenetic modification is cytosine methylation on DNA, which is in general associated with gene silencing. Epigenetic modifications of chromatin regroup post-translational alteration of histones including phosphorylation, acetylation, methylation, ubiquitination, and SUMOylation, in combination with the dynamic replacement of core histone by histone variants, such as the deposition of histone H3.3 on transcriptionally active promoters [11, 12]. In addition to epigenetic modifications, the positioning of transcriptional activators, transcriptional repressors, other chromatin remodeling enzymes, and small interfering RNAs on target genes also regulate gene expression. This review highlights our current view of the epigenetic landscape of ESCs and MSCs. This landscape is likely to provide a molecular basis for gene activation and multilineage differentiation potential.
DNA Methylation and Gene Expression
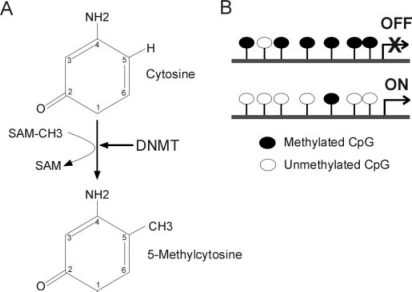
DNA methylation consists of the addition of a methyl group to position 5 of a cytosine in a cytosine-phosphate-guanine (CpG) dinucleotide (fig. 1 A). CpG methylation is symmetrical (it occurs on both DNA strands) and targets isolated CpGs, clustered CpGs, or even clustered CpGs within a CpG island. A CpG island is defined as a sequence in which the observed/expected CpG frequency is greater than 0.6 with a G+ C content greater than 50%. According to Gardiner-Garden and Frommer [13], the expected number of CpG dimers in a given 200-bp window is calculated as the number of Cs in the window multiplied by the number of Gs in the window, divided by window length. This 200-bp window is moving across the sequence of interest at 1 bp intervals. CpG islands are often found in the 5’ regulatory regions of vertebrate housekeeping genes. CpG islands are often protected from methylation, enabling constitutive expression of these genes. CpG islands in the promoter of tumor suppressor genes, for instance, are unmethylated in normal cells, whereas a hallmark of cancer is de novo methylation of these CpG islands, resulting in repression of tumor suppressor genes and triggering of an uncontrolled cell cycle. DNA methylation of tumor suppressor genes constitutes the basis of a number of anti-cancer therapies relying on the inhibition of DNA methyl transferases [14].
Fig. 1.
The basics of DNA methylation: A mechanism of DNA methylation; B textbook view of the relationship between DNA methylation and gene expression.
CpG methylation is catalyzed by DNA methyltransferases (DNMTs). The maintenance DNA methyltransferase DNMT1 specifically recognizes hemi-methylated DNA after replication and methylates the daughter strand, ensuring fidelity in the methylation profile after replication [15]. In contrast to DNMT1, DNMT3a and DNMT3b are implicated in de novo DNA methylation that takes place during embryonic development and cell differentiation [16], as a means of shutting down genes whose activity is no longer required as cells differentiate (e.g., that of pluripotency-associated genes). The 4th DNMT, DNMT2, has to date no clear ascribed function in DNA methylation [17, 18, 19, 20, 21] but has been shown to have cytoplasmic transfer RNA methyltransferase activity [22, 23].
DNA methylation is a hallmark of long-term gene silencing (fig. 1 B). The methyl groups create target sites for methyl-binding proteins which induce transcriptional repression by recruiting co-repressors such as histone deacetylases [24]. So, DNA methylation largely contributes to long-term gene silencing [25, 26], and as such it is essential for development [27, 28, 29, 30], × chromosome inactivation [31], and genomic imprinting [32, 33, 34, 35]. The relationship between DNA methylation and gene expression is complex [36], and recent evidence based on genome-wide CpG methylation profiling highlights promoter CpG content as a component of this complexity [37]. In vitro differentiation of ESCs and embryonal carcinoma (EC) cells also correlates with changes in DNA methylation notably on the promoter of developmentally regulated genes expressed in pluripotent ESCs such as the transcription factors OCT4 and NANOG [38, 39, 40]. However, to date, only sporadic indications of CpG methylation changes have been reported during differentiation of MSCs or precursor cells [9, 10, 41].
DNA Methylation on Promoters of Lineage Specification Genes in Mesenchymal Stem Cells
The epigenetic landscape of somatic stem cells remains largely unraveled. This section highlights recent published and unpublished findings on the relationship between DNA methylation of lineage specification genes, gene expression, and potential for differentiation of MSCs. We focus on ASCs on which most epigenetic studies have been reported to date. An emerging concept is that a CpG methylation pattern ‘pre-programs’ ASCs for adipogenic differentiation preferentially over other lineages. Adipose tissue constitutes an rich source of MSCs [7, 8, 42, 43]. ASCs with a CD34+ CD105+ CD45-CD31- phenotype have been isolated by negative selection against CD45 and CD31 with high purity (approximately 99%) from the stromal vascular fraction of human liposuction material [7]. Notably, cultured ASCs display a gene expression profile and surface antigen phenotype similar to bone marrow-derived MSCs [7, 44, 45, 46], suggesting a common mesodermal ancestor. ASCs exhibit primarily mesodermal differentiation abilities in vitro, and can promote neuronal functions, osteogenic repair, and reconstitution of the immune system in vivo [41, 42]. ASCs can also differentiate toward the endothelial cell lineage in vitro and contribute to re-vascularization of ischemic tissue; nonetheless, whether their contribution is direct or indirect remains debated [9, 47]. Transcriptional profiling of freshly isolated, uncultured ASCs reveals expression of genes extending across the 3 germ layers, suggestive of a differentiation potential toward non-mesodermal lineages [7]. However, whether ASCs form functional tissues of these lineages in vivo also remains under debate.
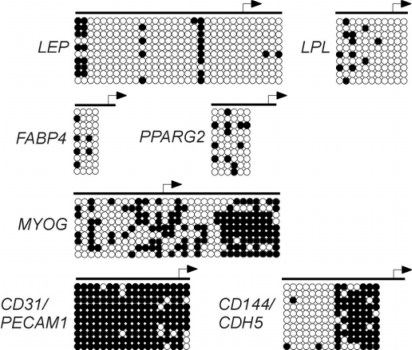
Recent studies relying on bisulfite genomic sequencing analyses have started to unveil the DNA methylation profile of tissue-specific genes in human ASCs. Of note, bisulfite sequencing consists of the bisulfite-mediated conversion of unmethylated (i.e., unprotected) cytosines in CpG dinucleotides to uracil, while methylated cytosines are protected from conversion and remain as cytosines [48]. Polymerase chain reaction (PCR) replaces the uracils with thymidines, and subsequent sequencing determines, by reading a thymidine or cytosine, the methylation state of the cytosines in the original sequence. Bacterial cloning of the PCR products generates several sequences, and thereby provides a quantitative assessment of the extent of methylation of a given CpG. As such, bisulfite genomic sequencing represents the state-of-the-art of DNA methylation analysis. Bisulfite sequencing analysis of 4 adipogenic specification promoters -namely, leptin (LEP), peroxisome proliferaior activated receptor gamma 2 (PPARG2), fatty acid-binding protein 4 (FABP4), and lipoprotein lipase (LPL) – reveals several DNA methylation features in freshly isolated ASCs [10] (fig. 2). Firstly, these promoters are globally hypomethylated, with a mere 5–30% of CpGs being methylated. Secondly, CpG methylation profiles are mosaic between ASC donors and within donors. This mosaicism is consistent with that observed in stem cells from intestinal crypts [49, 50, 51]. Mosaicism is believed to result from stochastic methylation which accumulates independently in different cells as a result of exposure to environmental, age, and health factors [14, 51, 52, 53, 54], together with a propensity of certain CpGs to be hypermethylated [55, 56]. Indeed, it is clear that each gene contains cytosines more susceptible to methylation than others [10].
Fig. 2.
Bisulfite genomic sequencing analysis of CpG methylation in the promoter regions of lineage-specific genes in undifferentiated human ASCs. Genes indicative of the adipogenic lineage (LEP, PPARG2, FABP4, LPL), endothelial cell lineage (CD31/PECAM1, CD144/CDH5), and myogenic lineage (MYOG) are shown. Each circle represents a CpG (white: unmethylated; black: methylated). Each row of circles represents the sequence of 1 bacterial clone of PCR product (i.e., 1 genomic fragment). 9–10 clones (i.e., sequences) are represented to provide a quantitative indication of the extent of methylation of each CpG in the ASC population examined for each locus. Note the mosaicism in the methylation pattern for each locus.
In contrast to adipogenic promoters, myogenic (MYOG) and endothelial cell (CD31/PECAM1 and CD144/CDH5) regulatory regions display more methylation [9, 10] (fig. 2). Housekeeping genes such as GAPDH and LMNB1 are unmethylated (not shown), as expected from their constitutive expression. So, in ASCs, adipogenic genes are un- or hypomethylated while non-adipogenic lineage-specific promoters seem to be more methylated. This raises the hypothesis of an epigenetic programming of ASCs for adipogenic differentiation by a CpG methylation pattern on critical promoters. An extension of these analyses to several genes and MSCs will bring more insight on this hypothesis.
Long-term culture of human ASCs does not significantly alter methylation states. Few CpGs in the LEP, FABP4, and LPL promoters become methylated upon culture of ASCs while even fewer are demethylated; however, the significance of these methylation changes remains uncertain. Indeed, increased mosaicism in CpG methylation is detected between cell clones relative to that detected between individual ASC donors [10], but culture to senescence does not enhance mosaicism [57]. In contrast to a previous report on CpG methylation in hESCs [58], we have no evidence of heritable methylation changes in cultured ASCs, suggesting randomness in the (de)methylation events occurring during culture. In addition to the presumed defects in DNMT1 function, different cells in the initial ASC population display mosaic CpG methylation. Moreover, asymmetric cell division, a characteristic of pluripotent stem cells, is expected to generate a different epigenetic pattern in each daughter cell within a clonal population. Studies available to date, therefore, argue that hypomethylation of adipogenic promoters, in contrast to other lineage-specific promoters, constitutes an epigenetic signature of human ASCs. A working hypothesis, then, is that MSCs are pre-programmed by DNA methylation of lineage-specific genes to preferentially differentiate into the cell type(s) of the tissues in which they reside. Hypomethylation of adipogenic promoters in undifferentiated ASCs raises the issue of how DNA methylation correlates with transcription. All genes examined in the above study are expressed (at low level) in freshly isolated ASCs, and a fraction of these genes become inactivated upon culture despite the maintenance of a hypomethylated state [10, 57]. Conversely, DNA methylation does not preclude expression of a gene, as exemplified by transcription of the methylated CD31 and CD144 loci in ASCs [9] (see also below). So gene expression in ASCs does not correlate with a specific methylation pattern in any of the genes examined to date, an observation not restricted to pluripotent cells [36, 59].
Relationship between Promoter DNA Methylation and Transcription
A genome-wide DNA methylation profiling in several somatic cell types and in sperm, by methyl-DNA immunoprecipitation (MeDIP) and hybridization of the precipitated DNA on genomic (promoter) arrays, shows that the relationship between promoter DNA methylation and promoter activity depends on the CpG content of the promoter [37]. Promoters with low CpG content show no correlation between activity (determined by RNA polymerase II occupancy) and abundance of methylated CpGs; therefore, transcriptionally active low CpG promoters (LCPs) are not necessarily un- or hypomethylated [37]. It seems in fact that most low CpG promoters are methylated regardless of their activity status. On the contrary, the activity of intermediate CpG content promoters (ICPs) and high CpG content promoters (HCPs) is inversely correlated to the extent of methylation [37]. In these categories, the proportion of transcriptionally active promoters decreases with increasing DNA methylation, arguing that methylation of ICPs and HCPs is incompatible with transcription. Further analysis, however, shows that inactive ICPs and HCPs differ in their DNA methylation status: most inactive HCPs are unmethylated, whereas a high proportion of inactive ICPs are methylated. So, collectively, the work of Weber et al. [37] argues that inactive HCPs globally remain unmethylated, inactive ICPs are often methylated, whereas LCPs are frequently methylated irrespective of their activation status. A genome-wide analysis of CpG methylation profiling in different MSC populations will be welcome to assess the relationship between CpG content, methylation state, and transcriptional status in these cells.
DNA Methylation Patterns in Embryonic Stem Cells
Limited evidence suggests that the DNA methylation signature of ESCs is distinct from that of MSCs and of differentiated cells; however, whether this reflects differences in gene expression or the true pluripotent nature of ESCs is unclear. Restriction enzyme digestion-mediated analyses of global DNA methylation show that mouse ESC genomes are less methylated than those of differentiated somatic cells [60, 61]. XX mouse ESCs are hypomethylated relative to XY ESCs. Hypomethylation affects both repetitive and unique sequences including differentially methylated regions which regulate expression of paternally imprinted loci [61]. Increased hypomethylation of XX ESCs has been attributed to the presence of 2 active × chromosomes (active × is hypomethylated relative to inactive X) and to reduced levels of DNMT3a and 3b. However, in DNMT-deficient (Dnmt3a-/- Dnmt3b-/-] mouse ESCs, only 0.6% of CpGs are demethylated [60], so the extent to which DNMT3a and 3b contribute to global DNA methylation in mouse ESCs remains uncertain. DNMT1 deficiency, in contrast, reduces global methylation levels from 65 to 20%, a condition which blocks differentiation potential [60]. Unfortunately, no indication currently exists on the methylation status of regulatory regions of lineage-specific genes in mouse ESCs, which could account for their potential for expression upon differentiation.
DNA methylation analyses of hESCs have been promoted by in vitro fertilization data on the unexpectedly high incidence of imprinting and other epigenetic abnormalities in embryos [62], suggesting that hESCs may also display variation in their epigenetic makeup. A restriction analysis-based methylation profiling of over 1,500 CpG sites from over 370 genes in 14 hESC lines [63] revealed an average of 35% methylation, a value substantially lower than that reported for mouse ES cells [61]. hESC methylation profiles were segregated from those of normal and cancer cell lines, normal tissue, and somatic stem cells, reflecting an epigenetic distance between hESCs and other cell types [63]. Interestingly, less than 50 CpGs within 40 genes contributed to this difference. Another 25 CpG sites from over 20 genes distinguished hESCs from normal differentiated cells and somatic stem cells; these sites were found to represent markers of developmental potential [63]. Other genes differentially methylated in hESCs relative to somatic cells are markers of pluripotency such as OCT4 and NANOG, which are unmethylated in undifferentiated hESCs [64], while being partially methylated in human MSCs in which they are not expressed (S. Timoskainen and PC, unpublished data). So on the basis of these analyses, it appears that the methylation pattern of a relatively small number of developmentally controlled genes may constitute an epigenetic mark unique to hESCs.
The need for large scale expansion of hESCs for any therapeutic use raises the question of epigenetic stability of hESCs in long-term culture. The consensus from published reports is that extended culture of hESCs alters DNA methylation. Restriction landmark genome scanning analysis of approximately 2,000 loci has identified epigenetic variations between hESC lines at loci important for differentiation [58]. Most changes occur shortly after hESC derivation and are heritable, whereas some alterations are maintained even after in vitro differentiation. This study is supported by a similar methylation drift at a small number of promoters examined in late passage cultures of other hESC lines [63, 65]. In contrast, however, stable methylation patterns have been reported by bisulfite genomic sequencing in a small number of imprinted loci in 4 different hESC lines [66]. Thus, epigenetic variation occurs during extended culture of hESCs, however, timing and degree of this epigenetic drift are likely to be cell line-dependent. A picture missing from the ESC epigenetics is a high-resolution genome-wide DNA methylation profiling across regulatory and coding regions. MeDIP assays are particularly well suited for whole-genome and promoter investigations [37, 67]. Such data can be superimposed onto transcription factor binding [37, 67] and histone modification maps to elaborate a multi-layered epigenetic profile characteristic of pluripotent cells. New data on the DNA methylation landscape of ESCs will be enlightening.
Whether unscheduled CpG methylation occurs upon in vitro differentiation of hESCs remains to be established but appears as a possibility. Analysis of over 4,600 CpG islands revealed that 1.4% undergo unexpected hypermethylation upon neurogenic differentiation of hESCs, in regulatory regions of genes involved in metabolism, signal transduction, and differentiation [68]. Although distinct from tumor suppressor CpG island methylation, this hypermethylation leads to the down-regulation of the affected genes, and as such has been suggested to have implications in the development of metabolic diseases [68]. Thus, the risk of aberrant CpG island methylation upon hESC differentiation should be considered when optimizing differentiation protocols, in particular if they are going to be used in therapeutic applications.
Post-Translational Modifications of Histones
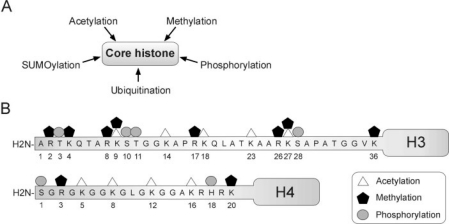
The eukaryotic genome is packaged by interactions of DNA with proteins into chromatin. The core element of chromatin is the nucleosome which consists of 147 base pairs of DNA wrapped around 2 sub units of each of hist one H2A, H2B, H3, and H4. Nucleosomes are spaced by the linker histone H1. The amino-terminal tails of histones are post-translationally modified to confer physical properties that affect their interactions with DNA on gene regulatory sequences. Histone modifications not only influence chromatin packaging but are also read by adaptor molecules, chromatin modifying enzymes, transcription factors, and transcriptional repressors, and thereby contribute to the regulation of transcription [69, 70, 71, 72, 73]. Epigenetic histone modifications have been best characterized so far for H3 and H4 and include combinatorial phosphorylation, ubiquitination, SUMOylation, acetylation, and methylation (fig. 3). In particular, di- and trimethylation of H3 lysine 9 (H3K9m2/m3) and trimethylation of H3K27 (H3K27m3) elicit the formation of repressive heterochromatin through the recruitment of heterochromatin protein 1 (HP1) [74] and polycomb group (PcG) proteins, respectively [75, 76, 77]. However, whereas H3K9m3 marks constitutive heterochromatin [78], H3K27m3 characterizes facultative heterochromatin, or chromatin domains harboring transcriptionally repressed genes that can be activated upon stimulation [79, 80]. In contrast, acetylation of histone tails loosens their interaction with DNA and creates a chromatin conformation suitable for targeting of transcriptional activators. Thus, acetylation on H3K9 (H3K9ac) and H4K16 (H4K16ac), together with di- or trimethylation of H3K4 (H3K4m2/3), are exclusively found in euchromatin, often in association with transcriptionally active genes [80, 81, 82, 83, 84]. In addition to altering histone-DNA interactions, H3K4m3 and H3K9ac mediate the recruitment and tethering of transcriptional activators [85, 86]. Mapping of the positioning of histone modifications throughout the genome or on given promoters has been enabled by chromatin immunoprecipitation (ChIP) assays whereby a specific histone modification is immunoprecipitated and associated DNA sequences are identified by PCR or by labeling and hybridization onto genomic arrays [87].
Fig. 3.
Posttranslational histone modifications: A core histories can be methylated, acetylated, phosphorylated, ubiquitinated, or SUMOylated to modulate gene expression; B site and nature of known posttranslational modifications on the amino-terminal tails of the core histones H3 and H4.
Histone Modifications Associated with Differentiation-Regulated Genes in Mesenchymal Stem Cells
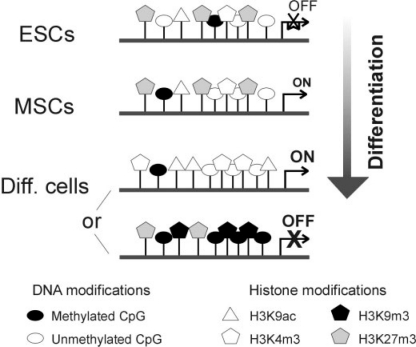
The nature of histone modifications marking promoters regulated by differentiation in MSCs remains at present largely unknown. Analyses have up to now been limited to normal differentiated cultured cells, cancer cell lines, and mouse ESCs. The availability of ChIP assays suitable for chromatin from small cell numbers [39, 88], however, opens avenues for investigating limiting cell samples such as embryonic cells [88]. Observations from our laboratory point to the presence of H3K4m3 and of the repressive H3K27m3 modification on lineage-specific promoters in undifferentiated ASCs (AN, L. Lindeman, and PC, manuscript submitted) (fig. 4, MSCs). So, together with the hypomethylated state of these promoters [10], these presumably bivalent histone marks on the same promoter (although not necessarily on the same nucleosome) reinforce the view of adipogenic promoters pre-programmed for activation upon adipogenic stimulation. Upon adipogenic differentiation, activation of adipogenic genes is expected to be accompanied by acetylation of H3K9 and demethylation of H3K27 (fig. 4, differentiated somatic cells). Inactivation of a promoter upon lineage-specific differentiation would, conversely, be expected to lead to deacetylation and trimethylation of H3K9 and maintenance of trimethylated H3K27 (fig. 4, differentiated somatic cells).
Fig. 4.
Epigenetic landscape of genes associated with lineage specification as function of differentiation: undifferentiated embryonic stem cells (ESCs), undifferentiated mesenchymal stem cells (MSCs), differentiated somatic cells. Two scenarios are presented for lineage-specific genes in differentiated cells, depending on whether the gene is activated (ON) or repressed (OFF).
Mapping of Histone Modifications in the Embryonic Stem Cell Genome
Recent mapping of histone modifications has shown that lineage-specific genes, which are either silent or active in differentiated somatic cells, are in a potentially active state in pluripotent ESCs. Genome-wide and locus-specific ChIP analyses reveal that repressed but potentially active promoters are associated with so-called ‘bivalent’ histone modifications characterized by H3K4m3, a mark of active genes, and H3K27m3 which associates with inactive genes [79, 80] (fig. 5 A). Azuara et al. [79] have shown that several transcription factors essential for lineage specification are not expressed in mouse ESCs but are marked on their promoter by H3K4m3, H3K27m3, and by H3K9ac. Unscheduled expression of these genes is induced in ESCs deficient for embryonic ectoderm development protein, a component of the polycomb repressor complex PRC2 which harbors H3K27 methyltransferase activity [89], demonstrating the essential role of trimethylation of H3K27 in maintaining a transcriptional brake in a context of transcriptionally permissive chromatin. At the genome-wide level, these ‘bivalent domains’ consist of large regions of H3K27 trimethylation embedding smaller areas of H3K4 trimethylation [80]. Consistent with the findings of Azuara et al. [79], these domains include transcription factor encoding genes that are repressed or expressed at low levels. Intriguing-ly, the correlation between histone methylation marks and ge-nomic sequence in ESCs raises the hypothesis that DNA sequence may prime the epigenetic landscape in pluripotent cells [80]. Nevertheless, not all lineage control genes in ESCs are associated with bivalent histone modifications; rather, they are marked by H3K4m3 only or do not display H3K4m3 or H3K27m3 [80]. The critical role of these genes in lineage determination suggests that they are also in a transcriptionally poised state and await, through yet unknown epigenetic mechanisms, permission for transcription.
Fig. 5.
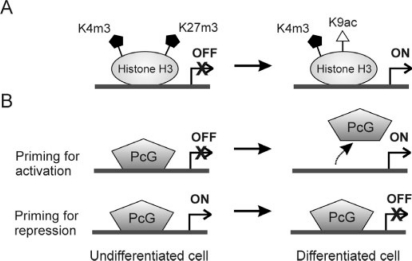
Control of lineage-specific gene expression by histone H3K27 methylation and PcGs. A In undifferentiated cells, repressed lineage-specific genes are marked by trimethylation of K4 and K27 (the bivalent marks). These marks are believed to prime genes for activation. Upon differentiation, acetylation of H3K9 and demethylation of H3K27 results in transcriptional activation of the gene. B In undifferentiated cells, repressed lineage-specific genes can be either primed for activation by occupancy of PcGs on the promoter; differentiation coincides with removal of the PcGs and activation of the gene. However, genes expressed in undifferentiated cells can also be primed for transcriptional repression by PcG binding to the promoter.
Evidence for Hyperdynamic Chromatin in Embryonic Stem Cells
A dynamic reorganization of chromatin domains is essential for setting up heritable transcriptional programs in the context of differentiation [90]. Many structural chromatin proteins such as heterochromatin protein 1 (HP1) and histones have been shown to bind more loosely to chromatin of ESCs than differentiated or somatic cells [91]. These proteins are also hyperdynamic in ESCs relative to differentiated cells. Fluorescence recovery after photobleaching studies have shown that all 3 isoforms of HP1 fused with green fluorescence protein exchange faster in heterochromatic foci of undifferentiated mouse ESCs than after differentiation [91]. Likewise, exchange rates of fluorescently tagged histones H1, H2B, and H3 are significantly higher in pluripotent ESCs than in differentiated counterparts. These studies unravel the existence of a greater fraction of loosely bound HP1 as well as core and linker histones in ESCs. The hyperdynamic nature of chromatin-associated proteins in pluripotent ESCs reflects some plasticity in chromatin organization, and thereby provides a basis for pluripotency. The concept of hyperdynamic chromatin in ESCs is in line with an attractive yet highly speculative ‘histone modification pulsing’ model whereby developmentally regulated genes would be marked by transient histone modifications in pluripotent cells to enable the appropriate response upon differentiation [92].
Polycomb Group Proteins: A ‘Brake’ on Lineage Specification Genes
PcGs are transcriptional repressors [93, 94] found within 2 distinct and conserved polycomb repressor complexes (PRC1 and PCR2), working cooperatively [95]. Involvement of PRCs in pluripotency has been suggested by the requirement of PcG proteins for the patterning of gene expression during development, for establishing pluripotent ESCs, and for maintaining somatic stem cell cultures (reviewed in [96]).
In undifferentiated ESCs, PcGs preferentially occupy genes that are activated upon differentiation, consistent with the view that these genes are poised for transcription [97, 98, 99] (fig. 5). Histone methyltransferase activity of Eed and enhancer of zeste homologue 2 (Ezh2; another PRC2 component) is responsible for trimethylation of H3K27 on these target genes [75, 76]. In addition, trimethylation of H3K4 is mediated by Trithorax group (Trx) proteins [94]. Thus, the known interplay between PcG and Trx proteins is also likely to establish bivalent histone modifications on developmentally regulated genes in pluripotent cells. PcGs, however, are also dynamic and not always associated with transcriptionally repressed genes. For genes activated upon differentiation, PcGs are displaced from promoters [99]. Furthermore, genes that are repressed during differentiation have also paradoxically been found to be already occupied by PcG proteins in undifferentiated cells while in a state of activity. These findings suggest that PRCs constitute a ‘pre-programmed memory system’ established during embryogenesis [99]. This program would mark certain genes for transcriptional repression upon differentiation, while other genes would be primed for activation (fig. 5 B). It will be interesting to determine whether genes poised for transcriptional activation or repression by PcG proteins are marked by distinct histone modifications (e.g., different levels of the active H3K9ac mark) or by a specific CpG methylation status. An increasing body of evidence, therefore, suggests that unique combinations of CpG methylation, histone modifications, PcG occupancy, and nucleosome positioning [100, 101, 102, 103] on developmentally regulated gene promoters, in a context of hyperdynamic chromatin, define a pluripotent genomic organization in ESCs.
DNA Methylation, Histone Modifications, and Timing of DNA Replication
DNA methylation has long been implicated in the organization of the nuclear compartment, particularly in regions of constitutive heterochromatin (see [104] for an overview of the evidence). A recent study shed light on the nature of the relationship between global DNA methylation levels and chromatin organization [104]. Indeed, Dnmt3a-/- Dnmt3b-/- mouse ESCs lacking DNA methylation have been shown to exhibit enhanced clustering of pericentric heterochromatin and major changes in chromatin structure [104]. Levels of H3K9m2 are reduced (H3K9m3 remains surprisingly unaltered) while levels of acetylated H3K9, H4K5, and H4K16 increase, both globally and on major satellite repeats, suggesting a reorganization of heterochromatin in these cells. Mobility of the linker his-tones H1 and H5 is also reduced. In contrast, absence of DNA methylation does not seem to affect compaction of bulk and heterochromatin, on the basis of nuclease digestion, nucleosome spacing and chromatin fractionation [104]. Genes reactivated by elimination of DNMT1 in mouse ESCs become enriched in acetylated H3K9 and H3K14, H4ac and H3K4m3, while those not reactivated by removal of DNA methylation show no hyperacetylation [105]. Thus, some methylated genes in ESCs are subject to additional repressive mechanisms affecting histone H3 acetylation. These studies illustrate how DNA methylation affects global chromatin packaging and subsequently, organization of the nucleus, but in a manner that does not involve chromatin compaction. Despite these global changes, however, different classes of genes respond differently to the absence of DNA methylation.
Timing of DNA replication has been shown to be influenced by the state of chromatin (active vs. inactive), albeit not always by transcription per se [79, 106]. Replication timing has been introduced as an additional epigenetic component [107], although whether it qualifies as an ‘epigenetic’ component on the basis of the definition of epigenetics remains questionable (replication timing is not a modification of DNA or chromatin). Interestingly, in mouse ESCs, a number of genes not necessarily expressed but which may be important later during differentiation have been shown to replicate early in S phase [79]. Genes that are not needed, however, replicate later in S phase. Indeed, genes encoding key neuronal-specific transcription factors replicate early in undifferentiated ESCs, but late in hematopoietic stem cells in which these genes are not required [79]. Therefore, lineage specification genes are able to undergo modifications in chromatin organization and switch from early to late replication timing in the course of differentiation.
Early replication timing has been linked to enriched histone acetylation [108, 109], but how replication timing functionally relates to DNA methylation remains to be explored. Recent evidence indicates that genes whose expression is dependent on DNA demethylation in ESCs consistently replicate early in S phase, while half of those genes not reactivated by DNA demethylation replicate late [105]. Nonetheless, the overall replication timing pattern does not seem to be dependent on CpG methylation [110], and methylation is not necessarily affected by replication timing profile, suggesting that replication timing and DNA methylation are independently established [105].
Perspectives
Genome-wide technologies have provided a wealth of information on mechanisms regulating gene expression in the context of development, cell differentiation, and disease. These studies have also started to unravel the epigenetic landscape of ESCs and somatic stem cells, providing a molecular frame for the pluripotent state. Such approaches have in our opinion been welcome because defining pluripotency simply on the basis of gene expression in ESCs has been deceptive [111]. Multiple aspects of stem cell function remain nevertheless to be investigated. We are looking at the tip of the iceberg in the epigenetic landscape of stem cells. Mapping of DNA methylation marks, of novel histone modifications, and of novel transcriptional regulators [112], together with improved bioinformatics tools, will enhance the resolution of the current stem cell epigenetic map.
A totally unexplored area is in vivo epigenetics. The fate of ESCs after transplantation into animal models is being studied, but the extent of contribution of MSCs to various tissues remains debated. Our analyses of DNA methylation in ASCs after in vitro differentiation suggest that the cells retain an undifferentiated ASC epigenetic program despite phenotypic changes [41]. In the event MSCs do directly contribute to host tissue in vivo, a hypothesis is that the target tissue provides a beneficial environment for stem cell function. Intuitively, the in vivo environment may be more conducive to epigenetic commitment of MSCs than the cell culture flask. Broader application of imaging techniques to chromatin dynamics, gene expression and epigenetics [90, 91, 113, 114, 115] will undoubtedly contribute to our understanding of genome organization in stem cells. Ultimately, compilation of nucleus-wide four-dimensional imaging data and genome-wide biochemical and genetic data will provide an integrated representation of functional genome organization in stem cells.
The restricted differentiation potential of MSCs currently limits their application to regenerative medicine. Qualities of the ideal stem cell in a clinical setting are expected to be extensive ability to be expanded in culture without genetic and epigenetic abnormalities, ability to form functional cell types in vitro and in vivo, and immunocompatibility with the patient. Patient-derived somatic stem cells fulfill the latter requirement; however, they currently do not meet the first two. Attempts to alleviate limited differentiation potential of MSCs aim at enhancing differentiation plasticity through a nuclear reprogramming process. Current strategies for reprogramming somatic cells to pluripotency include nuclear transplantation [116, 117, 118], fusion with ESCs [119, 120, 121], treatment with extracts from eggs [122], ESCs or other pluripotent cells [40], and retroviral transduction of pluripotency-associated factors [123, 124, 125, 126, 127]. These approaches have been reviewed elsewhere [128]. Notably, recent attempts at reprogramming somatic stem cells have been reported, and results suggest that the reprogramming efficiency by nuclear transfer of progenitor cells compared to terminally differentiated cells is not improved [129]. Efforts are needed to determine whether somatic stem cells will one day be safely reprogrammed to a pluripotent state to enable their therapeutic use.
Acknowledgements
Our work is supported by the FUGE, STORFORSK, YFF and STAM-CELLE programs of the Research Council of Norway, and by the Norwegian Cancer Society.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol. 2005;6:872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman LM, Carpenter MK. Characterization and culture of human embryonic stem cells. Nat Biotechnol. 2005;23:699–708. doi: 10.1038/nbt1102. [DOI] [PubMed] [Google Scholar]
- 4.Jahagirdar BN, Verfaillie CM. Multipotent adult progenitor cell and stem cell plasticity. Stem Cell Rev. 2005;1:53–59. doi: 10.1385/SCR:1:1:053. [DOI] [PubMed] [Google Scholar]
- 5.Verfaillie C. Stem cell plasticity. Hematology. 2005;10(suppl 1):293–296. doi: 10.1080/10245330512331390113. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, Du J, Aldrich S, Lisberg A, Low WC, Largaespada DA, Verfaillie CM. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 7.Boquest AC, Shahdadfar A, Fronsdal K, Sigurjonsson O, Tunheim SH, Collas P, Brinchmann JE. Isolation and transcription profiling of purified uncultured human stromal stem cells: alteration of gene expression after in vitro cell culture. Mol Biol Cell. 2005;16:1131–1141. doi: 10.1091/mbc.E04-10-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 9.Boquest AC, Noer A, Sorensen AL, Vekterud K, Collas P. CpG methylation profiles of endothelial cell-specific gene promoter regions in adipose tissue stem cells suggest limited differentiation potential toward the endothelial cell lineage. Stem Cells. 2007;25:852–861. doi: 10.1634/stemcells.2006-0428. [DOI] [PubMed] [Google Scholar]
- 10.Noer A, Sørensen AL, Boquest AC, Collas P. Stable CpG hypomethylation of adipogenic promoters in freshly isolated, cultured and differentiated mesenchymal stem cells from adipose tissue. Mol Biol Cell. 2006;17:3543–3556. doi: 10.1091/mbc.E06-04-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 12.Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 13.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 14.Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;14:R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 15.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 16.Turek-Plewa J, Jagodzinski PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–647. [PubMed] [Google Scholar]
- 17.Jeltsch A, Nellen W, Lyko F. Two substrates are better than one: dual specificities for Dnmt2 methyltransferases. Trends Biochem Sci. 2006;31:306–308. doi: 10.1016/j.tibs.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Kunert N, Marhold J, Stanke J, Stach D, Lyko F. A Dnmt2-like protein mediates DNA methylation in Drosophila. Development. 2003;130:5083–5090. doi: 10.1242/dev.00716. [DOI] [PubMed] [Google Scholar]
- 19.Tang LY, Reddy MN, Rasheva V, Lee TL, Lin MJ, Hung MS, Shen CK. The eukaryotic DNMT2 genes encode a new class of cytosine-5 DNA methyltransferases. J Biol Chem. 2003;278:33613–33616. doi: 10.1074/jbc.C300255200. [DOI] [PubMed] [Google Scholar]
- 20.Hermann A, Schmitt S, Jeltsch A. The human Dnmt2 has residual DNA-(cytosine-C5) methyl-transferase activity. J Biol Chem. 2003;278:31717–31721. doi: 10.1074/jbc.M305448200. [DOI] [PubMed] [Google Scholar]
- 21.Liu K, Wang YF, Cantemir C, Muller MT. Endogenous assays of DNA methyltransferases: evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol Cell Biol. 2003;23:2709–2719. doi: 10.1128/MCB.23.8.2709-2719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007;21:261–266. doi: 10.1101/gad.1472907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyl-transferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 24.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman AR, Hu JF. Directing DNA methylation to inhibit gene expression. Cell Mol Neurobiol. 2006;26:425–438. doi: 10.1007/s10571-006-9057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14:R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 28.Young LE, Beaujean N. DNA methylation in the preimplantation embryo: the differing stories of the mouse and sheep. Anim Reprod Sci. 2004;82–83:61–78. doi: 10.1016/j.anireprosci.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 29.Mann JR. Imprinting in the germ line. Stem Cells. 2001;19:287–294. doi: 10.1634/stemcells.19-4-287. [DOI] [PubMed] [Google Scholar]
- 30.Razin A, Shemer R. DNA methylation in early development. Hum Mol Genet. 1995;4:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- 31.Hellman A, Chess A. Gene body-specific methylation on the active × chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 32.Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 33.Reik W, Howlett SK, Surani MA. Imprinting by DNA methylation: from transgenes to endogenous gene sequences. Dev Suppl. 1990:99–106. [PubMed] [Google Scholar]
- 34.Sapienza C, Peterson AC, Rossant J, Balling R. Degree of methylation of transgenes is dependent on gamete of origin. Nature. 1987;328:251–254. doi: 10.1038/328251a0. [DOI] [PubMed] [Google Scholar]
- 35.Reik W, Collick A, Norris ML, Barton SC, Surani MA. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature. 1987;328:248–251. doi: 10.1038/328248a0. [DOI] [PubMed] [Google Scholar]
- 36.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 37.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 38.Deb-Rinker P, Ly D, Jezierski A, Sikorska M, Walker PR. Sequential DNA methylation of the Nanog and Oct-4 upstream regions in human NT2 cells during neuronal differentiation. J Biol Chem. 2005;280:6257–6260. doi: 10.1074/jbc.C400479200. [DOI] [PubMed] [Google Scholar]
- 39.Dahl JA, Collas P. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells. 2007;25:1037–1046. doi: 10.1634/stemcells.2006-0430. [DOI] [PubMed] [Google Scholar]
- 40.Freberg CT, Dahl JA, Timoskainen S, Collas P. Epigenetic reprogramming of OCT4 and NANOG regulatory regions by embryonal carcinoma cell extract. Mol Biol Cell. 2007;18:1543–1553. doi: 10.1091/mbc.E07-01-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boquest AC, Noer A, Collas P. Epigenetic programming of mesenchymal stem cells from human adipose tissue. Stem Cell Rev. 2006;2:319–329. doi: 10.1007/BF02698059. [DOI] [PubMed] [Google Scholar]
- 42.Fraser JK, Wulur I, Alfonso Z, Hedrick MH. Fat tissue: an underappreciated source of stem cells for biotechnology. Trends Biotechnol. 2006;24:150–154. doi: 10.1016/j.tibtech.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, Alfonso ZC, Fraser JK, Benhaim P, Hedrick MH. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katz AJ, Tholpady A, Tholpady SS, Shang H, Ogle RC. Cell surface and transcriptional characterization of human adipose-derived adherent stromal (hADAS) cells. Stem Cells. 2005;23:412–423. doi: 10.1634/stemcells.2004-0021. [DOI] [PubMed] [Google Scholar]
- 45.Kern S, Eichler H, Stoeve J, Kluter H, Bieback K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood or adipose tissue. Stem Cells. 2006;24:1294–1301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- 46.Urs S, Smith C, Campbell B, Saxton AM, Taylor J, Zhang B, Snoddy J, Jones VB, Moustaid-Moussa N. Gene expression profiling in human preadipocytes and adipocytes by microarray analysis. J Nutr. 2004;134:762–770. doi: 10.1093/jn/134.4.762. [DOI] [PubMed] [Google Scholar]
- 47.Cousin B, Andre M, Arnaud E, Penicaud L, Casteilla L. Reconstitution of lethally irradiated mice by cells isolated from adipose tissue. Biochem Biophys Res Commun. 2003;301:1016–1022. doi: 10.1016/s0006-291x(03)00061-5. [DOI] [PubMed] [Google Scholar]
- 48.Clark SJ, Statham A, Stirzaker C, Molloy PL, Frommer M. DNA methylation: bisulphite modification and analysis. Nat Protoc. 2006;1:2353–2364. doi: 10.1038/nprot.2006.324. [DOI] [PubMed] [Google Scholar]
- 49.Kim JY, Beart RW, Shibata D. Stability of colon stem cell methylation after neo-adjuvant therapy in a patient with attenuated familial adenomatous polyposis. BMC Gastroenterol. 2005;5:19–25. doi: 10.1186/1471-230X-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim JY, Tavare S, Shibata D. Counting human somatic cell replications: methylation mirrors endometrial stem cell divisions. Proc Natl Acad Sci USA. 2005;102:17739–17744. doi: 10.1073/pnas.0503976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yatabe Y, Tavare S, Shibata D. Investigating stem cells in human colon by using methylation patterns. Proc Natl Acad Sci U S A. 2001;98:10839–10844. doi: 10.1073/pnas.191225998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- 53.Hoffman LM, Carpenter MK. Human embryonic stem cell stability. Stem Cell Rev. 2005;1:139–144. doi: 10.1385/SCR:1:2:139. [DOI] [PubMed] [Google Scholar]
- 54.Ushijima T, Okochi-Takada E. Aberrant methylations in cancer cells: where do they come from? Cancer Sci. 2005;96:206–211. doi: 10.1111/j.1349-7006.2005.00035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfeifer GP, Steigerwald SD, Hansen RS, Gartler SM, Riggs AD. Polymerase chain reaction-aided genomic sequencing of an → chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci U S A. 1990;87:8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva AJ, Ward K, White R. Mosaic methylation in clonal tissue. Dev Biol. 1993;156:391–398. doi: 10.1006/dbio.1993.1086. [DOI] [PubMed] [Google Scholar]
- 57.Noer A, Boquest AC, Collas P. Dynamics of adipogenic promoter DNA methylation during clonal culture of human adipose stem cells to senescence. BMC Cell Biol. 2007;8:18–29. doi: 10.1186/1471-2121-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Allegrucci C, Wu YZ, Thurston A, Denning CN, Priddle H, Mummery CL, Ward-van OD, Andrews PW, Stojkovic M, Smith N, Parkin T, Jones ME, Warren G, Yu L, Brena RM, Plass C, Young LE. Restriction landmark genome scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum Mol Genet. 2007;16:1253–1268. doi: 10.1093/hmg/ddm074. [DOI] [PubMed] [Google Scholar]
- 59.Kaneko KJ, Rein T, Guo ZS, Latham K, Depamphilis ML. DNA methylation may restrict but does not determine differential gene expression at the Sgy/Tead2 locus during mouse development. Mol Cell Biol. 2004;24:1968–1982. doi: 10.1128/MCB.24.5.1968-1982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson M, Krassowska A, Gilbert N, Chevassut T, Forrester L, Ansell J, Ramsahoye B. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol. 2004;24:8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, John R, Feil R, Brockdorff N. Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
- 62.Jacob S, Moley KH. Gametes and embryo epigenetic reprogramming affect developmental outcome: implication for assisted reproductive technologies. Pediatr Res. 2005;58:437–446. doi: 10.1203/01.PDR.0000179401.17161.D3. [DOI] [PubMed] [Google Scholar]
- 63.Bibikova M, Chudin E, Wu B, Zhou L, Garcia EW, Liu Y, Shin S, Plaia TW, Auerbach JM, Arking DE, Gonzalez R, Crook J, Davidson B, Schulz TC, Robins A, Khanna A, Sartipy P, Hyllner J, Vanguri P, Savant-Bhonsale S, Smith AK, Chakravarti A, Maitra A, Rao M, Barker DL, Loring JF, Fan JB. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lagarkova MA, Volchkov PY, Lyakisheva AV, Philonenko ES, Kiselev SL. Diverse epigenetic profile of novel human embryonic stem cell lines. Cell Cycle. 2006;5:416–420. doi: 10.4161/cc.5.4.2440. [DOI] [PubMed] [Google Scholar]
- 65.Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, Sui G, Cutler DJ, Liu Y, Brimble SN, Noaksson K, Hyllner J, Schulz TC, Zeng X, Freed WJ, Crook J, Abraham S, Colman A, Sartipy P, Matsui S, Carpenter M, Gazdar AF, Rao M, Chakravarti A. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- 66.Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Epigenetic status of human embryonic stem cells. Nat Genet. 2005;37:585–587. doi: 10.1038/ng1556. [DOI] [PubMed] [Google Scholar]
- 67.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 68.Shen Y, Chow J, Wang Z, Fan G. Abnormal CpG island methylation occurs during in vitro differentiation of human embryonic stem cells. Hum Mol Genet. 2006;15:2623–2635. doi: 10.1093/hmg/ddl188. [DOI] [PubMed] [Google Scholar]
- 69.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 70.Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–24. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 71.De la Cru X, Lois S, Sanchez-Molina S, Martinez-Balbas MA. Do protein motifs read the histone code? Bioessays. 2005;27:164–175. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- 72.Cosgrove MS, Wolberger C. How does the histone code work? Biochem Cell Biol. 2005;83:468–476. doi: 10.1139/o05-137. [DOI] [PubMed] [Google Scholar]
- 73.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 74.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 75.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 76.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 77.Pasini D, Bracken AP, Jensen MR, Lazzerini DE, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 79.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 80.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 81.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 82.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 83.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van LF, Gottschling DE, O'Neill LP, Turner BM, Delrow J, Bell SP, Groudine M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, Bourque G, Oh S, Ruan Y, Ng HH, Wei CL. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 85.Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 86.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, III, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 87.Collas P, Dahl JA. Chop it, ChIP it, check it: the current status of chromatin immunoprecipitation. Front Biosci. 2008;13:929–943. doi: 10.2741/2733. [DOI] [PubMed] [Google Scholar]
- 88.O'Neill LP, Vermilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet. 2006;38:835–841. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- 89.Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 90.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 91.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gan Q, Yoshida T, McDonald OG, Owens GK. Concise review: epigenetic mechanisms contribute to pluripotency and cell lineage determination of embryonic stem cells. Stem Cells. 2007;25:2–9. doi: 10.1634/stemcells.2006-0383. [DOI] [PubMed] [Google Scholar]
- 93.Kennison JA. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. 289–303. [DOI] [PubMed] [Google Scholar]
- 94.Ringrose L, Paro R. Polycomb/trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 95.Otte AP, Kwaks TH. Gene repression by Polycomb group protein complexes: a distinct complex for every occasion? Curr Opin Genet Dev. 2003;13:448–454. doi: 10.1016/s0959-437x(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 96.Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 97.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 98.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pusarla RH, Vinayachandran V, Bhargava P. Nucleosome positioning in relation to nucleosome spacing and DNA sequence-specific binding of a protein. FEBS J. 2007;274:2396–2410. doi: 10.1111/j.1742-4658.2007.05775.x. [DOI] [PubMed] [Google Scholar]
- 101.Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 102.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 104.Gilbert N, Thomson I, Boyle S, Allan J, Ramsahoye B, Bickmore WA. DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction. J Cell Biol. 2007;177:401–411. doi: 10.1083/jcb.200607133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lande-Diner L, Zhang J, Ben-Porath I, Amariglio N, Keshet I, Hecht M, Azuara V, Fisher AG, Rechavi G, Cedar H. Role of DNA methylation in stable gene repression. J Biol Chem. 2007;282:12194–12200. doi: 10.1074/jbc.M607838200. [DOI] [PubMed] [Google Scholar]
- 106.Azuara V, Brown KE, Williams RR, Webb N, Dillon N, Festenstein R, Buckle V, Merkenschlager M, Fisher AG. Heritable gene silencing in lymphocytes delays chromatid resolution without affecting the timing of DNA replication. Nat Cell Biol. 2003;5:668–674. doi: 10.1038/ncb1006. [DOI] [PubMed] [Google Scholar]
- 107.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 108.Schubeler D, Scalzo D, Kooperberg C, van SB, Delrow J, Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet. 2002;32:438–442. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- 109.Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 110.Gribnau J, Hochedlinger K, Hata K, Li E, Jaenisch R. Asynchronous replication timing of imprinted loci is independent of DNA methylation, but consistent with differential subnuclear localization. Genes Dev. 2003;17:759–773. doi: 10.1101/gad.1059603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fortunel NO, Otu HH, Ng HH, Chen J, Mu X, Chevassut T, Li X, Joseph M, Bailey C, Hatzfeld JA, Hatzfeld A, Usta F, Vega VB, Long PM, Libermann TA, Lim B. Comment on 'Sternness': transcriptional profiling of embryonic and adult stem cells' and 'a stem cell molecular signature'. Science. 2003;302:393. doi: 10.1126/science.1086384. [DOI] [PubMed] [Google Scholar]
- 112.Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 113.Heard E, Bickmore W. The ins and outs of gene regulation and chromosome territory organisation. Curr Opin Cell Biol. 2007;19:311–316. doi: 10.1016/j.ceb.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 114.Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- 115.Gilbert N, Bickmore WA. The relationship between higher-order chromatin structure and transcription. Biochem Soc Symp. 2006:59–66. doi: 10.1042/bss0730059. [DOI] [PubMed] [Google Scholar]
- 116.Gurdon JB, Byrne JA. The first half-century of nuclear transplantation. Proc Natl Acad Sci U S A. 2003;100:8048–8052. doi: 10.1073/pnas.1337135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wilmut I, Beaujean N, De Sousa PA, Dinnyes A, King TJ, Paterson LA, Wells DN, Young LE. Somatic cell nuclear transfer. Nature. 2002;419:583–586. doi: 10.1038/nature01079. [DOI] [PubMed] [Google Scholar]
- 118.Yang X, Smith SL, Tian XC, Lewin HA, Renard JP, Wakayama T. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- 119.Kimura H, Tada M, Nakatsuji N, Tada T. Histone code modifications on pluripotential nuclei of reprogrammed somatic cells. Mol Cell Biol. 2004;24:5710–5720. doi: 10.1128/MCB.24.13.5710-5720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ying QL, Nichols J, Evans EP, Smith AG. Changing potency by spontaneous fusion. Nature. 2002;416:545–548. doi: 10.1038/nature729. [DOI] [PubMed] [Google Scholar]
- 121.Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 122.Miyamoto K, Furusawa T, Ohnuki M, Goel S, Tokunaga T, Minami N, Yamada M, Ohsumi K, Imai H. Reprogramming events of mammalian somatic cells induced by Xenopus laevis egg extracts. Mol Reprod Dev. 2007;74:1268–1277. doi: 10.1002/mrd.20691. [DOI] [PubMed] [Google Scholar]
- 123.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 124.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2007 doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 125.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 126.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 127.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 128.Collas P, Gammelsaeter R. Novel approaches to epigenetic reprogramming of somatic cells. Cloning Stem Cells. 2007;9:26–32. doi: 10.1089/clo.2006.0080. [DOI] [PubMed] [Google Scholar]
- 129.Sung LY, Gao S, Shen H, Yu H, Song Y, Smith SL, Chang CC, Inoue K, Kuo L, Lian J, Li A, Tian XC, Tuck DP, Weissman SM, Yang X, Cheng T. Differentiated cells are more efficient than adult stem cells for cloning by somatic cell nuclear transfer. Nat Genet. 2006;38:1323–1328. doi: 10.1038/ng1895. [DOI] [PubMed] [Google Scholar]