Abstract
Background
The genome of serotype M28 group A Streptococcus (GAS) strain MGAS6180 contains a novel genetic element named Region of Difference 2 (RD2) that encodes seven putative secreted extracellular proteins. RD2 is present in all serotype M28 strains and strains of several other GAS serotypes associated with female urogenital infections. We show here that the GAS RD2 element is present in strain MGAS6180 both as an integrative chromosomal form and a circular extrachromosomal element. RD2-like regions were identified in publicly available genome sequences of strains representing three of the five major group B streptococcal serotypes causing human disease. Ten RD2-encoded proteins have significant similarity to proteins involved in conjugative transfer of Streptococcus thermophilus integrative chromosomal elements (ICEs).
Results
We transferred RD2 from GAS strain MGAS6180 (serotype M28) to serotype M1 and M4 GAS strains by filter mating. The copy number of the RD2 element was rapidly and significantly increased following treatment of strain MGAS6180 with mitomycin C, a DNA damaging agent. Using a PCR-based method, we also identified RD2-like regions in multiple group C and G strains of Streptococcus dysgalactiae subsp.equisimilis cultured from invasive human infections.
Conclusions
Taken together, the data indicate that the RD2 element has disseminated by lateral gene transfer to genetically diverse strains of human-pathogenic streptococci.
Background
We sequenced the genome of a strain (MGAS6180) of serotype M28 group A Streptococcus [1], a human-specific pathogen that is non-randomly associated with neonatal female urogenital infections [2]. The genome of strain MGAS6180 has a novel 37-kb element designated RD2 (Region of Difference 2) [1]. RD2 is one of seven elements integrated into the chromosome of this strain (4 phages, 3 ICE and ICE related elements) [1,3]. Subsequently we demonstrated that all serotype M28 strains studied contained RD2 integrated at the same chromosomal site [1,3]. RD2 encodes seven secreted extracellular proteins that are expressed in human infections. One of these proteins (M28_Spy1336) is also known as the R28 protein, and has been previously studied in GAS and group B Streptococcus (GBS) [4-7]. The R28 protein has been implicated in virulence based on its ability to mediate binding of GBS to human vaginal epithelial cells [6]. Protein M28_Spy1325 was recently studied extensively and shown to be a member of the antigen I/II family of proteins originally described in oral streptococcal species. M28_Spy1325 binds to salivary agglutinin, a 340-kDa protein abundantly found in human saliva. Zhang et al. [8] recently demonstrated that immunization of mice with recombinant purified M28_Spy1325 confers protection against invasive infection. Thus, two proteins encoded by RD2 likely contribute to host-pathogen interactions.
Several lines of evidence suggest that RD2 in GAS was acquired by horizontal gene transfer (HGT). First, the RD2 element is integrated into a tRNA-threonine gene and flanked by 16bp imperfect direct repeats ATTC(C/T)CGGTGGTGGCA [1,3]. The chromosomal location of RD2 is identical in the majority of RD2-positive strains suggesting a conserved mode of integration [1]. Second, the G+C content of RD2 (35%) is significantly lower than the average GAS genome (38%) and contains different di-nucleotide content and codon usage [1,3]. An RD2-like element also has been identified in the genome of a serotype M2 GAS strain [3]. The RD2 element in this strain is virtually identical at the nucleotide level to RD2 present in M28 strains. However, the genome sequence of strain MGAS6180 (M28) and MGAS10270 (M2) are otherwise quite divergent from one another. Based on single nucleotide polymorphism (SNPs), the average SNP difference between genomes is about 137 per 1 kb (total of 14096 SNPs), while only 8 nucleotide differences are found within 37 kb RD2 region [3,9]. The differences in SNP frequency within chromosome and RD2 region strongly suggests that the RD2 element in these strains has had a very different evolutionary history compared to the core chromosome, and was acquired via horizontal transfer [3].
The primary goal of the experiments described herein was to test the hypothesis that the RD2 element was laterally transferable in vitro under laboratory conditions, and we found that this was the case. Moreover, we identified an RD2-like element in multiple strains of Lancefield group C and G streptococci, indicating that this genetic element is more phylogenetically widespread than previously appreciated.
Methods
Bacterial strains and growth
Streptococcal strains of serotypes A, C, and G (Additional File 1, Table S1) were cultured routinely at 37°C in an atmosphere of 5% CO2 on Trypticase soy agar II plates containing 5% sheep blood (Becton Dickinson, Franklin Lakes, NJ) or in liquid Todd Hewitt medium supplemented with 0.5% yeast extract (THY medium). Antibiotics were used at the following concentrations: spectinomycin, 150 μg/ml; erythromycin, 1 μg/ml; and kżanamycin, 400 μg/ml.
Isolation of total DNA from streptococci
DNA was isolated from cultures grown overnight in THY medium using a modified phenol-chloroform procedure [10]. Briefly, 5 to 35 ml of overnight THY cultures were pelleted by centrifugation and suspended in TE, pH 7.5. Bacteria were treated with mutanolysin (500 U/ml) and lysozyme (2 mg/ml) (Sigma Aldrich, St. Louis, MO) with occasional mixing for 1 h at 37°C, followed by 2% wt/vol SDS (Gibco, Carlsbad, CA), and proteinase K (0.2 mg/ml) (Sigma Aldrich, St. Louis, MO) for 1 h at 37°C. DNA was extracted with phenol:chloroform:isoamyl alcohol, precipitated with two volumes of ice-cold ethanol, washed with 70% ice-cold ethanol, and suspended in TE, pH 8.0 buffer containing 0.2 mg/ml RNase A (Invitrogen, Carlsbad, CA).
DNA amplification by PCR
Primers used in PCR reactions are listed in Table 1 and Additional File 2 (Table S2). PCR was used to investigate the presence and organization of the RD2 element in streptococcal strains. The PCR primers #1-#4 detect a chromosomal and extrachromosomal circular form, and tile across the entire RD2. Confirmation of RD2 presence by tailing and detection of genes encoding extra-chromosomal proteins was performed as described previously [1,2]
Table 1.
PCR primers used in this study
| A. Primers used for detection of multiple RD2 genes, Q-PCR and tiling. | ||
|---|---|---|
| Primer name | Primer sequence | Source |
| emm sequencing | ||
| CDC emm1 | TATT(C/G)GCTTAGAAAATTAA | [19] |
| CDC emm2 | GCAAGTTCTTCAGCTTGTTT | [19] |
| Detection of circular form | ||
| #1 | GAAAACAAAAGTTTCTTCATGCGTTTGGCG | this work |
| #2 | CAATTAATAGAAACATATGGTCATTTG | this work |
| #3 | GGAATTAGCCCACTAGAATATAAGC | this work |
| #4 | TAGCAAGTAAACCCTAGATTGTCTATGTTC | this work |
| Detection of genes encoding extracellular RD2 proteins | ||
| M28_Spy1306F | ACTAAGCCAAGCGAGGACAA | [1] |
| M28_Spy1306R | CCAAAACCGTGTAGCCTGTA | [1] |
| M28_Spy 1307F | TCATCGTCAAAAGCCATCTC | [1] |
| M28_Spy 1307R | TTGCTCTGATAAACCTCAAG | [1] |
| M28_Spy1308F | TACGACAGAAGCAGGTGGAG | [1] |
| M28_Spy1308R | ACCGAGTTTCGCAGGATTG | [1] |
| M28_Spy1325F | TGAATGATGCGGGGACTTAT | [1] |
| M28_Spy1325R | TGTAAAAGGCTGCTGGGTCT | [1] |
| M28_Spy1326F | ACACCGACTGAGATTGCTGA | [1] |
| M28_Spy1326R | TTGGCTTGTGAGGTTTGAGA | [1] |
| M28_Spy1332F | ATGCCAAAAACCAAAGGAAG | [1] |
| M28_Spy1332R | TCATACTTTTCAGGTACACAAGCA | [1] |
| M28_Spy1336F | GATACTTCACAGACGAAACAACG | [1] |
| M28_Spy1336R | ATCACGACTCCCATCACTCC | [1] |
| Quantitative PCR (Taqman) | ||
| proS_F | TGAGTTTATTATGAAAGAGGCTATAGTTTC | [15] |
| proS_R | AATAGCTTCGTAAGCTTGACGATAAT | [15] |
| proS_P | TCGTAGGTCACATCTAATCTTCATAGTTG | [15] |
| M28_Spy1306 F | CGTTGTTCCTGCTACTGGATCTGCTAC | this work |
| M28_Spy1306 P | ACGATTGCAAGTATTGCTTTG | this work |
| M28_Spy1306 R | CAATCGGTGTCGTTGGTTG | this work |
| M28_Spy1325 F | ACCGTCGCAAGGACCTTGTCTTTCTG | [8] |
| M28_Spy1325 P | CAGCATACGCATGACCTC | [8] |
| M28_Spy1325 R | AGTGATAACACTACCATCTGATAAAG | [8] |
| M28_Spy1336 F | ACAGAAGCTGCACCAAACTTGAACTTCTTAATTGA | this work |
| M28_Spy1336 P | GTAGATGCAGCAACTATTGAC | this work |
| M28_Spy1336 R | ATGATACTTCACAGACGAAACAAC | this work |
| M28_Spy0784_RD0 F | AGCAGAGTATGAAGGCGGTTTT | this work |
| M28_Spy0784_RD0 P | ATATTCTATCTGAAACGGCTCG | this work |
| M28_Spy0784_RD0 R | AACATCTCTGCGAGTCGTTCTATACTT | this work |
| M28_Spy0980_6180.1 F | TCGTTAGGACTGGCGGTAGAG | this work |
| M28_Spy0980_6180.1 P | TGCAACTGCTGTCTTAA | this work |
| M28_Spy0980_6180.1 R | AACAGTCTTTGCCACCACCAT | this work |
| M28_Spy1087_RD1 F | TGTTTTTTGAATCTCTGACTTCTTTCC | this work |
| M28_Spy1087_RD1 P | AGAATTGCAGCTACTTGTATT | this work |
| M28_Spy1087_RD1 R | TGCAGACGAAAATAGCTGTAACTACTC | this work |
| M28_Spy1231_6180.2 F | GCAGTTGCTTGTTGCGTTGA | this work |
| M28_Spy1231_6180.2 P | TGCAACCCACTGATTT | this work |
| M28_Spy1231_6180.2 R | GCGCGTAGAGCTGGAGTCA | this work |
| M28_Spy1805_6180.3 F | AAAGGGCTATGGACGAACGA | this work |
| M28_Spy1805_6180.3 P | CAGACCAGCCTTTG | this work |
| M28_Spy1805_6180.3 R | GGTAAACCGATATTTTTCATCAATGA | this work |
| B. Primer combinations used for tiling across RD2 element, after [1]. | ||
| Tiling fragment | Amplified region | Primer sequence |
| 1 | M28_Spy1299-1304 | GGTTTCGACAAGGTCAGAGC |
| TGTGAGTGTTCCTGTACCAGATG | ||
| 2 | M28_Spy1304-1306 | ACGGCTACCTTTCCCCCTA |
| ACTAAGCCAAGCGAGGACAA | ||
| 3 | M28_Spy1306-1307 | CCAAAACCGTGTAGCCTGTA |
| TCATCGTCAAAAGCCATCTC | ||
| 4 | M28_Spy1307-1308 | TTGCTCTGATAAACCTCAAG |
| TACGACAGAAGCAGGTGGAG | ||
| 5 | M28_Spy1308-1310 | ACCGAGTTTCGCAGGATTG |
| GCTTGGAGGTGTTTCCTTTC | ||
| 6 | M28_Spy1310-1314 | CCTTGTTCTGCTTGATGTCC |
| ATCAAGCAAGCAACAAAACG | ||
| 7 | M28_Spy1314-1322 | TTTCCACCCATCAGTTCAGG |
| GACTGGTGGCGGTAAGACTG | ||
| 8 | M28_Spy1322-1325 | TTTCATCCCCAAAAAGCATC |
| TGAATGATGCGGGGACTTAT | ||
| 9 | M28_Spy1325-1326 | TGTAAAAGGCTGCTGGGTCT |
| ACACCGACTGAGATTGCTGA | ||
| 10 | M28_Spy1326-1331 | TTGGCTTGTGAGGTTTGAGA |
| TCATACTTTTCAGGTACACAAGCA | ||
| 11 | M28_Spy1331-1336 | ATGCCAAAAACCAAAGGAAG |
| GATACTTCACAGACGAAACAACG | ||
| 12 | M28_Spy1336-1338 | ATCACGACTCCCATCACTCC |
| CAAAGTTCCTGCCCCAAC | ||
Construction of isogenic mutant strain MGAS6180Δ1325-1326spcR
Allelic replacement was used to construct an isogenic mutant strain in which two contiguous genes (M28_Spy1325 and M28_Spy1326) encoded by RD2 were deleted and replaced by spectinomycin resistance cassette [11]. Upstream and downstream regions flanking the two-gene segment were cloned in pTOPO plasmid (Invitrogen) with spectinomycin resistance cassette between them. The gel purified PCR product encompassing both flanks with the spectinomycin cassette was electroporated into cells of strain MGAS6180 made competent as described before [12]. The resulting isogenic strain was confirmed to be the correct construct by PCR analysis, DNA sequencing, and Southern hybridization. Successful inactivation of the Spy1325 and Spy1326 genes also was confirmed by quantitative real-time PCR and Western immunoblot analysis. Detailed strain construction is presented as Additional File 3 and the confirmation of the proper construction as Additional File 4 (Figure S1).
Filter mating
Filter mating procedure was performed according to modified method described previously [13]. The MGAS6180Δ1325-1326spcR strain was used as a donor of the RD2 element in filter mating experiments. Strains MGAS2221ΔcovRS (M1, kanamycin resistance, RD2neg; P. Sumby unpublished), and MGAS10750 (M4 serotype, natural erythromycin resistance, RD2neg; [9]) were used as recipient strains. Overnight donor and recipient cultures (750 μl of each) were mixed and collected on the surface of a 0,45 μm pore size sterile nitrocellulose filter (Millipore). The filter was transferred to the surface of TSA plate without antibiotics and incubated for 3 h, 6 h, or 16 h. After the incubation, the filter was washed with sterile PBS and the bacteria collected in the wash fluid were plated on THY agar plates with appropriate antibiotics. Randomly selected colonies resistant to both antibiotics were screened by PCR for the size of emm gene amplicon that is characteristic for M28 or M4 type and presence of RD2 region genes.
Induction of genetic elements with mitomycin C and hydrogen peroxide
Genetic elements were induced by treating bacterial cultures with mitomycin C as described previously [14]. Briefly, 750 ml of pre-warmed THY medium was inoculated with an overnight culture of MGAS 6180 (1:50 dilution) and grown until the OD reached 0.15 (early log phase). The culture was divided into three aliquots, and one aliquot was treated with mitomycin C (Sigma, final concentration 0.2 μg/ml), second with hydrogen peroxide (final concentration 0,5 mM) and one aliquot was left untreated as a control sample. The concentration of mitomycin C and hydrogen peroxide used for induction of mobile genetic elements was tested for the ability to induce mobile elements and inhibit growth (Additional File 3). The concentrations used in the experiment were sufficient to induce mobile elements in MGAS6180 and were above the minimal inhibitory concentration (Additional File 5, Figure S2). Samples (35 ml) collected at 1 h, 2 h, and 3 h intervals and after overnight incubation and were used for total DNA isolation as described above.
Quantitative analysis of changes in gene copy number
Total DNA isolated from control GAS or cells treated with mitomycin C or hydrogen peroxide was used as a template in quantitative PCR (Taqman) reactions. Diluted DNA was amplified in multiplex reactions. The primers used amplified the chromosomal gene proS (internal calibrator [15]) and the target test gene of interest. Gene copy number was presented as the difference in amplified copies between control gene proS and the gene of interest (2ΔCt) at each experimental condition. The increase in copy number between start (T0, sample collected immediately prior splitting the cultures and the induction) and time point of interest (Texp; e.g. 1 h after the induction) was calculated according to the equation 2ΔCt TExp/2ΔCt T0.
Results
Comparative analysis of RD2 gene content and organization in GAS and GBS
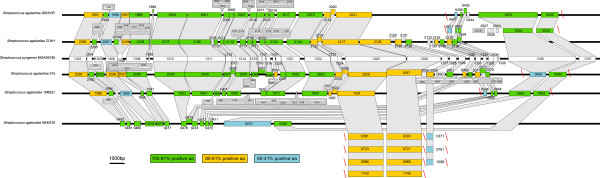
Sequences homologous to RD2 were initially reported to be present in strains of serotype III and V Group B Streptococcus (GBS) [1]. By analyzing the available GBS genomic sequences a number of sequences homologous to RD2 can be identified (Figure 1) [16,17]. The RD2 region in GAS is integrated into gene encoding tRNA for threonine, while elements found in GBS genomes carrying RD2 gene homologs are integrated into gene encoding tRNA for threonine as in GAS, but also tRNA for lysine [17]. Interestingly, the organization of RD2 like element in GBS is strain dependent. In case of strains 515 and COH1, almost the entire element is organized as in MGAS6180 with the exception of rib and its regulator (homologs of M28_Spy1336 and M28_Spy1337), whereas in other strains only fragments homologous to RD2 are detected. The regions in S. agalactiae genomes homologous to genes M28_Spy1303- M28_Spy1325 are located in majority of analyzed GBS strains within single chromosomal location, while genes M28_Spy1326-M28_Spy1337 are located in other chromosomal location or locations (Figure 1 and Additional File 6, Table S3).
Figure 1.
Comparison of the organization of RD2 ORFs in diverse GAS and GBS strains. Slanted red lines indicate discontinuity between fragments homologous to RD2 element present in MGAS6180. RD2 open reading frames are marked as white rectangles, open reading frames of GBS are color coded according to their homology to RD2 on the protein level. Numbers within each rectangle represent ORF designation number in particular GBS strain
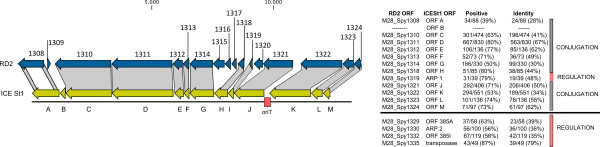
RD2 encodes a putative conjugation module
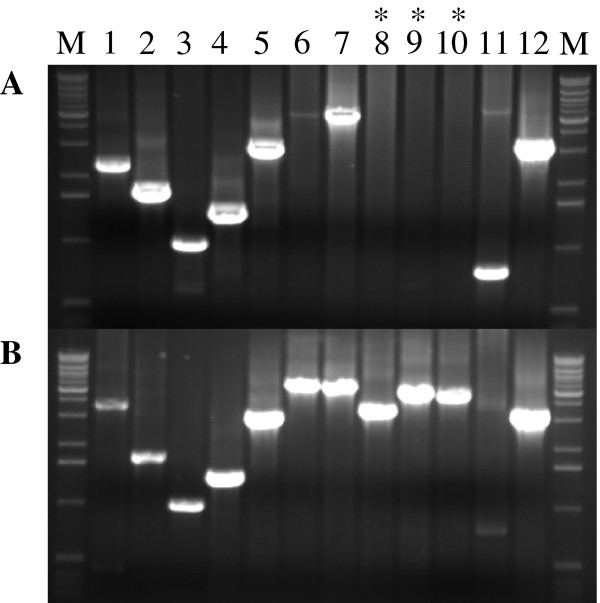
Based on DNA sequence analysis, RD2 does not appear to encode genes involved in replication as a circular plasmid. GAS is not considered to be naturally transformable under standard laboratory growth conditions, suggesting that other mechanisms must be used to transfer RD2-related genes between cells. DNA sequence analysis identified a putative transfer module encoded by RD2 with similarity to the ICESt1 and ICESt3 conjugation modules present in Streptococcus thermophilus (Figure 2) [18]. Thus, we hypothesized that RD2 uses a conjugation-like mechanism to transfer from donor to recipient strains. To test this hypothesis, we performed filter mating using donor strain MGAS6180Δ1325-1326spcR, which contains a spectinomycin resistance cassette integrated into the chromosomal copy of RD2. The recipient strain used was strain MGAS10750, a type emm4 organism that is naturally resistant to erythromycin. After filter mating of strain MGAS6180Δ1325-1326spcR and strain MGAS10750 for 3 h, 6 h, and 16 h, we obtained 1, 3, and 202 colonies, respectively (transfer frequency ~10-6 of transconjugants per donor cell), which were resistant to both antibiotics (spectinomycin 150 μg/ml and erythromycin 1 μg/ml). Eight putative transconjugant colonies were tested for the presence of the RD2 element and characterized for the emm gene sequence. In group A Streptococcus, emm gene is highly polymorphic in sequence and encodes for major surface protein M that is responsible for GAS serotype. Amplification of hyper-variable region of emm gene with primers CDCemm1 and CDCemm2 yields products that differ in size depending on the M serotype [19]. RD2 positive transconjugants were first screened based on the emm amplicon size (data not shown), and the amplified product was sequenced to confirm that transconjugants belong to M4 serotype, the same as the recipient. Successful transfer of the RD2 element from the emm28 strain to the emm4 strain was confirmed by PCR tiling across the entire RD2 element (Figure 3). Moreover, based on amplification of the junction region between RD2 and the chromosome, the RD2 element in the transconjugate was integrated at the same locus as in the donor emm28 strain (Figure 3). Based on analysis of 43 colonies resistant to both spectinomycin and kanamycin, similar results were obtained using strain serotype M1 strain as the recipient strain (MGAS2221ΔcovRS, resistant to kanamycin).
Figure 2.
RD2 encodes homologues of conjugative transfer genes present in the ICESt1 and ICESt3 elements of S. thermophilus.
Figure 3.
Detection of RD2 transfer from donor strain MGAS6180 (emm28) to recipient strain MGAS10750 (emm4). Amplicons 1-12 generated by PCR tiling across the RD2 element. A. transconjugant; * denote amplicons encompassing deleted M28_Spy1325-1326 region that is replaced by spectinomycin resistance cassette; B. control with chromosomal DNA isolated from strain MGAS6180. M - 1 kb ladder (Invitrogen)
RD2 is present in multiple, likely extrachromosomal, copies in GAS
Many gene transfer processes, including conjugation, require circular form of the transferred molecule or that more than one copy of the element exists during at least one point in the transfer cycle [20-22]. Therefore, we tested the hypothesis that multiple copies of the RD2 are present in the bacterial cell. PCR primers were used that allow detection of a circular form of RD2, and permit assessment of the orientation of chromosomal integration of multiple copies of RD2 (Figure 4A). Primers #1 and #4 recognize chromosomal sequences, whereas primers #2 and #3 recognize RD2 element sequences. Depending on the direction and/or arrangement of multiple copies of RD2 (i.e., head-to-head, tail-to-tail, head-to-tail), the different primer combinations would yield distinct amplicons. Based on the genome sequence of strain MGAS6180 [1] primer pairs #1-#2 and #3-#4 would amplify the junction region between the chromosome and RD2 on the left and right flank, respectively (positive control reactions). Using total DNA isolated from an overnight culture of MGAS6180 as template, PCR analysis yielded products amplified with primers #1-#2 and #3-#4, as expected. However, we also observed that primers #2 and #3 amplified a product, a result suggesting the presence of either multiple integrated copies of RD2 or a circular form of RD2 (Figure 4B). Next, we analyzed nine other GAS strains of multiple M protein serotypes using primers #2-#3 to determine if this was a general phenomenon. Regardless of emm type, all RD2-positive strains yielded an amplicon with the primer #2-#3 combination whereas RD2-negative organism did not (Figure 4C). Further, DNA sequence analysis revealed that all PCR amplicons generated with primers #2-#3 contained the sequence CGGTGGTGGCA, corresponding to a junction between the left and right flanking regions of RD2 (Figure 4).
Figure 4.
PCR screen detects multiple or circular copy of RD2. A. Primer combinations used for detection of seven potential arrangements of RD2. Thick black arrows represent RD2 element; thin gray line represents the chromosome. Red, purple, blue, and green arrows represent locations of primer #1, #2, #3, and #4, respectively. Potential arrangement variants are: arrangement as determined by genome sequencing [1], at least two head-to-tail copies of RD2, tail-to-tail, single copy arranged in reverse orientation than the integrative copy determined by genome sequencing, head-to-tail arrangement of copies in reverse orientation than the integrative copy determined by genome sequencing, head-to-head, and circular form. B. PCR screen detects product amplified with primer pairs #1+#2, #3+#4, and #2+#3, corresponding to arrangement variant (head-to-tail) or (circular form), and #1+#4 detecting chromosomal integration site lacking RD2. C. Primers #2+#3 detect arrangement variant 2 or 7 in multiple RD2 positive strains [1]. Serotype M1 strain MGAS5005 (lacks RD2) was used as a negative control of amplification.
To further investigate the putative presence of multiple extrachromosomal copies of RD2 in GAS cells, we performed quantitative real time PCR using total DNA isolated from MGAS6180 strain. Performed analysis revealed that RD2 is present in 6-9 copies per chromosome (Figure 5B, see below). Also, the amplification of chromosomal junction (primers #1+#4) suggests that RD2 can be excised from the site of integration.
Figure 5.
Mitomycin C treatment results in amplification of RD2. A Rapid decrease in O.D. of a liquid culture of strain MGAS6180 after mitomycin C addition. The decreased O.D. is likely due to prophage induction followed by lytic cycle phage release. Smaller drop in OD is observed after treatment with hydrogen peroxide. B. The RD2 element is present in 6-9 copies per chromosome in the absence of inducer. C. The RD2 element is not induced by oxidative stress. Bars in each group represent the RD2 copy number after 1 h, 2 h, 3 h, and 16 h after treatment with hydrogen peroxide. D. RD2 is induced by DNA damage. Bars in each group represent the increase in copy number at 1 h, 2 h, 3 h, and 16 h after treatment with mitomycin C. The statistical significance of the increase in RD2 copy number was determined by t-test, *** on the graph denotes p value below 0.001.
Taken together, these results indicate that a circular form of RD2 is present in strain MGAS6180.
Response of strain MGAS6180 to mitomycin C and hydrogen peroxide treatment
We hypothesized that the putative circular form detected in overnight cultures (see above) is a transient form involved in DNA transfer. DNA damaging factors as ultraviolet light, hydrogen peroxide, or mitomycin C can induce mobilization of genetic elements such as prophages or pathogenic islands as part of a response to DNA damage or oxidative stress [23]. To test hypothesis that RD2 was induced/excised by DNA damage and oxidative stress, we examined induction of RD2 and five other integrative elements present in the genome of strain MGAS 6180 by mitomycin C and hydrogen peroxide treatment.
In the absence of mitomycin C, no significant increase in target gene copy number of any of tested element occurred, even after overnight incubation (Figure 5B), and the copy number of the element remained constant. However, RD2 copy number increased by 1 h, 2 h, 3 h, and 16 h-post mitomycin C treatment (Figure 5D). Of note, we also detected increases in the copy number of genes encoded by several other integrative elements present in the genome of strain MGAS6180. For example, all three tested prophages were induced. In the most dramatic case of prophage 6180.2 (encoding SpeK, a superantigen, and SlaA, a secreted phospholipase A2 virulence factor) we observed a increase in relative copy number over 700 times compared with the pre-induction level (Additional File 7, Figure S3). Consistent with phage induction, mitomycin C treatment resulted in a rapid decrease in optical density of the culture, presumably corresponding to cell lysis (Figure 5A).
Treatment with hydrogen peroxide did not increase RD2 copy number (Figure 5C), however we observed induction of phages such as 6180.1 and 6180.2 (Additional File 7 Figure S3).
An RD2-like element is present in group C and G Streptococcus strains
Inasmuch as genome sequence information (Figure 1) and filter-mating data presented herein suggested that RD2 or an RD2-like element can spread between streptococcal species and multiple serotypes, we tested the hypothesis that the RD2 element has a phylogenetic distribution broader than GAS and GBS. To test the hypothesis, we screened 20 group C (GCS) and G (GGS) streptococci causing human infections by PCR for the presence of seven RD2 genes encoding putative extracellular secreted proteins. The primers and conditions we used were based on the sequence of RD2 found in GAS strain MGAS6180, and have been used previously to study the distribution of RD2 in GAS strains [1]. Because specific primers were used, this PCR analysis tests for the presence of genes with high homology to the RD2 element in GAS. The majority of the 20 GCS and GGS strains tested have homologs of RD2 element genes (Table 2A). DNA sequencing of all PCR products confirmed that the amplified gene fragments were homologues of RD2 element genes (data not shown). To test the hypothesis that the amplified genes were organized in an RD2-like genetic element, we used PCR primers described previously to tile across the entire RD2 element found in GAS strains [1]. The results (Table 2B) show that two GGS strains had an intact RD2 element, and one GCS strain had large segments of an intact RD2. The analysis also revealed a similar organization to RD2 in MGAS6180, as amplicons of the same size were generated (data not shown). Missing products of tiling PCR of GCS encompass homologs of M28_Spy1325 and M28_Spy1326 (fragments 9-10) which genes detected in single PCR reactions (Table 2A). The failure to amplify PCR products corresponding to the junction sites between the chromosome and RD2 suggests that the element is located in a different chromosomal location than in GAS. However, we cannot rule out the possibility that DNA sequence divergence at the primer pairing site was responsible for the lack of amplification of the target amplicon.
Table 2.
Detection of RD2 element genes in Lancefield group C and G streptococci by PCR.
| A. Detection of genes encoding putative extracellular proteins | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | M28_ Spy1306 | M28_ Spy1307 | M28_ Spy1308 | M28_ Spy1325 | M28_ Spy1326 | M28_ Spy1332 | M28_ Spy1336 | ||||||
| GCS | |||||||||||||
| 15169 | + | + | - | + | + | - | + | ||||||
| 15170 | + | + | - | - | + | - | - | ||||||
| 15172 | + | + | + | + | + | - | + | ||||||
| 15173 | + | + | - | + | + | - | + | ||||||
| 15178 | + | + | + | + | + | + | + | ||||||
| 15181 | + | + | - | + | + | - | + | ||||||
| GGS | |||||||||||||
| 15163 | + | + | - | + | + | - | + | ||||||
| 15164 | + | + | - | + | + | - | + | ||||||
| 15165 | + | + | - | + | + | - | + | ||||||
| 15166 | + | + | - | + | + | - | + | ||||||
| 15167 | + | + | - | + | + | - | + | ||||||
| 15168 | + | + | - | + | + | - | + | ||||||
| 15171 | + | + | + | + | + | - | + | ||||||
| 15174 | + | + | + | + | + | + | + | ||||||
| 15175 | + | + | + | - | - | - | - | ||||||
| 15176 | + | + | - | + | + | - | + | ||||||
| 15177 | + | + | - | + | + | - | + | ||||||
| 15179 | + | + | - | + | + | - | + | ||||||
| 15180 | + | + | + | + | + | - | + | ||||||
| 15182 | + | + | + | + | + | + | + | ||||||
| B. PCR-tiling across the entire RD2 element. Example of the tiling across RD2 is presented in Figure 3. (+) PCR product present, (-) no product, * amplified fragment of different size than for strain MGAS6180 | |||||||||||||
| PCR-tiling fragment no. | |||||||||||||
| Strain | group | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| 6180 | A | + | + | + | + | + | + | + | + | + | + | + | + |
| 15178 | C | - | + | + | + | + | - | - | + | - | - | + | - |
| 15174 | G | +(*) | + | + | + | + | + | - | + | + | + | + | - |
| 15182 | G | - | + | + | + | + | + | + | + | + | + | + | - |
Discussion and Conclusions
Analysis of multiple genomes of GAS shows that about 10% of the genome can be attributed to genetic material acquired horizontal gene transfer [3]. Multiple mobile genetic elements as prophages, ICE elements and ancient pathogenicity islands are part of GAS metagenome [3,24]. Lack of detected natural transformation of GAS, despite proposed mechanism mediated via quorum sensing mechanism, [25] stresses the importance of transduction and conjugation processes in HGT.
Since late 1970s multiple authors were studying plasmid conjugal transfer between various streptococcal species [26-28]. Later, based on sequence analyses and experimental rationale, horizontal transfer of genes/regions between GAS and GGS was implied [29-31]. Finally, recent publications report conjugative transfer of ICE elements in human and animal isolates of GAS, GBS, GGS, GCS and Streptococcus uberis [32,33].
Our work demonstrates that genetic element RD2 from GAS strain MGAS6180 (serotype M28) can be horizontally transferred in the laboratory to other GAS strains by filter mating. The transfer frequency is comparable with inter-species transfer of ICESt3 [34]. However, we cannot exclude that the transfer frequency was influenced by the inactivation of M28_Spy1325-1326 genes. The genes encode putative extracellular proteins and can act as aggregation factors, in particular, M28_Spy1325 has homology to enterococcal conjugative plasmid pAM373 aggregation factor [35]. However, because we used filter mating technique that can at least partially circumvent the need of aggregation factor in the conjugation process, the lack of M28_Spy1325-1326 genes does not have to affect transfer frequency during filter mating.
Presented study provides experimental support for the idea that the presence of RD2 in GAS strains of very diverse phylogenetic backgrounds that have not shared a recent common ancestor have acquired this element by lateral gene flow occurring in nature. Our results also show that RD2-like regions are present in multiple Lancefield group C and group G strains, additional evidence for horizontal dissemination of RD2 in natural populations of streptococci. Of note, the detection of an RD2-like element in group B [16], C and G streptococci (this work) is consistent with early reports of the production of the R28 antigen in these organisms [5,36].
We believe that RD2 has spread and been maintained in genetically diverse organisms in part because proteins encoded by this genetic element confer a survival advantage to the recipient organism. RD2 encodes at least seven proteins that are secreted into the extracellular environment, including several likely to participate in host-pathogen interactions such as cell adhesion. It is plausible that at least two of these proteins confer a survival premium. The best characterized is protein R28 encoded by M28_Spy1336. The RD2 protein has been shown to promote adhesion of GAS to human epithelial cells grown in vitro and confer protective immunity in a mouse model of invasive disease, together providing evidence that the R28 protein is a virulence factor [5,6]. Another RD2 encoded gene involved in virulence is M28_Spy1325. The protein is a member of the antigen I/II family of adhesions made by oral streptococci. It is made in vivo during invasive GAS infection, and binds GP340, a heavily glycosylated protein present in human saliva [8]. Similar to the R28 protein, immunization with recombinant purified M28_Spy1325 protect mice from experimental invasive infection, and the protein is made during human invasive infections [1,8]. Although far less is known about the other secreted extracellular proteins made by RD2, serologic analysis indicates that M28_Spy1306, M28_Spy1326 and M28_Spy1332 also are made during human invasive infections [1].
Although our work did not define the exact molecular mechanism(s) mediating horizontal gene transfer of RD2, the structure of the element and its transfer by filter mating point toward conjugation as a key process. Parts of RD2 share substantial homology with ICESt1 [37] and ICESt3 [38] conjugative elements from S. thermophilus. ICESt1 and ICESt3 elements have homology in sequence and organization with conjugative transposon Tn916 from Enterococcus faecalis [39]. Interestingly, a large intergenic region between M28_Spy1321 and M28_SpyM28_Spy1322 ORFs contains multiple palindromic sequences and might function as origin of transfer (oriT) as the equivalent region of Tn916 has been shown [40] or has been suggested to function as such [18].
We note that the copy number of RD2 increased rapidly and substantially following treatment of strain MGAS6180 with mitomycin C, a DNA damaging agent known to induce the SOS response and stimulate horizontal transfer of ICEs [23,41-43].
Interestingly, Ubeda et al. have reported that other factors as antibiotic treatment can mediate SOS response in staphylococci and promote horizontal dissemination of pathogenicity island-encoded virulence factor genes [44]. The postulated mechanism of SOS-induced induction and transfer of ICESt1/3 elements involves autoproteolysis of cI type repressor Arp1 [23,45]. As the RD2 element encodes multiple cI type repressors [1] it is plausible that the mechanism of RD2 induction is mediated by SOS-induced proteolysis or autoproteolysis of one of the RD2 cI regulators. The induction of RD2 was not observed after treatment with hydrogen peroxide i.e. in the condition of oxidative stress that is known to induce phages [46-48]. That suggests rather LexA dependent mechanism induced by DNA damage.
In conclusion, RD2 is a medium host range mobile element that is shared between multiple unrelated serotypes of GAS and other pathogenic streptococcal species. As a consequence of several extracellular secreted proteins encoded by RD2, the element may confer a selective advantage on organisms that acquire this element by horizontal gene transfer.
Authors' contributions
IS conceived, designed, coordinated the study and wrote the manuscript; performed the bioinformatics analysis of RD2 region, filter mating experiments and analysis of gene copy number. NMG participated in the design of the study, analysis of the results and wrote the manuscript; performed the bioinformatics analysis of RD2 region; screened GCS and GGS strains for the presence of RD2 element and constructed the RD2 mutant.
NG detected multiple RD2 copies. LM participated in data analysis, and screened GCS/GGS strains for the presence of RD2 element. JMM analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Table S1: Streptococcal strains used in the study
Table S2: Primers used for the mutant construction
Supplemental Methods
Figure S1: Conformation of proper mutant construction
Figure S2: Determination of MIC values for mitomycin C and hydrogen peroxide
Table S3: Homologs of RD2 genes found in GBS
Figure S3: Induction of prophages and ICE elements in MGAS6180 after treatment with mitomycin C and hydrogen peroxide.
Contributor Information
Izabela Sitkiewicz, Email: iza.sitkiewicz@gmail.com.
Nicole M Green, Email: Nicole.Green@cdph.ca.gov.
Nina Guo, Email: nguo@tmhs.org.
Laurent Mereghetti, Email: mereghetti@univ-tours.fr.
James M Musser, Email: jmmusser@tmhs.org.
Acknowledgements
We thank S. Beres and P. Sumby for advice and K. Stockbauer for critical reading of the manuscript.
References
- Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, Beres SB, LeFebvre RB, Musser JM. Genome sequence of a serotype M28 strain of group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis. 2005;192(5):760–770. doi: 10.1086/430618. [DOI] [PubMed] [Google Scholar]
- Green NM, Beres SB, Graviss EA, Allison JE, McGeer AJ, Vuopio-Varkila J, LeFebvre RB, Musser JM. Genetic diversity among type emm28 group A Streptococcus strains causing invasive infections and pharyngitis. J Clin Microbiol. 2005;43(8):4083–4091. doi: 10.1128/JCM.43.8.4083-4091.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Musser JM. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One. 2007;2(8):e800. doi: 10.1371/journal.pone.0000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancefield RC. Differentiation of group A streptococci with a common R antigen into three serological types, with special reference to the bactericidal test. J Exp Med. 1957;106(4):525–544. doi: 10.1084/jem.106.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancefield RC, Perlmann GE. Preparation and properties of a protein (R antigen) occurring in streptococci of group A, type 28 and in certain streptococci of other serological groups. J Exp Med. 1952;96(1):83–97. doi: 10.1084/jem.96.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalhammar-Carlemalm M, Areschoug T, Larsson C, Lindahl G. The R28 protein of Streptococcus pyogenes is related to several group B streptococcal surface proteins, confers protective immunity and promotes binding to human epithelial cells. Mol Microbiol. 1999;33(1):208–219. doi: 10.1046/j.1365-2958.1999.01470.x. [DOI] [PubMed] [Google Scholar]
- Stalhammar-Carlemalm M, Areschoug T, Larsson C, Lindahl G. Cross-protection between group A and group B streptococci due to cross-reacting surface proteins. J Infect Dis. 2000;182(1):142–149. doi: 10.1086/315693. [DOI] [PubMed] [Google Scholar]
- Zhang S, Green NM, Sitkiewicz I, Lefebvre RB, Musser JM. Identification and characterization of an antigen I/II family protein produced by group A Streptococcus. Infect Immun. 2006;74(7):4200–4213. doi: 10.1128/IAI.00493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, DeLeo FR, Musser JM. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc Natl Acad Sci USA. 2006;103(18):7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Current protocols in molecular biology. Vol. 1. John Wiley and Sons, Inc; 1994. [Google Scholar]
- Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Woischnik M, Podbielski A, Musser JM. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Invest. 1997;99(11):2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitkiewicz I, Musser JM. Expression microarray and mouse virulence analysis of four conserved two-component gene regulatory systems in group A Streptococcus. Infect Immun. 2006;74(2):1339–1351. doi: 10.1128/IAI.74.2.1339-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW. Conjugal transfer of plasmid pAM beta 1 in Lactobacillus reuteri and between lactobacilli and Enterococcus faecalis. Appl Environ Microbiol. 1987;53(11):2693–2695. doi: 10.1128/aem.53.11.2693-2695.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks DJ, Lei B, Musser JM. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect Immun. 2003;71(12):7079–7086. doi: 10.1128/IAI.71.12.7079-7086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, Sumby P, Sitkiewicz I, Granville C, DeLeo FR, Musser JM. Central role of a bacterial two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc Natl Acad Sci USA. 2005;102(44):16037–16042. doi: 10.1073/pnas.0505839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS. et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc Natl Acad Sci USA. 2005;102(39):13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet M, Couve E, Glaser P, Guedon G, Payot S. Integrative conjugative elements and related elements are major contributors to the genome diversity of Streptococcus agalactiae. J Bacteriol. 2008;190(20):6913–6917. doi: 10.1128/JB.00824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B, Guedon G. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid. 2002;48(2):77–97. doi: 10.1016/S0147-619X(02)00102-6. [DOI] [PubMed] [Google Scholar]
- Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34(4):953–958. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67(2):277–301. doi: 10.1128/MMBR.67.2.277-301.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, Babic A, Grossman AD. Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol. 2010;75(2):268–279. doi: 10.1111/j.1365-2958.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd EF, Almagro-Moreno S, Parent MA. Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 2009;17(2):47–53. doi: 10.1016/j.tim.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Bellanger X, Morel C, Decaris B, Guedon G. Derepression of excision of integrative and potentially conjugative elements from Streptococcus thermophilus by DNA damage response: implication of a cI-related repressor. J Bacteriol. 2007;189(4):1478–1481. doi: 10.1128/JB.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchaud A, Guy L, Collyn F, Haenni M, Nakata M, Podbielski A, Moreillon P, Roten CA. M-protein and other intrinsic virulence factors of Streptococcus pyogenes are encoded on an ancient pathogenicity island. BMC Genomics. 2009;10:198. doi: 10.1186/1471-2164-10-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol. 2010;78(3):589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buu-Hoi A, Bieth G, Horaud T. Broad host range of streptococcal macrolide resistance plasmids. Antimicrob Agents Chemother. 1984;25(2):289–291. doi: 10.1128/aac.25.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershfield V. Plasmids mediating multiple drug resistance in group B streptococcus: transferability and molecular properties. Plasmid. 1979;2(1):137–149. doi: 10.1016/0147-619X(79)90012-X. [DOI] [PubMed] [Google Scholar]
- Ravdonikas LE. The genetic control of virulence in group A streptococci. I. Conjugal transfer of plasmids and their effect on expression of some host cell properties. Acta Pathol Microbiol Immunol Scand B. 1983;91(1):55–60. [PubMed] [Google Scholar]
- Simpson WJ, Musser JM, Cleary PP. Evidence consistent with horizontal transfer of the gene (emm12) encoding serotype M12 protein between group A and group G pathogenic streptococci. Infect Immun. 1992;60(5):1890–1893. doi: 10.1128/iai.60.5.1890-1893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers RJ, Gal D, McMillan D, Sriprakash KS, Currie BJ, Walker MJ, Chhatwal GS, Fagan PK. Fibronectin-binding protein gene recombination and horizontal transfer between group A and G streptococci. J Clin Microbiol. 2004;42(11):5357–5361. doi: 10.1128/JCM.42.11.5357-5361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken C, Haase G, Brandt C, Weber-Heynemann J, Martin S, Lammler C, Podbielski A, Lutticken R, Spellerberg B. Horizontal gene transfer and host specificity of beta-haemolytic streptococci: the role of a putative composite transposon containing scpB and lmb. Mol Microbiol. 2001;41(4):925–935. doi: 10.1046/j.1365-2958.2001.02563.x. [DOI] [PubMed] [Google Scholar]
- Haenni M, Saras E, Bertin S, Leblond P, Madec JY, Payot S. Diversity and Mobility of Integrative and Conjugative Elements in Bovine Isolates of Streptococcus agalactiae, S. dysgalactiae subsp. dysgalactiae, and S. uberis. Appl Environ Microbiol. 2010;76(24):7957–7965. doi: 10.1128/AEM.00805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MR, Shera J, Van Domselaar GH, Sriprakash KS, McMillan DJ. A novel integrative conjugative element mediates genetic transfer from group G Streptococcus to other {beta}-hemolytic Streptococci. J Bacteriol. 2009;191(7):2257–2265. doi: 10.1128/JB.01624-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger X, Roberts AP, Morel C, Choulet F, Pavlovic G, Mullany P, Decaris B, Guedon G. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J Bacteriol. 2009;191(8):2764–2775. doi: 10.1128/JB.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boever EH, Clewell DB, Fraser CM. Enterococcus faecalis conjugative plasmid pAM373: complete nucleotide sequence and genetic analyses of sex pheromone response. Mol Microbiol. 2000;37(6):1327–1341. doi: 10.1046/j.1365-2958.2000.02072.x. [DOI] [PubMed] [Google Scholar]
- Maxted WR. Occurrence of the M. substance of type 28 group A in streptococci of Lancefield groups B, C and G. J Gen Microbiol. 1949;3(1):1–6. doi: 10.1099/00221287-3-1-1. [DOI] [PubMed] [Google Scholar]
- Burrus V, Roussel Y, Decaris B, Guedon G. Characterization of a novel integrative element, ICESt1, in the lactic acid bacterium Streptococcus thermophilus. Appl Environ Microbiol. 2000;66(4):1749–1753. doi: 10.1128/AEM.66.4.1749-1753.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic G, Burrus V, Gintz B, Decaris B, Guedon G. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology. 2004;150(Pt 4):759–774. doi: 10.1099/mic.0.26883-0. [DOI] [PubMed] [Google Scholar]
- Franke AE, Clewell DB. Evidence for conjugal transfer of a Streptococcus faecalis transposon (Tn916) from a chromosomal site in the absence of plasmid DNA. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):77–80. doi: 10.1101/sqb.1981.045.01.014. [DOI] [PubMed] [Google Scholar]
- Jaworski DD, Clewell DB. A functional origin of transfer (oriT) on the conjugative transposon Tn916. J Bacteriol. 1995;177(22):6644–6651. doi: 10.1128/jb.177.22.6644-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci USA. 2005;102(35):12554–12559. doi: 10.1073/pnas.0505835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427(6969):72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- McGrath BM, O'Halloran JA, Pembroke JT. Pre-exposure to UV irradiation increases the transfer frequency of the IncJ conjugative transposon-like elements R391, R392, R705, R706, R997 and pMERPH and is recA+ dependent. FEMS Microbiol Lett. 2005;243(2):461–465. doi: 10.1016/j.femsle.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Ubeda C, Maiques E, Knecht E, Lasa I, Novick RP, Penades JR. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol Microbiol. 2005;56(3):836–844. doi: 10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- Bellanger X, Morel C, Decaris B, Guedon G. Regulation of excision of integrative and potentially conjugative elements from Streptococcus thermophilus: role of the arp1 repressor. J Mol Microbiol Biotechnol. 2008;14(1-3):16–21. doi: 10.1159/000106078. [DOI] [PubMed] [Google Scholar]
- Glinkowska M, Los JM, Szambowska A, Czyz A, Calkiewicz J, Herman-Antosiewicz A, Wrobel B, Wegrzyn G, Wegrzyn A, Los M. Influence of the Escherichia coli oxyR gene function on lambda prophage maintenance. Arch Microbiol. 2010;192(8):673–683. doi: 10.1007/s00203-010-0596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los JM, Los M, Wegrzyn A, Wegrzyn G. Hydrogen peroxide-mediated induction of the Shiga toxin-converting lambdoid prophage ST2-8624 in Escherichia coli O157:H7. FEMS Immunol Med Microbiol. 2010;58(3):322–329. doi: 10.1111/j.1574-695X.2009.00644.x. [DOI] [PubMed] [Google Scholar]
- Los JM, Los M, Wegrzyn G, Wegrzyn A. Differential efficiency of induction of various lambdoid prophages responsible for production of Shiga toxins in response to different induction agents. Microb Pathog. 2009;47(6):289–298. doi: 10.1016/j.micpath.2009.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Streptococcal strains used in the study
Table S2: Primers used for the mutant construction
Supplemental Methods
Figure S1: Conformation of proper mutant construction
Figure S2: Determination of MIC values for mitomycin C and hydrogen peroxide
Table S3: Homologs of RD2 genes found in GBS
Figure S3: Induction of prophages and ICE elements in MGAS6180 after treatment with mitomycin C and hydrogen peroxide.