Abstract
Background
Deletion of the glycosyltransferase bgsA in Enterococcus faecalis leads to loss of diglucosyldiacylglycerol from the cell membrane and accumulation of its precursor monoglucosyldiacylglycerol, associated with impaired biofilm formation and reduced virulence in vivo. Here we analyzed the function of a putative glucosyltransferase EF2890 designated biofilm-associated glycolipid synthesis B (bgsB) immediately downstream of bgsA.
Results
A deletion mutant was constructed by targeted mutagenesis in E. faecalis strain 12030. Analysis of cell membrane extracts revealed a complete loss of glycolipids from the cell membrane. Cell walls of 12030ΔbgsB contained approximately fourfold more LTA, and 1H-nuclear magnetic resonance (NMR) spectroscopy suggested that the higher content of cellular LTA was due to increased length of the glycerol-phosphate polymer of LTA. 12030ΔbgsB was not altered in growth, cell morphology, or autolysis. However, attachment to Caco-2 cells was reduced to 50% of wild-type levels, and biofilm formation on polystyrene was highly impaired. Despite normal resistance to cationic antimicrobial peptides, complement and antibody-mediated opsonophagocytic killing in vitro, 12030ΔbgsB was cleared more rapidly from the bloodstream of mice than wild-type bacteria. Overall, the phenotype resembles the respective deletion mutant in the bgsA gene. Our findings suggest that loss of diglucosyldiacylglycerol or the altered structure of LTA in both mutants account for phenotypic changes observed.
Conclusions
In summary, BgsB is a glucosyltransferase that synthesizes monoglucosyldiacylglycerol. Its inactivation profoundly affects cell membrane composition and has secondary effects on LTA biosynthesis. Both cell-membrane amphiphiles are critical for biofilm formation and virulence of E. faecalis.
Background
The properties of the bacterial cell envelope are pivotal for the interaction of bacteria and the host organism [1]. Enterococcus faecalis expresses several cell-wall glycopolymers that make up the cell envelope, including capsular polysaccharides [2], cell-wall carbohydrates [3], cell-wall teichoic acid, lipoteichoic acid (LTA) [4], and glycolipids [5]. We have recently constructed a deletion mutant of the glycosyltransferase bgsA in E. faecalis [5]. Deletion led to a profound shift of the equilibrium of the two main cell wall glycolipids: monoglucosyldiacylglycerol (MGlcDAG) accumulated in the cell membrane of the bgsA mutant, while the production of diglucosyldiacylglycerol (DGlcDAG) was completely abrogated [5]. The bgsA mutant displayed normal cell morphology and growth characteristics but was impaired in attachment to colonic epithelial cells, and biofilm formation was almost completely abolished [5]. Remarkably, the LTA content of the mutant was higher due to the increased length of the glycerol-phosphate polymer.
The role of glycolipids in membrane physiology has been investigated in the cell wall-less bacterium Acholeplasma laidlawii, which produces glycolipids that are chemically identical to MGlcDAG and DGlcDAG of E. faecalis [6,7]. In Acholeplasma, the ratio of DGlcDAG to MGlcDAG governs the lipid bilayer's elasticity, curvature, and surface-charge density [6-8]. Interestingly, the pathway of glycolipid synthesis is highly conserved, and the type 4 family of NDP-glucose glycosyltransferases contains 107 UDP-sugar glycosyltransferases of bacterial, fungal, and plant origin [9]. Aside from their role as cell membrane components, glycolipids are also involved in the synthesis of LTA in bacteria with low G+C content [10]. LTA has a number of important functions in bacterial physiology including cation homeostasis, resistance to antimicrobial peptides, autolysin activity, non-covalent anchoring of cell-surface proteins, attachment to host tissues, and biofilm formation [1,11]. Glycolipids also function as acceptors of the glycerol-phosphate polymer during LTA synthesis, although the exact mechanism underlying this process is still under investigation [10]. If the processive glycosyltransferase YpfP is inactivated in Staphylococcus aureus, DAG instead of DGlcDAG is utilized as a building block in LTA synthesis, suggesting that glycolipids are not essential acceptors of the LTA polymer [12,13].
A second glycosyltransferase (EF 2890) is located immediately downstream of bgsA. To our knowledge, the function of this gene locus of E. faecalis or its homologues in streptococci is still unknown. In the current study, we report the construction of a deletion mutant of EF_2890 that we designated bgsB and studied the role of glycolipid metabolism in LTA biosynthesis and bacterial physiology.
Results
Construction of a deletion mutant of the glycosyltransferase bgsB
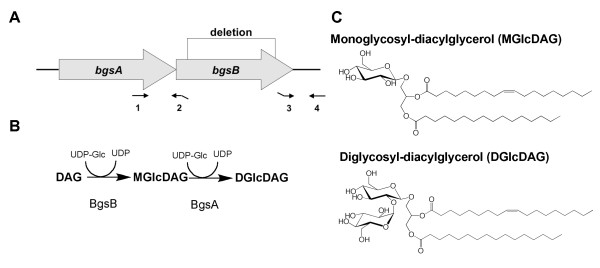
Immediately downstream from bgsA, we identified a putative 1,2-diacylglycerol 3-glucosyltransferase (TIGR number EF2890) by basic local alignment search tool (BLASTP) search (Figure 1). This glycosyl-transferase shows homology to YP_001620482.1 of Acholeplasma laidlawii (identity 34%, similarity 55%) [14] and to Lmo2555 of Listeria monocytogenes (identity 23%, similarity 41%) [15]. We designated this gene bgsB. To study the requirement of bgsB for glycolipid production, LTA synthesis, and bacterial physiology, we constructed a deletion mutant by targeted mutagenesis using the strategy previously applied for the bgsA deletion mutant. Unmarked deletions were created by allelic exchange, and all gene deletions were confirmed by PCR. In the resulting mutant, an internal fragment of 790 bp was deleted from the bgsB gene (Figure 1). Single gene reconstitution of bgsB in E. faecalis 12030ΔbgsB completely restored the wild-type phenotype, including the glycolipid expression profile in cell membrane extracts (Figure 2) and biofilm formation (Figure 3).
Figure 1.
Biosynthesis of glycolipids in E. faecalis. A Genetic organization of the bgs-locus in E. faecalis. The numbers refer to the primers described in Table 2. bgsB has a length of 1224 bp. A putative transcriptional terminator is found 10 bases downstream of bgsB. B Putative biosynthetic pathway of glycolipid synthesis in E. faecalis. C Structure of E. faecalis glycolipids. The position of 18:1 and 16:0 fatty acids has not been determined [5].
Figure 2.
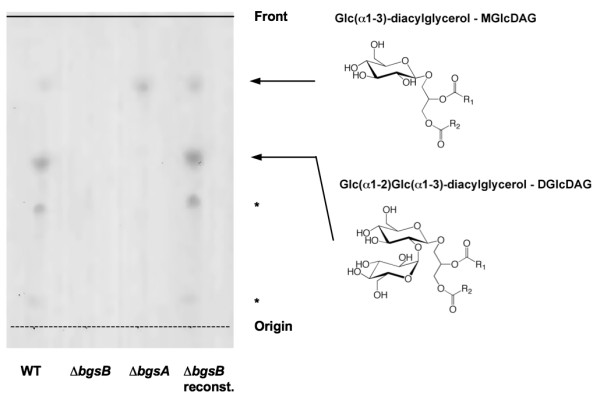
Thin-layer chromatography of cell-membrane total lipid extracts of E. faecalis strains. Bacterial cells were grown overnight, disintegrated, and stirred with butanol. Membrane lipids were extracted from butanol by phase partition according to Bligh and Dyer. 50 μg of lipid extract was applied to the TLC plate, separated using a solvent system of CHCl3/MeOH/H2O (65:25:4, v/v/v), and visualized with α-naphtol/sulfuric acid. The identity of the bands has been confirmed previously [5]. The glycolipids marked with an asterisk have not been analyzed.
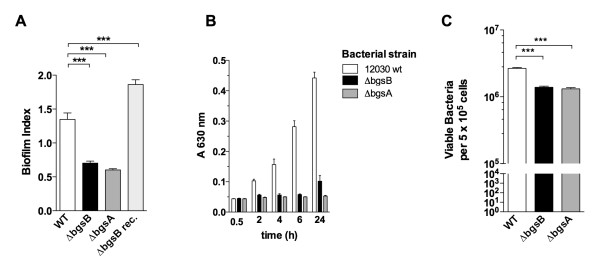
Figure 3.
Role of bgsB in biofilm formation and bacterial adherence to Caco-2 cells. A Biofilm formation on polystyrene. Microtiter plates were incubated with bacteria for 18 h, non-adherent bacteria removed by washing with PBS, and biofilms stained with crystal violet. Data represent the means ± SEM. *** P < Tukey's multiple comparison test. B Development of biofilm on polystyrene of E. faecalis 12030 wt, 12030ΔbgsB, and 12030ΔbgsA over time. After incubation periods of ≥ 4 h, E. faecalis 12030 wt elaborated significantly more biofilm than the deletion mutants (P < 0.001, Tukey's multiple comparison test). Bars represent means ± SEM. C Bacterial adherence to Caco-2 cells. Caco-2 cells were incubated at a multiple of infection of 100:1 for 2 h with the respective strain grown to mid-log phase. Data represent the means ± SEM. *** P < 0.001, Dunn's multiple comparison test.
Deletion of bgsB leads to a complete loss of glycolipids from the cell membrane and to expression of LTA with increased chain length
We hypothesized that, because it is located immediately downstream from bgsA and has high homology to ALmgs in Acholeplasma laidlawii, the gene product of bgsB glycosylates diacylglycerol to yield MGlcDAG. To test this hypothesis, we extracted the total lipids of the cell membrane, separated them by thin layer chromatography (TLC), and stained glycolipids with α-naphthol (Figure 2). As shown previously, inactivation of bgsA resulted in accumulation of MGlcDAG in the cell membrane (Figure 2). In contrast, no glycolipids were visualized in 12030ΔbgsB extracts, suggesting that bgsB encodes for a glycosyltransferase that glycosylates DAG to form MGlcDAG. MGlcDAG is the substrate of BgsA, which adds a second glucose to yield DGlcDAG (Figure 1). Since BgsA does not accept DAG as a substrate, inactivation of BgsB results in the loss of all glycolipids from the cell membrane (Figure 2).
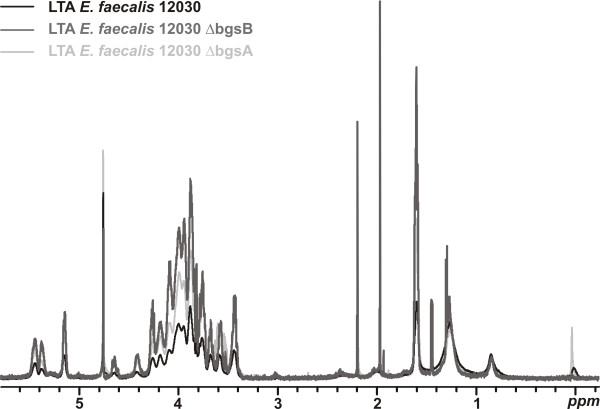
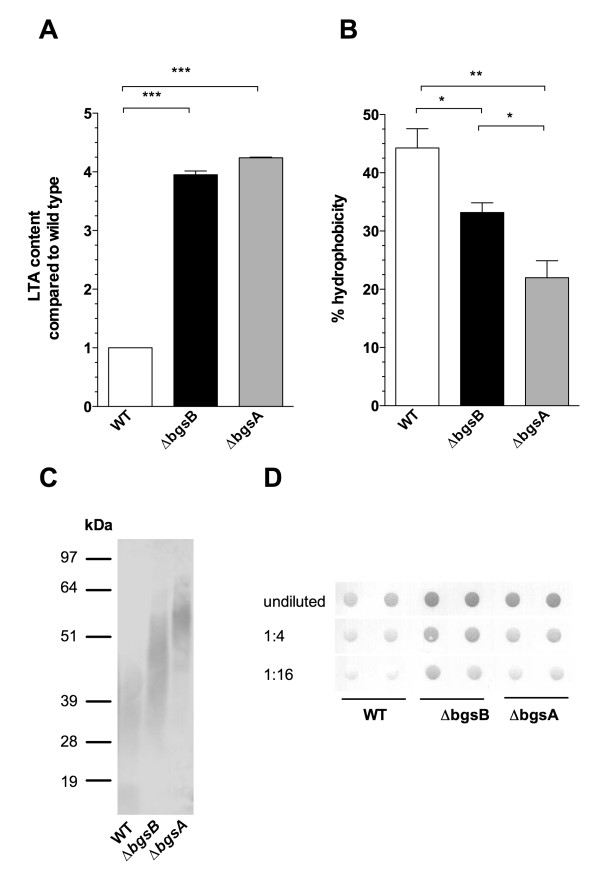
We recently showed that inactivation of bgsA also affects LTA synthesis, increasing the chain length of the glycerol-phosphate polymer [5]. Inactivation of bgsB has a similar effect on the LTA chain length (Figure 4). To estimate the chain length of the glycerol-phosphate chain by 1H-NMR analysis, we used the fatty acid signals of the molecule as an internal reference and compared the integration values of H1 of glucose and -CH3 of alanine to the -CH3 and -CH2- signals (δ 1.26-1.29, and 0.88) of the fatty acids [5]. The integral ratios yielded higher amounts of glucose and alanine incorporated into the LTA of 12030ΔbgsB and 12030ΔbgsA compared to the wild type, suggesting an increased length of the glycerol-phosphate polymer (Figure 4). These results are supported by quantification of LTA from butanol extracts by ELISA (Figure 5). Approximately fourfold more LTA was recovered from butanol extracts of cell walls of 12030ΔbgsB and 12030ΔbgsA than from wild-type bacteria. To determine whether increased amounts of LTA were also released into the culture medium, we blotted the culture supernatant onto PVDF membranes and performed semi-quantitative immuno-dot blot analysis (Figure 5). For both mutants, 12030ΔbgsB and 12030ΔbgsA, increased amounts of LTA in the liquid medium were detected, indicating a higher turnover of LTA in the cell envelope. Previous studies in S. aureus and Listeria monocytogenes have shown that substitution of DGlcDAG by MGlcDAG or DAG as the glycolipid anchor of LTA retards the migration of the molecule in SDS-PAGE [13,15]. LTA extracted from both mutants displayed a slower mobility in SDS PAGE than wild-type LTA, with LTA from 12030ΔbgsB migrating faster than LTA from 12030ΔbgsA (Figure 5). This suggests that both mutants express different lipid anchors from those in the wild type. As DAG is the only substrate available for LTA synthesis in 12030ΔbgsB, it likely serves as lipid anchor in this strain.
Figure 4.
Comparison of 1H-NMR spectra of LTA isolated from E. faecalis 12030 wt, 12030ΔbgsB, and 12030ΔbgsA. Comparison of integration values of fatty acid (FA) signals (-CH2- and -CH3) as an internal reference and anomeric proton signal of glucose (H1 Glc A and H1 Glc B) indicated that the glycerolphosphate polymer of LTA from 12030ΔbgsB and 12030ΔbgsA contains approximately four times more kojibiose. Comparison of the resonance signal of total alanine (-CH3 Ala) and fatty acid signals (-CH2- (FA) and -CH3 (FA)) revealed that LTA extracted from either mutant also contains more alanine residues. Gro - glycerol.
Figure 5.
Impact of bgsB on the synthesis and anchoring of LTA in the cell wall and on hydrophobicity of E. faecalis cells. A The total amount of butanol-extracted LTA from cell-wall extracts as determined by ELISA. For the quantification of LTA tethered to the cell wall, bacteria were grown overnight and adjusted to the same OD600. Cell walls were disrupted by shaking with glass beads, and LTA was mobilized by stirring bacterial cells with butanol/water. ELISA plates were incubated with various concentrations of the respective water phase of the extraction, and LTA was detected using a polyclonal rabbit anti-LTA antibody. Data points represent means ± SEM, *** P < 0.001, Tukey's multiple comparison test. B Cell-surface hydrophobicity of E. faecalis strains determined by adherence of bacterial cells to a mixture of dodecane and aqueous phase. Bars represent the percentage of bacteria remaining in the organic phase after partitioning of the solvent system. Data represent the means ± SEM, **P < 0.01, *P < 0.05, Tukey's multiple comparison test. C Western blot detection of LTA from 12030 wild type and deletion mutants. LTA was extracted from disrupted bacterial cells after shaking with glass beads by boiling in SDS. LTA was loaded onto 4-12% SDS PAGE gels and detected using a polyclonal rabbit antibody against enterococcal LTA. D Estimation of LTA shed into the culture medium. After overnight culture, bacterial density was adjusted to the same OD600, and bacteria were removed by centrifugation. 100 μl of supernatant was blotted onto PVDF membrane. Bound LTA was detected using the same antibody used in the ELISA. Dilution steps of culture supernatant are indicated in the legend.
LTA and glycolipids are also major determinants of cell-surface charge density. Therefore, hydrophobicity of wild-type and mutant bacteria was determined by measuring the adherence to dodecane. Reduced adherence was observed for both 12030ΔbgsA and 12030ΔbgsB (Figure 5). However, 12030ΔbgsB had higher hydrophobicity than 12030ΔbgsA (44% wild type versus 33% 12030ΔbgsB and 22% 12030ΔbgsA).
Bacterial physiology is not significantly impaired in a bgsB deletion mutant
Previous studies have shown that LTA and glycolipids play important roles in growth, cell envelope integrity, and cell division [11]. However, despite the complete lack of glycolipids in the cell membrane and increased production of LTA, important characteristics of 12030ΔbgsB did not differ from wild-type bacteria: Mutants did not differ from wild-type bacteria in their growth kinetics in broth culture (data not shown). Cell morphology of 12030ΔbgsB determined by transmission electron microscopy was not affected (Additional file 1). Likewise, autolysis was not affected in 12030ΔbgsB (Additional file 2). Since phosphatidylglycerol from the cell membrane is used as a substrate for polyglycerolphosphate synthesis by LtaS [10], we investigated whether increasing chain length of LTA affects cell membrane content of phosphatidylglycerol in the mutant. However, the semi-quantitative analysis of extracts of total membrane lipids by TLC and staining with molybdenum blue did not reveal differences in phospholipid composition (Additional file 3). The composition and total amount of aminophospholipids as assessed semi-quantitatively by TLC also did not differ between the wild type and 12030ΔbgsB (Additional file 3). Neither did analysis of non-covalently bound surface proteins by SDS-PAGE reveal major differences between the bgsB deletion mutant and the parental strain (Additional file 3).
Deletion of the glucosyltransferase bgsB has no effect on resistance to complement, antimicrobial peptides, and opsonophagocytic killing
LTA has been shown to be critical for resistance against killing by cationic antimicrobial peptides [1] and has been identified as a target of opsonic antibodies against E. faecalis [4]. To characterize the sensitivity of 12030ΔbgsB to host defense mechanisms, we assessed its resistance to antimicrobial peptides nisin, polymyxin B, and colistin. For nisin, no difference was found between the wild-type and the bgsB deletion mutant (Additional file 4). A two-fold lower concentration of polymyxin B and colistin was required for killing of 12030ΔbgsB compared to the isogenic wild type strain. At a serum concentration of 1.7%, sensitivity to complement-mediated phagocytosis did not differ between the 12030 wild type and 12030ΔbgsB (Additional file 2). Furthermore, rabbit antibodies raised against whole bacterial cells of E. faecalis 12030 mediated opsonophagocytic killing of 12030ΔbgsB comparable to levels obtained for the wild-type strain (Additional file 2).
The loss of glycolipids from the cell membrane is associated with reduced adherence to Caco-2 cells and impaired biofilm formation
We recently showed that deletion of bgsA leads to loss of biofilm formation on polystyrene and to reduced adherence to Caco-2 cells [5]. Partial deletion of bgsB also strongly impaired biofilm formation, reducing production by 50% (Figure 3). This defect in biofilm formation was not a result of decreased initial attachment (i.e., bacteria attached in ≤ 30 min of incubation); rather, it was due to defective accumulation of biofilm mass after initial attachment (Figure 3). Over a period of 24 h, biofilm mass of wild-type bacteria on polystyrene grew in a linear fashion. In contrast, the amount of biofilm produced by bgsB and bgsA mutants remained constant at the level of initial attachment. Adhesion to colonic epithelial cells (Caco-2 cells) was also impaired in 12030ΔbgsB, reaching only 50% of the adhesion of wild-type bacteria (Figure 3).
bgsB contributes to virulence during bacteremia in mice
Previous experiments with a bgsA deletion mutant in E. faecalis showed that it leads to an attenuation of virulence in a mouse bacteremia model [5]. To assess whether cell membrane glycolipids or glycolipid anchoring of LTA is required for the pathogenesis of enterococcal infections, we employed the same model to investigate the bgsB mutant. As mentioned above, 12030 wild-type and respective mutants had comparable growth characteristics. For virulence studies, we infected BALB/c mice 6 - 8 weeks old by i.v. injection, sacrificed the animals after 3 days, and enumerated the viable bacteria. Pilot experiments indicated that, with a high inoculum of 2 × 109 bacteria, infected mice are bacteremic up to 4 days without succumbing to the infection. Compared to the wild type, mice infected with 12030ΔbgsB or 12030ΔbgsA cleared significantly more bacteria from the bloodstream (Figure 6). No difference in virulence between 12030ΔbgsB and 12030ΔbgsA was detected in this model.
Figure 6.
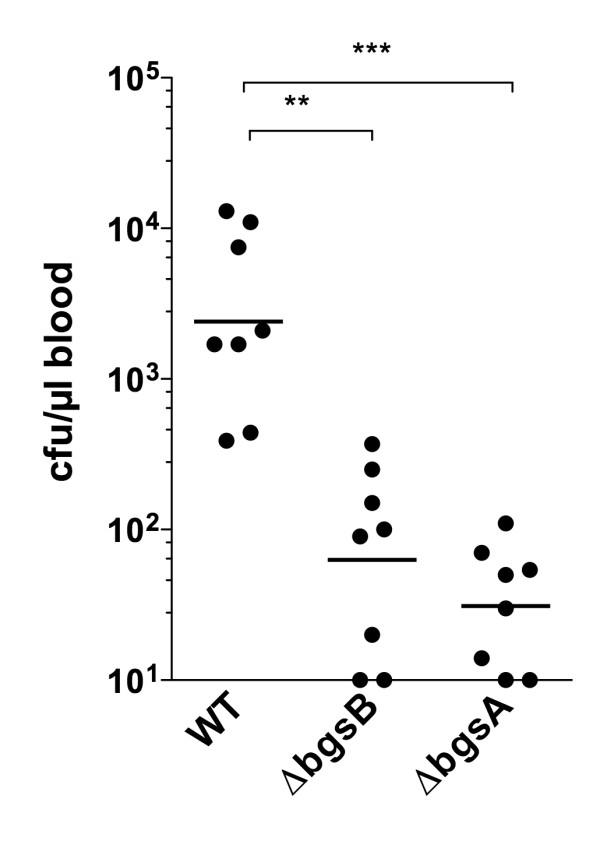
Virulence of E. faecalisΔbgsB in a mouse bacteremia model. Female BALB/c mice 6-8 weeks old were infected via the tail vein with stationary-phase E. faecalis strains (2.0 × 109 cfu). After 72 h mice were sacrificed and bacterial counts in the blood were enumerated. Data represent the individual bacterial counts and the geometric mean. ** P < 0.01, *** P < 0.001, Dunn's multiple comparison test. The lower limit of detection of the assay was 10 CFU/ml blood.
Discussion
Although MGlcDAG and DGlcDAG are the major glycolipids in many Gram-positive bacteria, distinct differences are found between species in biosynthetic pathways. In staphylococci and Bacillus, a single processive glucosyltransferase YpfP adds two glucose residues to DAG to synthesize DGlcDAG [12,16,17]. Depending on the bacterial species and strain background, the deletion of this enzyme may result in an increased LTA content and turnover [16], or loss of LTA from the cell membrane, associated with a reduced rate of autolysis and impaired biofilm formation [12]. In listeria, streptococci, and enterococci, genome analysis revealed two putative glycosyltransferases involved in the biosynthetic pathway of glycolipids [7,14,15,18]. Homologues of a (1→2) glucosyltransferase have been investigated in listeria (LafA), group B streptococci (IagA), and E. faecalis (BgsA) [5,15,18]. In group B streptococci, deletion of iagA results in the absence of capsule expression, reduced retention of LTA on the bacterial cell surface, and increased release of LTA into the culture medium [18]. Inactivation of lafA in L. monocytogenes strongly depletes LTA from both the cell wall and the culture medium [18]. In contrast to these findings, deletion of bgsA in E. faecalis results in an increased concentration of LTA in the bacterial cell envelope, most likely related to the longer glycerol-phosphate polymer. The different makeup of glycolipids and LTA in this mutant strongly impaired biofilm-formation and affected virulence in vivo [5].
In the current study, we constructed a deletion mutant by targeted mutagenesis of the putative glycosyltransferase bgsB located immediately downstream of bgsA. After inactivation of bgsB in E. faecalis 12030, no glycolipids or glycolipid-derivatives were recovered from the cell envelope of the 12030ΔbgsB mutant, indicating that BgsB is a 1,2-diacylglycerol 3-glucosyltransferase. BgsA cannot take the place of BgsB, which suggests that BgsA has higher substrate specificity than YpfP in S. aureus and B. subtilis [13,17]. The putative function assigned to BgsA and BgsB by this work is in agreement with data obtained for their homologues LafA and LafB in L. monocytogenes [15]. Although the lipid anchor of LTA from 12030ΔbgsB was not characterized chemically, indirect evidence suggests that DAG instead of DGlcDAG anchors LTA to the cell membrane in this mutant. LTA extracted from 12030ΔbgsB migrated more slowly than wild-type LTA in SDS PAGE, a feature that has been described for homologous LTA molecules substituted with DAG instead of DGlcDAG in S. aureus and L. monocytogenes [13,15]. In staphylococci and listeria it has been also demonstrated that, in the absence of glycolipids, the enzyme that transfers glycerolphosphate residues to the glycolipid anchor (LtaS) can utilize DAG as glycerolphosphate acceptor for the synthesis of the LTA backbone [13,15]. Deletion mutants of the glucosyltransferases bgsB and bgsA enabled us to study the individual roles of the two major glycolipids MGlcDAG and DGlcDAG in the physiology and virulence of E. faecalis. To our surprise, the complete loss of glycolipids from the cell membrane in 12030ΔbgsB had only minor effects on bacterial morphology, cell growth, and autolysis.
In contrast, MGlcDAG and DGlcDAG are critical for cell membrane elasticity and fluidity and important for the function of membrane-bound proteins in Acholeplasma laidlawii [6,7,14]. It is possible, however, that up-regulation of other cell membrane amphiphiles may compensate for the lack of glycolipids in the bgsB mutant [6]. In fact, the concentration of LTA was increased in 12030ΔbgsB and possibly compensates for the loss of phosphoglycolipid derivatives of MGlcDAG and DGlcDAG in the 12030ΔbgsB mutant [19]. A characteristic feature of both mutants is the increased chain length of the glycerol-phosphate polymer. However, the mechanism underlying this alteration in LTA structure remains unclear and deserves further attention.
The most notable feature of 12030ΔbgsB is its impairment in biofilm formation and adherence to colonic cells. As observed previously in the bgsA mutant, initial attachment to polystyrene was not impaired in 12030ΔbgsB, but the accumulation of bacteria in the growing biofilm was impaired. This is in contrast to other biofilm-defective mutants in E. faecalis, in which attachment to the foreign surface is the feature primarily affected and underlines the importance of cell envelope amphiphiles in the retention of bacteria within the biofilm architecture [20,21]. Several mechanisms may explain the biofilm phenotype of the mutants. As in the bgsA mutant, impaired biofilm formation in 12030ΔbgsB was associated with reduced hydrophobicity, a well-known determinant of biofilm formation in bacteria [22,23]. Also, increased LTA concentration in the cell envelope of the bgsB-mutant may impair biofilm formation by increasing the net negative charge of the cell envelope. The impact of the higher negative charge of the LTA molecule on biofilm formation has been demonstrated by mutants in the D-alanine-D-alanyl-carrier protein ligase DltA [24,25]. Finally, the increased amount of LTA released into the biofilm matrix (as observed with 12030ΔbgsB and 12030ΔbgsA) may act as a biosurfactant, promoting detachment of bacterial cells from the biofilm and thereby impeding its growth [26]. In contrast to our results the inactivation of the glycosyltransferase YpfP in S. aureus leads to depletion of LTA from the cell surface and to a reduced ability to form biofilm [12].
Aside from its effects on biofilm formation, the increased density of negative charges of the LTA molecule of the mutant may also explain the slight increase in sensitivity of 12030ΔbgsB to the antimicrobial peptides colistin and polymyxin B. If this difference explains the significantly impaired virulence in our mouse bacteremia model, however, is unclear. On balance, we observed a 2-log reduction in the number of CFU recovered for both mutants, suggesting that glycolipids, either as a cell membrane component or as an anchor of LTA, play a critical role in the cell envelope of enterococci during infection. In general, mutation of the glycosyl-transferase bgsA and bgsB yielded similar phenotypes, suggesting that the phenotypic changes observed for both mutants are mainly the result of the depletion of DGlcDAG or altered LTA structure. On the other hand, MGlcDAG seems to play a minor role in bacterial physiology and virulence.
Conclusions
We have shown that the bgsB gene is responsible for the glycosylation of DAG to form MGlcDAG, the first step in glycolipid synthesis in E. faecalis. bgsB deletion led to reduced biofilm formation and attachment to colonic cells, and to impaired virulence in vivo.
Methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are shown in Table 1. Enterococci were grown at 37°C without agitation in tryptic soy broth (TSB; Merck), M17 broth (Difco Laboratories), or TSB plus 1% glucose (TSBG) as indicated. In addition, tryptic soy agar or M17 agar plates were used. Escherichia coli DH5α and TOP10 (Invitrogen) were cultivated aerobically in LB-broth. Kanamycin was added for enterococci (1 mg/ml) and for E. coli (50 μg/ml); tetracycline was used at 12.5 μg/ml for E. coli and at 10 μg/ml for enterococci.
Table 1.
E. faecalis strains and plasmids used in this study.
| strain or plasmid | characterization | reference |
|---|---|---|
| strains | ||
| E. faecalis 12030 | Clinical isolate, strong biofilm producer | [33] |
| E. faecalis ATCC 29212 | Reference strain | |
| E. faecalis 12030ΔbgsA | (EF2891) bsgA mutant | [5] |
| E. faecalis 12030ΔbgsB | bgsB deletion mutant | This study |
| E. faecalis 12030ΔbgsB_rec. | Reconstituted mutant | This study |
| Escherichia coli DH5α | Gram-negative cloning host | |
| Escherichia coli TOP10 | Gram-negative cloning host | Invitrogen |
| plasmids | ||
| pCASPER | Gram-positive, temperature-sensitive mutagenesis vector | [34] |
| pCRII-TOPO | Gram-negative cloning vector | Invitrogen |
| pCASPER/ΔbgsB | This study | |
| pMAD/bsgB | This study | |
| pMAD | oripE194ts, EmR, AmpR, bgaB | [35] |
Construction of a nonpolar deletion mutant of bgsB
Molecular techniques used in this study have been described previously [5]. In brief, the bgsB mutant was constructed in E. faecalis 12030 by homologous recombination. The deletion of a portion of the gene bgsB (790 bp) (EF_2890 in the E. faecalis V583 genome, GenBank accession no. AAO82579.1) was created as described elsewhere [5]. Primers 1 and 2 (Table 2) were used to amplify a 581-bp fragment downstream, and primers 3 and 4 were used to amplify a 563-bp fragment upstream of the target gene. Primers 2 and 3 contain a 21-bp complementary sequence (underlined in Table 2). Overlap extension PCR was performed to generate a PCR product lacking a fragment of 790 bp in the center of bgsB (Figure 1). The resulting construct was cloned into the Gram-positive shuttle vector pCASPER containing a temperature-sensitive replicon; the resulting plasmid, pCASPER-ΔbgsB, was transformed into E. faecalis 12030 by electroporation. Integrants were selected at the non-permissive temperature (42°C) on TSA plates with kanamycin. Colonies were passaged between 6 and 10 times in liquid cultures without antibiotics at the permissive temperature (30°C) and subsequently screened by replica-plating for loss of kanamycin resistance. Kanamycin-sensitive clones were analyzed by PCR for the deleted sequence, and the deletion mutant was designated E. faecalis 12030ΔbgsB.
Table 2.
Primers used in this study.
| Name | Sequence (5'-3') | |
|---|---|---|
| 1 | EF2890 delF | CAAACTGCTCCTTCAGCAACT |
| 2 | EF2890 OEL | ACTAGCGCGGCCGCTTGCTCCCTATTTTGTCAGCGCCTCAAC |
| 3 | EF2890 OER | GGAGCAAGCGGCCGCGCTAGTTAGAAGTCGCTACCCCACTCA |
| 4 | EF2890 delR | GCGCGACAGTTACCAGAGTAT |
Complementation of the 12030ΔbgsB mutant has been done by a knocking in strategy as described previously [27]. Briefly, the bgsB gene (1224 bp) plus 212 bp upstream and 502 bp downstream was amplified using primers 1 and 4, cloned into pCRII-TOPO (Table 1) and digested with EcoRI. The resulting fragment was inserted into plasmid pMAD (Table 1). E. faecalis 12030ΔbgsB was transformed with the recombinant plasmid (pMAD-bgsB) and incubated at 37°C for 4 d on TSB plates supplemented with Xgal (40 μg/ml) and erythromycin (Erm, 100 μg/ml). Dark blue colonies were picked and incubated overnight on fresh plates supplemented with Xgal and Erm at the non-permissive temperature (44°C). Presence of the wild-type and mutated alleles was determined by PCR, and for each construct the positive clones were cultured in TSB medium supplemented with Erm (150 μg/ml) at 44°C over-night. This last step was repeated once, using the overnight culture to inoculate a fresh culture tube. To delete the erythromycin resistance gene, overnight cultures were inoculated in TSB medium without Erm and incubated for 12 h at 30°C, followed by 18 h at 44°C without shaking. This step was repeated until white colonies were obtained on Xgal-supplemented TSA plates incubated overnight at 37°C. Erm sensitivity of the white colonies was verified, and sensitive clones were tested by PCR for the presence of the intact bgsB gene.
Biofilm plate assay
Enterococci were tested for production of biofilm using a polystyrene microtiter assay [5,24]. Briefly, bacteria were grown at 37°C in TSB for 18 h. Polystyrene tissue-culture plates (Brandt, Germany) were filled with 180 μl of TSB plus 1% glucose and 20 μl of this culture, and the plates were then incubated at 37°C for 18 h. The plates were read in an ELISA reader (Bio-Rad Microplate reader) at an optical density of 630 nm to assess homogenous growth. The culture medium was discarded, and the wells were washed 3 times with 200 μl of PBS without disturbing the biofilm on the bottom of the wells. The plates were dried at 60°C for 1 h and stained with 2% Hucker's crystal violet for 2 min. Excess stain was removed by rinsing the plates under tap water, and the plates were dried at 60°C for 10 min. The optical density at 630 nm was determined. Biofilm formation was normalized to growth with the biofilm index, which was calculated as OD of the biofilm × (0.5/OD of growth) [24].
Adherence to Caco-2 cells
Adherence to Caco-2 cells was investigated using methods described previously [28]. In brief, cells were cultivated in DMEM medium supplemented with 10% fetal bovine serum and 1% non-essential amino acids under a 5% CO2 atmosphere. All the experiments were performed on cells between the 15th and 25th passage. Caco-2 cells were cultivated in 24-well plates to a density of 1 × 105 cells/well for 3-5 days. Bacteria were grown to mid-log phase at 37°C without agitation in tryptic soy broth; Caco-2 cells were incubated with bacteria for 2 h at a multiplicity of infection of 100:1. After infection of the monolayer, epithelial cells were washed and lysed with 0.25% Triton-X at 37°C for 20 min and adherent bacteria enumerated by quantitative bacterial counts. Pilot experiments had shown no significant bacterial invasion under the outlined conditions.
Isolation and analysis of glycolipids and LTA
Bacterial cells were resuspended in 0.1 M citrate buffer pH 4.7 and cell walls disrupted by shaking with an equal volume of glass beads (0.1 mm glass beads, 3 × 1 min intervals using a BeadBeater, Glenn Mills, Clifton, NJ). Glass beads were removed by sedimentation, and disrupted cells were stirred with an equal volume of n-butanol for 30 min. After phase separation by centrifugation, the aqueous layer was removed, dialyzed against 0.1 M ammonium acetate (pH 4.7) and lyophilized. LTA was purified from the aqueous phase by hydrophobic interaction chromatography [4]. The butanol phase was evaporated under a vacuum, and cell membrane lipids were extracted according to the method of Bligh and Dyer and separated by TLC (0.2 mm Silica gel 60 F254 Merck, Darmstadt) using a solvent system of CHCl3/MeOH/H2O (65:25:4, v/v/v) and detection with α-naphthol (3.2%). For detection of phospholipids, TLC plates were stained with molybdenum blue; amino phospholipids were stained with ninhydrin, as previously described [29]. LTA was also analyzed by SDS-PAGE as described previously [5]. Briefly, bacterial cell walls were disrupted by shaking with glass beads as described above, boiled in sample buffer containing SDS, and subjected to SDS-PAGE in gradient gels containing acrylamide (4/12% w/v, Invitrogen). Separated LTA was transferred onto PVDF membrane and blocked at 4°C in Tris-buffered saline (TBS) containing skim milk (5% w/v) for 18 h, then incubated at 20-22°C for 2 h with rabbit antibody raised against E. faecalis LTA (see below) diluted 1:200 in TBS/skim milk. After washing in TTBS (Tween 20 0.05% v/v in TBS), the sheets were incubated at 20-22°C for 1 h with a goat anti-rabbit IgG (whole cell) alkaline phosphatase conjugate (Sigma), diluted 1:1000 with TBS/skim milk, and then washed again in TTBS. Binding of the enzyme-conjugated antibodies was detected with the NBI/BCIP (Biorad). For visualization of proteins, SDS PAGE gels were stained with Coomassie blue.
Characterization of LTA
Lipoteichoic acid of E. faecalis strain 12030ΔbgsB was analyzed by NMR spectroscopy as described previously [5].
Rabbit antiserum against LTA
A female New Zealand White rabbit was immunized s.c. with 100 mg of LTA purified from E. faecalis strain 12030 suspended in complete Freund adjuvant (Sigma), followed by the same dose s.c. suspended in incomplete Freund adjuvant (Sigma) on day 7. The rabbit was boosted intravenously with three 10-mg doses over the following 3 weeks. After the last vaccination, the rabbit was sacrificed and exsanguinated to obtain the serum.
Autolysis assay and sensitivity to antimicrobial peptides
Cell autolysis was determined as described by Qin et al. [30]. The MIC of polymyxin B, nisin, and colistin against wild-type and 12030ΔbgsB were determined by a modified NCCLS broth dilution method [24].
Determination of hydrophobicity
Hydrophobicity was determined by measuring adherence to dodecane [31]. Briefly, bacteria were grown to logarithmic phase and resuspended in sodium phosphate to yield an OD600 of 0.4-0.5. The same volume of dodecane was added, and phases were vigorously vortexed for 1 min, then for 10 min to allow phase separation. Absorbance of the water-phase was measured. The proportion of cells in the dodecane phase was calculated according to the formula: % hydrophobicity = [1-(A/A0)] × 100.
Mouse bacteremia model
The virulence of E. faecalis strain 12030ΔbgsB was evaluated in a mouse bacteremia model [5,32]. In summary, eight female BALB/c mice 6-8 weeks old were challenged by i.v. injection of E. faecalis strains grown to stationary phase (2.0 × 109 cfu) via the tail vein. Seventy-two hours after infection, the mice were sacrificed and exsanguinated, and bacterial counts in the blood were enumerated by serial dilutions. All animal experiments were performed in compliance with the German animal protection law (TierSchG). The mice were housed and handled in accordance with good animal practice as defined by FELASA and the national animal welfare body GV-SOLAS. The animal welfare committees of the University of Freiburg (Regierungspräsidium Freiburg Az 35/9185.81/G-07/15) approved all animal experiments.
Transmission electron microscopy (TEM)
Bacterial cells were prepared for TEM as described previously [24].
Opsonophagocytic killing assay
An opsonophagocytic killing assay was used as previously described [5]. In summary, white blood cells (WBC) were prepared from fresh human blood collected from healthy adult volunteers. Using trypan blue staining to differentiate dead from live leukocytes, the final cell count was adjusted to 2.5 × 107 WBC per ml. Baby rabbit serum (Cedarlane Laboratories, Hornby, Ontario, Canada), diluted 1:15 in RPMI plus 15% fetal bovine serum (FBS) and absorbed with the target strain, was used as complement source. Bacteria cultured on agar plates were resuspended in TSB to an OD600 of 0.1 and then grown to an OD of 0.4. A final 1:100 dilution was made in RPMI-FBS. Equal amounts of PMN and bacteria (multiple of infection 1:1), complement, and heat-inactivated rabbit immune serum were incubated at 37°C for 90 min. For the controls, antibody, complement, or PMN were replaced by RPMI-FBS. For enumeration of surviving bacteria, the content of tubes was diluted in TSB, and samples were plated onto tryptic soy agar plates. The percentage of opsonophagocytic killing was calculated by determining the ratio of the CFU surviving in the tubes with bacteria, leukocytes, complement, and antibody to the CFU surviving in the tubes with all these components but lacking leukocytes.
Quantification of LTA
The LTA content of bacterial cell walls was measured according to the method of Fedtke et al. [12]. In summary, wild-type and mutant bacteria were grown for 18 h in TSB, adjusted to an equal OD600, and bacteria from equal volumes were collected by centrifugation. Bacterial were disrupted by shaking with glass beads as described above, and LTA was extracted from the cell walls by stirring them in an equal volume of butanol and 0.1 M Na-acetate buffer (pH 4,7). The aqueous phase of the extract was dialyzed, lyophilized, and resuspended in the same volume of phosphate buffer (pH 7.0). ELISA plates (Brandt) were coated with a range of LTA dilutions at 4°C for 18 h, and adherent LTA was detected using a rabbit antiserum specific for E. faecalis LTA as primary antibody (see above). A goat anti-rabbit IgG whole-molecule alkaline phosphatase conjugate (Sigma) served as secondary antibody [5]. LTA from E. faecalis 12030, purified by hydrophobic-interaction chromatography, was used as a standard. The amount of LTA shed into the culture medium was measured semi-quantitatively by immuno-dot-blot analysis. To this end, bacteria were grown in TSB at 37°C for 18 h and adjusted to the same OD600. Bacterial cells were removed by centrifugation, culture supernatant was passed through a 0.45 μm membrane filter, and 100 μl of supernatant was spotted in various dilutions onto PVDF membrane using a dot-blot microfiltration apparatus (Bio-Dot, Biorad Laboratories, Munich, Germany). The membranes were allowed to dry overnight. Staining of immuno-dot-blots was performed using the same protocol as described for western blot analysis of LTA.
Statistical Methods
Comparisons were made by one-way ANOVA and Tukey's multiple comparison test (parametric data) or Kruskal-Wallis test and Dunn's multiple comparison test (nonparametric data) as indicated using the Prism Graphpad 4 software package. A p-value of < 0.05 was considered statistically significant.
Authors' contributions
CT participated in the isolation and TLC analysis of glycolipids and LTA, the design and interpretation of the experiments, made the statistical analysis, and drafted the manuscript. IS performed the cell culture assays, autolysis assay and hydrophobicity assay. YB carried out the biofilm assay and participated in the molecular genetic studies. AK performed the opsonophagocytic killing assay and the mouse infection model. PSC performed the biochemical analysis of glycolipids and LTA. EG participated in the draft of the manuscript. OH participated in the biochemical analysis of the glycolipids and LTA and the draft of manuscript. JH participated in the design, coordination and interpretation of the study, and the draft of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Transmission electron microscopy of E. faecalis strains. E. faecalis 12030 wild type (A) and 12030ΔbgsB (B). Bar represents 500 nm.
Autolysis and opsonization of E. faecalis 12030ΔbgsB. A Spontaneous bacterial autolysis. Cells were grown to mid-log phase, resuspended in 10 mM sodium phosphate buffer containing 5% Triton X-100 and the decrease of the OD 600 at 30°C was recorded over time. B Bacterial killing in vitro after 90 min in the presence of 6.5% rabbit complement (white bar), 2 × 107 human PMN plus complement (gray bar) and rabbit antiserum raised against whole bacterial cells (serum dilution 1:2500) plus PMN and complement (black bar). Bars represent means ± SEM.
Characterization of E. faecalis ΔbgsB cell walls. A Thin-layer chromatography of cell membrane total lipid extracts of E. faecalis 12030 wild type (lane 1 and 4), 12030ΔbgsB (lane 2 and 5), 12030ΔbgsA (lane 3 and 6). TLC plates were developed using a solvent system of CHCl3/MeOH/H20 (65:25:4, v/v/v). Staining lane 1 - 3 molybdenum blue, lane 4 - 6 ninhydrin. B SDS PAGE of bacterial whole protein extracts. The material was extracted by disrupting the cells with glass-beads, boiling in Laemmli buffer, separated by 4-12% Bis-Tris gels and stained with Coomassie blue.
Minimal bactericial concentration of E. faecalis strains against antimicrobial peptides. Concentrations are expressed as μg/ml.
Contributor Information
Christian Theilacker, Email: christian.theilacker@uniklinik-freiburg.de.
Irina Sava, Email: irina1129@googlemail.com.
Patricia Sanchez-Carballo, Email: psanchez@fz-borstel.de.
Yinyin Bao, Email: baoyinyin1982@googlemail.com.
Andrea Kropec, Email: andrea.kropec@uniklinik-freiburg.de.
Elisabeth Grohmann, Email: elisabeth.grohmann@uniklinik-freiburg.de.
Otto Holst, Email: oholst@fz-borste.de.
Johannes Huebner, Email: johannes.huebner@uniklinik-freiburg.de.
Acknowledgements
The authors thank Dr. Friedrich Feuerhake for help with electron microscopy, Ioana Toma and Dominique Wobser for excellent technical assistance. J.H. was supported by a grant of the German Ministry of Science and Education (ERA-Net PathoGenoMics 0313933).
References
- Weidenmaier C, Peschel A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 2008;6(4):276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- Theilacker C, Kaczynski Z, Kropec A, Sava I, Ye L, Bychowska A, Holst O, Huebner J. Serodiversity of Opsonic Antibodies against Enterococcus faecalis -Glycans of the Cell Wall Revisited. PLoS ONE. 2011;6(3):e17839. doi: 10.1371/journal.pone.0017839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Singh KV, Bourgogne A, Zeng J, Murray BE. Further characterization of the epa gene cluster and Epa polysaccharides of Enterococcus faecalis. Infect Immun. 2009;77(9):3759–3767. doi: 10.1128/IAI.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theilacker C, Kaczynski Z, Kropec A, Fabretti F, Sange T, Holst O, Huebner J. Opsonic antibodies to Enterococcus faecalis strain 12030 are directed against lipoteichoic acid. Infect Immun. 2006;74(10):5703–5712. doi: 10.1128/IAI.00570-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theilacker C, Sanchez-Carballo P, Toma I, Fabretti F, Sava I, Kropec A, Holst O, Huebner J. Glycolipids are involved in biofilm accumulation and prolonged bacteraemia in Enterococcus faecalis. Mol Microbiol. 2009;71(4):1055–1069. doi: 10.1111/j.1365-2958.2008.06587.x. [DOI] [PubMed] [Google Scholar]
- Wikström M, Xie J, Bogdanov M, Mileykovskaya E, Heacock P, Wieslander A, Dowhan W. Monoglucosyldiacylglycerol, a foreign lipid, can substitute for phosphatidylethanolamine in essential membrane-associated functions in Escherichia coli. J Biol Chem. 2004;279(11):10484–10493. doi: 10.1074/jbc.M310183200. [DOI] [PubMed] [Google Scholar]
- Edman M, Berg S, Storm P, Wikstrom M, Vikstrom S, Ohman A, Wieslander A. Structural features of glycosyltransferases synthesizing major bilayer and nonbilayer-prone membrane lipids in Acholeplasma laidlawii and Streptococcus pneumoniae. J Biol Chem. 2003;278(10):8420–8428. doi: 10.1074/jbc.M211492200. [DOI] [PubMed] [Google Scholar]
- Vikström S, Li L, Wieslander A. The nonbilayer/bilayer lipid balance in membranes. Regulatory enzyme in Acholeplasma laidlawii is stimulated by metabolic phosphates, activator phospholipids, and double-stranded DNA. J Biol Chem. 2000;275(13):9296–9302. doi: 10.1074/jbc.275.13.9296. [DOI] [PubMed] [Google Scholar]
- Campbell J, Davies G, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1998;329(Pt 3):719. doi: 10.1042/bj3290719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman O, Dover LG, Sutcliffe IC. Lipoteichoic acid biosynthesis: two steps forwards, one step sideways? Trends Microbiol. 2009;17(6):219–225. doi: 10.1016/j.tim.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Neuhaus FC, Baddiley J. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67(4):686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedtke I, Mader D, Kohler T, Moll H, Nicholson G, Biswas R, Henseler K, Götz F, Zähringer U, Peschel A. A Staphylococcus aureus ypfP mutant with strongly reduced lipoteichoic acid (LTA) content: LTA governs bacterial surface properties and autolysin activity. Mol Microbiol. 2007;65(4):1078–1091. doi: 10.1111/j.1365-2958.2007.05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundling A, Schneewind O. Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J Bacteriol. 2007;189(6):2521–2530. doi: 10.1128/JB.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg S, Edman M, Li L, Wikstrom M, Wieslander A. Sequence properties of the 1,2-diacylglycerol 3-glucosyltransferase from Acholeplasma laidlawii membranes. Recognition of a large group of lipid glycosyltransferases in eubacteria and archaea. J Biol Chem. 2001;276(25):22056–22063. doi: 10.1074/jbc.M102576200. [DOI] [PubMed] [Google Scholar]
- Webb AJ, Karatsa-Dodgson M, Grundling A. Two-enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes. Mol Microbiol. 2009;74(2):299–314. doi: 10.1111/j.1365-2958.2009.06829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriukhin MY, Debabov DV, Shinabarger DL, Neuhaus FC. Biosynthesis of the glycolipid anchor in lipoteichoic acid of Staphylococcus aureus RN4220: role of YpfP, the diglucosyldiacylglycerol synthase. J Bacteriol. 2001;183(11):3506–3514. doi: 10.1128/JB.183.11.3506-3514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorasch P, Wolter FP, Zähringer U, Heinz E. A UDP glucosyltransferase from Bacillus subtilis successively transfers up to four glucose residues to 1,2-diacylglycerol: expression of ypfP in Escherichia coli and structural analysis of its reaction products. Mol Microbiol. 1998;29(2):419–430. doi: 10.1046/j.1365-2958.1998.00930.x. [DOI] [PubMed] [Google Scholar]
- Doran KS, Engelson EJ, Khosravi A, Maisey HC, Fedtke I, Equils O, Michelsen KS, Arditi M, Peschel A, Nizet V. Blood-brain barrier invasion by group B Streptococcus depends upon proper cell-surface anchoring of lipoteichoic acid. J Clin Invest. 2005;115(9):2499–2507. doi: 10.1172/JCI23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W. In: Handbook of Lipid Research. Hanahan DJ, editor. Vol. 6. New York: Plenum Press; 1990. Bacterial phosphoglycolipids and lipoteichoic acids; pp. 123–234. [Google Scholar]
- Mohamed JA, Huang DB. Biofilm formation by enterococci. J Med Microbiol. 2007;56(Pt 12):1581–1588. doi: 10.1099/jmm.0.47331-0. [DOI] [PubMed] [Google Scholar]
- Mohamed JA, Huang W, Nallapareddy SR, Teng F, Murray BE. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect Immun. 2004;72(6):3658–3663. doi: 10.1128/IAI.72.6.3658-3663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos R, van der Mei HC, Busscher HJ. Physico-chemistry of initial microbial adhesive interactions--its mechanisms and methods for study. FEMS Microbiol Rev. 1999;23(2):179–230. doi: 10.1111/j.1574-6976.1999.tb00396.x. [DOI] [PubMed] [Google Scholar]
- Courtney HS, Ofek I, Penfound T, Nizet V, Pence MA, Kreikemeyer B, Podbielski A, Podbielbski A, Hasty DL, Dale JB. Relationship between expression of the family of M proteins and lipoteichoic acid to hydrophobicity and biofilm formation in Streptococcus pyogenes. PLoS ONE. 2009;4(1):e4166. doi: 10.1371/journal.pone.0004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabretti F, Theilacker C, Baldassarri L, Kaczynski Z, Kropec A, Holst O, Huebner J. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect Immun. 2006;74(7):4164–4171. doi: 10.1128/IAI.00111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M, Cramton SE, Götz F, Peschel A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun. 2001;69(5):3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu TR. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol Rev. 1996;60(1):151–166. doi: 10.1128/mr.60.1.151-166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Carbona S, Sauvageot N, Giard JC, Benachour A, Posteraro B, Auffray Y, Sanguinetti M, Hartke A. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol Microbiol. 2007;66(5):1148–1163. doi: 10.1111/j.1365-2958.2007.05987.x. [DOI] [PubMed] [Google Scholar]
- Sava IG, Zhang F, Toma I, Theilacker C, Li B, Baumert TF, Holst O, Linhardt RJ, Huebner J. Novel interactions of glycosaminoglycans and bacterial glycolipids mediate binding of enterococci to human cells. J Biol Chem. 2009;284(27):18194–18201. doi: 10.1074/jbc.M901460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A. et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med. 2001;193(9):1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Singh KV, Xu Y, Weinstock GM, Murray BE. Effect of disruption of a gene encoding an autolysin of Enterococcus faecalis OG1RF. Antimicrob Agents Chemother. 1998;42(11):2883–2888. doi: 10.1128/aac.42.11.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Cuperus PL, Bruce AW, van der Mei HC, Tomeczek L, Khoury AH, Busscher HJ. Comparison of contact angles and adhesion to hexadecane of urogenital, dairy, and poultry lactobacilli: effect of serial culture passages. Appl Environ Microbiol. 1992;58(5):1549–1553. doi: 10.1128/aem.58.5.1549-1553.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufnagel M, Koch S, Creti R, Baldassarri L, Huebner J. A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of biofilm and prolonged bacteremia in mice. J Infect Dis. 2004;189(3):420–430. doi: 10.1086/381150. [DOI] [PubMed] [Google Scholar]
- Huebner J, Wang Y, Krueger WA, Madoff LC, Martirosian G, Boisot S, Goldmann DA, Kasper DL, Tzianabos AO, Pier GB. Isolation and chemical characterization of a capsular polysaccharide antigen shared by clinical isolates of Enterococcus faecalis and vancomycin-resistant Enterococcus faecium. Infect Immun. 1999;67(3):1213–1219. doi: 10.1128/iai.67.3.1213-1219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegan MC, Jett BD, Hancock LE, Gilmore MS. Role of hemolysin BL in the pathogenesis of extraintestinal Bacillus cereus infection assessed in an endophthalmitis model. Infect Immun. 1999;67(7):3357–3366. doi: 10.1128/iai.67.7.3357-3366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70(11):6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transmission electron microscopy of E. faecalis strains. E. faecalis 12030 wild type (A) and 12030ΔbgsB (B). Bar represents 500 nm.
Autolysis and opsonization of E. faecalis 12030ΔbgsB. A Spontaneous bacterial autolysis. Cells were grown to mid-log phase, resuspended in 10 mM sodium phosphate buffer containing 5% Triton X-100 and the decrease of the OD 600 at 30°C was recorded over time. B Bacterial killing in vitro after 90 min in the presence of 6.5% rabbit complement (white bar), 2 × 107 human PMN plus complement (gray bar) and rabbit antiserum raised against whole bacterial cells (serum dilution 1:2500) plus PMN and complement (black bar). Bars represent means ± SEM.
Characterization of E. faecalis ΔbgsB cell walls. A Thin-layer chromatography of cell membrane total lipid extracts of E. faecalis 12030 wild type (lane 1 and 4), 12030ΔbgsB (lane 2 and 5), 12030ΔbgsA (lane 3 and 6). TLC plates were developed using a solvent system of CHCl3/MeOH/H20 (65:25:4, v/v/v). Staining lane 1 - 3 molybdenum blue, lane 4 - 6 ninhydrin. B SDS PAGE of bacterial whole protein extracts. The material was extracted by disrupting the cells with glass-beads, boiling in Laemmli buffer, separated by 4-12% Bis-Tris gels and stained with Coomassie blue.
Minimal bactericial concentration of E. faecalis strains against antimicrobial peptides. Concentrations are expressed as μg/ml.