Abstract
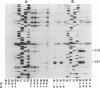
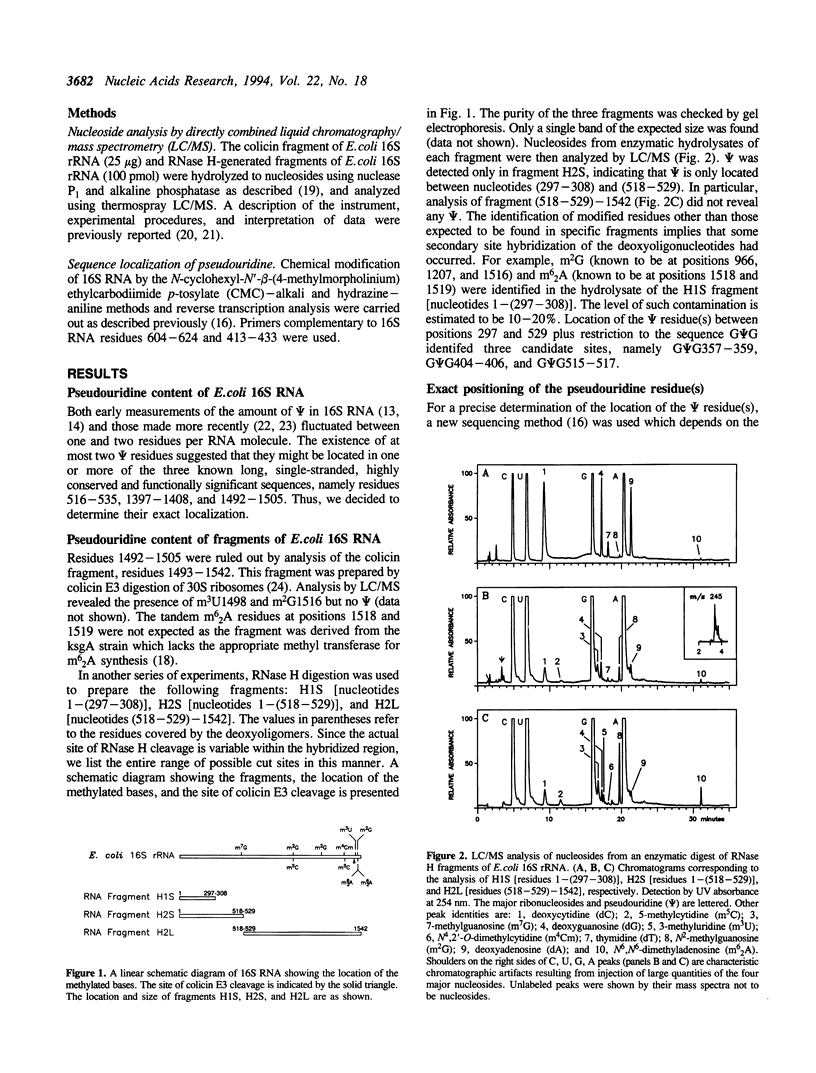
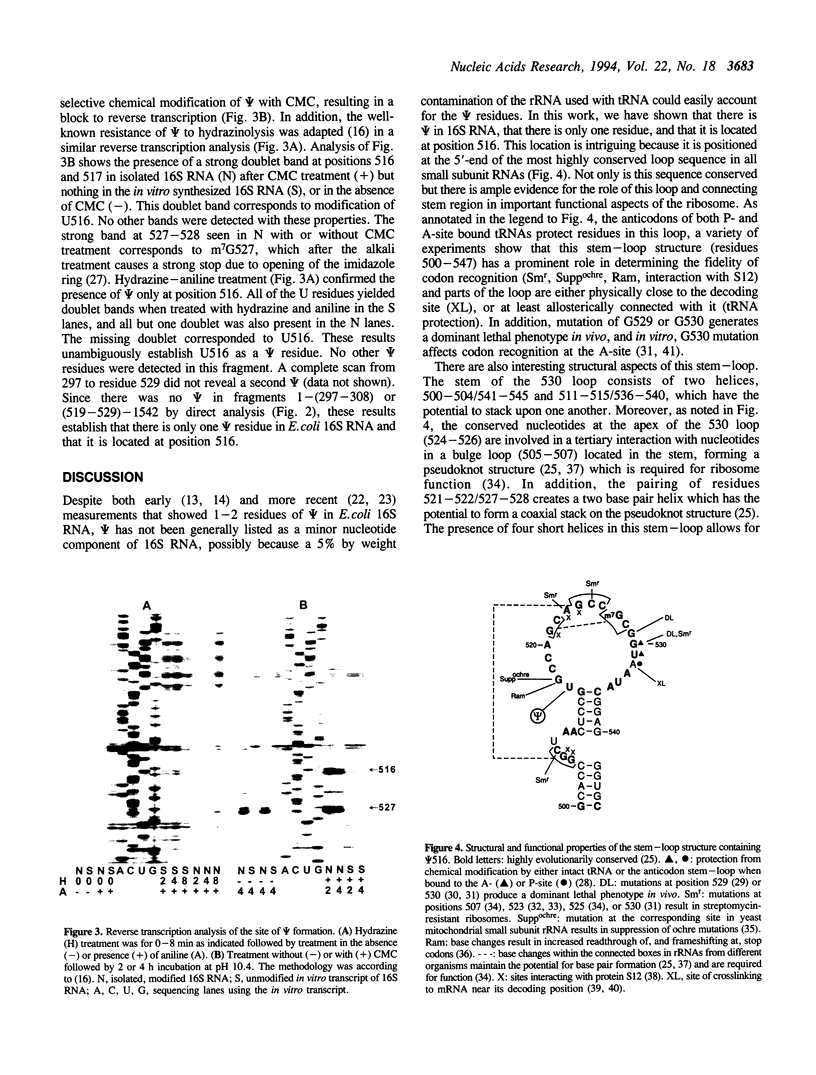
The number and location of pseudouridine residues in Escherichia coli 16S ribosomal RNA has been determined by a combination of direct and indirect methods. Only one residue was found, at position 516. This site is at the 5'-end of one of the three most highly conserved long sequences of this RNA molecule. A number of experimental findings have strongly implicated this loop in the fidelity of codon recognition by A-site bound tRNA. By virtue of its location, we suggest that psi 516 may also play a role in maintaining the fidelity of protein synthesis.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bakin A., Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993 Sep 21;32(37):9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- Bowman C. M., Dahlberg J. E., Ikemura T., Konisky J., Nomura M. Specific inactivation of 16S ribosomal RNA induced by colicin E3 in vivo. Proc Natl Acad Sci U S A. 1971 May;68(5):964–968. doi: 10.1073/pnas.68.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe R., Mitchell P., Osswald M., Stade K., Bochkariov D. Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J. 1993 Jan;7(1):161–167. doi: 10.1096/fasebj.7.1.8422963. [DOI] [PubMed] [Google Scholar]
- Bruenger E., Kowalak J. A., Kuchino Y., McCloskey J. A., Mizushima H., Stetter K. O., Crain P. F. 5S rRNA modification in the hyperthermophilic archaea Sulfolobus solfataricus and Pyrodictium occultum. FASEB J. 1993 Jan;7(1):196–200. doi: 10.1096/fasebj.7.1.8422966. [DOI] [PubMed] [Google Scholar]
- Crain P. F. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- Cunningham P. R., Nurse K., Weitzmann C. J., Ofengand J. Functional effects of base changes which further define the decoding center of Escherichia coli 16S ribosomal RNA: mutation of C1404, G1405, C1496, G1497, and U1498. Biochemistry. 1993 Jul 20;32(28):7172–7180. doi: 10.1021/bi00079a014. [DOI] [PubMed] [Google Scholar]
- Cunningham P. R., Weitzmann C. J., Nurse K., Masurel R., Van Knippenberg P. H., Ofengand J. Site-specific mutation of the conserved m6(2)A m6(2)A residues of E. coli 16S ribosomal RNA. Effects on ribosome function and activity of the ksgA methyltransferase. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):18–26. doi: 10.1016/0167-4781(90)90135-o. [DOI] [PubMed] [Google Scholar]
- Dontsova O., Dokudovskaya S., Kopylov A., Bogdanov A., Rinke-Appel J., Jünke N., Brimacombe R. Three widely separated positions in the 16S RNA lie in or close to the ribosomal decoding region; a site-directed cross-linking study with mRNA analogues. EMBO J. 1992 Aug;11(8):3105–3116. doi: 10.1002/j.1460-2075.1992.tb05383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds C. G., Vestal M. L., McCloskey J. A. Thermospray liquid chromatography-mass spectrometry of nucleosides and of enzymatic hydrolysates of nucleic acids. Nucleic Acids Res. 1985 Nov 25;13(22):8197–8206. doi: 10.1093/nar/13.22.8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier A., Turmel M., Lemieux C. Mapping of chloroplast mutations conferring resistance to antibiotics in Chlamydomonas: evidence for a novel site of streptomycin resistance in the small subunit rRNA. Mol Gen Genet. 1988 Oct;214(2):192–197. doi: 10.1007/BF00337710. [DOI] [PubMed] [Google Scholar]
- Gehrke C. W., Kuo K. C. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J Chromatogr. 1989 Jun 2;471:3–36. doi: 10.1016/s0021-9673(00)94152-9. [DOI] [PubMed] [Google Scholar]
- Helser T. L., Davies J. E., Dahlberg J. E. Mechanism of kasugamycin resistance in Escherichia coli. Nat New Biol. 1972 Jan 5;235(53):6–9. doi: 10.1038/newbio235006a0. [DOI] [PubMed] [Google Scholar]
- Johnson J. D., Horowitz J. Characterization of ribosomes and RNAs from Mycoplasma hominis. Biochim Biophys Acta. 1971 Oct 14;247(2):262–279. doi: 10.1016/0005-2787(71)90675-7. [DOI] [PubMed] [Google Scholar]
- Kowalak J. A., Pomerantz S. C., Crain P. F., McCloskey J. A. A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Res. 1993 Sep 25;21(19):4577–4585. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzosiak W., Denman R., Nurse K., Hellmann W., Boublik M., Gehrke C. W., Agris P. F., Ofengand J. In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into a functional 30S ribosome. Biochemistry. 1987 Apr 21;26(8):2353–2364. doi: 10.1021/bi00382a042. [DOI] [PubMed] [Google Scholar]
- Lane B. G., Ofengand J., Gray M. W. Pseudouridine in the large-subunit (23 S-like) ribosomal RNA. The site of peptidyl transfer in the ribosome? FEBS Lett. 1992 May 4;302(1):1–4. doi: 10.1016/0014-5793(92)80269-m. [DOI] [PubMed] [Google Scholar]
- Limbach P. A., Crain P. F., McCloskey J. A. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994 Jun 25;22(12):2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Forbes J., de Jonge P., Klootwijk J. Presence of a hypermodified nucleotide in HeLa cell 18 S and Saccharomyces carlsbergensis 17 S ribosomal RNAs. FEBS Lett. 1975 Nov 1;59(1):60–63. doi: 10.1016/0014-5793(75)80341-3. [DOI] [PubMed] [Google Scholar]
- Maden B. E. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- Melançon P., Lemieux C., Brakier-Gingras L. A mutation in the 530 loop of Escherichia coli 16S ribosomal RNA causes resistance to streptomycin. Nucleic Acids Res. 1988 Oct 25;16(20):9631–9639. doi: 10.1093/nar/16.20.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16 S rRNA. J Mol Biol. 1990 Jan 5;211(1):135–145. doi: 10.1016/0022-2836(90)90016-F. [DOI] [PubMed] [Google Scholar]
- Nichols J. L., Lane B. G. In vivo incorporation of methyl groups into the ribose of Escherichia coli ribosomal RNA. J Mol Biol. 1967 Dec 28;30(3):477–489. doi: 10.1016/0022-2836(67)90363-4. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Ribosomal RNA and translation. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- Noller H. F. tRNA-rRNA interactions and peptidyl transferase. FASEB J. 1993 Jan;7(1):87–89. doi: 10.1096/fasebj.7.1.8422979. [DOI] [PubMed] [Google Scholar]
- O'Connor M., Göringer H. U., Dahlberg A. E. A ribosomal ambiguity mutation in the 530 loop of E. coli 16S rRNA. Nucleic Acids Res. 1992 Aug 25;20(16):4221–4227. doi: 10.1093/nar/20.16.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz S. C., McCloskey J. A. Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol. 1990;193:796–824. doi: 10.1016/0076-6879(90)93452-q. [DOI] [PubMed] [Google Scholar]
- Powers T., Noller H. F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991 Aug;10(8):2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T., Noller H. F. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1042–1046. doi: 10.1073/pnas.87.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T., Noller H. F. Evidence for functional interaction between elongation factor Tu and 16S ribosomal RNA. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1364–1368. doi: 10.1073/pnas.90.4.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T., Noller H. F. The 530 loop of 16S rRNA: a signal to EF-Tu? Trends Genet. 1994 Jan;10(1):27–31. doi: 10.1016/0168-9525(94)90016-7. [DOI] [PubMed] [Google Scholar]
- SPECTOR L. B., KELLER E. B. Labile acetylated uracil derivatives. J Biol Chem. 1958 May;232(1):185–192. [PubMed] [Google Scholar]
- Santer M., Santer U., Nurse K., Bakin A., Cunningham P., Zain M., O'Connell D., Ofengand J. Functional effects of a G to U base change at position 530 in a highly conserved loop of Escherichia coli 16S RNA. Biochemistry. 1993 Jun 1;32(21):5539–5547. doi: 10.1021/bi00072a007. [DOI] [PubMed] [Google Scholar]
- Saponara A. G., Enger M. D. The isolation from ribonucleic acid of substituted uridines containing alpha-aminobutyrate moieties derived from methionine. Biochim Biophys Acta. 1974 Apr 27;349(1):61–77. doi: 10.1016/0005-2787(74)90009-4. [DOI] [PubMed] [Google Scholar]
- Shen Z. H., Fox T. D. Substitution of an invariant nucleotide at the base of the highly conserved '530-loop' of 15S rRNA causes suppression of yeast mitochondrial ochre mutations. Nucleic Acids Res. 1989 Jun 26;17(12):4535–4539. doi: 10.1093/nar/17.12.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Powers T., Changchien L. M., Noller H. F. Interaction of ribosomal proteins S5, S6, S11, S12, S18 and S21 with 16 S rRNA. J Mol Biol. 1988 Jun 20;201(4):683–695. doi: 10.1016/0022-2836(88)90467-6. [DOI] [PubMed] [Google Scholar]
- Thomas G., Gordon J., Rogg H. N4-Acetylcytidine. A previously unidentified labile component of the small subunit of eukaryotic ribosomes. J Biol Chem. 1978 Feb 25;253(4):1101–1105. [PubMed] [Google Scholar]
- Woese C. R., Gutell R. R. Evidence for several higher order structural elements in ribosomal RNA. Proc Natl Acad Sci U S A. 1989 May;86(9):3119–3122. doi: 10.1073/pnas.86.9.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]