Abstract
The regulatory spine is a set of conserved residues that are assembled and disassembled upon activation and inactivation of kinases. We recently identified the regulatory spine within the immunologically important Tec family kinases and have shown that in addition to the core spine residues within the kinase domain itself, contributions from the SH2-kinase linker region result in an extended spine structure for this kinase family. Disruption of the regulatory spine, either by mutation or removal of the amino-terminal SH2-kinase linker region or mutation of core spine residues, leads to inactivation of the Tec kinases. With a focus on the Tec family members, Itk and Btk, we now show that the gatekeeper residue is also critical for the assembly of the regulatory spine. Mutation of the bulky Itk F434 gatekeeper residue to alanine or glycine inactivates Itk. The activity of the Itk F434A mutant can be recovered by a secondary-site mutation within the N-terminal lobe, specifically L432I. The Itk L432I mutation likely rescues the activity of the gatekeeper F434A mutation by promoting the assembly of the regulatory spine. We also show that mutation of the Itk and Btk gatekeeper residues to methionine is sufficient to activate the isolated kinase domains of Tec kinases in the absence of the amino-terminal SH2-kinase linker. Thus, shifting the conformational equilibrium between the assembled and disassembled states of the regulatory spine by changing the nature of the gatekeeper residue is key to regulating the activity of Tec kinases.
INTRODUCTION
Protein kinases are enzymes that control key processes in the cell by catalyzing the transfer of a phosphate group from ATP to the hydroxyl moiety of Ser/Thr or Tyr (1, 2). The Tec family kinases (Itk, Btk, Tec, Txk and Bmx) are non-receptor tyrosine kinases that are expressed predominantly in hematopoietic cells and function downstream of important immune receptors (3-6). The domain architecture of the Tec kinases is similar to that of the Src family kinases; both kinase families are characterized by the SH3-SH2-kinase domain arrangement (7). In addition, the Tec kinases (with the exception of Txk) also contain a PH domain at the amino-terminus making the overall domain architecture distinct from the Src kinases (7-10). While extensive biochemical and structural analyses of full-length Src kinases have provided a mechanistic understanding of how the catalytic activity of this kinase family is regulated (11, 12), an understanding of Tec kinase regulation lags significantly behind due in large part to the lack of a high resolution structure for any of the full length Tec kinases.
Despite the relative dearth of structural information for the Tec kinase family, biochemical analyses have shown that the regulation of the Tec kinases differs considerably from that of Src kinases (10). One of the most notable differences lies in the fact that the isolated kinase domains of the Src family kinases are active, while the isolated kinase domains of the Tec kinases are catalytically inert (9, 10, 13). The catalytic activity of the Src kinases is negatively controlled by the amino-terminal domains (14, 15). In contrast, the catalytic activity of the Tec kinases is positively dependent on the presence of the amino-terminal regulatory domains, more specifically the 17-residue linker region between the SH2 domain and the kinase domain (10). A conserved tryptophan residue (Itk W355) in the linker between the SH2 and kinase domains of the Tec kinases, along with a conserved methionine residue (Itk M410) within the C-helix of the kinase domain, play a critical role in stabilizing the active state of Tec kinases (10, 16). Like other kinase families, the activity of the Tec kinases is also positively regulated by phosphorylation on a conserved tyrosine (Itk Y511 or Btk Y551) within a large flexible loop within the kinase domain called the activation loop (17).
Recently, a conserved structure called the ‘regulatory spine’ has been identified as a key component in controlling the activity of kinases (18, 19). The regulatory spine consists of a set of residues that span the three-dimensional structure of the kinase domain from the carboxy-terminal lobe to the amino-terminal lobe. The regulatory spine residues are organized into an assembled structure in active kinases and are disrupted in inactive kinases (18). We have previously identified the regulatory spine residues within the Tec family kinases (20). In that work we demonstrated that, in addition to the core regulatory spine residues present within the kinase domain itself, the Tec kinases contain an extended regulatory spine that includes the conserved tryptophan (Itk W355) from within the SH2-kinase linker and the conserved methionine (Itk M410) on the C-helix of the kinase domain.
Mutation of the residues that constitute the extended regulatory spine in the context of full-length Itk results in a drastic reduction in the activity of the kinase (20). Moreover, the presence of the SH2-kinase linker is critical for the assembly of a stable regulatory spine in Tec kinases. In fact, the crystal structures of the isolated kinase domain of Itk (without the SH2-kinase linker) in the ‘active’ (Y511 phosphorylated) and ‘inactive’ (Y511 unphosphorylated) states are identical (8). In both structures the distance between the crucial ion pair (Itk K390 and E405) is ~8 Å; significantly greater than the 3-4 Å that is typically observed in structures of the active states of kinases. Thus, in the absence of the SH2-kinase linker the regulatory spine of Tec kinases fails to assemble despite activation loop phosphorylation (20). The extended regulatory spine within Tec kinases therefore defines an allosteric mechanism by which the regulatory SH2-kinase linker region impinges on the kinase domain in a positive manner and stabilizes the active conformation of the kinase domain.
In addition to the regulatory spine and non-catalytic regulatory domains/regions, there are other features that modulate the activity of kinases. These include the gatekeeper residue, a key residue within the amino-terminal lobe of the kinase that controls access to a deep pocket within the kinase domain. Structural studies have shown that the gatekeeper residue lies at the edge of the regulatory spine and introduction of a bulky hydrophobic residue such as isoleucine or methionine at the gatekeeper position has been demonstrated to stabilize the regulatory spine and activate Src, Abl and EGFR kinases (21).
In this manuscript, we set out to explore the interplay between the regulatory spine and the gatekeeper residue in the Tec family kinases. The data demonstrate that mutation of the Itk gatekeeper residue F434 to alanine or glycine inactivates full-length Itk. In the context of this result, we identify a secondary mutation that activates the Itk F434A mutant. Introduction of a β-branched hydrophobic residue at position 432 (L432I) rescues the activity of the Itk F434A mutant. We suggest that the β-branched isoleucine at position 432 potentially compensates for the loss in hydrophobicity of the F434A mutation, and rescues the activity of Itk by promoting the assembly of the regulatory spine. Furthermore, we show that the isolated kinase domains of Itk and Btk can be activated (regulatory spine assembled) in the absence of the amino-terminal regulatory domains by mutation of the gatekeeper residue to methionine. Thus, the gatekeeper residue, in conjunction with the regulatory spine, are key modulators of Tec kinase activity.
EXPERIMENTAL PROCEDURES
Constructs
The baculoviral expression constructs for full-length (mouse) Itk has been described previously (10). The bacterial expression constructs for the Itk kinase domain have been described elsewhere (22). The mouse wild-type Btk kinase domain (396-659) was PCR amplified and cloned into the pET 28b (Novagen) vector to create the His-tagged Btk kinase domain. All mutations were introduced by using the site directed mutagenesis (SDM) kit (Stratagene). All constructs were verified by sequencing at the Iowa State DNA synthesis and sequencing facility.
Protein expression and purification
Baculoviral constructs were expressed and purified from Sf9 cells as described previously (10). The bacterial expression constructs for the His-tagged Itk, Btk kinase domains were expressed and purified from ArcticExpress cells (Stratagene) as described previously (22). Briefly, the Itk or Btk kinase domains were expressed in ArcticExpress bacteria at 12°C for 23 hours. The cell pellets were re-resuspended in lysis buffer (0.5 mg/ml lyzozyme, 50 mM KH2PO4 pH 8.0, 150 mM NaCl, 20 mM imidazole) and stored overnight at −80°C. The cell pellets were thawed after the addition of 1 mM PMSF and 3000 Units DNase I (Sigma). The lysate was spun at 14K for 1 hour at 4°C. The supernatant was incubated with Nickel NTA resin (Qiagen). The resin was washed with wash buffer (50 mM Hepes pH 7.5, 150 mM NaCl, 40 mM imidazole) and then eluted with elution buffer (50 mM Hepes pH 7.5, 150 mM NaCl, 250 mM imidazole, 10% glycerol). The proteins were aliquoted, flash frozen with liquid nitrogen and stored at −80°C.
Kinase assays and western blotting
In vitro kinase assays were performed by incubating the isolated kinase domain of Itk or Btk in a kinase assay buffer (50 mM Hepes pH 7.0, 10 mM MgCl2, 1 mM DTT, 1 mg/ml BSA, 1 mM Pefabloc and 200 μM ATP) at RT for one hour. The samples were boiled, separated by SDS-PAGE and western blotted with the Anti Btk phosphoY551 antibody (BD Biosciences), anti-FLAG (Sigma) antibody or anti-His (Upstate) antibody as described previously (23). The Anti Btk phosphoY551 antibody is also used for detection of phosphorylation on Itk Y511 . Kinetic parameters for the full-length wild-type and mutant Itk are derived using radioactive assays that have been described previously (10).
Activity measurements of pY551 level normalized Btk kinase domain wild-type and mutants were carried out by pre-incubating the Btk kinase domain with ATP at RT for varying lengths of time (Btk WT and T474I: 45 min; Btk T474A: 90 min; Btk T474M: 30 seconds). The autophosphorylated Btk kinases were subsequently tested for activity using radioactive assays as described previously (10).
RESULTS
The Itk F434A gatekeeper mutation inactivates full-length Itk
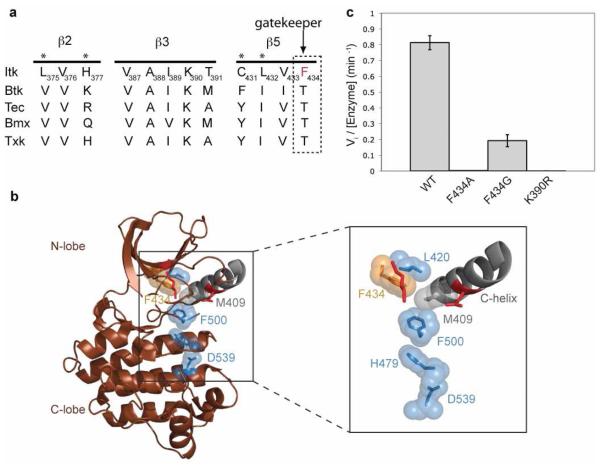
There is ample precedence for the activation of kinase activity by mutation of the gatekeeper residue to a bulky hydrophobic residue (21). In all kinases, the gatekeeper residue is located on the β5 strand at the edge of the regulatory spine within the N-terminal lobe of kinases and introduction of a large hydrophobic side chain is thought to promote kinase activity by favoring the active, assembled conformation of the regulatory spine (21). Interestingly, alignment of the β5 strand of the Itk kinase domain with other members of the Tec kinase family shows that Itk is unique within the Tec kinases as its gatekeeper residue is a phenylalanine instead of the threonine that is present in other Tec kinases (Fig. 1a). Thus, for wild type Itk, the gatekeeper residue (F434) is already large and hydrophobic and inspection of the available Itk structures suggests that F434 is adjacent to the spine and therefore might be expected to favor assembly of the regulatory spine (Fig. 1b). Given the bulky, hydrophobic nature of the Itk gatekeeper residue we first tested the prediction that mutation of the Itk gatekeeper residue to a smaller alanine or glycine residue would shift the conformational equilibrium to the disassembled state of the regulatory spine and result in inactive Itk. Indeed, mutation of the bulky Itk gatekeeper residue F434 in full-length Itk to either alanine or glycine leads to a dramatic loss in activity (Fig. 1c, Table 1). The Itk F434A mutant reduces the activity to that of the kinase inactive K390R mutant of Itk, whereas the Itk F434G mutant retains about 25% of the wild-type kinase activity (Fig. 1c, Table 1). Thus, mutation of the Itk gatekeeper residue to alanine or glycine inactivates full-length Itk presumably by destabilizing the assembled conformation of the regulatory spine.
Figure 1. The Itk gatekeeper residue F434 promotes the assembled state of the regulatory spine.
(a) Alignment of the β2, β3 and β5 strands of the N-terminal lobe of the kinase domain of Tec kinases. The gatekeeper residue of Itk is a phenylalanine (F434), all other Tec kinases contain a threonine at the gatekeeper position (indicated with arrow). Asterisks above the sequences indicate targets for the second-site mutagenesis. (b) The crystal structure of the isolated Itk kinase domain in the absence of the SH2-kinase linker (brown cartoon, PDB ID 1SNX) is shown with the disassembled regulatory spine residues: M409 (grey sticks and spheres), L420, H479, F500 and D539 (blue sticks and spheres). The boxed region is expanded to show the Itk gatekeeper F434 (orange sticks and spheres), and the C-helix is labeled. The crucial ion pair (Itk K390 and E405) that is assembled in active kinase structures is shown as red sticks and is not labeled. All structures in this and other figures were generated using PyMOL (33). (c) The gatekeeper residue of Itk (F434) was mutated to either alanine or glycine in the context of full-length Itk and tested for its in vitro kinase activity as described in materials and methods. Mutation of F434 to alanine or glycine reduces the activity of full-length Itk to almost that of the kinase inactive mutant (K390R) of full-length Itk.
Table 1.
| Itk full-length | Vi/[Enzyme] (min−1) |
|---|---|
| Itk wild-type | 0.81 ± 0.04 |
| Itk K390R | 0.001 ± 0.00 |
| Itk F434A | 0.003 ± 0.00 |
| Itk F434G | 0.19 ± 0.04 |
| Itk L432I/F434A | 0.60 ± 0.06 |
| Itk C431V/F434A | 0.009 ± 0.007 |
| Itk L375V/F434A | 0.021 ± 0.018 |
| Itk H377V/F434A | 0.030 ± 0.029 |
|
| |
| Itk isolated kinase domain | |
|
| |
| Background | 0.004 ± 0.0005 |
| Itk wild-type | 0.004 ± 0.0003 |
| Itk F434T | 0.003 ± 0.0002 |
| Itk F434I | 0.005 ± 0.0001 |
| Itk F434M | 0.008 ± 0.0003 |
| Itk L432I/F434M | 0.010 ± 0.0006 |
| Btk isolated kinase domain | (Without pY551 level normalization) |
(pY551 level normalized) |
|---|---|---|
| Background | 0.001 ± 0.000 | 0.001 ± 0.001 |
| Btk wild-type | 0.09 ± 0.001 | 0.10 ± 0.001 |
| Btk T474A | 0.06 ± 0.001 | 0.03 ± 0.015 |
| Btk T474M | 0.56 ± 0.060 | 0.53 ± 0.084 |
| Btk T474I | 0.06 ± 0.000 | 0.12 ± 0.008 |
Activity measurements were carried out at room temperature using 5 μCi of [32P] ATP and Peptide B (aminohexanoyl biotin-EQEDEPEGIYGVLF-NH2) as a substrate in a kinase assay buffer (50 mM Hepes pH 7.0, 10 mM MgCl2, 1 mM DTT, 1 mg/mL BSA, and 1 mM Pefabloc) as described previously (10).
The L432I is a second-site mutation that rescues the activity of the Itk F434A mutant
Gatekeeper mutations for many kinases have been extensively examined in the context of generating kinase variants that are sensitive to modified ATP analogs (24, 25). In the context of that work it has been found that certain kinases lose their activity upon mutation of the gatekeeper residue to alanine or glycine. Such kinases are referred to as ‘intolerant’ kinases, whereas the kinases that retain activity upon mutation of the gatekeeper residue to alanine or glycine are defined as ‘tolerant’ kinases (26). Previous work on ‘intolerant’ kinases has shown that their loss in activity due to the gatekeeper mutation can be rescued by a second site mutation at one of several specific locations within the amino-terminal lobe of the kinase (26). These activating secondary mutations always involve introduction of a β-branched amino acid residue at sites that do not contain a β-branched residue in the wild type sequence and cluster to the β2, β3 and β5 strands of the N-terminal kinase lobe (26). Since we have demonstrated that Itk is an ‘intolerant’ kinase (Fig. 1c), we next set out to rescue the activity of the Itk F434A mutant using this second-site mutagenesis strategy.
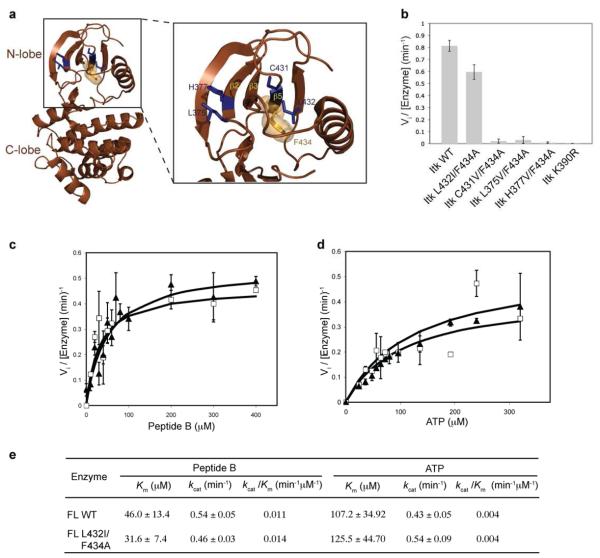
Alignment of the β2, β3 and β5 strands of Itk with other members of Tec kinases shows that within the β3 strand of Itk, V387 and I389 are already β-branched amino acids and hence are not candidates for a secondary mutation (Fig. 1a). The Itk β2 and β5 strands have four potential sites for second-site mutations: L375, H377, C431 and L432 (Fig. 1a and 2a). Each of these residues was therefore mutated to a β-branched amino acid within the context of full-length Itk F434A and tested for activity using a peptide substrate. Three of the double mutants: C431V/F434A, L375V/F434A and H377V/F434A do not exhibit significant activity (Fig. 2b, Table 1). However, the Itk L432I/F434A double mutant has significant kinase activity when compared to full-length wild-type Itk (Fig. 2b, Table 1). We further characterized the Itk L432I/F434A double mutant by measuring its kinetic parameters. As shown in Figure 2c, d and e, the kinetic parameters (Km and kcat) for the full-length Itk L432I/F434A double mutant are nearly identical to that of wild-type full-length Itk. Hence, the Itk L432I mutation is a second-site mutation that rescues the activity of the Itk F434A gatekeeper mutation.
Figure 2. The Itk L432I/F434A double mutation rescues the activity of the Itk gatekeeper F434A mutation.
(a) Location of potential second-site activating mutations within the N-terminal lobe of the Itk kinase domain. The crystal structure of the Itk kinase domain (brown cartoon) with the β2, β3 and β5 strands of the N-terminal lobe are indicated. The location of the four potential activating sites (L375, H377, C431 and L432) are shown as blue sticks and the gatekeeper (F434) residue is shown in orange (sticks and spheres). (b) The L432I mutation rescues the activity of the F434A gatekeeper mutation in Itk. The L375V, H377V, C431V and L432I mutations were made in the context of full-length Itk F434A, and the double mutants were tested for kinase activity as described in materials and methods. The Itk L432I/F434A double mutation recovers the activity of the Itk F434A mutant. (c, d and e) Kinetic parameters of the full-length Itk L432I/F434A double mutant. Substrate (peptide B or ATP) curves of wild-type full-length Itk (FL WT, filled triangles) or Itk L432I/F434A double mutant (FL L432I/F434A, open squares), were fit to the Michaelis-Menten equation using GraphFit to obtain the kinetic parameters reported in (e). The values for full-length wild-type Itk have been reported previously (10).
Stabilization of the assembled state of the regulatory spine activates Tec kinases in the absence of the N-terminal regulatory domains
Previous studies have demonstrated that kinases can be activated by the introduction of a bulky hydrophobic residue such as isoleucine or methionine at the gatekeeper position (21). In fact, the gatekeeper residue has been identified as a hotspot for oncogenic transformation of several kinases (27). An activating T315I gatekeeper mutation within the Abl kinase domain and a T790M gatekeeper mutation within EGFR have been detected in chronic myeloid leukemia (CML) patients and lung cancer patients, respectively (21, 28). Structural analysis of the Src gatekeeper T341I mutant showed that the gatekeeper residue lies at the tip of the hydrophobic spine. Since mutation of the gatekeeper residue to isoleucine or methionine stabilizes the hydrophobic spine in other kinase families, we next tested whether an analogous mutation would activate Itk and Btk (all Tec kinases contain Thr at the gatekeeper position with the exception of Itk that contains Phe as the gatekeeper residue). The Tec kinases pose an additional challenge; the isolated kinase domains exhibit very poor activity in the absence of the amino-terminal regulatory region (9, 10). Our previous work has demonstrated that the amino-terminal region (in particular the SH2-kinase linker region preceding the kinase domain) is required for assembly of the regulatory spine and activation of kinase actvity (10). We therefore hypothesized that mutation of the gatekeeper residue within Btk (and possibly Itk) to isoleucine or methionine might not only activate these kinases by pre-organizing the regulatory spine but might also overcome the requirement for the positive regulatory N-terminal region.
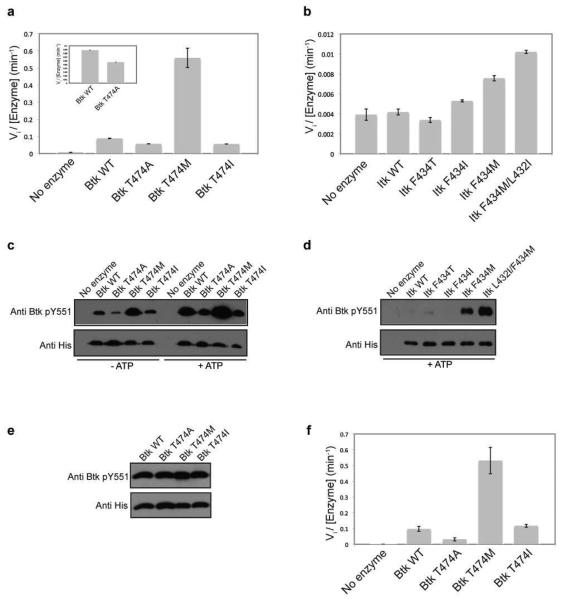
The gatekeeper residues of both Itk (F434) and Btk (T474) were mutated to either isoleucine or methionine in the context of the isolated Itk or Btk kinase domains. In vitro kinase activity measurements show that mutation of the Itk or Btk gatekeeper residue to methionine in the context of the isolated kinase domain results in an increase in activity compared to wild-type Btk and Itk, respectively (Fig. 3a and b, Table 1). Interestingly, mutation of the gatekeeper residue to an isoleucine does not activate either the Btk or Itk kinase domains (Fig. 3a and b, Table 1). This is in contrast to the findings that Src kinases are activated by mutation of the gatekeeper threonine to isoleucine (21). The activating effect of the methionine mutation versus the isoleucine mutation likely reflects the better accommodation of the methionine side chain over the β-branched isoleucine in the structure of Itk and Btk. Additionally, consistent with our earlier results that show the inactivating effect of the Itk F434A mutation (Fig. 1c), mutation of the Btk gatekeeper residue T474 to alanine also decreases the activity of Btk, albeit to a lesser extent than Itk (Fig 3a, Table 1). The CD spectra of the wild-type Itk and Btk isolated kinase domains overlay well with their respective gatekeeper mutants, suggesting that the observed changes in activity reflect the effect of mutation and are not due to misfolding in the kinase domain (Supplemental Figure S1a and b). Overall, these results suggest that both Itk and Btk are ‘intolerant’ kinases and mutation of the gatekeeper residue to methionine, but not isoleucine, activates these Tec kinases in the absence of the amino-terminal regulatory domains that normally activate kinase activity.
Figure 3. Mutation of the gatekeeper residue to methionine activates Tec kinases in the absence of the N-terminal regulatory domains.
(a and b) The gatekeeper residues of Itk and Btk were mutated to alanine, methionine, isoleucine and threonine in the context of the isolated kinase domains and tested for in vitro kinase activity as described in Experimental Procedures. Mutation of the Itk and Btk gatekeeper residues, F434 and T474 respectively, to methionine activates the isolated kinase domains of Itk and Btk. The inset in (a) shows the Btk T474A mutant is approximately half as active as wild type Btk kinase domain. (c and d) Activity of the Tec kinase gatekeeper mutations correlates with the level of phosphorylation on the activation loop of the kinase. Purified 250 nM kinase domain of Btk wild-type, T474A, T474I or T474M enzymes or Itk wild-type, F434T, F434I, F434M or L432I/F434M enzymes were incubated in a kinase assay buffer at RT for one hour without or with ATP, separated by SDS-PAGE, transferred onto polyvinylidene difluoride (PVDF) membrane, and probed with either an anti-Btk pY551 (also used to detect Itk pY511) or anti-His antibody. The exposure time for the Itk panel is ten times longer than that for the Btk panel. (e) Normalization of phosphorylation level on Btk Y551. Purified 250 nM kinase domain of Btk wild-type, T474A, T474I or T474M enzymes were pre-incubated with ATP in a kinase assay buffer as described in Experimental Procedures. The Btk enzymes were then probed with an anti-Btk pY551 and anti-His antibody as before. (f) The Btk kinase domain enzymes in (e) that were normalized for phosphorylation on Btk Y551 were tested for in vitro kinase activity as before.
In addition to measuring the initial velocity associated with peptide substrate phosphorylation, we can exploit the correlation between activation loop phosphorylation and activity for these enzymes. The activity of the wild type and variant Itk and Btk enzymes correlates with the level of phosphorylation on the activation loop tyrosine (Y551 in Btk and Y511 in Itk). Phosphorylation on Y551 is observed for bacterially expressed Btk kinase domains in the absence of incubation with ATP, with the Btk T474M mutant showing maximum Y551 phosphorylation (Fig 3c). Incubation of the purified Btk kinase domains with ATP shows that the Btk T474M mutant autophosphorylates itself on Y551 more efficiently when compared to wild-type Btk (Fig 3c). Unlike Btk, phosphorylation on Itk Y511 is only detected after incubation of the purified Itk kinases with ATP (Fig. 3d). The Itk F434M mutant shows greater phosphorylation on Y511 compared to wild-type Itk (Fig. 3d). Given the results described above that show the L432I mutation activates Itk, we tested whether this mutation would further activate the Itk gatekeeper F434M mutant. Indeed it does, the Itk L432I/F434M double mutant within the isolated kinase domain of Itk shows greater phosphorylation on Y511 compared to Itk F434M (Fig. 3d) and brings about a 1.5 fold increase in activity toward a peptide substrate over that of Itk F434M (Fig 3b, Table 1). As suggested above for the rescue of the intolerant Itk kinase, it is possible that the Itk L432I mutation promotes the assembly of the regulatory spine within Itk leading to the more active kinase.
While there is a direct correlation between the level of phosphorylation on the activation loop and activity of Itk and Btk, phosphorylation on the activation loop (Itk Y511 or Btk Y551) has been previously shown to activate Itk and Btk (17). While the Itk kinase domains purified from bacteria are uniformly unphosphorylated on Y511, the Btk kinase domain mutants display differential phosphorylation on the activation loop Y551 (Fig. 3c). To ensure that the activity measurements of the Btk kinase domain mutants (using a peptide substrate) are not skewed due to this differential phosphorylation on Y551, the Btk kinase domain mutants were pre-incubated for various times with ATP to normalize the pY551 levels and these pY551-normalized enzymes were then tested for kinase activity using a peptide substrate as before. As shown in Fig 3, the relative activities of the Btk kinases with normalized levels of pY551 (Fig. 3e and f, Table 1) is similar to that of the Btk kinases prior to pY551 level normalization (Fig. 3c, Table 1). Hence, the observed changes in activity due to the gatekeeper mutations are reflective of the changes in intrinsic kinase activity of Btk.
We note that the activity of Itk (wild-type and mutants) is consistently less than that of Btk (Table 1). One significant difference already noted is the identity of the gatekeeper residue. Despite the fact that the less active Itk contains the bulkier hydrophobic gatekeeper, we mutated Itk F434 to threonine, the corresponding gatekeeper residue in Btk, to assess whether this difference is responsible for the observed activity differences. This mutation, F434T, has no effect on Itk kinase activity (Fig 3b and d, Table 1) and so the differences in the basal activity of the wild-type isolated kinase domains of Itk and Btk cannot be accounted for by the difference in the identity of the gatekeeper residue. Future work will explore the basis for the observed differences in activity among the Tec family kinases.
DISCUSSION
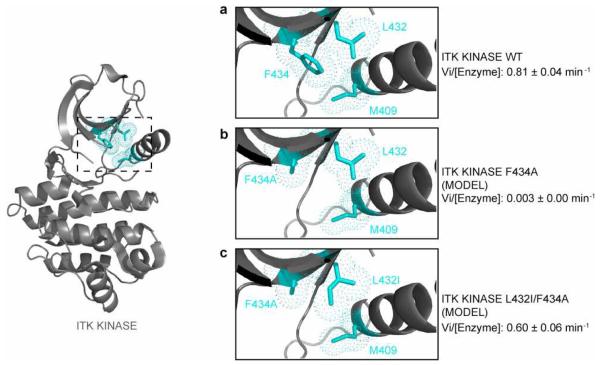
The regulatory spine is a key modulator of kinase activity (18, 19) and our recent work identified an extended spine structure that controls the activity of the Tec family kinases (20). The gatekeeper residue sits at the edge of the regulatory spine structure and so it is logical that, for Itk, the loss of the large phenylalanine gatekeeper by mutation to alanine inactivates Itk (Fig. 1c). The crystal structure of Itk (8) shows that the second-site activating mutation, L432I, is adjacent to the gatekeeper residue F434 and the regulatory spine residue M409 (Fig. 4). Models of the Itk kinase domain with the F434A single mutation and the L432I/F434A double mutation show that the substitution of a β-branched Ile residue at position 432 likely compensates for the loss in hydrophobicity of the F434A mutation favoring assembly of the regulatory spine within Itk (Fig. 4a, b and c). It is generally accepted that β-branched residues are preferred over non β-branched residues within β-sheets (29, 30) and so it is also possible that the β-branched isoleucine in the Itk L432I/F434A double mutant simply stabilizes the overall fold of the kinase. It should be noted, however, that we introduced β-branched residues at a number of different locations within the amino-terminal lobe of the kinase (all on β-strand structures) and none of these other substitutions rescued the activity of the Itk F434A mutant (Fig. 2a). Thus, while introduction of β-branched residues might increase the overall stability of the β-sheet, the fact that the L432I mutation is the only one among all the β-branched residues that rescued the activity of Itk points to its proximity to the regulatory spine as a key factor in its ability to act as a second-site activating mutation.
Figure 4. The Itk L432I mutation likely restores the regulatory spine in the Itk F434A mutant.
The crystal structure of the Itk kinase domain, PDB ID 3MIY, (grey cartoon) is shown at left with the location of the regulatory spine residue: M409, the gatekeeper residue: F434 and the site of the secondary activating mutation: L432 (cyan sticks and dots) indicated. The boxed region is expanded in panels (a), (b) and (c). Comparison of the side chain packing within (a) wild-type Itk (PDB ID 3MIY); (b) model of the Itk F434A single mutant; and (c) model of the Itk L432I/F434A double mutant. The introduction of the β-branched isoleucine residue at position 432 is likely more effective than leucine in compensating for the loss in hydrophobicity of the gatekeeper F434A mutation. The leucine rotamer about chi1 shown in (b) is unchanged from the Itk crystal structure (3MIY) shown in (a). While alternative leucine rotamers might be favored in the F434A single mutant, inspection of the alternative rotamers suggests that none appear to show increased packing with the residues at position 434 and 409 in the context of the 3MIY structure than that shown in (b). Kinetic data (see Table 1) are indicated for each structural model.
Interestingly, there are differences in regulation even within the Tec family kinases. The isolated wild-type Btk kinase domain is significantly more active than the isolated Itk kinase domain (Fig. 3a and b, Table 1). Swapping the gatekeeper residue in Itk to that of Btk did not generate a more active Itk enzyme (Fig. 3b and d, Table 1). One possibility for the observed difference in activity between Itk and Btk might be related to the nature of the residue at the activating second-site identified in this work. Comparison of the Itk and Btk kinase domain sequences shows that wild type Btk contains a β-branched residue (isoleucine) at the position corresponding to L432 in Itk (Fig. 1a). Mutation of L432 to isoleucine in Itk increased catalytic activity and so the isoleucine already present at this site in Btk might lead to the higher basal activity observed for the wild-type Btk kinase domain. However, while the Itk L432I mutation is activating, this substitution did not increase Itk activity to the level of Btk and its variants (Fig. 3a and b, Table 1) and so additional parameters must also affect the relative activities of each enzyme.
The non-catalytic regulatory domains of the Tec kinases serve a positive role in controlling kinase activity. Disconnecting the regulatory region from the catalytic kinase domain (either by mutation of W355 in the Itk SH2-kinase linker region or by truncation of the enzyme to yield the isolated, independent kinase domain) results in an enzyme with very poor catalytic activity (9, 10, 16). In the current work we have found that mutation of residues that surround the regulatory spines of Itk and Btk results in increased catalytic activity of the independent kinase domains. In particular, the Itk F434M and Btk T474M mutations in the context of the free kinase domain are significantly activating and partially overcome the requirement for the SH2-kinase linker (Fig. 3, Table 1). These findings suggest that for the wild type kinase domains of the Tec kinases, the conformational preference of the regulatory spine is the inactive, disassembled state and the active, assembled conformation is energetically disfavored. The active conformation is only stabilized upon additional interactions between the kinase domain and the amino-terminal regulatory region (20). Gain of function mutations for the Tec kinases have to date been elusive and we suggest that the Itk F434M and Btk T474M mutants might find utility as a research reagent.
In addition to the N-terminal regulatory domains, phosphorylation on the activation loop has also been shown to activate kinases (17). For Itk, activation by phosphorylation on the activation loop is completely dependent on the presence of the SH2-kinase linker region. Indeed, while phosphorylation on the activation loop has been shown to activate full-length Itk or deletion constructs of Itk kinase containing the SH2-kinase linker region (10, 17), the isolated kinase domain of Itk (without the SH2-kinase linker) exhibits poor catalytic activity even when completely phosphorylated on the activation loop (figure 1 in (8)). Additionally, activity assays that directly compare two equally phosphorylated fragments of Itk that differ only with respect to the presence of the linker region indicate that the linker-containing fragment of Itk is almost as active as full-length Itk while the smaller fragment that contains only the kinase domain of Itk (lacking the SH2-kinase linker) exhibits poor activity regardless of phosphorylation state (10). These data suggest that the poor activity of the isolated kinase domain of Itk is not due to the lack of phosphorylation on the activation loop but rather it is the SH2-kinase linker region that is essential for the assembly of the regulatory spine within the isolated kinase domain of Itk (10, 20). This is also consistent with the observation that the Itk regulatory spine is not assembled in the crystal structures of the phosphorylated Itk kinase domain lacking the linker region (8, 20). Thus, in the context of wild type full length Itk, phosphorylation on the activation loop certainly drives the conformational equilibrium of the regulatory spine to that of the assembled state, but the SH2-kinase linker region is an equal, if not more critical component of the activating machinery of this kinase.
Identification of legitimate in vivo substrates of kinases has been challenging (31). The gain of function mutant described above may be one approach to advance our understanding of Tec family kinase signaling. Another approach that has been developed and used widely for the identification of protein kinase substrates involves mutation of the gatekeeper residue to a residue with a small side chain (32). Mutation of the gatekeeper residue to alanine or glycine creates a ‘hole’ within the kinase domain active site that can accommodate larger modified ATP analogues or tailored inhibitors that can be used to identify kinase specific substrates in vivo. In the current work we have shown that Itk is ‘intolerant’ to this type of gatekeeper mutation (Itk F434A is inactive) but can be rescued by mutation at a second site (L432I). The location of this second-site activating mutation suggests a role for the kinase regulatory spine; loss of hydrophobic packing interactions upon mutation of the gatekeeper phenylalanine to alanine might be balanced by favorable contacts to the isoleucine sidechain at position 432. In addition to side chain packing effects, changes in backbone dynamics/flexibility could also contribute to the observed activating effect of these mutations. Regardless of the precise mechanism by which these combinations of mutations promote Itk activity, the Itk L432I/F434A double mutant is a useful tool for probing Itk specific signaling.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Owen Witte for providing the Btk cDNA template. This work is supported by grants from the National Institutes of Health (National Institute of Allergy and Infectious Diseases, AI043957 and AI075150) to A.H.A.
Abbreviations
- Itk
Interleukin-2 tyrosine kinase
- Btk
Bruton's tyrosine kinase
- SH2
Src homology 2
- SH3
Src homology 3
Footnotes
SUPPLEMENTAL INFORMATION
Comparison of the CD spectra of the Btk and Itk isolated kinase domains. S1 (a) The CD spectra of wild-type isolated Btk kinase domain superimposed with spectra of the isolated Btk kinase domain mutants (T474A, T474M and T474I). S1 (b) The CD spectra of wild-type isolated Itk kinase domain superimposed with spectra of the isolated Itk kinase domain mutants (F434M, F434I, F434T single mutants and L432I/F434M double mutant). Supplemental materials may be accessed free of charge online at http://pubs.acs.org.
REFERENCES
- 1.Bradshaw JM. The Src, Syk, and Tec family kinases: distinct types of molecular switches. Cell Signal. 2010;22:1175–1184. doi: 10.1016/j.cellsig.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 3.Andreotti AH, Schwartzberg PL, Joseph RE, Berg LJ. T-cell signaling regulated by the Tec family kinase, Itk. Cold Spring Harb Perspect Biol. 2010;2:a002287. doi: 10.1101/cshperspect.a002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg LJ, Finkelstein LD, Lucas JA, Schwartzberg PL. Tec family kinases in T lymphocyte development and function. Annu Rev Immunol. 2005;23:549–600. doi: 10.1146/annurev.immunol.22.012703.104743. [DOI] [PubMed] [Google Scholar]
- 5.Schwartzberg PL, Finkelstein LD, Readinger JA. TEC-family kinases: regulators of T-helper-cell differentiation. Nat Rev Immunol. 2005;5:284–295. doi: 10.1038/nri1591. [DOI] [PubMed] [Google Scholar]
- 6.Smith CI, Islam TC, Mattsson PT, Mohamed AJ, Nore BF, Vihinen M. The Tec family of cytoplasmic tyrosine kinases: mammalian Btk, Bmx, Itk, Tec, Txk and homologs in other species. Bioessays. 2001;23:436–446. doi: 10.1002/bies.1062. [DOI] [PubMed] [Google Scholar]
- 7.Joseph RE, Andreotti AH. Conformational snapshots of Tec kinases during signaling. Immunol Rev. 2009;228:74–92. doi: 10.1111/j.1600-065X.2008.00740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown K, Long JM, Vial SC, Dedi N, Dunster NJ, Renwick SB, Tanner AJ, Frantz JD, Fleming MA, Cheetham GM. Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors. J Biol Chem. 2004;279:18727–18732. doi: 10.1074/jbc.M400031200. [DOI] [PubMed] [Google Scholar]
- 9.Hawkins J, Marcy A. Characterization of Itk tyrosine kinase: contribution of noncatalytic domains to enzymatic activity. Protein Expr Purif. 2001;22:211–219. doi: 10.1006/prep.2001.1447. [DOI] [PubMed] [Google Scholar]
- 10.Joseph RE, Min L, Andreotti AH. The Linker between SH2 and Kinase Domains Positively Regulates Catalysis of the Tec Family Kinases. Biochemistry. 2007;46:5455–5462. doi: 10.1021/bi602512e. [DOI] [PubMed] [Google Scholar]
- 11.Boggon TJ, Eck MJ. Structure and regulation of Src family kinases. Oncogene. 2004;23:7918–7927. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- 12.Cowan-Jacob SW, Fendrich G, Manley PW, Jahnke W, Fabbro D, Liebetanz J, Meyer T. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure. 2005;13:861–871. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Veillette A, Caron L, Fournel M, Pawson T. Regulation of the enzymatic function of the lymphocyte-specific tyrosine protein kinase p56lck by the non-catalytic SH2 and SH3 domains. Oncogene. 1992;7:971–980. [PubMed] [Google Scholar]
- 14.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of cSrc reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 15.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 16.Guo S, Wahl MI, Witte ON. Mutational analysis of the SH2-kinase linker region of Bruton's tyrosine kinase defines alternative modes of regulation for cytoplasmic tyrosine kinase families. Int Immunol. 2006;18:79–87. doi: 10.1093/intimm/dxh351. [DOI] [PubMed] [Google Scholar]
- 17.Heyeck SD, Wilcox HM, Bunnell SC, Berg LJ. Lck phosphorylates the activation loop tyrosine of the Itk kinase domain and activates Itk kinase activity. J Biol Chem. 1997;272:25401–25408. doi: 10.1074/jbc.272.40.25401. [DOI] [PubMed] [Google Scholar]
- 18.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci U S A. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kornev AP, Taylor SS, Ten Eyck LF. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci U S A. 2008;105:14377–14382. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joseph RE, Xie Q, Andreotti AH. Identification of an allosteric signaling network within Tec family kinases. J Mol Biol. 2010;403:231–242. doi: 10.1016/j.jmb.2010.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat Struct Mol Biol. 2008;15:1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joseph RE, Andreotti AH. Bacterial expression and purification of Interleukin-2 Tyrosine kinase: Single step separation of the chaperonin impurity. Protein Expr Purif. 2008 doi: 10.1016/j.pep.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joseph RE, Severin A, Min L, Fulton DB, Andreotti AH. SH2-Dependent Autophosphorylation within the Tec Family Kinase Itk. J Mol Biol. 2009;391:164–177. doi: 10.1016/j.jmb.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alaimo PJ, Shogren-Knaak MA, Shokat KM. Chemical genetic approaches for the elucidation of signaling pathways. Curr Opin Chem Biol. 2001;5:360–367. doi: 10.1016/s1367-5931(00)00215-5. [DOI] [PubMed] [Google Scholar]
- 25.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C, Kenski DM, Paulson JL, Bonshtien A, Sessa G, Cross JV, Templeton DJ, Shokat KM. A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat Methods. 2005;2:435–441. doi: 10.1038/nmeth764. [DOI] [PubMed] [Google Scholar]
- 27.Dixit A, Yi L, Gowthaman R, Torkamani A, Schork NJ, Verkhivker GM. Sequence and structure signatures of cancer mutation hotspots in protein kinases. PLoS One. 2009;4:e7485. doi: 10.1371/journal.pone.0007485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mian AA, Schull M, Zhao Z, Oancea C, Hundertmark A, Beissert T, Ottmann OG, Ruthardt M. The gatekeeper mutation T315I confers resistance against small molecules by increasing or restoring the ABL-kinase activity accompanied by aberrant transphosphorylation of endogenous BCR, even in loss-of-function mutants of BCR/ABL. Leukemia. 2009;23:1614–1621. doi: 10.1038/leu.2009.69. [DOI] [PubMed] [Google Scholar]
- 29.Otzen DE, Fersht AR. Side-chain determinants of beta-sheet stability. Biochemistry. 1995;34:5718–5724. doi: 10.1021/bi00017a003. [DOI] [PubMed] [Google Scholar]
- 30.Minor DL, Jr., Kim PS. Measurement of the beta-sheet-forming propensities of amino acids. Nature. 1994;367:660–663. doi: 10.1038/367660a0. [DOI] [PubMed] [Google Scholar]
- 31.Manning BD, Cantley LC. Hitting the target: emerging technologies in the search for kinase substrates. Sci STKE. 2002;2002:pe49. doi: 10.1126/stke.2002.162.pe49. [DOI] [PubMed] [Google Scholar]
- 32.Bishop AC, Shah K, Liu Y, Witucki L, Kung C, Shokat KM. Design of allele-specific inhibitors to probe protein kinase signaling. Curr Biol. 1998;8:257–266. doi: 10.1016/s0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]
- 33.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.