Abstract
A hallmark of mammalian brain evolution is cortical expansion, which reflects an increase in the number of cortical neurons established by the progenitor cell subtypes present and the number of their neurogenic divisions. Recent studies have revealed a new class of radial glia-like (oRG) progenitor cells in the human brain, which reside in the outer subventricular zone. Expansion of the subventricular zone and appearance of oRG cells may have been essential evolutionary steps leading from lissencephalic to gyrencephalic neocortex. Here we show that oRG-like progenitor cells are present in the mouse embryonic neocortex. They arise from asymmetric divisions of radial glia and undergo self-renewing asymmetric divisions to generate neurons. Moreover, mouse oRG cells undergo mitotic somal translocation whereby centrosome movement into the basal process during interphase preceeds nuclear translocation. Our finding of oRG cells in the developing rodent brain fills a gap in our understanding of neocortical expansion.
Introduction
One of the most remarkable features in the evolution of the neocortex is the enormous increase in neuron number that reaches its peak in the human brain1–4. Although the laminar organization of the cortex is relatively similar in all mammals, an enormous expansion in cortical surface area underlies the transformation from smooth cortex to the highly folded primate neocortex, and the associated alteration of cortical architecture that is the substrate for the “higher” cortical functions that distinguish Homo sapiens from other mammalian species5. This transition underscores the importance of understanding the process of neurogenesis in the developing neocortex. Recent studies have identified two subtypes of neuronal progenitor cells in the developing rodent embryonic neocortex, radial glia (RG) and intermediate or basal progenitors (IP) 6–11. Neuroepithelial cells located in the apical-most region, the ventricular zone (VZ), transform to RG cells at the onset of neurogenesis. In addition to their well-characterized function as a scaffold supporting neuronal migration11, 15, RG cells constitute a major population of neural progenitor cells in the developing mammalian neocortex12–14. Radial glia display interkinetic nuclear migration (INM) and proliferate extensively at the luminal surface of the VZ (i.e. the apical VZ surface) 16, 17. The nuclei of newborn RG cells move away from the apical surface toward the basal lamina during G1, undergo S phase at a basal location, and return to the apical surface during G2 to undergo mitosis at the ventricular lumen9, 10, 12. Thus, INM is responsible for the pseudostratified appearance of the ventricular zone. Importantly, by moving interphase nuclei of RG cells away from the apical surface during G1, INM reserves the apical space for mitosis, and thereby may promote an expansion of RG cell number 18, 19. During the peak phase of neurogenesis (around embryonic day 13 to 18, [E13-E18], in mice), RG cells predominantly undergo asymmetric division to self-renew while simultaneously giving rise to either a neuron, or to an intermediate progenitor (IP) cell, the latter of which subsequently divides symmetrically to produce two neurons. IP cells appear to lack apical-basal polarity9, 11, 20.
An evolutionary increase in size and functional complexity of the cerebral cortex has culminated in the modern human brain that diverged from a rodent lineage ~ 100 million years ago4, 18, 21–23. Recent studies suggest that the development of oRG cells and their transit amplifying daughter cells (i.e. IP-like cells) may be the cellular mechanism underlying expansion in primate corticogenesis24. DiI-coated beads applied to the pial surface of fixed human cortical tissue have revealed oRG cells with RG-like morphology but lacking apical processes, and time-lapse imaging of fluorescently labelled human fetal brain slices show that oRG cells can self-renew and produce neuronal precursors24. Unlike RG cells, oRG cells show distinctive mitotic somal translocation behaviour instead of interkinetic nuclear migration (INM). It has been suggested that the OSVZ may be a primate specific feature and a hallmark of primate corticogenesis18, 25. But recent studies have shown that OSVZ progenitors (i.e. oRG cells) also exist in a non-primate species with a gyrencephalic brain, the ferret26, 27, which raises the question of whether oRG cells exist in lisencephalic species such as rodent, even though they have no cytoarchitectonically distinct OSVZ. While the RG cells and IP cells of the VZ and SVZ, respectively, are responsible for generating the majority of cortical neurons in rodent8, 10, additional sites of progenitor cell activity have been suggested including the subplate (SP, the first layer of cortical neurons produced in the mammalian cerebral cortex), the cortical plate (CP, future grey matter), the marginal zone (MZ), and the extra-ventricular zone28–30, which prompted us to ask whether oRG-like cells exist in the developing mouse neocortex. Furthermore, the origin of this distinct cell type remains unknown because of the challenge of experimental manipulation in primates and carnivores. oRG-like cells would be much easier to manipulate and study in the developing rodent brain.
To address these issues, we investigated whether progenitor cells resembling oRG cells exist in the rodent brain during periods of neocortical neurogenesis. We found cells in the superficial region of the subventricular zone (SVZ) in the developing mouse cortex that morphologically resemble oRG cells. Time-lapse imaging revealed that these cells undergo mitotic somal translocation and asymmetric division where one daughter cell inherits the basal process. Our long-term imaging revealed that oRG cells are generated directly from RG cells, and that they produce neurons directly, without an intervening intermediate progenitor cell. Furthermore, we found that during interphase, the centrosome moves into the basal process to maintain polarity prior to mitotic somal translocation. These results suggest that oRG cells are not a specialization of a larger brain with increased cortical area. Instead, oRG-like cells are probably present in all mammals and an evolutionary increase in the number of oRG cells likely amplified neuronal production and contributed to cortical expansion.
oRG-like cells exist in mouse neocortex
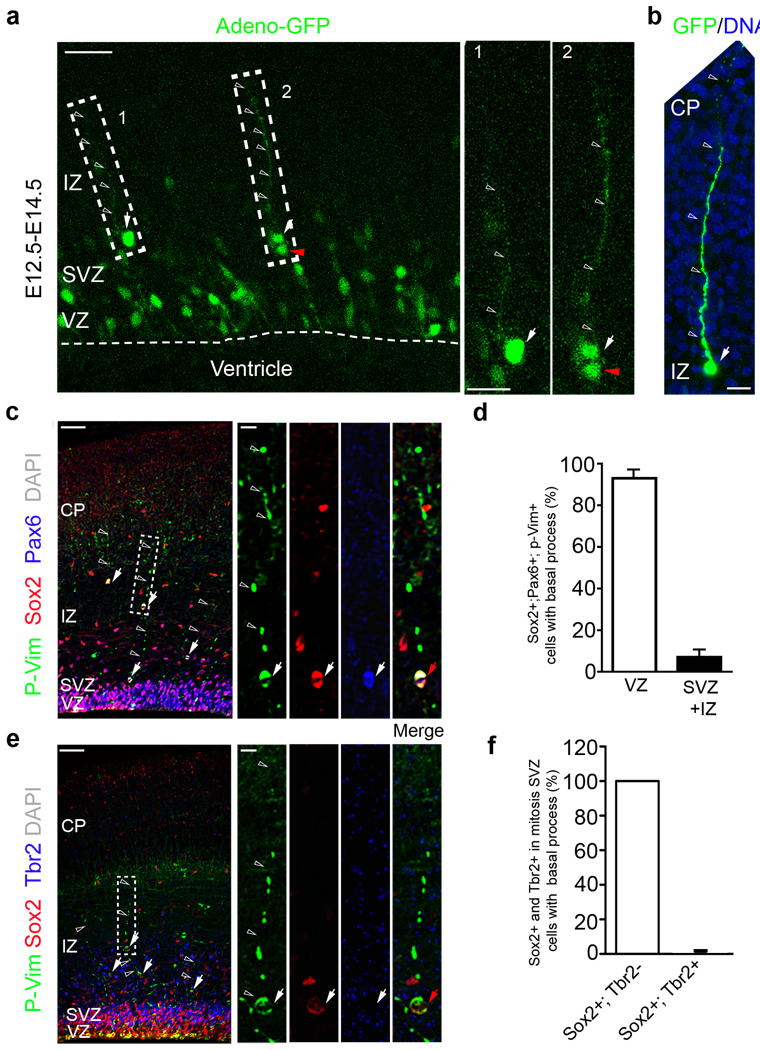
Recent studies have shown that radial glia-like cells referred to as oRG cells, abundantly populate the OSVZ of the fetal human brain and account for between 40% – 75% of all proliferating cells24. They are also present in the cortex of the ferret, a carnivore that also has an expanded gyrencephalic brain26, 27. These findings raise the question of whether OSVZ progenitors are specific to gyrencephalic species or whether they also exist in lissencephalic mammals. To examine this possibility, we introduced GFP-expressing adenovirus (Adeno-GFP) into the lateral ventricle of developing neocortex of E12.5 mouse embryos by in utero injection10. We searched for GFP+ cells with oRG morphology two days after infection. Interestingly, we observed GFP+ monopolar cells located in the superficial or outer region of the SVZ (Fig. 1a, cell 1 and 2 arrow). Similar to oRG cells in human fetal brain24, 26, 27, the mouse cells have a long basal process but not an apical process (Fig. 1a, c and e open arrowhead) so that they do not make contact with the ventricle (Fig. 1a cell 1, 2 and 1b). The morphology of these cells suggested that oRG-like cells might exist in the outer SVZ of the rodent brain. Moreover, in sparsely labelled slices we occasionally observed pairs of GFP+ cells (4 out of 25) close to each other in the region between the SVZ and intermediate zone (IZ), where one cell often had a long basal process and the adjacent cell was rounded, suggesting that the oRG-like cell might have divided (Fig. 1a, cell 2).
Figure 1. oRG cells in the developing mouse neocortex.
(a) Labelling of RG and oRG-like cells with Adeno-GFP. Note the oRG-like cell (box 1) that has a long basal process (open arrowhead) but no apical process. High magnification images are shown to the right (1 and 2). Scale bars: 50 µm and 15 µm. (b) Representative oRG-like cell (arrow), open arrowheads indicate the basal process. Scale bars: 25 µm. (c) Phosphovimentin (P-Vim, green) labels oRG cells in mitosis. The basal process has varicosities characteristic of M-phase oRG cells. The oRG-like cells co-stain with radial glial progenitor markers Pax6 (blue) and Sox2 (red). Arrows indicate triple positive oRG-like cells; open arrowheads indicate the basal process. Scale bars: 50 µm and 10 µm. (d) Quantification of the percentage of mitotic, basal process bearing oRG-like cells identified by P-Vim+, Pax6+, and Sox2+ immunostaining in the VZ (92.95 ± 5.90%) and superficial SVZ (7.05 ± 3.70%) (Total 78 cells from six animals). (e) Phosphovimentin+ (green) oRG cells at E16.5 co-stain for Sox2 (red) but are Tbr2− (blue, an intermediate progenitor marker). High magnification images of a representative outlined cell are shown to the right. Arrows indicate oRG-like cells co-stained for P-Vim (green) and Sox2 (red), open arrowheads indicate the basal process. Scale bars: 50 µm and 10 µm. (f) Quantification of the percentage of oRG-like cells identified by P-Vim+/Sox2+/Tbr2+ (0 %) or P-Vim+/Sox2+/Tbr2− (100%) in the superficial SVZ (Total 46 cells from six animals).
We used immunohistochemistry to examine whether cells with oRG-like morphology also express the transcription factors Pax6 and Sox2, neural stem/progenitor cell markers that are expressed by human and ferret oRG cells24, 26, 27. We also used phosphovimentin (P-Vim) which marks the cytoplasm of neural progenitor cells in M-phase of the cell cycle28 and helps to reveal their morphology. We observed Pax6+/Sox2+/P-Vim+ cells located in the IZ and the region adjacent to the SVZ (Fig. 1c, arrow for the cell). Among triple positive cells, 93% were located in the VZ, and only 7% were located in the superficial SVZ (Fig. 1d). Most of the triple positive cells (67 out of 78, E16) had a basal process revealed by the P-Vim antibody (Fig. 1c and e, open arrowheads). Furthermore, P-Vim+ cells with a basal process always expressed the RG cell markers Sox2 and Pax6 (67 out of 67, E16) indicating that the monopolar cells are progenitor cells. Tbr2 (t box brain 2) is a T-zone transcription factor that is specifically expressed by IP cells during development. IP cells are transit-amplifying progenitors arising from radial glia that can be characterized by expression of Tbr2 and by concurrent down-regulation of Pax6. IP cells divide symmetrically within the VZ or SVZ and generate a strictly neuronal population. We used Sox2, Tbr2 and P-Vim expression to further classify the cortical progenitors we observed. All (46/46) of the P-Vim+ cells with oRG-like morphology in the outer region of the SVZ were Sox2+/Tbr2− (Fig. 1e and f), indicating that oRG-like cells in the mouse resemble RG cells and not intermediate progenitors.
To examine the divisions of oRG-like cells directly, we used a low titre GFP-expressing retrovirus (Retro-GFP) to only transfect dividing cells. We injected the retrovirus into the lateral ventricle of E12.5 mouse embryos by in utero injection as described10, 11. Observations of division of the GFP-labelled oRG-like cells with long basal but no apical process confirmed that they were actively dividing progenitor cells (Fig. 1b). Moreover, a low density of dividing progenitors located in the IZ and outer SVZ were revealed by phospho-histone (pH3) immunostaining co-labelled with Sox2 and Pax6 (Supplementary Fig. 1). These results indicate that the developing mouse cortex contains a subtype of actively dividing progenitor cell that maintains process contact with the pia but not the ventricle. Based on their oRG-like morphology and expression of molecular markers characteristic of RG cells, we tentatively identified these cells as mouse oRG cells to set them apart from traditional RG cells in the VZ and IP cells in the SVZ.
Mouse oRG cells undergo mitotic somal translocation (MST)
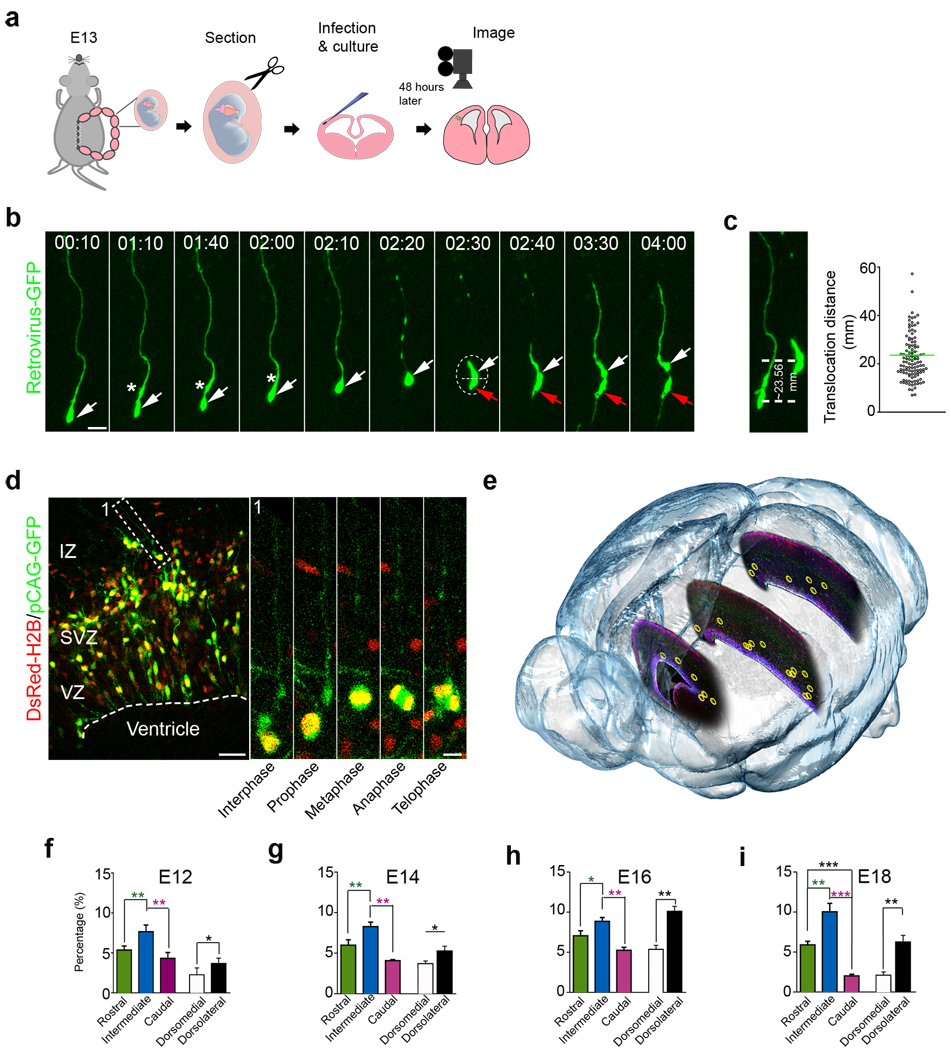
A characteristic feature of neuroepithelial and RG cells is interkinetic nuclear migration (INM). The apical anchoring of the centrosome within the ventricular endfoot serves as the cellular mechanism for INM by maintaining cell polarity; orienting the microtubule minus ends apically to direct dynein motor protein migration and asymmetric division of RG cells16, 17. Recent studies in human fetal brain show that oRG cells exhibit a distinctive behaviour prior to mitosis where the cell body moves rapidly up the basal fibre, referred to as mitotic somal translocation (MST). The similar morphological and molecular characteristic of mouse oRG and human oRGs prompted us to ask whether mouse oRG cells behave similarly. To test this, we first developed a new assay to specifically visualize oRG cells in the mouse (Fig. 2a). We delivered a GFP-expressing retrovirus directly into organotypic brain slices from E13 embryos, and started to perform confocal time-lapse imaging 48 hours later to monitor cellular behaviour.
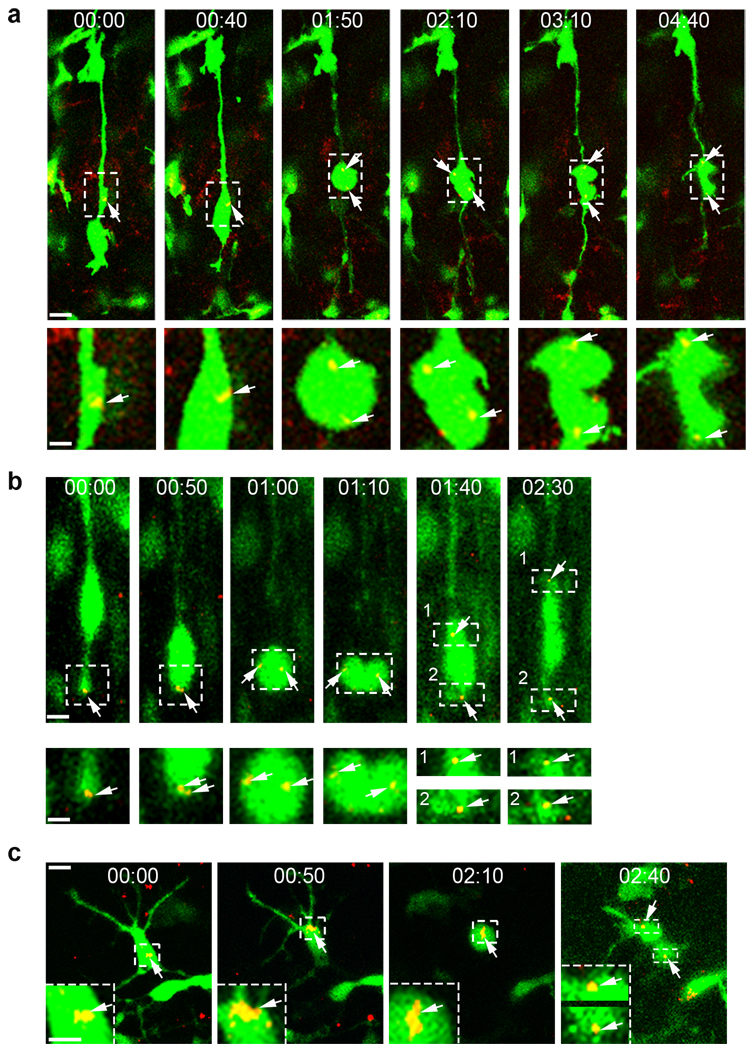
Figure 2. oRG cells undergo mitotic somal translocation.
(a) Experimental procedure for time-lapse. (b) oRG-like cells undergo mitotic somal translocation before mitosis (see supplementary Movie 1). Arrows indicate oRG-like cells (white) and a non-oRG daughter (red). An asterix indicates the characteristic swelling in the proximal basal process. A dashed line indicates the cleavage plane. Scale bar: 20 µm. (c) Quantification of mitotic somal translocation distances. Average distance 23.56 ± 1.56 µm (from 114 time-lapse sequences). (d) Dual-labelled oRG cell (broken box, cell 1) 1d after electroporation in utero at E13.5. High magnification images of mitotic cell behaviour from the outlined regions (right). Scale bar: 50 µm and 10 µm. (e) 3D illustration of oRG cell distribution pattern in E16 brain. Yellow donut-shape circles indicate the locations of mouse oRG cells. (f–i) Quantification of the percentage of Pax6+/P-Vim+/Sox2+ triple positive cells located in the outer region of the VZ and SVZ versus total triple positive cells located in the entire developing neocortex. *, p<0.05; **, p<0.005; ***, p<0.001.
We found that GFP retrovirus-labelled oRG cells spontaneously divided (Fig. 2b and Supplementary movie 1) and exhibited the same distinct behaviours as human oRG cells24. The cell body moved rapidly along the basal process led by a swelling within the process (Fig. 2b). The pattern of somal translocation was reminiscent of behaviour observed in migrating neurons31. The duration of oRG cell translocation was usually less than an hour, which is similar to the duration observed in human oRG cells. However, the translocation distance of oRG cells was around 25 µm (Fig. 2c), which is shorter compared to human oRG cells which average 57µm24. Of ~ 114 oRG cell divisions we observed, all but two cells divided with a horizontal cleavage plane (parallel to the ventricular surface). This plane of division ensured that the basal daughter cell inherited the basal fibre and maintained oRG morphology while the apical daughter did not, consistent with asymmetric oRG cell division (Fig. 2b). The two cells that did not divide horizontally gave rise to two similar daughter cells, both of which had oRG morphology and basal fibres (Supplementary Fig.2). The mitotic somal translocation of oRG cells contrasts with the INM of RG cells, in which the nucleus moves apically and mitosis occurs at the ventricular surface (Supplementary Fig. 3a and supplementary Movies 2), and with the mitotic behaviour of IP cells, which divide in place without nuclear translocation (Supplementary Fig.3b and supplementary Movies 3). To explore this further, we used a plasmid encoding human histone H2B, a protein that enables sensitive analysis of the nucleus in living mammalian cells, fused with DsRedexpress (DsRedex-H2B) and co-electroporated this with a plasmid encoding pCAG-GFP32, which diffuses throughout cells and thereby reveals their morphology. These constructs were electroporated into the developing mouse neocortex at E13.5 (Fig. 2d). We were able to observe oRG cells in brain slices at E14.5 (Fig. 2d, cell 1). Moreover, serial microscopic visualization revealed that these cells progressed through the cell cycle, including interphase and M phase (i.e. prophase, metaphase, anaphase and telophase) (Fig. 2d) after somal translocation, demonstrating that oRG cells are actively cycling cells. Together, these data support the conclusion that a subtype of oRG progenitor cell exists in the mouse brain that undergoes mitotic somal translocation and asymmetric division.
oRG cells distribute sparsely in developing neocortex
We next asked how numerous are oRG cells during mouse neocortical development and where are they located. To address these questions we mapped the location of Pax6+/P-Vim+/Sox2+ triple positive cells with basal processes at four embryonic ages (E12, E14, E16 and E18) in the rostro-caudal and medio-lateral axes (Fig. 2e–i and supplementary Fig. 4). We compared the relative distribution of oRG cells located in the superficial SVZ and IZ versus total triple positive cells located in the entire developing neocortex (Fig. 2e–i and Supplementary Fig.4b–d). At E12, the SVZ was present throughout the ventro-medial extent of the cortical wall, although no CP was detected medially. At this age we observed oRG cells (indicated by Pax6+/P-Vim+/Sox2+ with basal process) located in the superficial SVZ and inner IZ (Fig. 2f and Supplementary Fig.4b). oRG cells represented 5.35 ± 1.25% (n=200), 7.64 ± 2.06% (n=157) and 4.31 ± 1.80% (n= 209) of mitotic progenitors, respectively, in the rostral, intermediate, and caudal cortex. More oRG cells were observed in the dorso-lateral (3.65 ±1.70%, n= 411) than dorsal medial cortex (2.24 ± 2.15%, n=402). At E14, the proportions of oRG cells increased compared to E12, with 6.09 ± 1.65% (n=115) in rostral, 8.27 ±1.36% (n=145) in the intermediate, and 4.12 ±0.32% (n=97) in caudal cortex (Fig. 2g and Supplementary Fig. 4c). The number of oRG cells reached a peak at E16, consisting of 7.02 ±1.54% (n=228) in the rostral, 8.89 ±1.15% (n=180) in the intermediate and 5.26 ±0.95% (n=152) in caudal cortex (Fig. 2e and h). Concurrent with the depletion of RG cells in the VZ2, 12, the proportion of oRG cells increased to 10.03 ±2.60% (n=279) in the E18 intermediate cortex. Consistent with the dramatic decrease of neurogenesis at E18, the proportion of oRG cells compared to E16 dropped to 5.90±1.10% (n= 84) in the rostral cortex, and 2.04± 0.51% (n=49) in caudal cortex (Fig. 2i and supplementary Fig.4d).
At all stages, a similar distribution pattern of oRG cells was observed, with a lateral to medial developmental gradient (Fig. 2f–i). At E12, the proportion of oRG cells in lateral cortex comprised 3.65±1.70% (15 out of 411) of all progenitors, and 2.24± 2.15% (9 out of 402) in the medial cortex (fig. 2f). At E14, lateral oRG cells accounted for 5.06± 0.85% (18 out of 356) compared to 3.58± 1.24% of medial oRG cells (12 out of 335) (fig. 2g). At E16, oRG cells exhibited a more lateral (10.06± 1.54%, 32 out of 318) than medial (5.03± 1.10% 15 out of 298) location (fig. 2h). We still detected 6.11± 1.78% (8 out of 131) of oRG cells laterally, but a dramatic reduction medially to 2.03± 1.02% (6 out of 296) at E18 (Fig. 2i). These results suggest that oRG cells are relatively rare at all ages, but exhibit a general lateral to medial spatial gradient with the highest density of oRG cells in intermediate cortex, less in rostral cortex, and lowest in caudal cortex at all stages of embryonic neocortical development.
oRG cells generate neurons
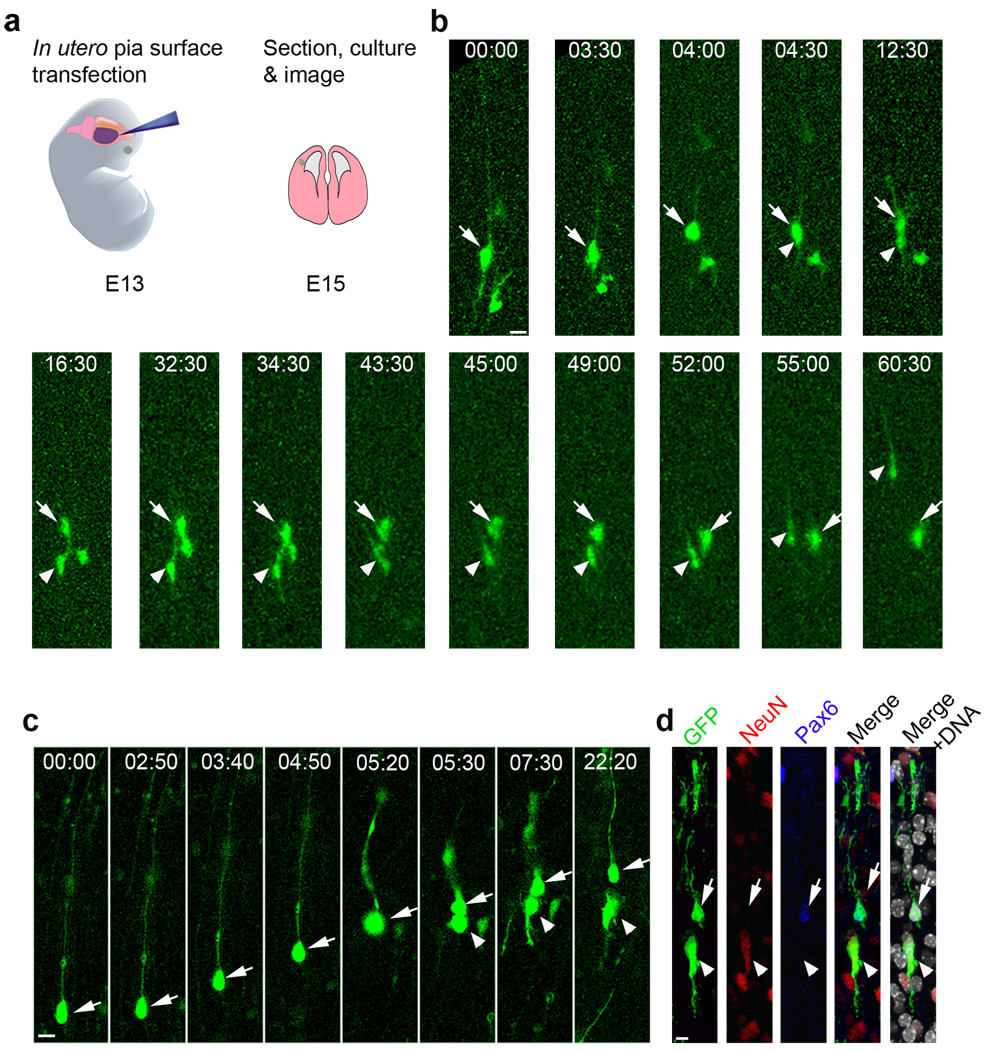
We next examined whether oRG cells generate neurons. Taking advantage of the basal processes of oRG cells, we developed a new assay using in utero pial surface injection of Adeno-GFP to retrogradely label oRG cells close to the cortical plate, as illustrated schematically in figure 3a, and started long-term (days long) time-lapse imaging two days after injection. Successfully labelled oRG cells were identified by their monopolar morphology (Supplementary Fig. 5a). We observed asymmetric, self-renewing, oRG cell divisions, where the apical daughter cell acquired neuronal morphology over the subsequent ~ 40 hours, including a leading process oriented towards the pia (Fig. 3b and supplementary Movies 4). After acquiring a short trailing process, the bipolar daughter cells migrated radially to the cortical plate with speeds (0.11 ± 0.02 µm/min) similar to those of neighbouring migrating neurons (0.12 ± 0.03µm/min)31.
Figure 3. oRG cells generate neurons.
(a) Experimental procedure for time-lapse analysis of oRG-like cell behaviour by in utero pial surface injection of EGFP expressing adenovirus (Adeno-GFP). (b) Time-lapse images of daughter neuron migration following oRG division. The apical daughter cell becomes bipolar after ~ 30h and migrates radially (see supplementary Movie 4). White arrows indicate the oRG cell and yellow arrows indicate the daughter oRG cell after division. Yellow arrowheads indicate the neuron daughter. Scale bar: 15 µm. (c) Asymmetric division of an oRG cell (arrows) generates a self-renewed oRG cell (arrows) and a daughter neuron (arrowheads, see supplementary Movie 5). (d) The oRG daughter was Pax6+ (a neuronal stem cell marker, blue), and the non-oRG daughter was NeuN+ (a neuronal marker, red) after 12h more in culture. Scale bar: 10 µm.
To further explore whether oRG cells produce neurons, we monitored oRG cell divisions in real time, and then determined daughter cell fate 12 or more hours later by immunostaining with cell-type specific markers. A commitment to the neuronal lineage was indicated by the expression of NeuN or βIII-tubulin (Tuj1). In most of the oRG cell divisions (13 out of 17), the apical daughter cell expressed NeuN or Tuj1 (Fig. 3c,d and supplementary Fig. 5b). In all cases, the basal daughter cells inherited the basal fibre and expressed Pax6 (Fig. 3c, d). In four cases, daughter cells were unlabeled, possibly due to limitation of antibody penetration. These results demonstrate that oRG cells divide asymmetrically to self-renew and give rise to neurons.
oRG Cells originate from RG cells
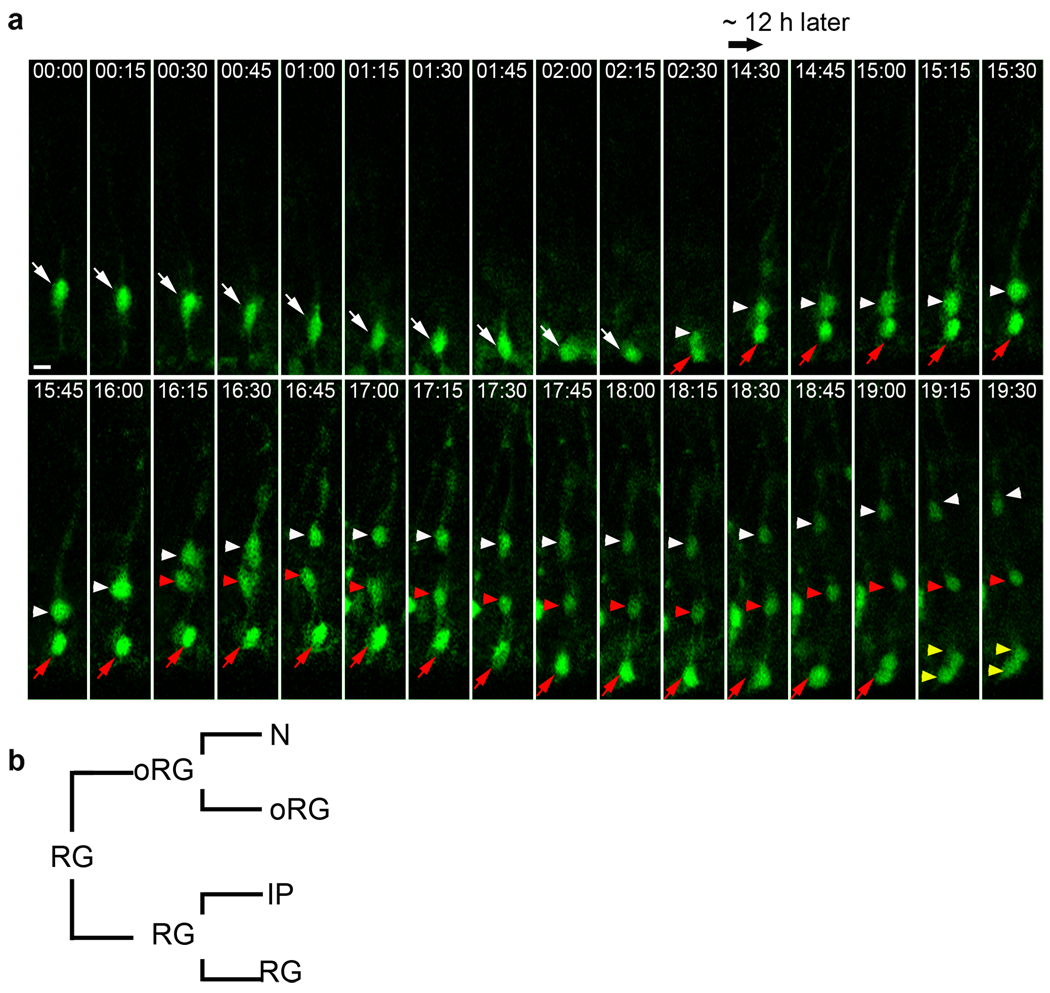
We previously hypothesized that human oRG cells originate in the VZ and use mitotic somal translocation to move into the SVZ24. However, due to technical limitations this has not been demonstrated in the human fetal brain. Having identified oRG cells in the developing mouse neocortex, we next examined their origin. We used Retro-GFP to label RG cells in E11.5 embryos by in-utero intra-ventricular injection as described10, 11. At E13.5 we removed the embryos and prepared organotypic brain slice cultures for time-lapse imaging11, 17, 33. As expected, GFP+ cells were radial glial cells with bipolar morphology located in the VZ (Fig. 4a). We monitored GFP+ RG cell at 15-minute intervals over 80 hours. Most (45 out of 52) dividing radial glial cells we analysed appeared to undergo asymmetric self-renewing division based on morphologically distinct differences between the daughter cells10, 11. We could confirm that some of the divisions we observed (4 out of 52) generated RG and IP cells (indicated by mutlipolar morphology and/or subsequent migration to the SVZ and symmetric division). However, a few divisions we monitored (3 out of 52) generated a self-renewed RG cell and a daughter cell with oRG cell morphology. Moreover, the oRG-like daughter underwent characteristic mitotic somal translocation associated with cell division in the SVZ, while the parent RG cell underwent INM and division at the ventricular surface as shown in the representative time-lapse figure (Fig. 4a and supplementary Movie 6). These data support a model of oRG cell origins (Fig. 4b and and supplementary Fig. 6) in which oRG cells are generated directly from asymmetric divisions of RG cells.
Figure 4. oRG cells originate from RG cells.
(a) Time-lapse image of RG cell division. A GFP-labelled RG cell was monitored at 15m intervals (white arrow) 2d after in-utero intra-ventricular retrovirus infection in E11.5. Asymmetric RG cell division generates a self-renewed RG cell (red arrow), which divides again (yellow arrowheads). The first RG daughter cell (white arrowhead) undergoes mitotic somal translocation and divides (white and red arrowheads follow the two daughter cells after oRG division) (see supplementary Movie 6). Scale bar: 10 µm. (b) Lineage tree of RG and oRG cell divisions. RG cells can divide asymmetrically to self-renew and generate oRG cells. Both progenitors can divide again to self-renew and generate daughter cells including neurons (N) and intermediate progenitors (IP).
Distinct centrosome dynamics of oRG cells
Given that the centrosome is required for the maintenance of RG cell progenitors in the VZ16, 17, we explored the cellular mechanism of oRG self-renewing asymmetric division by analyzing the distribution and dynamics of centrosome behavior in oRG cells. We electroporated pCAG-YFP plasmid or injected Retro-GFP to reveal cell morphology. To examine centrosome behaviour, we introduced a plasmid DsRedex-Centrin17, encoding a central component of the centrosome, into the developing neocortex of E13.5 mouse embryos by in utero electroporation17, 33. Six dividing oRG cells from independent experiments proceeded through mitosis in the outer SVZ and reached the two-cell stage. In all cases we found movement of the centrosome into a varicosity in the basal process at interphase, followed by movement of the nucleus in M phase (Fig. 5a and supplementary Movie 7), suggesting that the centrosome is involved in the maintenance of oRG cell polarity.
Figure 5. Distinct behaviour of centrosomes in different progenitor cells.
(a–c) Time-lapse images of centrosome dynamics in oRG cells (a, see supplementary Movie 7), RG cells (b, see supplementary Movie 8) and IP cells (c, see supplementary Movie 9). High magnification images from the outlined regions are shown (a, b, c). Arrows indicate centrosomes. Scale bars: a–c, 10 µm and 2.5 µm.
Our data indicate that mitotic somal translocation is one of the defining features of oRG cells in mouse as well as in human24, with the centrosome situated in a varicosity in the basal process. This feature is reminiscent of centrosome behaviour in migrating neurons31 in which the centrosome also moves into a varicosity in the leading process followed by the saltatory movement of the nucleus34. We hypothesized that a subcellular mechanism of centrosome positioning regulates mitotic somal translocation (Fig. 5a and Supplementary Movie 7). To study centrosome behaviour in all three subtypes of progenitors in mouse neocortex, we monitored centrosome positioning and behaviour in RG and IP cells. The positioning of the centrosome in the ventricular endfoot at the VZ surface helps explain why RG cells go through interkinetic nuclear migration. In all the seven dividing RG cells with centrosome labeling, the centrosome divides, and one daughter centrosome migrates back to the ventricular endfoot of the self-renewed RG cell, while the other daughter centrosome migrates away from the VZ with the non-RG daughter cell as previously reported17 (Fig. 5b and supplementary Movie 8). Furthermore, we found that centrosomes remain within the IP cell body throughout the cell cycle of IP cells (n=7); consistent with the observation that IP cells remain stationary when they divide (Fig. 5c and Supplementary Movie 9). Taken together, these data strongly suggest that specific centrosome positioning underlies mitotic somal translocation. The dynamic spatial arrangement of the centrosome in oRG cells is distinct from the other cortical precursor cells as the centrosome is located in the ventricular endfoot of the RG cell during interphase, and remains in the cell body of dividing IP cells17.
Discussion
Our finding of mitotic oRG cells provides a new lineage for neurogenesis in the rodent cortex. Real-time imaging data show that OSVZ progenitors are generated directly from RG cells in the VZ, and daughter oRG cells migrate away to the superficial SVZ by mitotic somal translocation. Since the number of oRG cells is very low, the contribution of oRG cells to neurogenesis and cortical layer formation in rodents is small. This may help explain why genetic mutations causing strong microcephaly phenotypes in human (aspm for example) do not necessary cause the same dramatic phenotype in rodents (Pulvers, et al 2010) 35. Moreover, although mouse oRG cells share two defining features (i. e. morphology and mitotic somal translocation) with human oRG cells, time-lapse imaging reveals an important difference between those two cells, namely that oRG cells undergo self-renewing asymmetric division to generate neurons directly, while human oRG cells generate transit-amplifying cells (i. e. IP cells) that in turn generate neurons.
Our data also demonstrate that during mitotic somal translocation, the oRG centrosome moves into a varicosity in the basal process at interphase, and the nucleus follows prior to mitosis. In this way oRG cell mitosis shares features of both neuronal migration and the G2 phase of interkinetic nuclear migration characteristic of neuroepithelial cell division. Given that centrosomes anchored in the ventricular (apical) endfeet of RG cells are a defining feature of the asymmetric division of RG cells16, 17, these data imply an important role of the centrosome in establishing oRG cell polarity. oRG cells go through asymmetric division with the cleavage plane perpendicular to the apico-basal axis, while RG cells undergo asymmetric division with a cleavage plane parallel to the apico-basal axis, implying that it is not cleavage plane orientation but possibly centrosome positioning that determines asymmetric division of oRG cells. Lack of apical polarity proteins (i.e. Par3) in human and ferret OSVZ progenitors26 raises an intriguing question of whether the asymmetric inheritance of mother versus daughter centrosome is necessary for oRG cell asymmetric division and fate specification as it is in RG cells17.
The results presented here suggest that a specific new subtype of progenitor cell exists in the superficial SVZ of mouse neocortex. Expression of cell fate markers, morphological features, and mitotic behavior link this progenitor with an OSVZ-like progenitor cell, the oRG cell, recently described in the developing human and ferret neocortex24, 26. Progenitors in this region of developing mouse cortex were observed a decade ago but had not been well-characterized28. It has been suggested that OSVZ-like cells may be primate specific25, and contribute to expansion of the neocortex in human and gyrencephalic species24, 26. However, the observation of oRG-like cells in mouse, a lissencephalic species, suggests that oRG cells are not unique to gyrencephalic brain development. The appearance of oRG cells in rodents may nonetheless foreshadow human cortical expansion. It is attractive to speculate that oRG-like cells arose in an ancestral rodent-like animal about 100 million years ago,4 and selective pressures led to an enormous expansion in their numbers in the lineages leading to primates. Additional features such as an increase in the number of progenitor cell cycles and the appearance of an intervening transit amplifying-type cell may have provided additional mechanisms to massively increase neuron number during neocortical evolution. It would be interesting to study the cortical development of more species with gyrencephalic or lissencephalic brains to obtain a more complete understanding of how progenitor cell behavior contributes to the architecture of the developing neocortex.
Methods
Plasmids and in utero electroporation
DsRedexpress (DsRedex) cDNAs were obtained by PCR and cloned into pEGFP-H2B-C1 to replace EGFP in generating the DsRedex-H2B plasmids.
In utero electroporation was performed as previously described. In brief, a timed pregnant Swiss webster mouse at 13.5 days of gestation (E13.5) was anesthetized, the uterine horns were exposed, and ~1 µl of plasmid DNA (1–3 µg/µl) mixed with Fast Green (Sigma,) was manually microinjected through the uterus into the lateral ventricle, using a bevelled and calibrated glass micropipette (Drummond Scientific). For electroporation, five 50 ms pulses of 40–50 mV with a 950 ms interval were delivered across the uterus with two 9-mm electrode paddles positioned on either side of the head (BTX, ECM830). After the procedure, the uterus was placed back in the abdominal cavity and the wound was surgically sutured. The animal was then placed in a 28°C recovery incubator under close monitoring until it recovered and resumed normal activity. All procedures for animal handling and usage were approved by our institutional research animal resource centre (RARC).
Retroviral and adenovirus in-utero infection
Replication-incompetent EGFP-expressing retrovirus was produced from a stably transfected packaging cell line (293gp NIT-GFP; a kind gift F. H. Gage). CMV-GFP adenovirus was acquired from Vector Biolabs (1×106 c.f.u). Animals were maintained according to protocols approved by the Institutional Animal Care and Use Committee at the University of California at San Francisco. Uterine horns of E11.5 - E13.5 gestation stage pregnant swiss webster mice (Charles River Laboratories) were exposed in a clean environment. Retrovirus or Adenovirus (~1.0 µl) with Fast green (2.5 mg ml−1, Sigma) was injected into the embryonic cerebral ventricle or pia surface through a bevelled, calibrated glass micropipette (Drummond Scientific). After injection, the peritoneal cavity was lavaged with ~10 ml warm PBS (pH 7.4) containing antibiotics, the uterine horns were replaced, and the wound was closed.
Brain sectioning, cortical slice culture, virual infection on slice and time-lapse imaging
Brains were dissected out into ice-cold artificial cerebro-spinal fluid (ACSF) containing (in mM): 125 NaCl, 5 KCl, 1.25 NaH2PO4, 1 MgSO4, 2 CaCl2, 25 NaHCO3 and 20 glucose; pH 7.4, 310 mOsm/L. Brains were embedded in 4% low-melting agarose in ACSF and sectioned at 400 µm using a vibratome (Leica microsystems). Brain slices that were transferred onto a slice culture insert (Millicell) in a glass-bottom petri dish (MatTek Corporation) with culture medium containing (by volume): 66% BME, 25% Hanks, 5% FBS, 1% N-2, 1% Penicillin/Streptomycin/Glutamine (all from Gibco) and 0.66% D-(+)-glucose (Sigma). Cultures were maintained in a humidified incubator at 37°C with constant 5% CO2 supply. For Fig 2a and b, GFP-expressing retrovirus (1× 106 colony forming units (c.f.u.) was manually microinjected into intermediate zone of the cultured brain slice, using a bevelled and calibrated glass micropipette (Drummond Scientific). All the time-lapse images were acquired using an inverted Leica TCS SP5 with an on-stage incubator (while streaming 5% CO2, 95% O2) and a 40X air objective lens.
Immunohistochemistry and confocal imaging
Mouse brains or cultured slices were fixed in 4% PFA in PBS at 4°C overnight. Incubated for one hour at room temperature in a blocking solution (10% normal goat or donkey serum as appropriate, 0.1% Triton X-100, and 0.2% gelatin in PBS), followed by incubation with the primary antibodies 3 days at 4°C. Sections were then washed in 0.1% Triton X-100 in PBS and incubated with the appropriate secondary antibody for one to two hours at room temperature.
The primary antibodies used were: mouse monoclonal anti-β-III Tubulin (clone TUJ1) (Covance, 1:500), rabbit polyclonal anti-Pax6 (Covance, 1:500), rabbit polyclonal anti-Tbr2 (Millipore/Chemicon, 1:500), goat anti-Sox2 (Santa Cruz sc-17320, 1:200) and mouse anti-phospho-vimentin (MBL International D076-3s (Ser55), or D095-s (Ser82), 1:500). Secondary antibodies used were: Alexafluor 488 (1:1000), 546 (1:100), or 647 (1:1000) conjugated donkey anti-mouse, anti-rabbit or goat (Invitrogen). DNA was stained with 4',6-diamidino-2-phenylindole (DAPI) (Molecular Probes). Images were acquired with a Leica TCS SP5 broadband laser confocal microscope, and analyzed with LCS Leica confocal software (Leica), Imaris imaging software (Bitplane), Volocity (ImproVision) and Photoshop (Adobe Systems). Data are presented as mean±s.e.m. and student’s t-test were used for statistical significance estimation.
Supplementary Material
Acknowledgement
We thank A. Alvarez-Buylla, D. Lim, C. Harwell, W. P. Ge and L. Fuentealba for comments on the manuscript and members of the Kriegstein laboratory for discussions. We thank W. Walantus and Y. Y. Wang for mouse surgery and technical support; D. V. Hansen and J. H. Lui for ideas and discussion. We thank F. Gage for GFP-retrovirus reagents. This work was supported by grants from the Bernard Osher Foundation and the NINDS (to A.R.K)
Footnotes
Author Contributions
X. W. conceived the project and carried out most of the experiments. J.-W.T. helped on some of the time-lapse imaging experiments and B. L. helped on the immunohistochemistry staining procedure. X. W. analyzed data, interpreted results and wrote the manuscript. A.R.K., as the principal investigator, provided conceptual and technical guidance for all aspects of the project. All authors edited the manuscript.
References
- 1.Fishell G, Kriegstein AR. Neurons from radial glia: the consequences of asymmetric inheritance. Curr Opin Neurobiol. 2003;13:34–41. doi: 10.1016/s0959-4388(03)00013-8. [DOI] [PubMed] [Google Scholar]
- 2.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kriegstein A, Noctor S, Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- 4.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 6.Englund C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 8.Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 9.Miyata T, et al. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- 10.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 11.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 12.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 13.Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Neural stem and progenitor cells in cortical development. Novartis Found Symp. 2007;288:59–73. discussion 73-58, 96-58. [PubMed] [Google Scholar]
- 14.Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Chenn A, Zhang YA, Chang BT, McConnell SK. Intrinsic polarity of mammalian neuroepithelial cells. Mol Cell Neurosci. 1998;11:183–193. doi: 10.1006/mcne.1998.0680. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fish JL, Dehay C, Kennedy H, Huttner WB. Making bigger brains-the evolution of neural-progenitor-cell division. J Cell Sci. 2008;121:2783–2793. doi: 10.1242/jcs.023465. [DOI] [PubMed] [Google Scholar]
- 19.Kosodo Y, et al. Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J. 2004;23:2314–2324. doi: 10.1038/sj.emboj.7600223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- 22.Rakic P. Elusive radial glial cells: historical and evolutionary perspective. Glia. 2003;43:19–32. doi: 10.1002/glia.10244. [DOI] [PubMed] [Google Scholar]
- 23.Abdel-Mannan O, Cheung AF, Molnar Z. Evolution of cortical neurogenesis. Brain Res Bull. 2008;75:398–404. doi: 10.1016/j.brainresbull.2007.10.047. [DOI] [PubMed] [Google Scholar]
- 24.Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 25.Smart IH, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex. 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fietz SA, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- 27.Reillo I, de Juan Romero C, Garcia-Cabezas MA, Borrell V. A Role for Intermediate Radial Glia in the Tangential Expansion of the Mammalian Cerebral Cortex. Cereb Cortex. doi: 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- 28.Kamei Y, et al. Visualization of mitotic radial glial lineage cells in the developing rat brain by Cdc2 kinase-phosphorylated vimentin. Glia. 1998;23:191–199. doi: 10.1002/(sici)1098-1136(199807)23:3<191::aid-glia2>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 29.Carney RS, Bystron I, Lopez-Bendito G, Molnar Z. Comparative analysis of extra-ventricular mitoses at early stages of cortical development in rat and human. Brain Struct Funct. 2007;212:37–54. doi: 10.1007/s00429-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 30.Cheung AF, Pollen AA, Tavare A, DeProto J, Molnar Z. Comparative aspects of cortical neurogenesis in vertebrates. J Anat. 2007;211:164–176. doi: 10.1111/j.1469-7580.2007.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walantus W, Castaneda D, Elias L, Kriegstein A. In utero intraventricular injection and electroporation of E15 mouse embryos. J Vis Exp. 2007;239 doi: 10.3791/239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elias LA, Wang DD, Kriegstein AR. Gap junction adhesion is necessary for radial migration in the neocortex. Nature. 2007;448:901–907. doi: 10.1038/nature06063. [DOI] [PubMed] [Google Scholar]
- 35.Pulvers JN, et al. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proc Natl Acad Sci U S A. 107:16595–16600. doi: 10.1073/pnas.1010494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–872. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.