Abstract
We present the 207 Mb genome sequence of the outcrosser Arabidopsis lyrata, which diverged from the self-fertilizing species A. thaliana about 10 million years ago. It is generally assumed that the much smaller A. thaliana genome, which is only 125 Mb, constitutes the derived state for the family. Apparent genome reduction in this genus can be partially attributed to the loss of DNA from large-scale rearrangements, but the main cause lies in the hundreds of thousands of small deletions found throughout the genome. These occurred primarily in non-coding DNA and transposons, but protein-coding multi-gene families are smaller in A. thaliana as well. Analysis of deletions and insertions still segregating in A. thaliana indicates that the process of DNA loss is ongoing, suggesting pervasive selection for a smaller genome.
Genome sizes in angiosperms range from 64 Mb in Genlisea1 to an enormous 149 Gb in Paris2-4. Two major processes increase genome size: polyploidization and transposable elements (TE) proliferation. Processes that counteract genome expansion include the loss of entire chromosomes, as well as deletion-biased mutations due to unequal homologous recombination and illegitimate recombination5-9. Recent work comparing two cereals, rice and sorghum, has begun to shed light on some of these processes10. However, these species are separated by 60 to 70 million years, making it difficult to disentangle the different evolutionary forces at work.
An exciting opportunity to understand what drives differences in genome size over shorter time scales is offered by the genus Arabidopsis in the Brassicaceae. The genome of the self-incompatible perennial A. lyrata is larger than 200 Mb, near the family average11,12, while the self-compatible annual A. thaliana has one of the smallest angiosperm genomes, at about 125 Mb, even though the two species diverged only about 10 million years ago13-15. Compared to the difference between the two species, variation within A. thaliana is much less11.
A high-quality genome sequence for the partially inbred A. lyrata strain MN47 was assembled from approximately 8.3x coverage of dideoxy sequencing reads, making use of information from genetic maps and chromosome painting16-19 (Online Methods). The final assembly included 206.7 Mb of sequence, 90% of which are included in eight large scaffolds covering the majority of each of the eight chromosomes, and another large scaffold of 1.9 Mb representing one of the centromeres. Based on cytological observations20, the centromeric gaps were estimated to span 17.2 Mb. A combination of de novo predictions, homology to A. thaliana features, and RNA sequencing was used to annotate the genome. In A. lyrata, we predicted 32,670 protein-coding genes, compared to 27,025 genes in A. thaliana21.
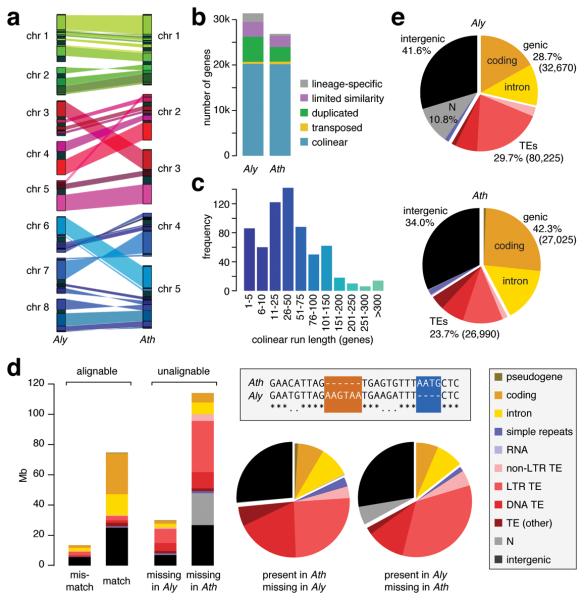
Since overall sequence identity between A. lyrata and A. thaliana is greater than 80% (Supplementary Fig. 1), the two genomes could be easily aligned (Fig. 1a). Genetic mapping16,18,19 has revealed 10 major rearrangements, including two reciprocal translocations and three chromosomal fusions, that led to the A. thaliana karyotype of five chromosomes, compared to the ancestral state of eight, as found in A. lyrata and other Brassicaceae. Although centromeric regions are difficult to assemble, we could identify the syntenic region in A. thaliana that corresponds to the chromosome 4 centromere of A. lyrata. The entire centromere has been lost, with only two remnants of satellite repeats in the 1.4 kb intergenic region between the genes At2g26570 and At2g26580 (Supplementary Fig. 2).
Figure 1.
Comparison of A. lyrata and A. thaliana genomes. (a) Alignment of A. lyrata (Aly) and A. thaliana (Ath) chromosomes. Genomes are scaled to equal size. Only syntenic blocks of at least 500 kb are connected. (b) Orthology classification of genes. (c) Distribution of run lengths of collinear genes. The mode at 1-5 reflects frequent single-gene transpositions. (d) Unalignable sites can be considered as present in one species and absent in the other, as shown in the boxed sequence diagram; matches are indicated by asterisks, and mismatches by periods. The histogram on the left indicates the absolute number of unalignable sites, and the pie charts in the middle compare their relative distribution over different genomic features. See also Supplementary Table 3. (e) Genome composition (number of elements in parentheses).
Apart from chromosomal-scale changes, approximately 90% of the two genomes have remained syntenic, with the great majority in highly conserved collinear arrangements (Fig. 1b and Supplementary Fig. 1d). The run length distribution of collinear gene pairs is bimodal, with a first peak of fragments of five or fewer collinear gene pairs (Fig. 1c), reflecting an abundance of small-scale rearrangements (<10 kb), including single gene transpositions. Windows containing a breakpoint in collinearity are enriched for TEs and other repeats (Supplementary Table 1), in agreement with repetitive elements often being associated with chromosomal rearrangements and transposed genes22-27, although they might not necessarily be causal28. Two thirds of the 154 inversions identified between the two species are flanked by inverted repeats (Supplementary Table 2).
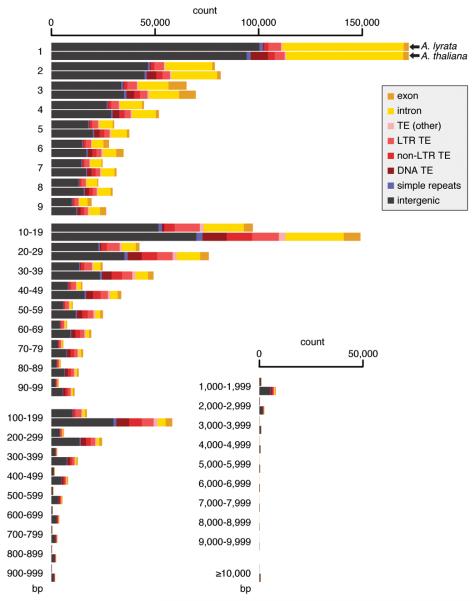
Despite this overall similarity in gene arrangement, the two genomes are strikingly different in size. A whole-genome alignment reveals that more than 50% (<114 Mb) of the A. lyrata genome appears to be missing from the A. thaliana reference genome. In contrast, only about 25% (<30 Mb) of the A. thaliana genome is absent from A. lyrata (Fig. 1d; Supplementary Fig. 1e; Supplementary Table 3). Nevertheless, the distribution across different sequence classes is similar: half of the unalignable sequences are in TEs, and a quarter in intergenic regions. The net effect of these changes is that the A. thaliana genome is ~80 Mb smaller than the A. lyrata genome, with a much higher fraction of genic sequences, 42% instead of 29%, even though the total gene count is smaller (Fig. 1e). The apparent shrinkage of the A. thaliana genome is not simply due to a few chromosome-scale changes: only 10% of the size difference is attributable to the three missing centromeres; the rest is due to hundreds of thousands of smaller insertions and deletions, spanning all classes of sites. Strikingly, while large differences much more often correspond to sequences only found in A. lyrata, this is not true for very small insertions and deletions (Fig. 2). This is in stark contrast to other genomes from other closely related species, but with similarly sized genomes, such as chimpanzee and human29.
Figure 2.
Apparent deletions by size and annotation. A. lyrata is always shown on top, A. thaliana on bottom.
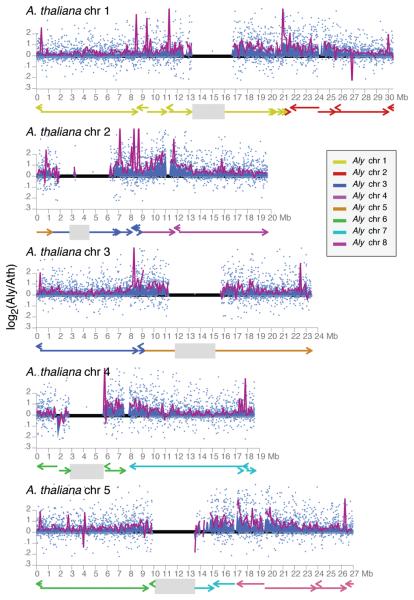
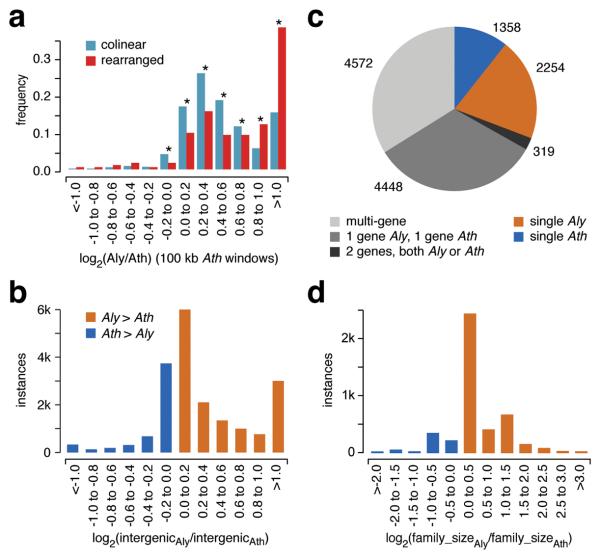
Although rearrangements are correlated with genome shrinkage (rearranged regions are on average shorter in A. thaliana than are collinear regions; Fig. 3 and Fig. 4a), unalignable sequences are found throughout the genome. An analysis of collinear gene pairs confirmed that in most cases, intergenic regions in A. lyrata are longer than their counterparts in A. thaliana (Fig. 4b). Introns behave similarly, although the difference is smaller13.
Figure 3.
Changes in genomic intervals along the A. thaliana genome. Mean ratios for all collinear gene pairs in each 100 kb window are shaded in blue, with individual values shown as light blue dots. The ratio of the absolute length of each non-overlapping 100 kb window is shown as a dark purple line. Centromeres are indicated as grey boxes.
Figure 4.
Change in size of collinear and rearranged regions, intergenic regions and gene families. (a) Size comparison of collinear regions, relative to 100 kb windows in A. thaliana. Asterisks indicate significant differences (binomial test, p<0.001). (b) Relative size of intergenic regions. (c) MCL clusters. (d) Relative size of gene families.
The gene content of A. thaliana is ~17% lower than that of A. lyrata, but without major differences in Gene Ontology (GO) distribution. Similarly, divergence patterns for different gene families between the two species mirror those of within-Arabidopsis thaliana polymorphism levels30,31. The combined gene sets of A. lyrata and A. thaliana result in 12,951 MCL32 clusters, with fewer singletons in A. thaliana (Fig. 4c). Among the 8,794 shared multi-gene MCL clusters (Fig. 4d), clusters that are smaller in A. thaliana outnumber those that are smaller in A. lyrata (1,797 to 612). F-box and NB-LRR genes are examples of gene families with particularly high birth and death rates in plants30,31,33-35. Arabidopsis lyrata has 596 F-box and 187 NB-LRR genes, compared to 502 and 159, respectively, in A. thaliana. The trend of fewer genes in A. thaliana is supported by a broader comparison of the Arabidopsis gene set with those of two other dicots36-38. Arabidopsis lyrata has 114 ortholog clusters39 shared with poplar and grapevine but not A. thaliana, while A. thaliana has only 45 clusters found in poplar and grapevine but not A. lyrata. Similarly, A. lyrata has 875 clusters not detected in any of the other three species, while A. thaliana has only 156 species-specific clusters (Supplementary Table 4 and Supplementary Fig. 3).
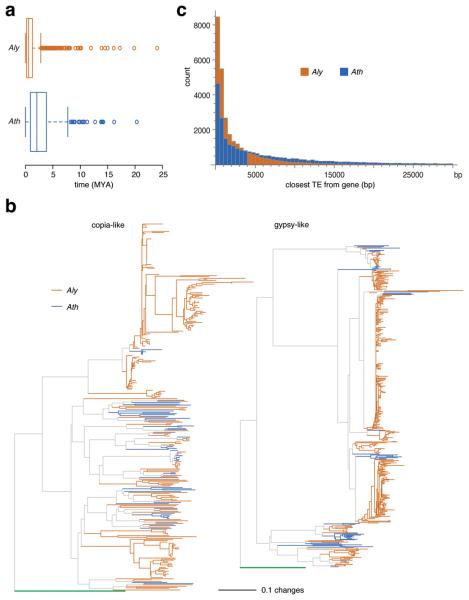
As in other taxa, TEs make an important contribution to the change in genome size (Fig. 1d), and TEs comprise a larger fraction of the A. lyrata genome (Fig. 1e). Without an outgroup, one cannot infer directly how much such patterns are shaped by different TE activity levels or the differential purging of ancestral TEs since speciation. To obtain an estimate of relative activity levels, one can exploit the molecular clock to estimate the average age of long terminal repeat (LTR) retrotransposons40 (Fig. 1e). Using the experimentally determined mutation rate in A. thaliana14, we calculated the mean and median age in A. thaliana to be 3.1 and 2.1 million years, respectively, compared to 1.1 and 0.6 million years in A. lyrata (Fig. 5a). In agreement with previous estimates41, this suggests that LTR retrotransposons have been recently more active in A. lyrata. A phylogenetic analysis also supports a greater expansion of specific LTR retrotransposon clades in A. lyrata (Fig. 5b). Coupled with higher activity levels of TEs in A. lyrata, we find that TEs are differently distributed in the two species, with A. lyrata having a higher proportion of genes with a TE nearby than A. thaliana (Fig. 5c), and this distance is skewed towards larger values in A. thaliana (Supplementary Table 5 and Supplementary Fig. 4). Together, these observations are consistent with a model under which selection purges TEs with deleterious effects on adjacent genes, such that TEs more distant from genes preferentially survive42, with TE elimination having been more efficient in A. thaliana. In addition, there is the possibility that TEs in A. lyrata have experienced less natural selection because they are on average younger.
Figure 5.
Comparison of transposable elements. (a) Estimated insertion times of LTR retrotransposons, based on the experimentally determined mutation rate for A. thaliana. The whiskers indicate values up to 1.5 times the interquartile range. The difference between the species is highly significant (Wilcoxon rank sum test, p<2.2×10−16). (b) Phylogeny of Ty1/copia-like and Ty3/gypsy-like LTR retrotransposons. S. cerevisiae Ty1 and Ty3 used as outgroups are indicated in green. (c) Distances of nearest TE from each gene. The difference between the two species is not simply due to fewer transposable elements in the A. thaliana genome (Supplementary Table 8 and Supplementary Fig. 7).
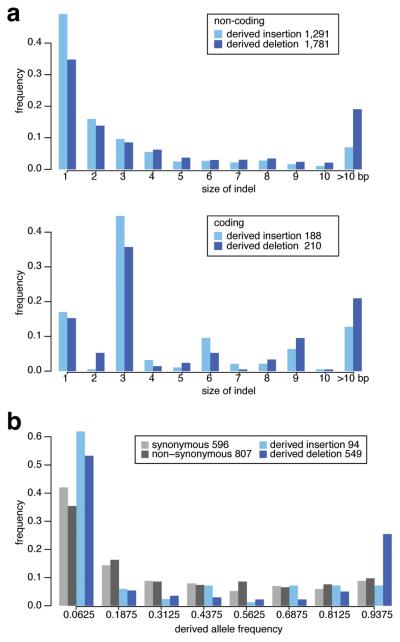
The evidence presented so far points to A. thaliana having suffered a large number of deletions throughout its genome. We can use within-species polymorphisms to shed light on the process by which this has happened. If the A. thaliana genome continues to shrink, we would expect fewer segregating insertions than deletions. Using the A. lyrata genome as a proxy in determining the derived state among a set of insertion and deletion polymorphisms found throughout the genome of 95 A. thaliana individuals43, we find a clear excess of deletions over insertions, with 2,685 fixed and 852 segregating deletions, compared to 1,941 fixed and 106 segregating insertions. Furthermore, among the fixed differences, deletions are on average longer than insertions (Fig. 6a). If selection were not involved, and if this pattern were only due to mutational bias favoring deletions44,45, deletion and insertion polymorphisms should have similar allele frequencies in the A. thaliana population. However, segregating insertions are on average found in fewer individuals than are deletions or single-nucleotide polymorphisms. Deletions are often found in the majority of individuals, and many are approaching fixation in A. thaliana (Fig. 6b). This pattern suggests that deletions are favored over insertions because of selection, rather than simple mutational bias, thus leading to a smaller genome.
Figure 6.
Sizes and allele frequency distribution of insertions and deletions that are either fixed or still segregating in 95 A. thaliana individuals43 and that are presumed to be derived based on comparison with the A. lyrata allele. (a) Size distribution of fixed insertions and deletions. Insertions and deletions that are multiples of a single codon (3 bp) are overrepresented in coding regions. (b) Allele frequency of segregating non-coding insertion and deletion frequencies compared to that of synonymous and non-synonymous polymorphisms.
The pattern of divergence between the two genomes supports this hypothesis. While more deletions have occurred on the A. thaliana than the A. lyrata lineage, the bias towards deletions becomes stronger the longer the missing sequence, and it is absent for sequences shorter than 5 bp or so (Fig. 2). This is consistent with a model where long deletions are selectively favored in A. thaliana, whereas short deletions are not. We acknowledge that without an outgroup to reconstruct the ancestral state shared by the ancestor of both A. lyrata and A. thaliana, one cannot accurately determine whether all changes are derived in A. thaliana.
In summary, we have presented a high-quality reference genome sequence for A. lyrata, which will be a valuable resource for functional, evolutionary and ecological studies in the genus Arabidopsis. Several processes contribute to the remarkable difference in genome size between the predominantly selfing A. thaliana and the outcrossing A. lyrata. In just a few million generations, numerous chromosomal rearrangements have occurred, consistent with theoretical predictions of rearrangements that reduce fitness in heterozygotes being fixed much more easily in strongly selfing species46. Though A. thaliana has 17% fewer genes than A. lyrata, much of the genome size difference seems to be due to reduced TE activity and/or more efficient TE elimination in A. thaliana, especially near genes, as well as shortening of non-TE intergenic sequences and introns in A. thaliana. Specifically, by making the reasonable assumption that the A. lyrata allele presents in the majority of cases the ancestral state, we find that segregating deletions at non-coding sites in A. thaliana are skewed towards higher allele frequencies, and that both fixed and polymorphic deletions are more common than insertions. Together, this suggests pervasive selection for a smaller genome in A. thaliana. Apart from apparent advantages for species with smaller genomes that have been inferred from meta analyses47, the transition to selfing might be an important factor in this process46. In addition, a shorter life span may allow a reduction of the genetic repertoire and thus contribute to the smaller genome of A. thaliana as well.
What role, if any, genome expansion might play in A. lyrata can be addressed once detailed A. lyrata polymorphism information as well as closely related outgroup genomes become available, such as the one from Capsella rubella, which is currently being assembled. A complete understanding of the processes behind genome contraction and expansion over short time scales will also require better knowledge of mutational events, and a deeper understanding of the distribution of, and selection on, non-coding regulatory sequences42. For both, high-quality whole genome sequences of additional Arabidopsis relatives will be an important tool.
METHODS
Sequencing and assembly
Arabidopsis lyrata strain MN47 was derived by forced selfing from material collected in Michigan, USA, by Dr. Charles Langley (UC Davis). It was inbred six times before extracting DNA for sequencing. Libraries with various insert sizes including fosmids and BACs were dideoxy sequenced on ABI 3730XL capillary sequencers. Reads were assembled with Arachne48, and collinearity information was integrated with marker information from genetic maps16,18,19 to reconstruct the eight linkage groups. Additional details and specifics are presented in the Supplementary Note.
Annotation
The genome was annotated using ab initio and homology-based gene predictors along with RNA-seq data (Supplementary Note). The complete details are described in the Supplementary Note.
MCL cluster analyses
MCL (mcl-06-058 package; http://micans.org/mcl/src/) was used with default parameters (−I 2, −S 6) based on clustering of hits with E-value ≤ 10−5. MCL uses a Markov cluster algorithm that attempts to overcome many of the difficulties with protein sequence clustering, such as the presence of multi-domain proteins, peptide fragments and proteins with very common domains. The method has been used for a variety of animal genomes49-51.
OrthoMCL analysis
Orthologous gene clusters were computed from OrthoMCL comparisons39 of four dicotyledonous species with finished genomes: A. thaliana and A. lyrata, Populus trichocarpa36 and Vitis vinifera37,38. A search for potentially missed genes in both Arabidopsis genomes resulted in minor adjustments of the OrthoMCL clusters. Instead of 10,573, 10,878 clusters now contained at least one gene of each the four species, and instead of 5,699, 5,800 clusters were Arabidopsis-specific. To determine deleted or newly generated orthologs (by OrthoMCL definition) between the two species, we focused on clusters specific for either A. lyrata or A. thaliana. For both species, there are two cluster types, those that are supported by members in P. trichocarpa and/or V. vinifera (supported specific cluster, SSC), and clusters exclusively found in one of the Arabidopsis species (exclusive specific cluster, ESC). We did not consider 2,939 and 6,103 unclustered genes (singletons) in A. thaliana and A. lyrata, respectively.
In our initial analysis, we detected 354 SSCs and 161 ESCs for A. thaliana, and 168 SSCs and 833 ESCs for A. lyrata. Whole genome projects, however, may contain false positive as well as missed or incomplete/partial gene calls that impose difficulties for OrthoMCL to detect orthologous relationships. To ensure that genes from the previously detected SSCs were indeed specific for one of the Arabidopsis species, we re-evaluated absence or presence of specific gene calls in the two genome sequences. Previously missed genes detected by GenomeThreader were added to each of the gene sets and the OrthoMCL analysis was repeated.
F-box and NB-LRR gene analysis
Using F-box PF00646.hmm as HMM profile with hmmsearch (E-value ≤ 10−5), 394 hits were found from in A. thaliana and 461 hits in A. lyrata. Alignment of these sequences was optimized with the PF00646 seed using ClustalX 2.052. The final alignment was produced by aligning with hmmalign against PF00646.hmm, to construct an Arabidopsis specific HMM F-box profile. With this HMM profile, 502 hits were found in A. thaliana, and 596 hits in A. lyrata. hmmalign was used to align all of these against PF00646.hmm.
A blastp search (E-value ≤ 10−10) performed with the NB domain (based on HMMEMIT, from http://niblrrs.ucdavis.edu/At_RGenes/). The NB domains of the retrieved proteins, 142 in A. thaliana and 162 in A. lyrata, were aligned using ClustalX52. This alignment was used to develop an Arabidopsis-specific HMM profile, which was used to search the complete set of proteins encoded by both the two genomes (cut off E ≤ 10−5).
PAUP* version 4.0b1053 was used to reconstruct phylogenetic trees with neighbor-joining method.
RepeatMasker analyses
To develop de novo repeat libraries for both species, we used RepeatModeler (version Beta 1.0.3, http://www.repeatmasker.org/RepeatModeler.html). To reduce false positives, unclassified repeats were compared to annotated genes, eliminating all that had at least 80% identity to annotated genes over at least 80 bp (GenBank: Green plant GB all [protein]; blastx with E-value ≤ 10−10). The remaining RepeatModeler predictions were classified with the 80-80-80 rule54, grouping repeats if they shared at least 80% identity over at least 80% of the aligned sequence, which had to be at least 80 bp long. The identified repeats were appended to RepBase (Arabidopsis library - RM database version 20080611), resulting in a final library with 1,152 repeat units. The final libraries were used to annotate TEs using RepeatMasker version 3.2.5.
LTR retrotransposons
Intact LTR retrotransposons were identified de novo using LTR_STRUC55 with default parameters. Based on the sequence divergence between the two LTRs of the same element, insertion times were estimated. All LTR pairs were aligned using MUSCLE56, and the distance K between them calculated with the Kimura two-parameter model using the distmat program implemented in the EMBOSS package (http://emboss.sourceforge.net/). The insertion time T was calculated as T = K/(2) (r), with r as the rate of nucleotide substitution. The molecular clock was set based on the observed mutation rate of 7 × 10−9 per site per generation (assumed to equal one year)14.
Classification and phylogeny of LTR retrotransposons
LTR retrotransposons can be classified into Ty1/copia-like and Ty3/gypsy-like elements57. We classified repeats using RepBase (version 13.08, http://www.girinst.org/server/RepBase/) and blastn (E-value ≤ 10−10), and by direct comparison against the JCVI/TIGR plant repeat database (http://blast.jcvi.org/euk-blast/index.cgi?project=plant). All intact LTR retrotransposons were compared with blastx (E-value ≤ 10−10) against a conserved 156 amino acid segment corresponding to the reverse transcriptase domain58 of Ty1/copia-like and Ty3/gypsy-like sequences, and this segment was then used for phylogenetic reconstruction, with PAUP* version 4.0b1053 and neighbor-joining method. As outgroup sequence, we used yeast the reverse transcriptase domain from yeast Ty1 and Ty3 elements, respectively58.
Detection and analysis of chromosomal breakpoints
Genome wide collinearity was detected by running i-ADHoRe59 on the core-orthologous genes, allowing the identification of breakpoints including inversions and nested inversions. For each inversion, 10 kb up- and downstream of the delimiting breakpoints were compared to each other using blastn (word size 4), tblastx (word size 1) and SSEARCH60-62. Tblastx outperforms blastn for coding regions. In non-coding regions, SSEARCH is more sensitive than blastn, but computationally less efficient, and hence most useful for comparison of shorter sequences. Only one hit per strand was reported. Therefore, for each pair of inversion flanking regions, all combinations of repeats and protein coding genes were evaluated. Default settings were used for gap penalties. An E-value of ≤ 0.01 was considered as indicating similarity between the up- and downstream regions.
Similarity of syntenic regions
To investigate nucleotide divergence of intergenic regions around coding genes (Supplementary Figure 1b), we extracted for each syntenic gene pair the 2 kb sequences 5′ of the start codon and 3′ from the stop codon. If the neighboring gene was closer than 2 kb, the extracted sequence was accordingly trimmed. Coding sequences of syntenic genes were also analyzed. Global alignments of syntenic sequences were generated using the Needleman-Wunsch algorithm as implemented in the EMBOSS package 5.0 (default parameters). Sequence identity of coding regions was measured over the full-length alignment. To investigate whether divergence of intergenic sequence is affected by relative orientation to neighboring genes, upstream sequences were split into head-to-tail and head-to-head groups, and downstream sequences into tail-to-head and tail-to-tail groups.
Fixed insertions and deletions
To identify fixed insertions and deletions among 1,238 fragments that had been amplified by PCR and sequenced in 95 A. thaliana individuals43, two representative sequences for each fragment were first constructed to represent the insertion and deletion states among all segregating indels. The representative sequence consisting of insertions was then queried against the A. lyrata genome with both BLAT63 (−maxGap=100 −extendThroughN −minIdentity=80) and BLAST64 (−e .00001 −F F −G −5 −E −1). Based on the longest hit from the union of hits obtained by both methods, the representative sequences for each alignment were profile-aligned with the A. lyrata allele with MAFFT65. Fixed insertions and deletions were identified in the resulting alignment.
Segregating insertions/deletions
A similar procedure to that described above was used to identify the A. lyrata allele (presumed ancestral state) for each polymorphic indel in A. thaliana. Instead of querying the entire fragment, we queried each insertion allele along with 25 bps flanking each side, against the A. lyrata genome using BLAT. For each polymorphic indel, we filtered for the best hit that spanned both sides of the indel site (by at least 3 bps) and reported each indel as either a derived insertion (if the A. lyrata allele was a deletion in the resulting profile alignment) or a derived deletion (if the A. lyrata allele was not a deletion).
Data and seed availability
The assembly and annotation (Entrez Genome Project ID 41137) are available from GenBank (accession number ADBK00000000) and from JGI's PHYTOZOME portal (http://www.phytozome.net/alyrata.php). Seeds of the MN47 strain have been deposited with the Arabidopsis Biological Resource Center under accession number CS22696.
Supplementary Material
ACKNOWLEDGMENTS
The U.S. Department of Energy Joint Genome Institute (JGI) provided sequencing and analyses under the Community Sequencing Program supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. We are particularly grateful to Dan Rokhsar and Kerrie Barry for providing leadership for the project at JGI. We thank Justin Borevitz, Anne Hall, Charles Langley, June Nasrallah, Barbara Neuffer, Outi Savolainen and Stephen Wright for contributing to the initial sequencing proposal submitted to the Community Sequencing Program at JGI, Christa Lanz and Kenneth Lett for technical assistance, and Peter Andolfatto and Rod Wing for comments on the manuscript. This work was supported by NSF DEB-0723860 (B.S.G.), NSF DEB-0723935 (M.N.), NSF MCB-0618433 (J.C.C.), NSF IOS-0744579 (M.E.N.), NIH GM057994 (J.B.), grant GABI-DUPLO 0315055 of the German Federal Ministry of Education and Research (K.F.X.M.), ERA-PG grant ARelatives from the Deutsche Forschungsgemeinschaft (D.W.) and Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT) and the Inter-University Network for Fundamental Research (P6/25, BioMaGNet) (Y.V.d.P.), a Gottfried Wilhelm Leibniz Award of the DFG (D.W.), the Austria Academy of Sciences (M.N.), and the Max Planck Society (D.W. and Y.-L.G.).
Footnotes
METHODS
Methods and any associated reference are available in the online version of the paper at http:/www.nature.com/naturegenetics/.
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
J.B., J.C.C., B.S.G., I.V.G., Y.-L.G., K.F.X.M., M.N., Y.V.d.P. and D.W. conceived the study; M.E.N. provided the biological material; J.C., J.-F.C., R.M.C., N.F., J.G. and Y.-L.G. performed the experiments; E.G.B., J.A.F., N.F., H.G., Y.-L.G., G.H., J.D.H., T.T.H., R.P.O., S.O., P.P., A.A.S., J.S., K.S., M.S., X.W., and L.Y. analyzed the data; and Y.-L.G., T.T.H., M.N. and D.W. wrote the paper with contributions from all authors.
COMPETING INTEREST STATEMENT
The authors declare no competing financial interests.
References
- 1.Greilhuber J, et al. Smallest angiosperm genomes found in Lentibulariaceae, with chromosomes of bacterial size. Plant Biol. 2006;8:770–7. doi: 10.1055/s-2006-924101. [DOI] [PubMed] [Google Scholar]
- 2.Gregory TR, et al. Eukaryotic genome size databases. Nucleic Acids Res. 2007;35:D332–8. doi: 10.1093/nar/gkl828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaut BS, Ross-Ibarra J. Selection on major components of angiosperm genomes. Science. 2008;320:484–6. doi: 10.1126/science.1153586. [DOI] [PubMed] [Google Scholar]
- 4.Pellicer J, Fay MF, Leitch IJ. The largest eukaryotic genome of them all? Botanical Journal of the Linnean Society. 2010;164:10–15. [Google Scholar]
- 5.Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Ann. Bot. 2005;95:127–32. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawkins JS, Proulx SR, Rapp RA, Wendel JF. Rapid DNA loss as a counterbalance to genome expansion through retrotransposon proliferation in plants. Proc Natl Acad Sci U S A. 2009;106:17811–6. doi: 10.1073/pnas.0904339106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piegu B, et al. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006;16:1262–9. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vitte C, Panaud O, Quesneville H. LTR retrotransposons in rice (Oryza sativa, L.): recent burst amplifications followed by rapid DNA loss. BMC Genomics. 2007;8:218. doi: 10.1186/1471-2164-8-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodhouse MR, et al. Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS Biol. 2010;8:e1000409. doi: 10.1371/journal.pbio.1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–6. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 11.Johnston JS, et al. Evolution of genome size in Brassicaceae. Ann. Bot. 2005;95:229–35. doi: 10.1093/aob/mci016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oyama RK, et al. The shrunken genome of Arabidopsis thaliana. Plant Systemat. Evol. 2008;273:257–271. [Google Scholar]
- 13.Wright SI, Lauga B, Charlesworth D. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol. Biol. Evol. 2002;19:1407–20. doi: 10.1093/oxfordjournals.molbev.a004204. [DOI] [PubMed] [Google Scholar]
- 14.Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–4. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2010;107:18724–8. doi: 10.1073/pnas.0909766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuittinen H, et al. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics. 2004;168:1575–84. doi: 10.1534/genetics.103.022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch MA, Kiefer M. Genome evolution among cruciferous plants: a lecture from the comparison of the genetic maps of three diplod species–Capsella rubella, Arabidopsis lyrata subsp. petraea, and A. thaliana. Am. J. Bot. 2005;92:761–767. doi: 10.3732/ajb.92.4.761. [DOI] [PubMed] [Google Scholar]
- 18.Yogeeswaran K, et al. Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 2005;15:505–15. doi: 10.1101/gr.3436305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lysak MA, et al. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA. 2006;103:5224–9. doi: 10.1073/pnas.0510791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berr A, et al. Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata. Plant J. 2006;48:771–83. doi: 10.1111/j.1365-313X.2006.02912.x. [DOI] [PubMed] [Google Scholar]
- 21.Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim JK, Simmons MJ. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. Bioessays. 1994;16:269–75. doi: 10.1002/bies.950160410. [DOI] [PubMed] [Google Scholar]
- 23.Stankiewicz P, et al. Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am. J. Hum. Genet. 2003;72:1101–16. doi: 10.1086/374385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–6. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J, Han K, Meyer TJ, Kim HS, Batzer MA. Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons. PLoS ONE. 2008;3:e4047. doi: 10.1371/journal.pone.0004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braumann I, van den Berg MA, Kempken F. Strain-specific retrotransposon-mediated recombination in commercially used Aspergillus niger strain. Mol. Genet. Genomics. 2008;280:319–25. doi: 10.1007/s00438-008-0367-9. [DOI] [PubMed] [Google Scholar]
- 27.Woodhouse MR, Pedersen B, Freeling M. Transposed genes in Arabidopsis are often associated with flanking repeats. PLoS Genet. 6:e1000949. doi: 10.1371/journal.pgen.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ranz JM, et al. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 2007;5:e152. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 30.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–42. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 31.Borevitz JO, et al. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2007;104:12057–62. doi: 10.1073/pnas.0705323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–84. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–30. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 34.Thomas JH. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 2006;16:1017–30. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, et al. The F-box gene family is expanded in herbaceous annual plants relative to woody perennial plants. Plant Physiol. 2008;148:1189–200. doi: 10.1104/pp.108.121921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 37.Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–7. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 38.Velasco R, et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Stoeckert CJ, Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat. Genet. 1998;20:43–5. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 41.Devos KM, Brown JK, Bennetzen JL. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002;12:1075–9. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19:1419–28. doi: 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrov DA, Sangster TA, Johnston JS, Hartl DL, Shaw KL. Evidence for DNA loss as a determinant of genome size. Science. 2000;287:1060–2. doi: 10.1126/science.287.5455.1060. [DOI] [PubMed] [Google Scholar]
- 45.Petrov DA, Lozovskaya ER, Hartl DL. High intrinsic rate of DNA loss in Drosophila. Nature. 1996;384:346–9. doi: 10.1038/384346a0. [DOI] [PubMed] [Google Scholar]
- 46.Charlesworth B. Evolutionary rates in partially self-fertilizing species. Am. Nat. 1992;140:126–48. doi: 10.1086/285406. [DOI] [PubMed] [Google Scholar]
- 47.Knight CA, Molinari NA, Petrov DA. The large genome constraint hypothesis: evolution, ecology and phenotype. Ann Bot. 2005;95:177–90. doi: 10.1093/aob/mci011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaffe DB, et al. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 2003;13:91–6. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW. The evolution of mammalian gene families. PLoS ONE. 2006;1:e85. doi: 10.1371/journal.pone.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prachumwat A, Li WH. Gene number expansion and contraction in vertebrate genomes with respect to invertebrate genomes. Genome Res. 2008;18:221–32. doi: 10.1101/gr.7046608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 52.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL-X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods): Version 4. Sinauer Associates; Sunderland, Massachusetts: 2003. [Google Scholar]
- 54.Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 55.McCarthy EM, McDonald JF. LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics. 2003;19:362–7. doi: 10.1093/bioinformatics/btf878. [DOI] [PubMed] [Google Scholar]
- 56.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–62. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Wessler SR. Genome-wide comparative analysis of the transposable elements in the related species Arabidopsis thaliana and Brassica oleracea. Proc. Natl. Acad. Sci. USA. 2004;101:5589–94. doi: 10.1073/pnas.0401243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simillion C, Vandepoele K, Saeys Y, Van de Peer Y. Building genomic profiles for uncovering segmental homology in the twilight zone. Genome Res. 2004;14:1095–106. doi: 10.1101/gr.2179004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–7. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 62.Pearson WR. Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics. 1991;11:635–50. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- 63.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 65.Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.